Abstract
Phosphatase of regenerating liver (PRL)-3 (PTP4A3) has gained much attention in cancer research due to its involvement in tumor promoting and metastatic processes. It belongs to the protein tyrosine phosphatase (PTP) superfamily and is thought to follow the catalytic mechanism shared by this family, which aside from the conserved active-site amino acids includes a conserved glutamic acid residue that is usually required for the integrity of the active site in PTPs. We noted that in structures of PRL-3, PRL-1, and PTEN these residues do not clearly align and therefore we sought to investigate if the glutamic acid residue fulfills its usual function in these proteins. Although this residue was essential for PTEN’s catalytic activity, it was nonessential for PRL-1 and PRL-3. Surprisingly, the mutation E50R increased PRL-3 activity against all tested in vitro substrates and also enhanced PRL-3-promoted cell adhesion and migration. We show that the introduction of Arg50 leads to an enhancement of substrate turnover for both PRL-3 and, to a lesser extent, PRL-1, and that the stronger gain in activity correlates with a higher structural flexibility of PRL-3, likely allowing for conformational adaptation during catalysis. Thus, in contrast to its crucial functions in other PTPs, this conserved glutamic acid can be replaced in PRL-3 without impairing the structural integrity. The variant with enhanced activity might serve as a tool to study PRL-3 in the future.
1. Introduction
Phosphatase of regenerating liver (PRL)-3 belongs to the dual-specificity phosphatases of the protein tyrosine phosphatase (PTP) superfamily. It has attracted attention through its involvement in cancerogenesis and metastatic events (for review, see refs (1, 2)). Still, although substrates and interaction partners were suggested by studies in various cell lines,1−4 molecular mechanisms of PRL-3 remain a challenge to address. Efforts to identify substrates are often hampered by insufficiently clear readouts and seemingly cell-line-specific phenotypes.1,2
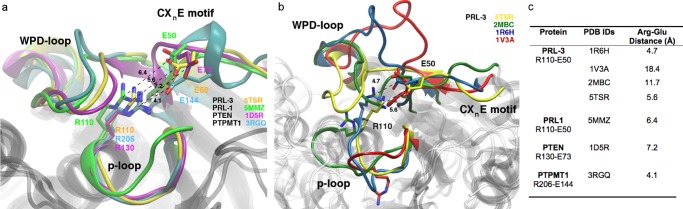
The catalytic mechanism of PTPs involves a cysteine residue whose thiolate attacks the scissile phosphate group in the first step of the two-step catalytic reaction, an arginine residue for transition-state stabilization of the phosphocysteine intermediate, and an aspartic acid functioning as general acid/base during both steps of catalysis. Both the cysteine and arginine are part of the conserved active-site p-loop of PTPs, and the aspartic acid is found in the flexible WPD-loop (Asp72 in PRL-3) that closes toward the active site for catalysis.5,6 A conserved serine/threonine residue stabilizes the thiolate before the nucleophilic attack on the phosphoester and assists in the hydrolysis of the phosphocysteine intermediate in the second catalytic step.5,7 Maintenance of a functional catalytic site is furthermore achieved by a glutamic acid residue positioning the catalytic p-loop arginine through hydrogen bonding.1,8 This conserved glutamic acid was proposed to be part of a structural CXnE motif in PTPs containing a regulatory (“backdoor”) cysteine,1 which regulates the enzyme’s activity through disulfide bond formation with the active-site cysteine.9 Alignment of a recent X-ray structure of PRL-3 (pdb entry 5TSR)10 with the closely related phosphatases PRL-1, PTEN, and PTPMT1 that contain the proposed motif suggests, however, that the glutamic acid may not position the catalytic arginine residue through hydrogen bonds in PRL-3, PRL-1, and PTEN given the further distance between the residues (Figure 1a,c). This suggests that the glutamic acid within the proposed CXnE motif could serve different purposes in these phosphatases, which is further indicated by the structural flexibility and variability shown in the available NMR structures of PRL-3 (pdb entries 2MBC, 1R6H, 1V3A) (Figure 1b,c).
Figure 1.
(a) Structural superposition of PTPs and the CXnE motif. The respective glutamic acids and the catalytic arginine residues are indicated for PRL-3 (orange: 5TSR), PRL-1 (green: 5MMZ), PTEN (magenta: 1D5R), and PTPMT1 (cyan: 3RGQ). Tight glutamate–arginine interactions as seen with PTPMT1 seem rather questionable for PRL-3, PRL-1, and PTEN. For PTPMT1, Glu144 from the “EEYE” motif aligns well with the Glu of the CXnE motif of the other PTPs. (b) Superposition of an X-ray (5TSR) and various NMR structures (others) of PRL-3 showing the protein’s flexibility for active-site conformations. The respective position of Arg110 and Glu50 is indicated. (c) Distances between Arg and Glu residues of the indicated phosphatases. For 5TSR, the reported value is an average calculated on both chains A and C; for 2MBC and 1R6H, the reported value is the average value calculated on the deposited 20 models.
Furthermore, PRL-3 shows a somewhat different active-site architecture from that of other PTPs. Still harboring the invariant catalytic cysteine, aspartic acid, and arginine residues, the conserved serine/threonine is replaced by an alanine residue in wild-type (wt) PRL-3 (Ala111).9,11 This is also the case in CDC25 and DUSP19, accompanied with inherently low phosphatase activity.12,13 For PRL-3, it was shown that the absence of a serine in position 111 leads to slower hydrolysis of the phosphocysteine intermediate involving two kinetic phases, a “burst-phase” followed by a slower steady-state phase, which can lead to a temporal blockage of the active site.9,10 Reconstituting this serine in PRL-3 through an A111S mutation led to the expected gain in catalytic activity against the unnatural substrates 3-O-methylfluorescein phosphate (OMFP)9 and 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP), where the steady-state kinetic phase was not observed anymore.10 Nevertheless, in a previous study, we observed less in vitro dephosphorylation of phosphatidylinositol (4,5)-bisphosphate (PI(4,5)P2) by the PRL-3 A111S variant compared to wt PRL-3. Also, PRL-3-like promotion of cell migration was lacking when overexpressing the A111S variant,14 showing that Ala111 is not replaceable without disturbing PRL-3’s integrity and that it appears to serve a function in the native enzyme.
In this study, we investigated how mutations of Glu50 in PRL-3 affect its catalytic activity, in comparison to the respective Glu73 in PTEN and Glu50 in PRL-1. We found that although for PTEN the glutamic acid is important for catalysis, it was nonessential for PRL-3’s and PRL-1’s catalytic activities. By contrast, the PRL-3 variant E50R led to a strong gain in PRL-3’s enzymatic activity, maintaining the specificity pattern against phosphatidylinositol phosphates (PIPs) and showing enhanced activity also in cells. Thus, as opposed to the respective conserved glutamic acid in other PTPs and to the Ala111 in PRL-3, Glu50 in PRL-3 is replaceable without disturbing PRL-3’s structural integrity. Kinetic and stability studies revealed that the unique flexibility of PRL-3 and ability of Arg50 to rescue the deletion of the catalytically important Arg110 when dephosphorylating PIPs are underlying features for the enhanced activity.
2. Results and Discussion
2.1. Different Roles of a Conserved Glutamic Acid in PRL-3 and PTEN
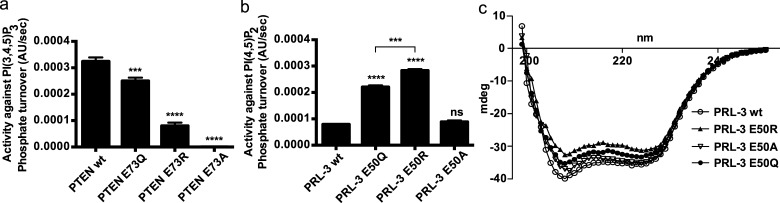
To investigate the role of glutamic acid as part of the proposed CXnE motif for PRL-3’s and PTEN’s catalytic activity, we carried out a mutagenesis study and compared enzymatic activities to the respective wt proteins. Mutations were selected according to loss of charge in the case of mutation E → Q, reversion of charge from negative to positive with mutation E → R, and replacement with a small, hydrophobic group with mutation E → A. PTEN was monitored for dephosphorylating its substrate PI(3,4,5)P3,15 and concomitantly, the proposed natural substrate PI(4,5)P214 was chosen for PRL-3. All three PTEN Glu73 variants showed a decrease or loss of activity (Figure 2a), suggesting an important role of this glutamic acid in PTEN’s catalytic mechanism in agreement with the proposed structural interaction with the catalytic arginine (see Figure 1).1 By contrast, the results with the corresponding PRL-3 variants indicated a different mechanism. Mutation E50A did not have any effect on the activity against PI(4,5)P2. Variants E50Q and especially E50R were significantly more active than wt PRL-3 (Figure 2b). A similar effect of the three PRL-3 mutations was found when comparing activities to the unnatural substrate DiFMUP, as shown by kinetic profiles in Table 1a. Although binding (reflected by Km values) was not altered, turnover rates were increased, with PRL-3 E50R again showing the highest activity, followed by E50Q and E50A. Introducing the mutations in PRL-3 did not significantly affect overall protein folding (Figure 2c). When comparing the hydrolysis of PRL-3 wt and E50R with DiFMUP, E50R accelerated both the burst and steady-state kinetics of the reaction (Figure S1), suggesting that both the formation of the phosphocysteine intermediate and its hydrolysis are facilitated in PRL-3 E50R compared to the wt enzyme. Taken together, these results suggest differences in the active-site architecture of PRL-3 and PTEN regarding the role of the glutamic acid in maintaining a catalytically competent active site. Although the decrease of activity of PTEN Glu73 variants suggests a similar mechanism to that seen with PTPMT1,5 PRL-3 activity was not affected by the E50A mutation and positively modulated by mutating residue Glu50 with polar amino acids, implying that Glu50 is nonessential for PRL-3’s catalytic activity.
Figure 2.
Different roles of a conserved glutamic acid for PRL-3 and PTEN. (a) PTEN Glu73 mutations decreased or abolished dephosphorylation of PI(3,4,5)P3 in vitro. (b) Mutations E50Q and E50R positively influenced PRL-3 activity against PI(4,5)P2. Results are shown as mean ± SD. Statistics, compared to respective wt enzyme or where indicated: ****p = 0.0001; ***p = 0.0002; ns: not significant. Enzymes were used at 6 μM, substrates at 100 μM concentration. (c) Circular dichroism (CD) spectrometry of PRL-3 Glu50 variants.
Table 1. Kinetic Parameters for PRL-3 (a) and PRL-1 (b) Variants (50 nM) Dephosphorylating the Unnatural Substrate DiFMUP, Obtained by Hyperbolic Curve Fittinga.
| Km (μM) | Kcat (s–1) | Kcat/Km (L mol–1 s–1) | p-value | |
|---|---|---|---|---|
| (a) | ||||
| PRL-3 wt | 21.40 ± 6.68 | 1.11 × 10–3 ± 6.83 × 10–4 | 53.17 ± 20.15 | |
| E50Q | 17.49 ± 4.52 | 4.70 × 10–3 ± 1.19 × 10–3 | 270.88 ± 47.58 | <0.0001 (***) |
| E50R | 22.24 ± 6.60 | 7.35 × 10–3 ± 1.56 × 10–3 | 336.08 ± 27.60 | <0.0001 (***) |
| E50A | 25.77 ± 5.10 | 3.15 × 10–3 ± 3.63 × 10–4 | 124.60 ± 20.09 | 0.0046 (**) |
| (b) | ||||
| PRL-1 wt | 32.22 ± 5.33 | 1.53 × 10–3 ± 3.02 × 10–5 | 48.46 ± 8.95 | |
| E50R | 37.11 ± 9.29 | 4.84 × 10–3 ± 4.18 × 10–4 | 136.31 ± 45.41 | 0.1153 (ns) |
Variants show similar affinities to the wild-type enzyme, but accelerated activity regarding turnover rates. Results are shown as mean ± SD. For the turnover parameters Kcat/Km, significance was calculated between variants and respective control, according to the statistical tests outlined in Experimental Section. The difference in Kcat/Km between PRL-3 E50R and E50Q is not statistically significant (not shown).
2.2. Biochemical Characterization of the PRL-3 E50R Variant
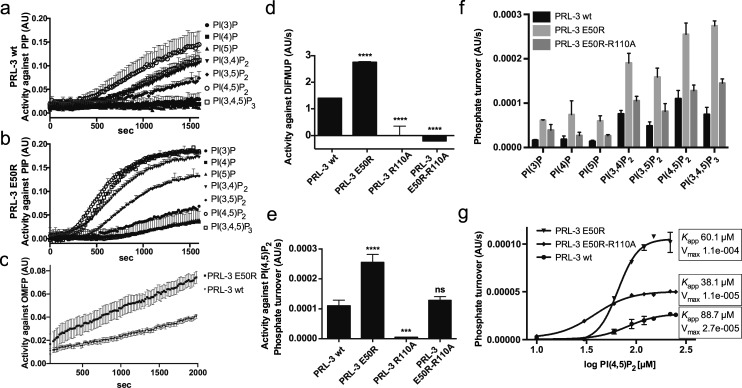
Given the common role of the conserved glutamic acid to position the arginine of the p-loop, replacement of Glu50 in PRL-3 without impairing the structural integrity of the protein would be a unique structural feature among the PTPs. To address if this is the case and to explain the observation of enhanced activity of PRL-3 E50R, we sought to gain insight into a potential role of this residue in catalysis and investigated the catalytic mechanism in detail. First, we compared PRL-3 E50R’s activity with wt PRL-3 against the naturally occurring PIP species. PRL-3 E50R was able to dephosphorylate all seven PIPs more efficiently than the wt enzyme (Figure 3a,b). The individual activity pattern against the various PIP species remained similar for both enzymes, suggesting that mutation E50R generally enhances activity without altering the enzyme’s specificity and that Glu50 can be replaced without impairing the protein’s structural integrity. In addition, the variant exhibited also increased activity against the unnatural substrate OMFP (Figure 3c). Overall, PRL-3 E50R shows similar substrate recognition preferences to wt PRL-3, as opposed to the previously reported hyperactive variant A111S.14 In addition, PRL-3 E50R was inhibited by our recently developed small-molecule inhibitor analog 3(16) with similar potency to wt PRL-3 (IC50 23.9 ± 0.2 vs 31.4 ± 3.5 μM, respectively, using DiFMUP as substrate). This suggests that the inhibitor does not interact with the Glu50-containing stretch in PRL-3.
Figure 3.
Kinetic characterization of PRL-3 variant E50R. (a, b) PRL-3 variant (b) E50R dephosphorylates all PIP species more efficiently than (a) wild-type PRL-3, with the individual activity curves shown. (c) E50R shows increased activity against the unnatural substrate OMFP. (d–f) PRL-3 variant E50R-R110A reveals different roles of Arg50 for catalysis, depending on the substrate used. In vitro activities of PRL-3 variants against (d) DiFMUP and (e) PI(4,5)P2 are shown. Results are shown as mean ± SD. Statistics, compared to respective wt enzyme: ***p = 0.0004; ****p = 0.0001. (f) E50R-R110A dephosphorylates all seven PIP species with turnover values between E50R and wild-type PRL-3. (g) PI(4,5)P2 titration profiles reveal non-Michaelis–Menten kinetics with sigmoidal curve shapes, showing altering apparent affinity (Kapp) and maximal turnover for the different variants. Data are shown as mean ± SD. The following concentrations were used: PIPs 100 μM with 6 μM enzyme; OMFP 600 μM with 6 μM enzyme; and DiFMUP 21 μM with 50 nM enzyme.
To address the mechanistic basis for the gain of activity of the PRL-3 E50R variant, we hypothesized that the E50R mutation would introduce a new arginine residue in close proximity to the active site that could stabilize the phosphocysteine in a similar manner to the catalytic Arg110.17,18 Also, having an additional positively charged arginine residue in place could facilitate binding of negatively charged phosphosubstrates or change the structure of the active site without taking active part in transition-state stabilization. To probe these hypotheses, we created PRL-3 variants R110A and E50R-R110A. The R → A variant, lacking the transition-state-stabilizing residue, was shown for similar phosphatases to be catalytically inactive.19 The double mutant possessed only the newly introduced Arg50 and lacked the catalytic Arg110. None of the newly introduced mutations disrupted protein folding (Figure S2). Interestingly, although both PRL-3 R110A and E50R-R110A were completely inactive against the unnatural substrate DiFMUP (Figure 3d), E50R-R110A was able to hydrolyze PI(4,5)P2, whereas R110A was, as expected, also inactive against the latter (Figure 3e). As shown in Figure 3f, PRL-3 E50R-R110A dephosphorylated all seven PIP species with a constant efficiency pattern between wt PRL-3 and E50R, again leaving the substrate specificity intact. Furthermore, using isothermal titration calorimetry, we observed that the PRL-3 variant E50R-R110A did not bind to DiFMUP anymore (Figure S3), explaining the absence of dephosphorylation activity.
To understand the influence of Arg50 on PIP catalysis in more detail, we carried out a kinetic analysis on PI(4,5)P2 hydrolysis with the PRL-3 E50R and E50R-R110A variants. The results are shown in Figure 3g. PI(4,5)P2 hydrolysis by PRL-3 and its variants followed non-Michaelis–Menten kinetics with sigmoidal curve shapes. The apparent affinity (Kapp, PI(4,5)P2 concentration at the inflection point) was similar in all cases and indicated only a slightly stronger PI(4,5)P2 binding for PRL-3 E50R-R110A than for E50R, and for both, binding was again slightly stronger than wt PRL-3. As shown by the maximal activity reached, PRL-3 E50R was more active than E50R-R110A and the least-active PRL-3 wt, showing a similar advantage for E50R in PI(4,5)P2 substrate turnover to that seen with DiFMUP. Because the overall phosphate turnover was enhanced for all of the PIP species when using the two PRL-3 variants (see Figure 3f), this effect of Arg50 seems to be independent of the number of negative charges present on the substrate. Together, these results show that for PI(4,5)P2 Arg50 is able to compensate for the catalytic Arg110, whereas substrate recognition is lost in the case of DiFMUP, and that it assists for both substrates in more efficient substrate turnover. To accomplish this, the additional arginine must be placed in a feasible position toward PI(4,5)P2, which could be enabled by the flexible nature of the protein20 allowing for structural rearrangements during catalysis. Furthermore, given its close proximity to both the p- and WPD-loops in some structural conformations (Figure 1), Arg50 might assist in the phosphocysteine hydrolysis by providing an additional hydrogen bond similarly to the conserved serine would, which is not present in PRL-3. Alternatively, it could interact with Asp71 or Asp72 and place the WPD-loop in PRL-3 in a favorable position for faster accessibility of Asp72 during catalysis, and this would lead to the accelerated steady-state kinetics observed above.
The non-Michaelis–Menten kinetics that we observed with PIP hydrolysis are not a general feature of the assay format, as the same kind of assay (EnzChek Phosphate Assay Kit) has been used in other types of studies with classical Michaelis–Menten profiles.21 The behavior is not specific to PRL-3 because the enzyme showed classical behavior when tested against DiFMUP (see Figure S1). Non-Michaelis–Menten behavior is typically known for oligomerizing enzymes.22 Because PRL-3 was reported to oligomerize,23 a protein concentration effect could be the reason for possible PRL-3 oligomerization23 as different enzyme concentrations were used in both assay types (see Experimental Section). Additionally, a more complex picture, including oligomerizing PIPs or the substrates inducing PRL-3 oligomerization, cannot be ruled out.
2.3. Mutation E50R Does Not Alter PRL-3’s Redox Properties
Redox regulation is an important mechanism to modulate PTP activity.24 Therefore, we further investigated if the enhanced activity of PRL-3 E50R resulted from altered redox properties compared to wt PRL-3. Similar to other PTPs, such as PTEN, CDC25, or LMW-PTP,25−27 PRL-3 contains a regulatory cysteine (Cys49) in close proximity to the active site9 that forms a reversible intramolecular disulfide bond with the catalytic cysteine (Cys104) under oxidative conditions, protecting the now inactive catalytic residue from further irreversible oxidation by reactive oxygen species.9,28−31Figure S4 confirms that mutation of residue Cys49 (PRL-3 C49A variant) abolishes the ability to be reduced after full oxidation with hydrogen peroxide and that the mutant is unable to form the oxidized disulfide. The regulatory Cys49 resides directly adjacent to Glu50 as part of the proposed CXnE motif (n = 0) in PRL-3. Therefore, mutations at position 50 might potentially alter the enzyme’s redox properties and thus influence its activity profile. We tested wt PRL-3 and E50R for their potential to be reduced or oxidized in the presence of varying amounts of dithiothreitol (DTT) or hydrogen peroxide, respectively. As shown in Figure S4, both enzymes behaved similarly with respect to the amount of DTT or H2O2 needed to fully reduce (activate) or oxidize (inactivate). Also, the same DTT plateau was reached for gaining full enzymatic activity. PRL-3 E50R was constantly about 2-fold more active than wt PRL-3 against the DiFMUP substrate. To conclude, altered redox properties do not seem to cause E50R’s enhanced activity. Thus, although both cysteine and glutamic acid are part of the same structural motif, only cysteine seems to be important for redox regulation in PRL-3 and the full motif is not a functional entity.
2.4. PRL-3 E50R Shows Enhanced Structural Flexibility and Decreased Stability
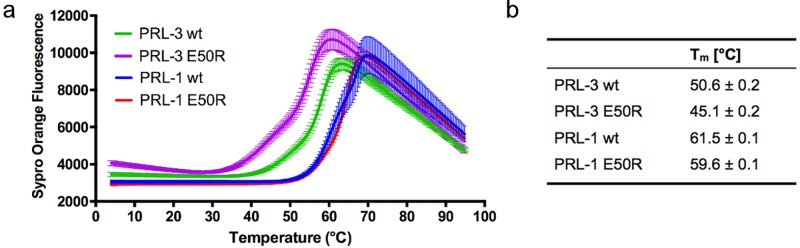
To address the hypothesis that the structurally flexible nature of PRL-3 could aid in substrate turnover, we investigated how the E50R mutation would influence structural stability and flexibility. Several NMR structures of PRL-3 have been reported,9,20,32 all showing variations in the structural models. The apo form of PRL-3, as well as PRL-3 bound to small ligands, has not yet been crystallized, and our own attempts have been futile. Recently, however, PRL-3 stabilized through the interaction with a large protein domain was successfully crystallized.10 In contrast, several consistent crystal structures of PRL-1, the closest homologue of PRL-3, are available in the apo form or bound to proteins, suggesting that PRL-1 is less flexible than PRL-3.1 We thus included PRL-1 in this study. We first measured the thermal stability of the respective variants. Our data clearly support trends from crystallography discussed above: PRL-3 is considerably less stable than PRL-1 (Figure 4a,b), suggesting together with the discussed NMR data increased conformational flexibility in PRL-3. For PRL-3, a further decrease in stability was observed upon introduction of the E50R mutation, whereas for PRL-1 E50R compared to PRL-1 wt, changes were minimal. On the basis of this finding, we hypothesized that if these shifts would contribute to enhancing the catalytic activity, we should observe a slight increase in catalytic activity of PRL-1 E50R compared to PRL-1 wt. However, this increase should not exceed the impact on the catalytic activity enhancement of PRL-3 E50R compared to PRL-3 wt. As listed in Table 1b, the data we observed for DiFMUP dephosphorylation follow exactly these expected trends: (1) the PRL-1 variants gave similar Km values toward DiFMUP, but the values are slightly lower than those for the respective PRL-3 variants; (2) the Kcat values of PRL-3 and PRL-1 wt are similar, but PRL-1 E50R showed a 3.2 times higher Kcat than PRL-1 wt, whereas PRL-3 E50R’s Kcat is 6.6 times higher than that of PRL-3 wt.
Figure 4.
PRL-3 is less stable than PRL-1, and the E50R mutation further destabilizes PRL-3. (a) Enzyme thermal stability determined by a ThermoFluor assay for PRL-3 wt and E50R, PRL-1 wt and E50R. (b) Melting temperatures of the respective proteins.
Finally, we asked if the change of the surface potential when introducing Arg50 in both PRL-1 and PRL-3 (Figure S5) could have an influence on the structural flexibility difference in both proteins. We observed a charge change in both proteins, as expected, from the wt to the E50R variant. This change is not only present locally in close proximity of Arg50 but also in the area around the CXnE motif. The two best comparable structures between PRL-1 and PRL-3, both bound to CNNM domains (5TSR and 5MMZ), suggest that the difference of the impact of this change on both proteins is similar and therefore this cannot clearly explain the difference in structural flexibility or reduction in PRL-3 stability.
Together, these data show that conformational flexibility and protein stability have an important influence on the catalytic turnover abilities of PRL-3. These features potentially enable the correct positioning of Arg50 in the PRL-3 E50R variant to aid catalysis, which offers an explanation for the enhanced catalytic activity of this variant. With a sequence identity of 79% between the two proteins,1 however, finding the reason for the differing structural stability will require introduction of each differing amino acid found scattered along the sequence of PRL-1 into PRL-3 together with thermostability analysis of the resulting variants.
2.5. PRL-3 Variant E50R Accelerates Cell Adhesion and Migration
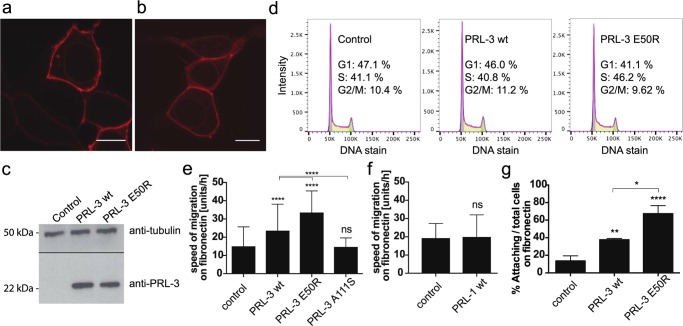
To investigate the biological relevance of our biochemical findings, we compared the migration abilities of stably overexpressing PRL-3 wt and E50R HEK293 cell lines. Enhanced cell migration is a well-established phenotype for PRL-3 that depends on its catalytic activity.14,16 We first controlled for correct protein localization of PRL-3 upon introducing the E50R point mutation (Figure 5a,b). After generation of inducible stable cell lines and control of protein/mutant expression by Western blot (Figure 5c), also potential strong side effects on the cell cycle were ruled out for PRL-3 variant E50R (Figure 5d).
Figure 5.
Cellular analysis of PRL-3 variant E50R. (a, b) E50R retains wt-like localization. HEK293 control cells were transiently transfected with (a) mKate-PRL-3 or (b) mKate-E50R constructs and visualized by confocal microscopy. The scale bar is 10 μm. Representative pictures are shown. (c) Immunoblots of HEK293 FlpIn T-Rex stable cell lines blotted with anti-tubulin- and anti-PRL-3-specific antibodies, indicating similar expression levels of the introduced PRL-3 wt/E50R protein. (d) Cell cycle profiles of HEK293 control, PRL-3 wt, and E50R stable cell lines showing similar G1, S, and G2/M phase distributions. (e) HEK293 PRL-3 wt and E50R cells show accelerated migration on fibronectin substratum compared to control cells, with enhanced activity of the E50R cell line. PRL-3 A111S (e) and PRL-1 (f) HEK cell lines do not show enhanced cell migration on this substratum. (g) PRL-3 wt and E50R cell lines show enhanced adhesion to fibronectin substratum. Cells were seeded on the respective support and analyzed by microscopy for percentage of adhering cells or speed of migration, respectively. Results are shown as mean ± SD of a representative experiment. Statistics, compared to respective control: *p = 0.0453; **p = 0.0062; ****p = 0.0001; ns: not significant.
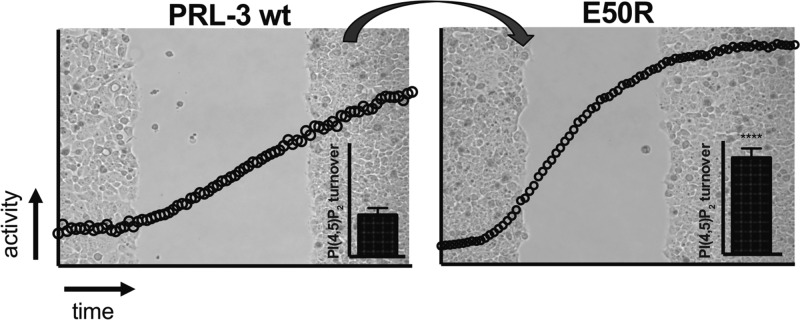
We next compared PRL-3 wt and E50R HEK293 stable cell lines for their migration behavior on different substrata. As biological migration processes in extracellular matrix (ECM) proceed through passing matrix layers containing fibronectin, laminin, and various forms of collagens,33−36 we investigated PRL-3 cell migration on these coatings. On laminin, collagen-IV, collagen-I, and gelatin (Figure S6a–d), both PRL-3 and E50R cell lines migrated equally fast as the control cell line. On fibronectin, however, PRL-3 cells showed about 1.5-fold accelerated migration compared to control (Figure 5e), suggesting enhanced interaction of PRL-3 HEK293 cells with this ECM component. The E50R cell line showed a more than 2-fold accelerated cell migration on fibronectin compared to control cells (Figure 5e), exceeding the migration speed of wt PRL-3. Thus, the mutation E50R enhances this activity-dependent cellular phenotype. Contrarily, we observed that cells expressing the previously suggested hyperactive A111S variant14 had a control-like phenotype on fibronectin (Figure 5e). This we had already observed for migration on uncoated dishes;14 however, when testing our cell lines here on standard uncoated dishes of various plastic types (Figure S7), we obtained contradictory and unpredictable migration results, bringing the relevance of migration studies on such uncoated support into question. Together, these results show that the PRL-3 E50R mutant, as opposed to PRL-3 A111S, accelerates PRL-3-dependent cell migration processes on fibronectin-coated slides in agreement with our biochemical data. Finally, we tested if the PRL-1 cell line would show faster migration on fibronectin substratum. PRL-1 wt enhances cell migration on uncoated support (Figure S8). However, PRL-1 did not enhance cell migration here (Figure 5f), suggesting that PRL-3 promotes cell migration on fibronectin through a mechanism that is not shared by PRL-1. Because both PRL-3 wt and E50R, but not PRL-1 wt and PRL-3 A111S,14 dephosphorylate PI(4,5)P2 in vitro and promote cell migration on fibronectin, this could be a potential pathway through which PRL-3 acts here. Future studies will need to address this question.
We next investigated if the observed effects in cell migration correlate with effects in cell adhesion to the respective ECM components, as cell–matrix adhesion is a known prerequisite for cell migration processes.33 In agreement with the observed migration results, no difference in adhesion of the cell lines was observed on laminin, collagen-I, collagen-IV, and gelatin (Figure S6e–h). Similarly, PRL-3 wt cells adhered about 2-fold faster on fibronectin compared to control, and the activity gain of the E50R variant was again reflected by a fourfold accelerated adhesion to fibronectin substratum compared to wt PRL-3 (Figure 5g, see also Figure S9 for original microscope images). Together, these results suggest that PRL-3-driven HEK293 cell migration relates to an adhesion advantage to the respective substratum and that the choice of substratum layer seems of importance for the promotion of cell migration by PRL-3. Further studies are required to corroborate this observation, with special attention given to integrin repertoires. Integrins are the major receptors responsible for engaging in specific cell–ECM contacts,37 and they were shown to rely on (phospho)lipid compositions.38 Accordingly, PRL-3/E50R might affect integrin composition and/or function by its action on PI(4,5)P2. In addition, cell adhesion to fibronectin was reported to correlate with tumorigenicity,39,40 and it is tempting to speculate that the observed PRL-3-dependent effect could represent a potential link to its tumorigenic potential. Of note, mutation E50R is not found among the so-far known PRL-3 cancer mutations summarized in the COSMIC database for PTP4A3.41
3. Conclusions
To conclude, we show here that PRL-3’s catalytic mechanism and active-site architecture differ from those of other PTPs regarding the role of the glutamic acid within the proposed structural CXnE motif, which in PRL-3 does not appear to be a functional entity. Glu50 in PRL-3, and also in PRL-1, can be replaced without structural and catalytic impairment of the phosphatases. However, PRL-3 is structurally more flexible and less stable than PRL-1, which explains at least in part the differences seen here in the catalytic behavior.
PRL-3 E50R’s structural flexibility demonstrated here could place Arg50 in a favorable position to enhance catalytic turnover. Although the detailed mechanism remains unclear, possible mechanisms could be Arg50 providing a hydrogen bond to aid in the second catalytic step of phosphate hydrolysis from the active-site cysteine or Arg50 positioning the WPD-loop favorably for catalysis, or given the flexibility of PRL-3, both these interactions could transiently be happening during catalysis. Future studies including mutational analyses will address these hypotheses.
A defined, moderate enhancement of enzymatic activity can be explored to study the respective enzyme by revealing a stronger but not arbitrary phenotype or increasing substrate turnover to allow its identification. Activity-enhancing variants have been successfully applied to study various enzymes in the past.42−44 Accordingly, PRL-3 E50R could represent a novel tool for potentiating PRL-3 activity in vitro and in cellular applications, as it shows enhanced phosphatase activity toward all tested in vitro substrates, similar substrate specificity to the wt protein with regard to phosphoinositides, and enhanced PRL-3-dependent cellular phenotypes.
4. Experimental Section
4.1. Materials
FBS was obtained from Gibco or Sigma (Sigma Lot #014M3395 for cell migration experiments). Di-C8:0-phosphoinositide phosphates were ordered from Avanti Polar Lipids or Echelon. PRL-3 inhibitor analog 3 was purchased from Enamine.19 6,8-Difluoro-4-methylumbelliferyl phosphate (DiFMUP) was obtained from Life Technologies, and all other chemicals were purchased from Sigma or Merck. Standard cell culture dishes were from Thermo Scientific/Nunclon, wound healing dishes with preinserted migration inserts were from ibidi, as well as separated migration inserts were used for self-insertion. Precoated cell culture plates with various matrix component coatings were purchased from Corning.
4.2. Bacterial Strains, Plasmids, and Cloning
Plasmid vector pETM-20 was used for bacterial overexpression of recombinant PRL phosphatase wt and variants. PTEN and variants were overexpressed as GST-tagged fusion proteins using pGEX-4T-1 plasmid vector. Escherichia coli Top10 strain was used for cloning of recombinant DNA, and BL21 DE3 strain was used for protein expression. The different mutations were constructed by site-directed mutagenesis polymerase chain reaction (PCR) using the following primer pairs. PRL-3 E50Q: 5′ gcgtgtgtgtcaagtgacctat 3′ and 5′ ataggtcacttgacacacacgc 3′; PRL-3 E50A: 5′ gcgtgtgtgtgcagtgacctat 3′ and 5′ ataggtcactgcacacacacgc 3′; PRL-3 E50R: 5′ gcgtgtgtgtcgagtgacctatg 3′ and 5′ cataggtcactcgacacacacgc 3′; and PRL-3 R110A and E50R-R110A were constructed from wt PRL-3 or E50R as template with the following primer pair: 5′ cctgggcgcggctccagtcc 3′ and 5′ cataggtcactcgacacacacgc 3′. PTEN mutants were generated using the following primers: E73Q: 5′ atctttgtgctcaaagacattatga 3′ and 5′ tcataatgtctttgagcacaaagat 3′; E73A: 5′ atctttgtgctgcaagacattatga 3′ and 5′ tcataatgtcttgcagcacaaagat 3′; E73R: 5′ aatctttgtgctagaagacattatga 3′ and 5′ tcataatgtcttctagcacaaagatt 3′; and PRL-1 E50R: 5′ agagtatgtcgagcaacttacg 3′ and 5′ cgtaagttgctcgacatactct 3′. All generated constructs were confirmed by DNA sequencing. pmKate-2N plasmid vector was used for fluorescently labeled constructs for transient mammalian expression. E50R mutation was generated from wt PRL-3 using the above-mentioned primer pair. pcDNA5/FRT/TO construct for creation of the stable E50R cell line was cloned from pcDNA5/FRT/TO[3Flag-PRL-3] plasmid obtained from G. Varsano (EMBL) using the same primer pair as above. The respective stable A111S cell line was created as reported previously.14
4.3. Protein Expression and Purification
Wt PRL-3 as well as the respective protein variants were prepared as previously described,16 yielding pure protein in all cases. PTEN and variants were prepared as GST-fusion proteins as previously described,14 with the following modifications. The purification was performed in two steps. From GSTrap column purification, the proteins were eluted in 50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 10 mM glutathione, and 5 mM DTT. To purify the proteins further, ion-exchange chromatography was performed.
4.4. CD Spectroscopy
Circular dichroism experiments were performed on a Jasco J-715 spectrophotometer, using Quartz cuvette 110-QS from Hellma Analytics. The experiments were performed at 20 °C. Proteins were measured in a buffer containing 20 mM Tris–HCl, pH 7.4, 50 mM NaCl, and 1 mM DTT. Protein concentrations were 0.17 mg mL–1 for all CD spectra recordings and 0.48 mg/mL for all thermal stability measurements. Far-UV spectra were recorded between 200 and 250 nm and normalized to the respective buffer control. Data were collected in triplicates and averaged for each protein. Thermal stability was observed at 223 nm, over a temperature range of 20–95 °C. Stability data were analyzed using GraphPad Prism 6.0 software and asymmetric sigmoidal curve fitting.
4.5. In Vitro Phosphatase Activity Assays
Dephosphorylation assays using DiFMUP as phosphosubstrate were performed as previously described for wt PRL-3.16 Mutant enzymes were treated accordingly, with enzyme concentrations of 50 nM and DiFMUP concentration of 21 μM used in all cases. Inhibition with analog 3 was performed as described previously.16 Assays using substrate 3-O-methylfluorescein phosphate (OMFP) were performed in an assay buffer containing 40 mM Tris–HCl, pH 7.4, 150 mM NaCl, and 10 mM DTT. OMFP concentration was 600 μM. Enzyme concentration was 6 μM. Absorbance at 450 nm was monitored over time. Dephosphorylation of soluble phosphoinositide phosphates was performed using the purine nucleoside phosphorylase-coupled EnzChek Phosphate Assay Kit (Molecular Probes), according to the provider’s instructions. DTT concentration was kept 10 mM in all cases to ensure full cysteine reduction and therefore maximal enzyme activity. Enzymes were used at 6 μM concentration and preincubated in DTT-containing assay buffer at least 10 min prior to starting the assay. Kinetic recordings were performed at 37 °C. For single measurements comparing various PIP species, PIPs were kept at 100 μM assay concentration. Initial velocities were calculated from the slope of the linear part of the proceeding reaction. For kinetic titrations, PI(4,5)P2 substrate was used in a concentration series range between 150 and 0.4 μM. Log (PIP concentration) was plotted against reaction velocity. All data were recorded on a Tecan multiwell plate reader and analyzed with GraphPad Prism 6 software. All measurements were performed in triplicate, with two (PIP titration) or three (all other) independent experiments.
4.6. Thermofluor Analysis
The thermostability of purified PRL-3 and PRL-1 in both wt and E50R variants was analyzed with ThermoFluor technology.45 In detail, 6 μM concentration of each purified protein was incubated with SYPRO orange solution (Thermo Fischer Scientific, S-6651) in 20 mM TRIS, pH 7.4, 50 mM NaCl, and 1 mM DTT. With a StepOne Plus Real-Time PCR system (Applied Biosystems), the samples were subjected to a temperature gradient of 4–95 °C (over a period of 3 h), simultaneously measuring the emitted fluorescence signal. The melting temperature was obtained using a two-state fitting model. The reported Tm in Figure 4b corresponds to the first melting temperature.
4.7. Cell Culture and Generation of Inducible Stable Cell Lines
Cells were cultured in Dulbecco’s modified Eagle’s medium containing 4.5 g L–1 glucose, 10% (v/v) FBS, 2 mM l-glutamine, and 1% (v/v) penicillin/streptomycin solution and grown in a humidified atmosphere containing 5% CO2. HEK FlpIn T-Rex 293 cells (Invitrogen) were used to create inducible stable cell lines expressing Flag-tagged phosphatase (variant), according to the manufacturer’s instructions and as previously reported for wt PRL-3.16 Stable cell lines were grown in selection medium supplemented with 15 μg mL–1 blasticidin and 100 μg mL–1 hygromycin. Protein expression was induced by the addition of tetracycline (1 μg mL–1) 20 h prior to experimentation.
4.8. Western Blot
HEK293 stable cell lines were lysed in standard HNTG lysis buffer, and cleared lysate was normalized to total protein amount and subjected to sodium dodecylsulfate-polyacrylamide gel electrophoresis and immunoblot analysis using anti-PRL-3-specific monoclonal antibody (mouse, EMBL in-house antibody facility, clone 1E7, used 1:1000). Blocking was performed in TBS-Tween (0.1% (v/v)) containing 5% (w/v) BSA, according to standard procedures. Anti-tubulin-specific antibody (mouse, Sigma, clone T-5168, used 1:5000) was used according to the manufacturer’s instructions. Detection of respective peroxidase-coupled secondary antibody (Sigma or GE Healthcare, used 1:10 000) was carried out with ECL prime reagents from Amersham.
4.9. Confocal Microscopy
Transient transfection of mKate-labeled protein cDNA was carried out using Fugene-HD (Roche; 5 μL/μg DNA). HEK293 control cells were transfected with 0.5 μg of DNA construct 18 h before imaging on a Leica SP2 confocal microscope equipped with a 63× oil lense. Imaging was performed at room temperature. The following imaging buffer was used: 20 mM HEPES, pH 7.4, 115 mM NaCl, 1.2 mM CaCl2, 1.2 mM MgCl2, 1.2 mM K2HPO4, and 2 g L–1 glucose. Pictures were taken using Leica confocal software.
4.10. Cell Adhesion Experiments
Cell adhesion assays were performed at room temperature on a Leica DMI 4000 B widefield microscope with a 20× objective. Tetracycline-induced HEK293 stable cell lines were collected and seeded into the respective precoated or uncoated cell culture plates at a density of 1 × 105 cells mL–1 at time point zero. At the respective time points, pictures of the adhering cells were taken (three pictures per condition) and counted according to cell morphology, distinguishing between round/unattached cells and adhering cells with visible protrusions and a flat shape. Fibronectin plates were monitored 20 min after seeding. Data are presented as percent of adhering cells over total cells. Independent experiments were performed at least in triplicate.
4.11. Cell Migration Experiments
Cell migration was monitored as wound healing assay on a Zeiss Cell Observer HS widefield microscope with a 10× objective. A total of 9 × 105 cells mL–1 of the respective HEK293 stable cell lines were seeded into fibronectin-coated plates (Corning) and allowed to settle for 20 h in the presence of tetracycline. The plates were manually equipped with migration inserts for self-insertion (Ibidi) by placing the insert with sterile tweezers and gently pressing it onto the plate. Cell culture dishes and matrix component-precoated plates were used according to the information in the text. On the day of experimentation, new growth medium was added containing 7.5% (v/v) FBS (Sigma), and the inserts were gently removed with sterile tweezers. Dishes were kept at 37 °C, and pictures of the wound gap were taken at 0 and 1.5 h time points, using Zeiss ZEN software. All pictures were analyzed with Fiji software according to length (unit) difference of the remaining wound gap and presented as speed of migration (unit/h). Independent experiments were performed at least in triplicate.
4.12. Statistical Analysis
Multiple samples were compared to respective control or wt enzyme by one-way ANOVA with Dunnett correction. Two individual samples were compared by unpaired two-sided Student’s t-test with Welch correction.
Acknowledgments
M.K. is grateful to the German Science Foundation (DFG) for support within the Emmy Noether Programme (KO 4013/1-1). Bi.H. acknowledges the Austrian Academy of Sciences (ÖAW) for a DOC fellowship (DOC 23592). A.B. acknowledges EMBL and Marie Curie Actions for an EMBL Interdisciplinary Postdoc (EIPOD) fellowship. Be.H., G.D., and P.R. were funded by EMBL. The authors thank G. Varsano (EMBL) for providing plasmid pcDNA5/FRT/TO[3Flag-PRL-3], the PRL-3 and A111S HEK293 stable cell lines, and for helpful discussions. They also thank P. Lujan (EMBL) for providing mKate constructs. Plasmid pGEX-4T-1[PTEN] was a kind gift from R. Pulido (BioCruzes Health Research Institute, Barakaldo, Spain). The authors thank I. Racke and J. Rossmanith for protein purification (Protein Expression and Purification Core Facility, EMBL), R. Opitz for ThermoFluor analysis (Protein Expression and Purification Core Facility, EMBL), and A. Perez Gonzales (Flow Cytometry Core Facility, EMBL), and acknowledge the Advanced Light Microscopy Facility, EMBL, for support.
Glossary
Abbreviations
- CD
circular dichroism
- CDC25
cell division cycle, phosphatase 25
- DiFMUP
6,8-difluoro-4-methylumbelliferyl phosphate
- DMSO
dimethyl sulfoxide
- DSP
dual-specificity phosphatase
- DTT
dithiothreitol
- DUSP19
dual-specificity phosphatase, 19
- ECM
extracellular matrix
- Kapp
apparent affinity
- MD
molecular dynamics
- LMW-PTP
low-molecular-weight protein tyrosine phosphatase
- PIP
phosphatidylinositol phosphate
- PRL
phosphatase of regenerating liver
- PTEN
phosphatase and tensin homologue
- PTP
protein tyrosine phosphatase
- PTPMT1
protein tyrosine phosphatase, mitochondrial-1
- OMFP
3-O-methylfluorescein phosphate
- wt
wild-type
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b01208.
Structural changes upon introducing mutation E50R; raw data of DiFMUP dephosphorylation; further CD spectroscopy data; isothermal titration calorimetry data; redox activity data; additional migration/adhesion data (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Rios P.; Li X.; Köhn M. Molecular mechanisms of the PRL phosphatases. FEBS J. 2013, 280, 505–524. 10.1111/j.1742-4658.2012.08565.x. [DOI] [PubMed] [Google Scholar]
- Campbell A. M.; Zhang Z.-Y. Phosphatase of regenerating liver: A novel target for cancer therapy. Expert Opin. Ther. Targets 2014, 18, 555–569. 10.1517/14728222.2014.892926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato Y.; Yamazaki D.; Mizukami S.; Du L.; Kikuchi K.; Miki H. Membrane protein CNNM4-dependent Mg2+ efflux suppresses tumor progression. J. Clin. Invest. 2014, 124, 5398–410. 10.1172/JCI76614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulerez I.; Funato Y.; Wu H.; Yang M.; Kozlov G.; Miki H.; Gehring K. Phosphocysteine in the PRL-CNNM pathway mediates magnesium homeostasis. EMBO Rep. 2016, 17, 1890–1900. 10.15252/embr.201643393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.-Y. Protein-tyrosine phosphatases: Biological function, structural characteristics, and mechanism of catalysis. Crit. Rev. Biochem. Mol. Biol. 1998, 33, 1–52. 10.1080/10409239891204161. [DOI] [PubMed] [Google Scholar]
- Barford D.; Das A. K.; Egloff M. P. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu. Rev. Biophys. Biomol. Struct. 1998, 27, 133–164. 10.1146/annurev.biophys.27.1.133. [DOI] [PubMed] [Google Scholar]
- Xiao J.; Engel J. L.; Zhang J.; Chen M. J.; Manning G.; Dixon J. E. Structural and functional analysis of PTPMT1, a phosphatase required for cardiolipin synthesis. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 11860–11865. 10.1073/pnas.1109290108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoret L. M.; Rader S. D.; Fletterick R. J.; Cohen F. E. Hydrogen bonds involving sulfur atoms in proteins. Proteins: Struct., Funct., Genet. 1991, 9, 99–107. 10.1002/prot.340090204. [DOI] [PubMed] [Google Scholar]
- Kozlov G.; Cheng J.; Ziomek E.; Banville D.; Gehring K.; Ekiel I. Structural insights into molecular function of the metastasis-associated phosphatase PRL-3. J. Biol. Chem. 2004, 279, 11882–11889. 10.1074/jbc.M312905200. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Kozlov G.; Li X.; Wu H.; Gulerez I.; Gehring K. PRL-3 phosphatase active site is required for binding the putative magnesium transporter CNNM3. Sci. Rep. 2017, 7, 48 10.1038/s41598-017-00147-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchetot C.; Chagnon M.; Dubé N.; Hallé M.; Tremblay M. L. Substrate-tracking techniques in the identification of cellular PTP targets. Methods 2005, 35, 44–53. 10.1016/j.ymeth.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Dunphy W. G.; Kumagai A. The cdc25 protein contains an intrinsic phosphatase activity. Cell 1991, 67, 189–196. 10.1016/0092-8674(91)90582-J. [DOI] [PubMed] [Google Scholar]
- Nakamura K.; Tanoue K.; Satoh T.; Takekawa M.; Watanabe M.; Shima H.; Kikuchi K. A novel low-molecular mass dual-specificity phosphatase, LDP-2, with a naturally occurring substitution that affects substrate specificity. J. Biochem. 2002, 132, 463–470. 10.1093/oxfordjournals.jbchem.a003244. [DOI] [PubMed] [Google Scholar]
- McParland V.; Varsano G.; Li X.; Thornton J.; Baby J.; Aravind A.; Meyer C.; Pavic K.; Rios P.; Köhn M. The metastasis-promoting phosphatase PRL-3 shows activity toward phosphoinositides. Biochemistry 2011, 50, 7579–7590. 10.1021/bi201095z. [DOI] [PubMed] [Google Scholar]
- Maehama T.; Dixon J. E. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1998, 273, 13375–13378. 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Hoeger B.; Diether M.; Ballester P. J.; Köhn M. Biochemical evaluation of virtual screening methods reveals a cell-active inhibitor of the cancer-promoting phosphatases of regenerating liver. Eur. J. Med. Chem. 2014, 88, 89–100. 10.1016/j.ejmech.2014.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. P.; Wang W. Q.; Yang H.; Liu S.; Liang F.; Fedorov A. A.; Almo S. C.; Zhang Z.-Y. Structure and biochemical properties of PRL-1, a phosphatase implicated in cell growth, differentiation, and tumor invasion. Biochemistry 2005, 44, 12009–12021. 10.1021/bi0509191. [DOI] [PubMed] [Google Scholar]
- Bessette D. C.; Wong P. C.; Pallen C. J. PRL-3: a metastasis-associated phosphatase in search of a function. Cells Tissues Organs 2007, 185, 232–236. 10.1159/000101324. [DOI] [PubMed] [Google Scholar]
- Hsu F.; Mao Y. The structure of phosphoinositide phosphatases: Insights into substrate specificity and catalysis. Biochim. Biophys. Acta 2015, 1851, 698–710. 10.1016/j.bbalip.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong K.-W.; Kang D.-I.; Lee E.; Shin A.; Jin B.; Park Y.-G.; Lee C.-K.; Kim E.-H.; Jeon Y. H.; Kim E. E.; Kim Y. Structure and backbone dynamics of vanadate-bound PRL-3: Comparison of 15N nuclear magnetic resonance relaxation profiles of free and vanadate-bound PRL-3. Biochemistry 2014, 53, 4814–4825. 10.1021/bi5003844. [DOI] [PubMed] [Google Scholar]
- Roberts S. J.; Stewart A. J.; Sadler P. J.; Farquharson C. Human PHOSPHO1 exhibits high specific phosphoethanolamine and phosphocholine phosphatase activities. Biochem. J. 2004, 382, 59–65. 10.1042/BJ20040511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins W. M. Non-Michaelis-Menten kinetics in cytochrome P450–catalyzed reactions. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 291–310. 10.1146/annurev.pharmtox.45.120403.100004. [DOI] [PubMed] [Google Scholar]
- Sun J. P.; Luo Y.; Yu X.; Wang W. Q.; Zhou B.; Liang F.; Zhang Z.–Y. Phosphatase activity, trimerization, and the C-terminal polybasic region are all required for PRL1-mediated cell growth and migration. J. Biol. Chem. 2007, 282, 29043–29051. 10.1074/jbc.M703537200. [DOI] [PubMed] [Google Scholar]
- Tonks N. K. Redox redux: Revisiting PTPs and the control of cell signaling. Cell 2005, 121, 667–670. 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Lee S. R.; Yang K. S.; Kwon J.; Lee C.; Jeong W.; Rhee S. G. Reversible inactivation of the tumor suppressor PTEN by H2O2. J. Biol. Chem. 2002, 277, 20336–20342. 10.1074/jbc.M111899200. [DOI] [PubMed] [Google Scholar]
- Savitsky P. A.; Finkel T. Redox regulation of Cdc25C. J. Biol. Chem. 2002, 277, 20535–20540. 10.1074/jbc.M201589200. [DOI] [PubMed] [Google Scholar]
- Chiarugi P. The redox regulation of LMW-PTP during cell proliferation or growth inhibition. IUMBM Life 2001, 52, 55–59. 10.1080/15216540252774775. [DOI] [PubMed] [Google Scholar]
- Skinner A. L.; Vartia A. A.; Williams T. D.; Laurence J. S. Enzyme activity of phosphatase of regenerating liver is controlled by the redox environment and its C-terminal residues. Biochemistry 2009, 48, 4262–4272. 10.1021/bi900241k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato Y.; Miki H. Reversible oxidation of PRL family protein-tyrosine phosphatases. Methods 2014, 65, 184–189. 10.1016/j.ymeth.2013.06.032. [DOI] [PubMed] [Google Scholar]
- Yu L.; Kelly U.; Ebright J. N.; Malek G.; Saloupis P.; Rickman D. W.; McKay B. S.; Arshavsky V. Y.; Bowes Rickman C. Oxidative stress-induced expression and modulation of Phosphatase of Regenerating Liver-1 (PRL-1) in mammalian retina. Biochim. Biophys. Acta 2007, 1773, 1473–1482. 10.1016/j.bbamcr.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner J. J.; Parsons Z. D.; Cummings A. H.; Zhou H.; Gates K. S. Redox regulation of protein tyrosine phosphatases: structural and chemical aspects. Antioxid. Redox Signaling 2011, 15, 77–97. 10.1089/ars.2010.3611. [DOI] [PubMed] [Google Scholar]
- Kim K. A.; Song J. S.; Jee J.; Sheen M. R.; Lee C.; Lee T. G.; Ro S.; Cho J. M.; Lee W.; Yamazaki T.; Jeon Y. H.; Cheong C. Structure of human PRL-3, the phosphatase associated with cancer metastasis. FEBS Lett. 2004, 565, 181–187. 10.1016/j.febslet.2004.03.062. [DOI] [PubMed] [Google Scholar]
- Huttenlocher A.; Horwitz A. R. Integrins in cell migration. Cold Spring Harb. Perspect. Biol. 2011, 3, a005074 10.1101/cshperspect.a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P.; Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat. Rev. Cancer 2003, 3, 362–374. 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- Hynes R. O. Integrins: Bidirectional, allosteric signaling machines. Cell 2002, 110, 673–687. 10.1016/S0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Nerenberg P. S.; Salsas-Escat R.; Stultz C. M. Collagen - a necessary accomplice in the metastatic process. Cancer Genomics Proteomics 2007, 4, 319–328. [PubMed] [Google Scholar]
- Hynes R. O. Integrins: Bidirectional, allosteric signaling machines. Cell 2002, 110, 673–687. 10.1016/S0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Gopalakrishna P.; Chaubey S. K.; Manogaran P. S.; Pande G. Modulation of alpha5beta1 integrin functions by the phospholipid and cholesterol contents of cell membranes. J. Cell. Biochem. 2000, 77, 517–528. . [DOI] [PubMed] [Google Scholar]
- Gourley C.; Paige A. J.; Taylor K. J.; Ward C.; Kuske B.; Zhang J.; Sun M.; Janczar S.; Harrison D. J.; Muir M.; Smyth J. F.; Gabra H. WWOX gene expression abolishes ovarian cancer tumorigenicity in vivo and decreases attachment to fibronectin via integrin alpha3. Cancer Res. 2009, 69, 4835–4842. 10.1158/0008-5472.CAN-08-2974. [DOI] [PubMed] [Google Scholar]
- Reyes-Reyes M. E.; George M. D.; Roberts J. D.; Akiyama S. K. P-selectin activates integrin-mediated colon carcinoma cell adhesion to fibronectin. Exp. Cell. Res. 2006, 312, 4056–4069. 10.1016/j.yexcr.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=PTP4A3#dist (accessed May 29, 2017).
- Yaakov G.; Bell M.; Hohmann S.; Engelberg D. Combination of two activating mutations in one HOG1 gene forms hyperactive enzymes that induce growth arrest. Mol. Cell. Biol. 2003, 23, 4826–4840. 10.1128/MCB.23.14.4826-4840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milano S. K.; Kwon W.; Pereira R.; Antonyak M. A.; Cerione R. A. Characterization of a novel activated Ran GTPase mutant and its ability to induce cellular transformation. J. Biol. Chem. 2012, 287, 24955–24966. 10.1074/jbc.M111.306514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinmayeh H.; Zimmerman D. M.; Cantrell B. E.; Schober D. A.; Bruns R. F.; Gackenheimer S. L.; Ornstein P. L.; Hipskind P. A.; Britton T. C.; Gehlert D. R. Structure-activity relationship of a series of diaminoalkyl substituted benzimidazole as neuropeptide Y Y1 receptor antagonists. Bioorg. Med. Chem. Lett. 1999, 9, 647–652. 10.1016/S0960-894X(99)00082-7. [DOI] [PubMed] [Google Scholar]
- Boivin S.; Kozak S.; Meijers R. Optimization of protein purification and characterization using ThermoFluor screens. Protein Expression Purif. 2013, 91, 192–206. 10.1016/j.pep.2013.08.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.