Abstract
SK3 channels are abnormaly expressed in metastatic cells, and Ohmline (OHM), an ether lipid, has been shown to reduce the activity of SK3 channels and the migration capacity of cancer cells. OHM incorporation into the plasma membrane is proposed to dissociate the protein complex formed between SK3 and Orai1, a potassium and a calcium channel, respectively, and would lead to a modification in the lipid environment of both the proteins. Here, we report the synthesis of deuterated OHM that affords the determination, through solid-state NMR, of its entire partitioning into membranes mimicking the SK3 environment. Use of deuterated lipids affords the demonstration of an OHM-induced membrane disordering, which is dose-dependent and increases with increasing amounts of cholesterol (CHOL). Molecular dynamics simulations comfort the disordering action and show that OHM interacts with the carbonyl and phosphate groups of stearoylphosphatidylcholine and sphingomyelin and to a minor extent with CHOL. OHM is thus proposed to remove the CHOL OH moieties away from their main binding sites, forcing a new rearrangement with other lipid groups. Such an interaction takes its origin at the lipid–water interface, but it propagates toward the entire lipid molecules and leads to a cooperative destabilization of the lipid acyl chains, that is, membrane disordering. The consequences of this reorganization of the lipid phases are discussed in the context of the OHM-induced inhibition of SK3 channels.
1. Introduction
The dysregulation of ion channel activity/expression emerged as a common feature of cancer cells. These dysregulations constitute new biomarkers1 and were deeply studied to better understand the physiology of cancer cells. Interestingly, the modulation of abnormaly and overexpressed ion channels opens new perspectives in cancer chemotherapy as illustrated, for instance, by antiproliferative actions triggering apoptosis2 or for the reduction of cancer cell spreading.3 The development of new modulators of ion channels based on an amphiphilic molecular structure4 requires knowledge of the mechanisms involved in channel gating and also their direct interaction with the membrane environment that is a very complex lipid mixture.5
Different mechanisms have been proposed to explain the regulation of ion channels by lipids. (i) Global change in the physical properties of the membrane. Analysis of the crystal structure of the KvAP channel revealed that a lipid bilayer is required to maintain the correct relative orientations of channel domains6 and the activity of this voltage-dependent potassium channel also depends on the negatively charged lipids.7 TRAAK and TREK1 mechano-gated potassium channels are mechanically gated by the lipid bilayer in the absence of any other cellular components.8 Moreover, inverted-conical shape lipids tend to favor a convex deformation of the plasma membrane which leads to the opening of the TREK-1/TRAAK channels.9 It is well-accepted that changes in cholesterol (CHOL) content in the plasma membrane result in a modification of membrane fluidity and also bilayer thickness. This bilayer thickness can indeed regulate the activity of the membrane protein10 as exemplified by a reduction of gramicidin channel activity11−13 induced by an increase of membrane thickness triggered by higher incorporation of CHOL. (ii) Specific lipid–protein interactions as exemplified with the interaction between CHOL and the KirBac1.1 channel.14 (iii) Interactions between channels and proteins localized in nanodomains of the plasma membrane. Caveolin which is present in CHOL- and sphingolipid-rich nanodomains was found to regulate potassium channels.15,16 This modulation occurred either through direct protein–lipid interactions17,18 or by influencing the physical characteristics of the bilayer as described above.
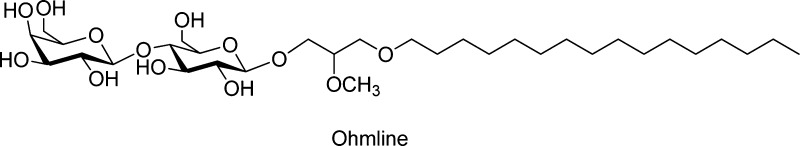
Ohmline, named hereafter OHM, 1-O-hexadecyl-2-O-methyl-rac-glycero-3-lactose (Figure 1), and belonging to a family of synthetic glyco–glycero ether lipids, has been demonstrated to specifically reduce the activity of SK3 potassium channels via an interaction that remains elusive. The SK3 channel is a potassium channel belonging to a small-conductance calcium-activated potassium channel family (with SK1 and SK2 channels). When expressed in a cancer cell, the SK3 channel increases twice the capacity of cells to migrate and invades a matrix that is similar to the physiological extracellular matrix.19,20 Using mouse models of metastatic breast cancers, the SK3 channel was found to promote the development of metastases, mainly the development of bone metastases—this observation has a direct link with the activation of SK3 by calcium and the high calcium concentrations found in the bone environment.21 OHM at 10 nM concentration was found to reduce the SK3 channel activity and the migration of SK3-expressing cancer cells with 50% efficiency in the bone metastasis development.21
Figure 1.
Chemical structure of OHM (1-O-hexadecyl-2-O-methyl-rac-glycero-3-lactose).
OHM is the first specific inhibitor of SK3 channels: it inhibits the SK3 channel and, in a minor extent, the SK1 channel, and it does not significantly affect the SK2 channel.22 Altogether, these results provide evidence that amphiphilic compounds such as OHM can selectively modulate the activity of SK3 channels likely due to the modification of the physicochemical properties of the plasma membrane and of the lipid environment even though a direct interaction of OHM with SK3 channel cannot be excluded.
Despite the outstanding action of OHM on the SK3 channel activity and its potent use to prevent the formation of metastases, much less is known about its interaction with a membrane and even less is known about the mechanism of action at the molecular level. In this study, we aimed to assess the behavior of OHM after its incorporation in a model membrane by answering two basic questions: Does an equilibrium exist between a location inside and outside the plasma membrane? Are there any specific interactions with the lipids present in the membrane that could explain some modification of the membrane biophysical properties?
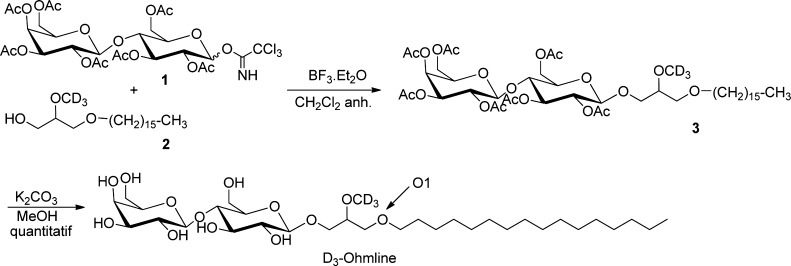
To answer the first question, we synthesized deuterated OHM at sn2 position (Figure 2) and used 2H NMR, which allows determining whether OHM, when placed in contact with a membrane model, features an isotropic signal (a water environment) or an anisotropic signal (a lipid environment). To answer the second question, we carried out molecular dynamics (MD) simulations on a model mimicking the SK3 lipid environment and assessed membrane dynamic via 2H NMR experiments that made use of deuterated POPC as molecular probe.
Figure 2.
Last two steps for the synthesis of C2H3O-OHM (2H3-OHM).
2. Results
2.1. NMR
2.1.1. OHM Partitioning into the Membrane
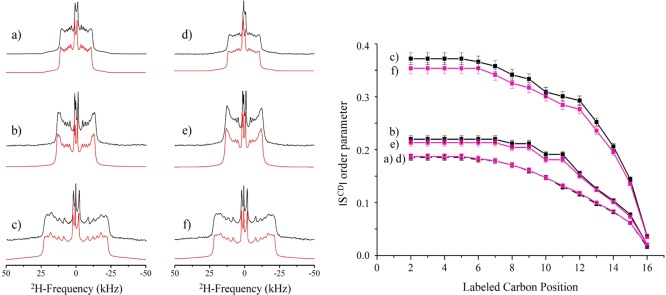
Wide-line 2H NMR of methyl-deuterated OHM was performed in water and in POPC/SM/CHOL and POPC membranes, Figure 3. The spectrum of OHM in solution (7.6 mM) (Figure 3a, black trace) is a small so-called “powder” pattern with two isotropic lines superimposed. One is assigned to residual deuterated water (centered at 4.7 ppm), and the other one is assigned to OHM in solution (at 2.3 ppm). Solid-state 2H NMR is indeed capable of distinguishing molecules in solution, in small isotropic micelles, or in larger entities. Fast molecular tumbling as in solution or for nanometric micelles leads to the so-called isotropic lines; the solid-state quadrupolar interaction is averaged to zero by fast (picosecond) Brownian motions.23 For larger entities, micrometric liposomes or large vesicles, the slow motions (microseconds to nanoseconds), mainly anisotropic, do not average the interaction to zero. One obtains residual “powder patterns” reflecting the onset of such slow motions. An intermediate situation may be obtained for edifices of a 50–200 nm hydrodynamic radius, such as wormlike micelles, which present “exchange” line shapes23 as observed for the very small residual powder pattern in Figure 3a. The experimental spectrum could be simulated (red traces and inset) using a quadrupolar splitting of 2.6 kHz and two isotropic lines of 29 and 13% contributing to water and OHM, respectively, in solution. The presence of such a powder pattern (58%) indicated that deuterated OHM is under the form of large micelles (possibly wormlike). When applied to liposomes, the spectrum of 2H OHM is composed of a central isotropic line at 4.7 ppm (deuterated water) and a well-defined axially symmetric powder pattern representative of entire partitioning of OHM into the membrane (Figure 3b); signals of OHM in micelles or in solution are no longer detected (less than 1% experimental error). Such patterns are characteristic of molecules embedded in liquid-disordered or liquid-ordered (lo) environments that maintain the axial symmetry of the motional processes occurring under such conditions. Simulations (red traces) report quadrupolar splittings of 9.2 and 7.4 kHz for OHM in POPC/SM/CHOL and POPC liposomes, respectively. This indicates that OHM sits in a more ordered environment when embedded in a CHOL-containing membrane (Figure 3b).
Figure 3.
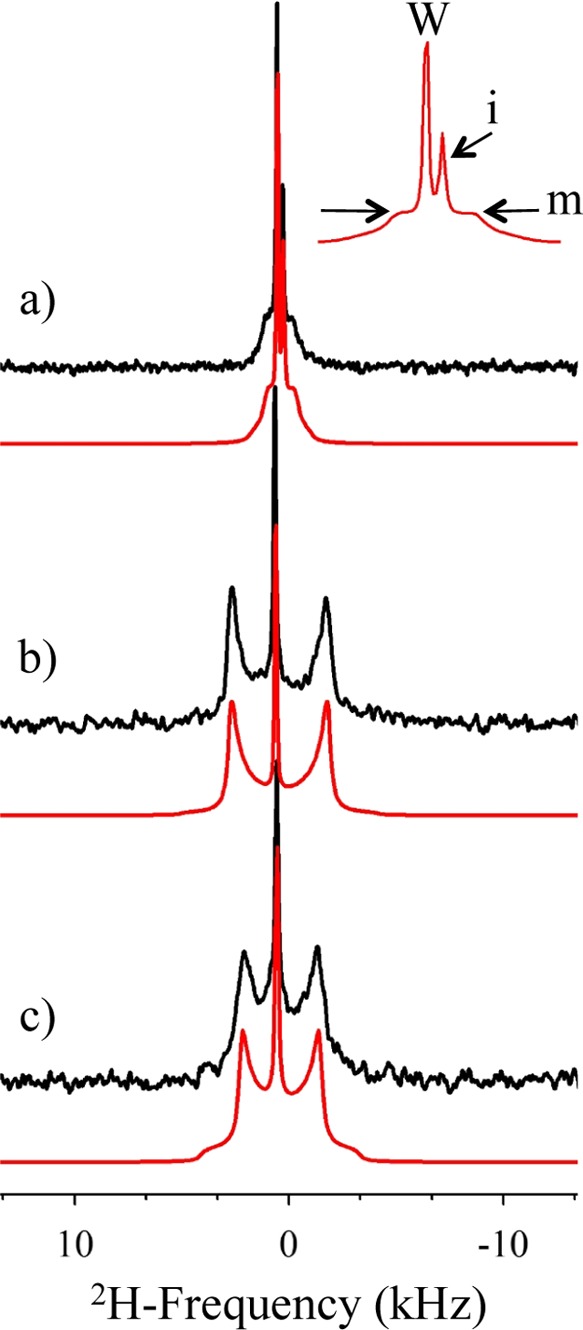
2H NMR spectra of deuterated OHM (a) in water (7.6 mM), (b) in POPC/SM/CHOL (10/60/30) at Ri = 15, and (c) in POPC at Ri = 15. Black traces stand for the experimental spectra, and red traces stand for the simulated ones. The top inset is the expansion of the simulation of (a), with W, i, and m representing water, isotropic, and micelle traces of OHM.
2.1.2. OHM Disorders Membranes
The effect of OHM on the dynamics of several membrane systems was monitored using POPC perdeuterated on the palmitic sn-1 chain. POPC was used either as a pure system or in the presence of major amounts of sphingomyelin (SGML), that is, POPC/SM/CHOL (10/85/5, mol %) or with a high concentration of CHOL, that is, POPC/SM/CHOL (10/60/30, mol %). The temperature was varied from 25 to 45 °C on all systems. Spectra are recorded at 45 °C in Figure 4 (left) and in the presence of OHM at lipid-to-OHM ratio, Ri, of 30 (right). All spectra display the axially symmetric shape, characteristic of the manifestation of several axially symmetric motional processes (bond or molecule rotation, anisotropic reorientations, etc.) known to be present in rather dynamic membranes.24,25 The pure POPC system is the most dynamic membrane (narrow spectrum). The presence of high amounts of SGML and 5 mol % CHOL widens the spectra (middle traces), which indicates a reduction in membrane dynamics. With higher amounts of CHOL (POPC/SM/CHOL, 10/60/30, mol %), the spectra become very wide, indicating further reduction of dynamics. In the presence of OHM, the spectral shapes are unchanged but become a little narrower. This is better perceived when performing spectral simulations (red traces below the experimental spectra) and reporting the SCD order parameter measured at each labeled carbon position, k, on the palmitic chain of POPC (Figure 4, right). All profiles display the well-known behavior: “plateau” of elevated order parameters near the very rigid glycerol backbone (positions 2–8) and monotonic decrease on moving toward the chain end at position 16, that is, at the bilayer center.24 The presence of OHM is almost undetectable on POPC, slightly decreases the whole profile of order parameters for POPC/SM/CHOL (10/85/5), and has more disordering action on that for POPC/SM/CHOL (10/60/30). By summing up all order parameters along the chain, the effect is of 1 ± 1% for POPC, 3 ± 1% for POPC/SM/CHOL (10/85/5), and 5 ± 1% for POPC/SM/CHOL (10/60/30). The disordering action is concentration-dependent: upon increasing the amount of OHM (Ri = 15, not shown) on the POPC/SM/CHOL (10/60/30) membrane, the whole disordering becomes 8%. Of interest is the observation that disordering is observed for temperatures in the range 35–45 °C; no effect on membrane dynamics is detected at ambient temperature. It is worth mentioning that membrane disordering is synonymous of membrane thinning. OHM thus leads to a reduction of membrane thickness; the more OHM, the greater the effect and the more CHOL in the membrane, the greater the effect.
Figure 4.
Left panel: solid-state 2H NMR spectra in the absence (a–c) and the presence (d–f) of OHM (Ri = 30), at 45 °C: (a) 2H31-POPC, (b) 2H31-POPC/SM/CHOL (10/85/5, mol %), and (c) 2H31-POPC/SM/CHOL (10/60/30, mol %). Black upper traces represent the experimental spectra, and red lower traces represent the simulated spectra. Right panel: order parameter profile |SkCD| as a function of kth labeled carbon position, of the palmitic chain of POPC in the absence (black symbols, a–c) and the presence (purple symbols, d–f) of OHM at Ri = 30. |Sk| is obtained from spectral simulations, with arbitrary decremental assignment for positions 2–10. The error bars represent the accuracy.
2.2. MD Simulation
2.2.1. OHM Position in the Bilayer and the 2D Order Parameter
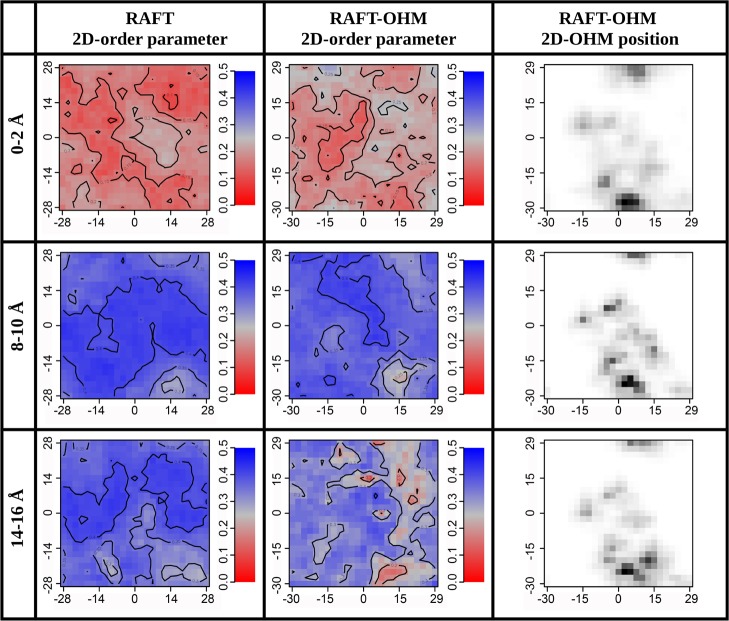
From MD simulations, information on membrane dynamics and the location of OHM in the bilayer can be obtained. OHM is located inside the bilayer, with its sugar groups pointing at the water interface and the hydrocarbon chain well-inserted in the bilayer aliphatic core. This is well-shown in the density profile for the final arrangement of the OHM molecules along the z axis of the bilayer (Figure S12).
Figure 5 shows the 2D lipid tail order parameters calculated at different distances from the center of the membrane (slices at different depths) and their correlation with the OHM molecule position. In the control bilayer (the first column), there is a clear difference in the surface order parameter between the interface at 14–16 Å and the hydrocarbon core region at 8–10 Å and the bilayer center at 0–2 Å. The center is clearly more disordered than the two other bilayer regions, in complete accordance with the above-mentioned NMR data. The bilayer with OHM (the second column) shows, for the regions at 8–10 and 14–16 Å, a decrease in the surface order parameter for regions that match with the OHM positions that are shown in the third column. Of interest is the fact that the region around 8–10 Å appears to be the most ordered, as was also noted in the NMR experimental order parameters (positions 2–10 plateau along the fatty acyl chains). SCD acyl chain order parameters can also be obtained from MD calculations: we have obtained results in the same line of the NMR experiments, and the presence of ≈10% of OHM also induces a decrease in order parameters of about 10% (Figure S13).
Figure 5.
Two-dimensional order parameter in the lipid model system. Total order parameter of the upper leaflet of the bilayer is represented without and with OHM. The position of the OHM molecules for the same leaflet is indicated in 2D-OHM position. The numbers in angstrom on the left indicate the distances from the center of the membrane at which the 2D order parameter was evaluated. The bilayer center is set at 0 Å.
2.2.2. Consequences of the OHM Partitioning: Interleaflet Mixing
The order parameter decrease is usually associated with an increase in the bilayer interdigitation and its related diminution in the thickness. The interleaflet distributions of the methyl group ends of the acyl chains are measures of the mixing between the opposing monolayers.26 The density profile of OHM (Figure S12, bottom row) shows the interdigitation of the acyl chains.
Distances between the peaks, which characterize the distributions of the terminal methyl groups in both leaflets, and the percentage of the terminal methyl groups from each monolayer shared by the opposite (overlap) are shown in Table 1.
Table 1. Interleaflet Mixinga.
| CH3 source | distance between peaks (Å) | overlap (%) | |
|---|---|---|---|
| RAFT | PC | 2.3 ± 0.4 | ∼59.6 |
| SGML | 5.3 ± 0.5 | ∼25.4 | |
| CHOL | 4.1 ± 0.4 | ∼30.4 | |
| RAFT + OHM | PC | 3.2 ± 0.7 | ∼60.7 |
| SGML | 5.2 ± 0.7 | ∼41.6 | |
| CHOL | 3.2 ± 0.6 | ∼55.8 |
The interleaflet mixing was calculated as the overlap area between the atomic density profiles of the terminal CH3 groups coming from both monolayers and by the distance between the peaks of such densities.
The overlap in the distributions of the CH3 terminal groups increases after OHM addition for all three lipids [1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), SGML, and CHOL]. For DSPC, a slight increase in the overlap is observed (1.1%); for SGML and CHOL molecules, instead the increase in the overlap is more significant (16.2 and 25.4%, respectively). The analysis confirms that the OHM molecules induce a change at the level of the interlayer space with small consequences in terms of average area per lipid and thickness, +0.2 Å2 and −1.6 Å, respectively.
2.2.3. Molecular Interactions at the Interface That Stabilize OHM
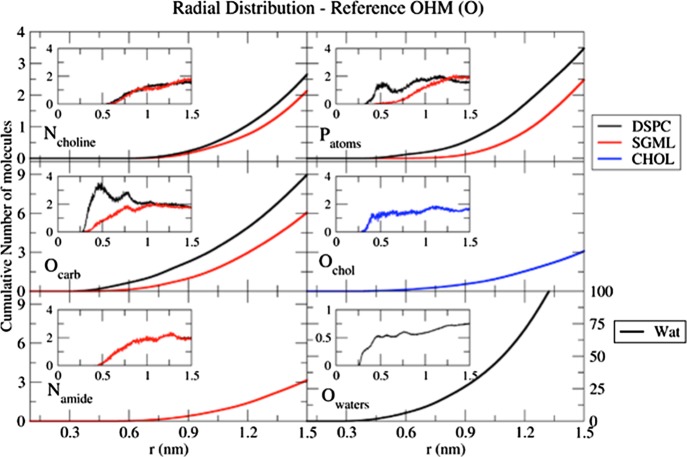
To characterize the intermolecular interactions that stabilize OHM at the interface level, the cumulative number of molecules located around the different atomic moieties of the lipids is plotted for each lipid group of the bilayer as well as the water molecules (Figure 6). The sn3 oxygen atom of the OHM glycerol group that links the lipid chain (noted O1, Figure 1) is located at the water–lipid interface, and we have chosen it as a reference (0 Å). In each inset, the radial distribution functions (RDFs) of the lipid groups considered are drawn.
Figure 6.
Cumulative number of molecules and RDFs of OHM oxygen glycerol associated with respect to the main lipid groups located at the interface. The RDFs for the different membrane groups around one OHM oxygen atom, between the tail and the sugar head [oxygen O1 of the glycerol moiety (Figure 1)], are shown in the inset. The colors indicate the lipid considered, and in each graph, the moiety involved is indicated. The origin of the graphs at 0 Å is not shown for clarity.
From the first rising region of the cumulative number and RDF curves, the first groups that appear nearest to the OHM oxygen atom O1 (at around 0.3 nm) are the carbonyl groups of DSPC and SGML; the CHOL hydroxyl and water molecules; and then phosphates, amides, and cholines. Nevertheless, only the carbonyl and phosphorus atoms from DSPC seem to be ordered at around 0.5 nm from the location of the OHM oxygen O1 because only both of these atoms show a well-defined peak in the RDF curve. Table 2 allows the comparison of the number of atoms that are located, on average, at a specific distance of 0.8 nm from the OHM oxygen O1.
Table 2. Summary of the Cumulative Number of Different Lipid Species and Their Corresponding Atomic Groups at 0.8 nm Distance for OHM Oxygen O1.
| DSPC | SGML | CHOL | |
|---|---|---|---|
| O carbonyl | 1.63 | 0.63 | |
| P atoms | 0.3 | 0.04 | |
| N choline | 0.09 | 0.05 | |
| N amide | 0.2 | ||
| O chol | 0.4 | ||
| waters around oxygen OHM | 18 | ||
The OHM molecules interact mostly with the carbonyl oxygen of DSPC molecules, followed by the carbonyl oxygen of SGML and finally the CHOL oxygen. The strong solvation of the OHM molecules, principally at the level of the sugar head, suggests that the water molecules could be playing a role in the stabilization of the head group through hydrogen interactions with other lipid head groups.
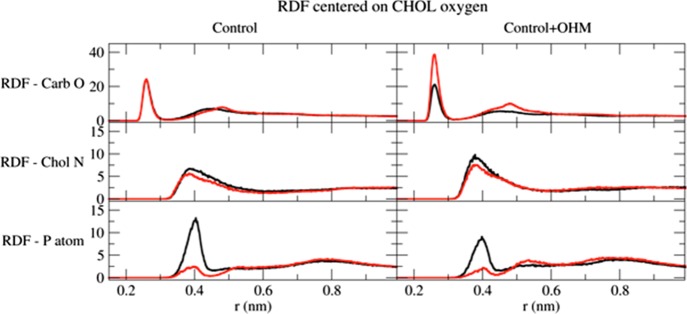
We also calculated the RDFs of the different lipid moieties around the CHOL oxygen (Figure 7). The RDFs show that CHOL is stabilized by the DSPC as well as SGML carbonyls (upper row). The presence of a sharp peak centered at 0.27 nm indicates that a strong H bond is formed between the hydroxyl and the carbonyl groups.
Figure 7.
RDFs of CHOL hydroxyl. The RDFs for the different membrane groups (black for DSPC and red for SGML) around the CHOL oxygen are shown.
On the basis of the peak heights, in the control bilayer, there are equal possibilities to find a carbonyl oxygen atom from DSPC or SGML at around 0.27 nm. However, in the presence of OHM, the possibility increases (around 50%) for the SGML carbonyl oxygen. For the interactions of the CHOL oxygen with the choline nitrogen (middle row), the OHM molecule has little effect and the possibilities to find it at around 0.4 nm are maintained after the addition of OHM. Finally, for the phosphorus atom, the possibility to find it at 0.4 nm from the CHOL oxygen is higher for the DSPC molecule than that for SGML (lower left row). Because of the addition of OHM, the CHOL hydroxyl again misses some of these interactions.
3. Discussion
Over the last 10 years, the role of ion channels in cancer development and cancer spreading was deeply assessed,4,27 which paved the way for the design of efficient inhibitors. In this direction, the implication of SK3 channels in cancer cell migration and metastasis development was already established.19 Edelfosine28 and OHM21 were the first amphiphilic compounds that exhibited strong SK3 inhibition. Although more works were reported to conceive heterocyclic compounds as inhibitors of SK3 channels,29 the fact that amphiphilic compounds can also act as efficient modulators offers new possibilities of assessment. These original results invited to study the mechanism of action of OHM that was first addressed by designing new analogues (structure/activity relationship). Accordingly, analogues of OHM featuring the incorporation of a phosphate group or other disaccharide unit30 gave new efficient inhibitors, but OHM remains the most promising compound. Accordingly, further insights into the mechanism of action of amphiphilic compounds on ion channels require the assessment of their interactions with model membranes, which is the aim of our study.
From the NMR experiments, two main pieces of information can be derived: OHM is partitioned entirely into the membrane and disorders the lipid bilayer. The first information is of interest because it implies that there is a greater affinity for the lipid bilayer compared to the self-association of OHM as a micelle in water. The OHM critical micelle concentration value of 12 μM31 is hundreds of times lower than the values used in the experiments. The composition of the membrane appears not to be of importance; pure phospholipids or mixtures with sphingolipids and CHOL still lead to 100% of OHM entering into liposomes both at Ri = 15 and 30. We have, however, to be careful because no accurate determination of binding constants has been performed. The second important observation is the disordering effect observed on membranes containing large amounts of CHOL. The system POPC/SM/CHOL (10/60/30) is indeed in a lo phase where molecular motions are clearly reduced by the condensing effect of CHOL. The action of OHM appears to limit the action of CHOL by reducing the condensing effect.
From MD (Figures 5 and S13), we apprehend that the disordering effect can be attributed to distinct mechanisms, one involving a direct interaction of the OHM molecules with both species (DSPC and SGML) or another involving a CHOL-mediated interaction, affecting the binding of those lipids with the CHOL molecules because it was already mentioned that CHOL molecules induce an ordering effect on the lipid tails. A combination of both mechanisms cannot be excluded.
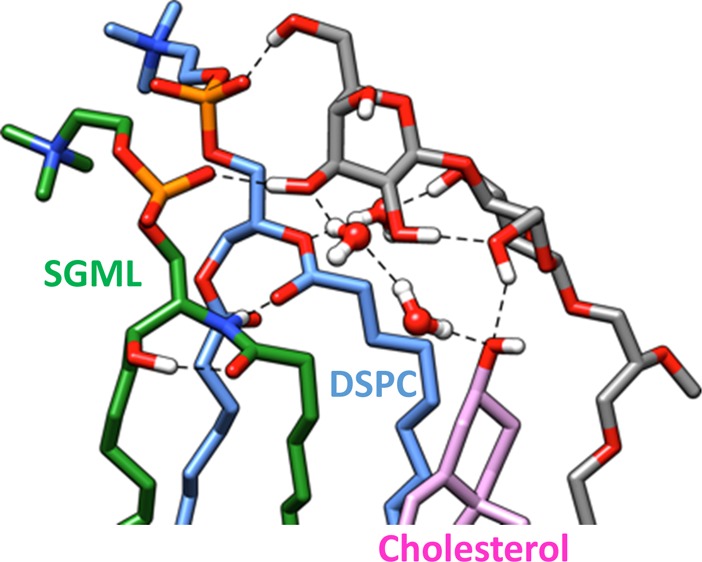
For edelfosine and miltefosine, a tendency was proposed to be associated with CHOL in certain lipid domains.4,32 Other ether lipids have been demonstrated to force the CHOL OH to interact with the phosphate lipid group due to the absence of carbonyl, to be stabilized.33 The consequence of this new stabilization promoted a tilt in the CHOL molecule that the same author has mentioned as a modulator of membrane-active proteins. In our model, the latter would seem to be the case (Figures 6 and 7).
CHOL controls the activity of a wide range of membrane proteins through specific interactions.34 Indeed, protein sequence analysis revealed that several sites of SK3 are CHOL-binding candidates (not shown). The OHM-induced modifications of CHOL availability in the bilayer could modify its interaction with regions that recognize CHOL, leading to an incorrect or inclusive misfolding of the SK3 structure with loss of activity. This mechanism of decrease/loss of activity (inactivation is a particular state for a channel that occurs after activation) does, however, not exclude the fact that OHM provokes a general destabilization of the acyl chains that would also affect the activity of the channel.
So far, it was not demonstrated that SK3 is a lipid-dependent channel, but the channel is activated by cell swelling and inhibited by shrinkage,35 which suggests that the channel activity is sensitive to the biophysical characteristics of the plasma membrane. A change in the global organization of the lipid environment could explain the modulation of the SK3 channel activity, but a very specific lipid interaction should not be excluded.
The results in Table 1 show that the presence of OHM induces an increase in the interleaflet mixing. It should be noticed that the length of the hydrophobic alkyl chain was described as a key characteristic for the activity of alkylphospholipid derivatives that have been used as precursors to design the OHM molecules. As shown previously for glycerol-derived phospholipid ether analogues, the decrease in the length from 12 carbons to 7 resulted in little or no anticancer activity, whereas increasing the length had the opposite effect.36 For the OHM case, this behavior is in complete agreement with our preliminary experimental data (unpublished results), meaning that as a result of the OHM internalization or as a driving force to pull the hydrophilic moiety at the interface, the interleaflet mixing (the mixing between methyl group ends of the acyl chains from opposite monolayers) should be considered in the mechanism of action of this kind of compound.
4. Conclusions
This work brings new insights into the mechanism of action of OHM that, as previously shown, can inhibit (in vitro and in vivo) the activity of SK3 ion channels. We demonstrate, for the first time, that deuterated OHM (2H3-OHM) is fully incorporated in a model of membrane as revealed by 2H NMR studies. Additional 2H NMR studies that make use of model membranes that incorporate perdeuterated POPC indicate that the addition of OHM has a weak effect on the ordering parameters within the bilayer. However, model membranes with a high concentration of CHOL (30%) or the recording around the physiological temperature (35–45 °C instead of 20 °C) shows a more pronounced effect of OHM on the behavior of the bilayer that becomes less ordered. These results indicate that OHM can slightly modify the physicochemical properties of the bilayer, especially the moieties rich in CHOL. The question of the location of OHM in this lipid bilayer is then addressed by MD simulations. From this study, it is concluded that OHM mainly interacts with the oxygen atoms of the carbonyl groups of DSPC and SGML and, in a less extent, with the oxygen atom of CHOL. The consequences of these interactions imply a reduction of the stabilization of CHOL within the membrane that, in the absence of OHM, interacts also with the oxygen atoms of the carbonyl groups of DSPC and SGML. These results show, for the first time, the molecular interactions between OHM and model membranes. The presence of direct interaction between OHM embedded in the lipid bilayer and ion channels cannot be so far excluded. This point still needs to be studied and is the focus of our current efforts.
5. Materials and Methods
5.1. Synthesis of Deuterated OHM at the sn2 Position
The full synthesis is reported in the Supporting Information. The last two steps (glycosylation and deprotection of the lactose unit) are reported below.
5.1.1. Glycosylation Reaction
A solution of 1 (459 mg, 0.59 mmol, 1.0 equiv) and 2 (200 mg, 0.60 mmol, 1.02 equiv) in dry CH2Cl2 (10 mL) was stirred with molecular sieves (4 Å) for 1 hour under a N2 atmosphere. At 0 °C, BF3·Et2O (29 μL, 0.235 mmol, 0.4 equiv) was added dropwise, and the mixture was stirred for 24 h at room temperature under inert atmosphere. The mixture was quenched by the addition of water (3 mL). The organic layer was washed twice with an aqueous saturated NaHCO3 solution (2 × 3 mL) and an aqueous saturated NaCl solution (3 mL). The organic layer was dried upon MgSO4, filtered, and concentrated to give the crude compound 3. The product was purified by chromatography on silica gel [eluent: petroleum spirit/ethyl acetate (6:4)] to give the pure compound 3 (42% yield). Rf [petroleum spirit/ethyl acetate (6:4)]: 0.12; 1H NMR (C2HCl3, 399.972): 5.32 (d, 1H, 3JHH = 3.2 Hz, H4′); 5.16 (t, 1H, 3JHH = 8.8 Hz, H3); 5.08 (dd, 1H, 3JHH = 10.0, 7.6 Hz, H2′); 4.94–4.86 (m, 2H, H2 + H3′); 4.52–4.44 (m, 3H, H1 + H1′ + H6a); 4.10–4.05 (m, 3H, H6′a + H6′b + H6b); 3.86–3.79 (m, 2H, H5′ + Ha CH2sn-3); 3.79 (t, 1H, 3JHH = 9.4 Hz, H4); 3.58–3.56 (m, 2H, Ha CH2sn-1 + Hb CH2sn-3); 3.43–3.35 (m, 5H, H5 + CH sn-2 + CH2 α fatty chain + Hb CH2sn-1); 2.12 (s, 3H, OCH3); 2.09 (s, 3H, OCH3); 2.03 (s, 3H, OCH3); 2.01 (s, 3H, OCH3); 2.01 (s, 3H, OCH3); 2.00 (s, 3H, OCH3); 1.93 (s, 3H, OCH3); 1.52 (m, 2H, β CH2 fatty chain); 1.22 (br s, 26H, CH2 fatty chain); 0.86 (t, 3H, 3JHH = 6.6 Hz, CH3 fatty chain); 13C NMR (CDCl3, 74.475): 170.3 (s, C=O); 170.1 (s, C=O); 170.0 (s, C=O); 169.7 (s, C=O); 169.5 (s, C=O); 169.0 (s, C=O); 101.0 (s, C1′); 100.9–100.8 (C1 two diastereoisomers); 79.2–78.8 (CH sn-2, two diastereoisomers); 76.2 (s, C4); 72.8 (s, C3); 72.6 (s, C5); 71.8 (CH2 α fatty chain); 71.7 (s, C2); 71.0 (C3′); 70.6 (C5′); 70.3–70.0 (CH2sn-3, two diastereoisomers); 69.8–68.8 (CH2sn-1, two diastereoisomers); 69.1 (s, C2′); 66.6 (s, C4′); 62.0 (s, C6); 60.8 (s, C6′); 31.9 (s, CH2 fatty chain); 29.4 (s, CH2 fatty chain); 29.3 (s, CH2 fatty chain); 26.0 (s, CH2 fatty chain); 22.6 (s, CH2 fatty chain); 20.8 (s, OCH3); 20.6 (s, OCH3); 20.5 (s, OCH3); 14.1 (s, CH3 fatty chain).
5.1.2. Deprotection of 3 to Produce 2H3-OHM
K2CO3 (2.8 mL, 0.02 mmol, 0.5 equiv) was added to a solution of 3 (40 mg, 0.10 mmol, 1.0 equiv) in MeOH (5 mL). The mixture was stirred at room temperature for 15 h. Then, Amberlyst IR-120 (H+) was added, and the mixture was stirred for 30 min at room temperature. The reaction mixture was warmed (reflux), quickly filtered, and concentrated to give the crude compound 2H3-OHM in a quantitative yield. 2H NMR (deuterium NMR probehead): 3.78 (s, C2H3); 1H NMR (DMSO-2H6, 500.133): 5.11–5.07 (m, 2H, 2OH); 4.77 (br s, 1H, OH); 4.66 (s, 1H, OH); 4.63 (br s, 1H, OH); 4.53–4.50 (m, 2H, H1′ + OH); 4.20–4.17 (m, 2H, H4 + H1′); 3.77–3.26 (m, 17H, H3 + H5 + H6 + H2′+ H3′ + H4′ + H5′ + H6′ + CH2 α fatty alkyl chain + CH2sn-1 + CH2sn-3 + CH sn-2); 3.00–2.99 (m, 1H, H2); 1.47 (m, 2H, β CH2 fatty alkyl chain); 1.22 (br s, 26H, CH2 fatty alkyl chain); 0.85 (t, 3H, 3JHH = 6.8 Hz, CH3 fatty alkyl chain); 13C NMR (C2HCl3, 74.475): 103.8 (s, C1′); 102.9–102.8 (C1 two diastereoisomers); 80.7 (C4); 78.6–78.5 (CH sn-2, two diastereoisomers); 75.5; 75.0; 74.8; 73.2; 73.1; 70.5; 68.1 (C2 + C3 + C5 + C2′ + C3′ + C4′ + C5′); 70.6 (CH2 α fatty alkyl chain); 69.9 (CH2sn-3); 68.7–68.5 (CH2sn-1, two diastereoisomers); 60.5–60.4 (2s, C6 + C6′); 31.3 (s, CH2 fatty alkyl chain); 29.2 (s, CH2 fatty alkyl chain); 29.0 (s, CH2 fatty alkyl chain); 28.8 (s, CH2 fatty alkyl chain); 28.7 (s, CH2 fatty alkyl chain); 25.6 (s, CH2 fatty alkyl chain); 22.1 (s, CH2 fatty alkyl chain); 13.9 (s, CH3 fatty alkyl chain).
5.2. Liposome Preparation
Appropriate amounts of lipids (ca. 10 mg) and unlabeled OHM (0.56 mg for lipid-to-OHM molar ratio, Ri, of 15 and 0.28 mg for Ri = 30) were weighted and cosolubilized in an organic solvent mixture [chloroform/methanol (2:1)] to ensure a complete mixing of the components, followed by solvent evaporation under a nitrogen gas flow, removal of solvent traces using a high-speed vacuum apparatus, and suspending in water and overnight freeze-drying. Deuterium-depleted water (100 μL) was added to obtain lipid hydration, h = 90% [h = mass of water over the total mass of the system (phospholipids and water)]. After shaking in a vortex mixer, the last step was carried out, which consisted of three freeze–thaw cycles; samples were frozen in liquid nitrogen for 30 s, heated at 45 °C for 10 min in a water bath, and shaken again for better sample homogeneity. Three different systems of lipids with and without OHM were prepared following this protocol: one with pure POPC-2H31 (PC, purchased from Avanti Polar Lipids) and two others containing PC, CHOL, and egg SGML. The molar ratios were the following for PC/SGML/CHOL: 10/60/30 and 10/85/05 (mwPOPC-2H31 = 760.08 g·mol–1; mwSGML = 710.96 g·mol–1; mwCHOL = 386.65 g·mol–1; mwOHM = 654 g·mol–1; and mwH2O = 18.01 g·mol–1). Samples with deuterated OHM and protonated lipids were prepared using the same procedure.
5.3. Solid-State and Liquid-State NMR
NMR experiments were carried out on
Bruker Avance III 800 MHz (18.8
T) and 400 MHz (9.4 T) spectrometers. Wide-line 2H NMR
spectra were acquired at 122.8 MHz (18.8 T) by means of a quadrupolar
echo pulse sequence37 with a π/2
pulse width of 4.5 μs, an interpulse delay of 40 μs, and
a recycle delay of 2 s. Typically, 3k to 6k scans were recorded depending
on the concentration and the temperature of the sample. The reference
for solid-state deuterium powder patterns was set to 4.7 ppm for 1HO2H. A Lorentzian noise filtering of 100–300
Hz was always applied prior to Fourier transformation from the top
of the echo signal. Quadrature detection was used in all cases. Samples
were allowed to equilibrate for at least 20 min for each temperature
before the NMR signal was acquired. Liquid-state 1H NMR
spectra (Supporting Information) were recorded
at 400.1 MHz using a simple pulse sequence, with a π/2 pulse
width of 4 μs and a recycle delay of 5 s. Tetramethylsilane
was used for reference. Nonoriented solid-state NMR spectra were recorded
in the time domain (as free induction decays) and then Fourier transformed.
Individual components were built from the experimental estimates of
quadrupolar splittings, isotropic chemical shifts, and individual
line widths (line width is considered to be constant throughout the
pattern). Small variations were allowed to match with sharp experimental
features of the spectra. For perdeuterated chains, weights are not
variable and depend on the number of deuterons per labeled carbon
position; the individual time-dependent signals were then added accordingly,
leading after Fourier transformation of the multicomponent spectrum.
Once the sharp features are well-defined in the simulation (ΔνQ), the only adjustable variable which remains is the ellipsoid c/a ratio that is bound to the orientation
dependence of the global solid-state spectrum,38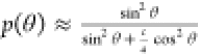 , where θ is the orientation of the
bilayer normal with respect to the magnetic field direction and c and a are the ellipsoid axes. SCD order parameters in the bilayer membranes
are proportional to quadrupolar splittings:39,40SkCD = 4ΔνQk/3AQ.
, where θ is the orientation of the
bilayer normal with respect to the magnetic field direction and c and a are the ellipsoid axes. SCD order parameters in the bilayer membranes
are proportional to quadrupolar splittings:39,40SkCD = 4ΔνQk/3AQ.
5.4. MD Simulations
MD simulations of the system containing the OHM molecules were performed using a newly developed force field for such molecules and a standard force field for lipids (see the Supporting Information).
A bilayer system mimicking the lipid environment of the SK3 channel was simulated with and without the OHM molecules as a control (Table 3). The lipid bilayer is made of molecules of DSPC, N-stearoyl-d-erythro-sphingosylphosphorylcholine (d18:1/18:0 SGML), and CHOL. The lipid composition was taken from previous work,41 where a RAFT domain with a totally saturated lipid, SGML, and CHOL molecules was analyzed. It is worth to notice that CHOL strongly interacts with saturated lipids and SGML, and therefore RAFT domains are enriched in saturated lipids.42,43
Table 3. Composition of the Simulated Systems.
| number of lipids (fractional composition) | water molecules | simulated time (ns) | |
|---|---|---|---|
| RAFT | DSPC: 54 (0.36) | 5586 | 200 |
| SGML: 44 (0.30) | |||
| CHOL: 50 (0.34) | |||
| RAFT–OHM | DSPC: 54 (0.33) | 5901 | 400 |
| SGML: 44 (0.27) | |||
| CHOL: 50 (0.31) | |||
| OHM: 14 (0.09) |
All MD simulations were performed using the Gromacs package version 4.5.44 A direct cutoff for nonbonded interactions of 1.6 nm and particle mesh Ewald for long-range electrostatics were applied.45 Berendsen46 baths were used to couple the simulation boxes with an isotropic pressure of 1 atm and to the reference temperature of 310 K. The OHM molecules together with the lipid membrane and water molecules were coupled to separate the Berendsen thermostats with a relaxation time of 0.1 ps. All bond lengths were constrained using the LINCS algorithm,47 whereas the SETTLE algorithm48 was used for water molecules. All systems were solvated with SPC water molecules.49 The time step in all simulations was set to 2 fs. The simulation time for each system is shown in Table 3.
5.4.1. Starting Conformations
The starting conformations for all systems were taken from previous publication.41 It is worth to notice that in that publication, the systems were simulated using another force field for the lipids, so the system without the OHM molecules was simulated again using the previously mentioned conditions. Most properties of this system were practically identical to those reported previously.41
The protocol to insert the OHM molecules into the membrane was described previously for the insertion of proteins into membranes.50 Basically, it consists of several steps of membrane and OHM molecule preparations, followed by series of scaling of the lipid position and energy minimization calculations (see the Supporting Information).
All trajectories were analyzed using the standard tools from the Gromacs package and custom tcl/tk scripts in the visual molecular dynamics environment.51 Different properties were calculated for each of the simulated systems along the trajectory (density profiles, RDFs, order parameters, etc.). Temporal averages were calculated in all cases along the production stage (the last 50 ns of the simulation).
Additionally, density profile and order parameters of each system were measured not only to quantify the overall organization of the lipid component but also to evaluate the production stage of each simulation. Therefore, a system was considered to be equilibrated when no further changes were found in those properties on at least three consecutive intervals of 20 ns.
5.4.2. Analysis
Most of the analysis on the trajectories was done using standard tools from the Gromacs package 4.5. The number density profile, the order parameter, the RDF, and the interleaflet mixing were calculated and averaged over the last 20 ns of each simulation. The number of hydrogen bonds was calculated based on an angle acceptor–donor-Hydrogen cutoff of 30° and a distance donor–acceptor cutoff of 3.5 Å.
The area per lipid was calculated from the P atom projection into a 2D surface using the Voronoi tessellation technique as it was previously used and described in the literature.52−54 It should be mentioned that this approach provides only an approximation to the real values because it tends to overestimate the area of smaller molecules and underestimate that of larger ones.55 Nevertheless, it allows us to obtain a good comparative estimation of the area per lipid for each component and its variation when the OHM molecules are present.
The thickness was measured as the average distance, in a direction normal to the membrane, between the centers of mass of the phosphorous atoms in each of the two layers to minimize the errors due to lateral fluctuations. The obtained errors are quite small in comparison to the experimental values because they came from an average value along the trajectory but are comparable to the experimental measurements of the thickness using the Luzzati method.56,57 The order parameters were calculated for the tail C carbon atoms located on a slice at different depths into the membrane, to check the OHM-induced variation of the lipid order along the membrane normal.
Acknowledgments
Financial support from the TGIR-RMN-THC Fr3050 CNRS for conducting the research and La Ligue contre le cancer is gratefully acknowledged. The Aquitaine Government is also thanked for providing grants to set up the NMR platform. This work was also funded by “Université François Rabelais de Tours,” “INSERM,” The Région Centre Val de Loire (LIPIDS Project ARD 2020 Biomédicaments), the committee Indre et Loire of “La Ligue Contre le Cancer,” the association CANCEN and Tours’ hospital oncology association ACORT and by Agencia Nacional de Promoción Científica y Tecnológica, project PICT 2014 - 1249.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b00936.
Synthesis of deuterated OHM at the sn2 position; NMR spectra of the intermediates and CD3-OHM in solution; and MD starting conformation and parameter development (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Lastraioli E.; Iorio J.; Arcangeli A. Ion Channel Expression as Promising Cancer Biomarker. Biochim. Biophys. Acta, Biomembr. 2015, 1848, 2685–2702. 10.1016/j.bbamem.2014.12.016. [DOI] [PubMed] [Google Scholar]
- Bortner C. D.; Cidlowski J. A. Ion Channels and Apoptosis in Cancer. Philos. Trans. R. Soc., B 2014, 369, 20130104. 10.1098/rstb.2013.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantome A.; Potier-Cartereau M.; Clarysse L.; Fromont G.; Marionneau-Lambot S.; Gueguinou M.; Pages J.-C.; Collin C.; Oullier T.; Girault A.; et al. Pivotal Role of the Lipid Raft SK3-Orai1 Complex in Human Cancer Cell Migration and Bone Metastases. Cancer Res. 2013, 73, 4852–4861. 10.1158/0008-5472.can-12-4572. [DOI] [PubMed] [Google Scholar]
- Jaffrès P.-A.; Gajate C.; Bouchet A. M.; Couthon-Gourvés H.; Chantôme A.; Potier-Cartereau M.; Besson P.; Bougnoux P.; Mollinedo F.; Vandier C. Alkyl Ether Lipids, Ion Channels and Lipid Raft Reorganization in Cancer Therapy. Pharmacol. Ther. 2016, 165, 114–131. 10.1016/j.pharmthera.2016.06.003. [DOI] [PubMed] [Google Scholar]
- Ingólfsson H. I.; Melo M. N.; van Eerden F. J.; Arnarez C.; Lopez C. A.; Wassenaar T. A.; Periole X.; de Vries A. H.; Tieleman D. P.; Marrink S. J. Lipid Organization of the Plasma Membrane. J. Am. Chem. Soc. 2014, 136, 14554–14559. 10.1021/ja507832e. [DOI] [PubMed] [Google Scholar]
- Lee S.-Y.; Lee A.; Chen J.; MacKinnon R. Structure of the KvAP Voltage-Dependent K+ Channel and Its Dependence on the Lipid Membrane. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 15441–15446. 10.1073/pnas.0507651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D.; Jiang Q.-X.; MacKinnon R. Phospholipids and the Origin of Cationic Gating Charges in Voltage Sensors. Nature 2006, 444, 775–779. 10.1038/nature05416. [DOI] [PubMed] [Google Scholar]
- Brohawn S. G.; Su Z.; MacKinnon R. Mechanosensitivity Is Mediated Directly by the Lipid Membrane in TRAAK and TREK1 K+ Channels. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, 3614–3619. 10.1073/pnas.1320768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maingret F.; Patel A. J.; Lesage F.; Lazdunski M.; Honoré E. Lysophospholipids Open the Two-Pore Domain Mechano-Gated K+ Channels TREK-1 and TRAAK. J. Biol. Chem. 2000, 275, 10128–10133. 10.1074/jbc.275.14.10128. [DOI] [PubMed] [Google Scholar]
- Gimpl G.; Burger K.; Fahrenholz F. Cholesterol as Modulator of Receptor Function. Biochemistry 1997, 36, 10959–10974. 10.1021/bi963138w. [DOI] [PubMed] [Google Scholar]
- Lundbæk J. A.; Birn P.; Girshman J.; Hansen A. J.; Andersen O. S. Membrane Stiffness and Channel Function. Biochemistry 1996, 35, 3825–3830. 10.1021/bi952250b. [DOI] [PubMed] [Google Scholar]
- Schagina L. V.; Blaskó K.; Grinfeldt A. E.; Korchev Y. E.; Lev A. A. Cholesterol-Dependent Gramicidin A Channel Inactivation in Red Blood Cell Membranes and Lipid Bilayer Membranes. Biochim. Biophys. Acta, Biomembr. 1989, 978, 145–150. 10.1016/0005-2736(89)90509-9. [DOI] [PubMed] [Google Scholar]
- Schagina L. V.; Korchev Y. E.; Grinfeldt A. E.; Lev A. A.; Blaskó K. Sterol Specific Inactivation of Gramicidin A Induced Membrane Cation Permeability. Biochim. Biophys. Acta, Biomembr. 1992, 1109, 91–96. 10.1016/0005-2736(92)90191-n. [DOI] [PubMed] [Google Scholar]
- Singh D. K.; Rosenhouse-Dantsker A.; Nichols C. G.; Enkvetchakul D.; Levitan I. Direct Regulation of Prokaryotic Kir Channel by Cholesterol. J. Biol. Chem. 2009, 284, 30727–30736. 10.1074/jbc.m109.011221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alioua A.; Lu R.; Kumar Y.; Eghbali M.; Kundu P.; Toro L.; Stefani E. Slo1 Caveolin-Binding Motif, a Mechanism of Caveolin-1-Slo1 Interaction Regulating Slo1 Surface Expression. J. Biol. Chem. 2008, 283, 4808–4817. 10.1074/jbc.m709802200. [DOI] [PubMed] [Google Scholar]
- Garg V.; Sun W.; Hu K. Caveolin-3 Negatively Regulates Recombinant Cardiac KATP Channels. Biochem. Biophys. Res. Commun. 2009, 385, 472–477. 10.1016/j.bbrc.2009.05.100. [DOI] [PubMed] [Google Scholar]
- Epshtein Y.; Chopra A. P.; Rosenhouse-Dantsker A.; Kowalsky G. B.; Logothetis D. E.; Levitan I. Identification of a C-Terminus Domain Critical for the Sensitivity of Kir2.1 to Cholesterol. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 8055–8060. 10.1073/pnas.0809847106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini J.; Barrantes F. J. Sphingolipid/cholesterol Regulation of Neurotransmitter Receptor Conformation and Function. Biochim. Biophys. Acta, Biomembr. 2009, 1788, 2345–2361. 10.1016/j.bbamem.2009.08.016. [DOI] [PubMed] [Google Scholar]
- Potier M.; Joulin V.; Roger S.; Besson P.; Jourdan M.-L.; LeGuennec J.-Y.; Bougnoux P.; Vandier C. Identification of SK3 Channel as a New Mediator of Breast Cancer Cell Migration. Mol. Cancer Ther. 2006, 5, 2946–2953. 10.1158/1535-7163.mct-06-0194. [DOI] [PubMed] [Google Scholar]
- Chantome A.; Girault A.; Potier M.; Collin C.; Vaudin P.; Pagès J.-C.; Vandier C.; Joulin V. KCa2.3 Channel-Dependent Hyperpolarization Increases Melanoma Cell Motility. Exp. Cell Res. 2009, 315, 3620–3630. 10.1016/j.yexcr.2009.07.021. [DOI] [PubMed] [Google Scholar]
- Chantome A.; Potier-Cartereau M.; Clarysse L.; Fromont G.; Marionneau-Lambot S.; Gueguinou M.; Pages J.-C.; Collin C.; Oullier T.; Girault A.; et al. Pivotal Role of the Lipid Raft SK3-Orai1 Complex in Human Cancer Cell Migration and Bone Metastases. Cancer Res. 2013, 73, 4852–4861. 10.1158/0008-5472.can-12-4572. [DOI] [PubMed] [Google Scholar]
- Girault A.; Haelters J.-P.; Potier-Cartereau M.; Chantome A.; Pinault M.; Marionneau-Lambot S.; Oullier T.; Simon G.; Couthon-Gourves H.; Jaffres P.-A.; et al. New Alkyl-Lipid Blockers of SK3 Channels Reduce Cancer Cell Migration and Occurrence of Metastasis. Curr. Cancer Drug Targets 2011, 11, 1111–1125. 10.2174/156800911798073069. [DOI] [PubMed] [Google Scholar]
- Douliez J. P.; Bellocq A. M.; Dufourc E. J. Effect of Vesicle Size, Polydispersity and Multilayering on Solid State 31P- and 2H-NMR Spectra. J. Chim. Phys. 1994, 91, 874–880. 10.1051/jcp/1994910874. [DOI] [Google Scholar]
- Dufourc E. J. Sterols and Membrane Dynamics. J. Chem. Biol. 2008, 1, 63–77. 10.1007/s12154-008-0010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufourc E. J.; Mayer C.; Stohrer J.; Althoff G.; Kothe G. Dynamics of Phosphate Head Groups in Biomembranes. Comprehensive Analysis Using phosphorus-31 Nuclear Magnetic Resonance Lineshape and Relaxation Time Measurements. Biophys. J. 1992, 61, 42–57. 10.1016/s0006-3495(92)81814-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capponi S.; Freites J. A.; Tobias D. J.; White S. H. Interleaflet Mixing and Coupling in Liquid-Disordered Phospholipid Bilayers. Biochim. Biophys. Acta, Biomembr. 2016, 1858, 354–362. 10.1016/j.bbamem.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.; Jan L. Y. Targeting Potassium Channels in Cancer. J. Cell Biol. 2014, 206, 151–162. 10.1083/jcb.201404136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potier M.; Chantome A.; Joulin V.; Girault A.; Roger S.; Besson P.; Jourdan M.-L.; LeGuennec J.-Y.; Bougnoux P.; Vandier C. The SK3/KCa2.3 Potassium Channel Is a New Cellular Target for Edelfosine. Br. J. Pharmacol. 2011, 162, 464–479. 10.1111/j.1476-5381.2010.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girault A.; Haelters J.-P.; Potier-Cartereau M.; Chantome A.; Jaffres P.-A.; Bougnoux P.; Joulin V.; Vandier C. Targeting SKCa Channels in Cancer: Potential New Therapeutic Approaches. Curr. Med. Chem. 2012, 19, 697–713. 10.2174/092986712798992039. [DOI] [PubMed] [Google Scholar]
- Sevrain C. M.; Haelters J.-P.; Chantôme A.; Couthon-Gourvès H.; Gueguinou M.; Potier-Cartereau M.; Vandier C.; Jaffrès P.-A. DiGalactosyl-Glycero-Ether Lipid: Synthetic Approaches and Evaluation as SK3 Channel Inhibitor. Org. Biomol. Chem. 2013, 11, 4479–4487. 10.1039/c3ob40634b. [DOI] [PubMed] [Google Scholar]
- Berthe W.; Sevrain C. M.; Chantôme A.; Bouchet A. M.; Gueguinou M.; Fourbon Y.; Potier-Cartereau M.; Haelters J.-P.; Couthon-Gourvès H.; Vandier C.; et al. New Disaccharide-Based Ether Lipids as SK3 Ion Channel Inhibitors. ChemMedChem 2016, 11, 1531–1539. 10.1002/cmdc.201600147. [DOI] [PubMed] [Google Scholar]
- De Sá M. M.; Sresht V.; Rangel-Yagui C. O.; Blankschtein D. Understanding Miltefosine–Membrane Interactions Using Molecular Dynamics Simulations. Langmuir 2015, 31, 4503–4512. 10.1021/acs.langmuir.5b00178. [DOI] [PubMed] [Google Scholar]
- Pan J.; Cheng X.; Heberle F. A.; Mostofian B.; Kučerka N.; Drazba P.; Katsaras J. Interactions between Ether Phospholipids and Cholesterol as Determined by Scattering and Molecular Dynamics Simulations. J. Phys. Chem. B 2012, 116, 14829–14838. 10.1021/jp310345j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini J.; Di Scala C.; Baier C. J.; Barrantes F. J. Molecular Mechanisms of Protein-Cholesterol Interactions in Plasma Membranes: Functional Distinction between Topological (Tilted) and Consensus (CARC/CRAC). Chem. Phys. Lipids 2016, 199, 52–60. 10.1016/j.chemphyslip.2016.02.009. [DOI] [PubMed] [Google Scholar]
- Jorgensen N. K.; Pedersen S. F.; Rasmussen H. B.; Grunnet M.; Klaerke D. A.; Olesen S.-P. Cell Swelling Activates Cloned Ca2+-Activated K+ Channels: A Role for the F-Actin Cytoskeleton. Biochim. Biophys. Acta, Biomembr. 2003, 1615, 115–125. 10.1016/s0005-2736(03)00237-2. [DOI] [PubMed] [Google Scholar]
- Weichert J. P.; Clark P. A.; Kandela I. K.; Vaccaro A. M.; Clarke W.; Longino M. A.; Pinchuk A. N.; Farhoud M.; Swanson K. I.; Floberg J. M.; et al. Alkylphosphocholine Analogs for Broad-Spectrum Cancer Imaging and Therapy. Sci. Transl. Med. 2014, 6, 240ra75. 10.1126/scitranslmed.3007646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J. H.; Jeffrey K. R.; Bloom M.; Valic M. I.; Higgs T. P. Quadrupolar Echo Deuteron Magnetic Resonance Spectroscopy in Ordered Hydrocarbon Chains. Chem. Phys. Lett. 1976, 42, 390–394. 10.1016/0009-2614(76)80392-2. [DOI] [Google Scholar]
- Pott T.; Dufourc E. J. Action of Melittin on the DPPC-Cholesterol Liquid-Ordered Phase: A Solid State 2H-and 31P-NMR Study. Biophys. J. 1995, 68, 965–977. 10.1016/s0006-3495(95)80272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J. H. The Description of Membrane Lipid Conformation, Order and Dynamics by 2H-NMR. Biochim. Biophys. Acta, Rev. Biomembr. 1983, 737, 117–171. 10.1016/0304-4157(83)90015-1. [DOI] [PubMed] [Google Scholar]
- Seelig J. Deuterium Magnetic Resonance: Theory and Application to Lipid Membranes. Q. Rev. Biophys. 1977, 10, 353–418. 10.1017/s0033583500002948. [DOI] [PubMed] [Google Scholar]
- Herrera F. E.; Pantano S. Structure and Dynamics of Nano-Sized Raft-like Domains on the Plasma Membrane. J. Chem. Phys. 2012, 136, 015103. 10.1063/1.3672704. [DOI] [PubMed] [Google Scholar]
- Pike L. J. Lipid Rafts: Bringing Order to Chaos. J. Lipid Res. 2003, 44, 655–667. 10.1194/jlr.r200021-jlr200. [DOI] [PubMed] [Google Scholar]
- Calder P. C.; Yaqoob P. Lipid Rafts–Composition, Characterization, and Controversies. J. Nutr. 2007, 137, 545–547. [DOI] [PubMed] [Google Scholar]
- Hess B.; Kutzner C.; van der Spoel D.; Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Darden T.; York D.; Pedersen L. Particle Mesh Ewald: An N log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. 10.1063/1.464397. [DOI] [Google Scholar]
- Berendsen H. J. C.; Postma J. P. M.; van Gunsteren W. F.; DiNola A.; Haak J. R. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684. 10.1063/1.448118. [DOI] [Google Scholar]
- Hess B.; Bekker H.; Berendsen H. J. C.; Fraaije J. G. E. M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. . [DOI] [Google Scholar]
- Miyamoto S.; Kollman P. A. Settle: An Analytical Version of the SHAKE and RATTLE Algorithm for Rigid Water Models. J. Comput. Chem. 1992, 13, 952–962. 10.1002/jcc.540130805. [DOI] [Google Scholar]
- Berendsen H. J. C.; Postma J. P. M.; van Gunsteren W. F.; Hermans J.. Interaction Models For Water In Relation To Protein Hydration. Intermolecular Forces; Springer, 1981; pp 331–342. [Google Scholar]
- Kandt C.; Ash W. L.; Tieleman D. P. Setting up and Running Molecular Dynamics Simulations of Membrane Proteins. Methods 2007, 41, 475–488. 10.1016/j.ymeth.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: Visual Molecular Dynamics. J. Mol. Graphics 1996, 14, 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Pandit S. A.; Vasudevan S.; Chiu S. W.; Mashl R. J.; Jakobsson E.; Scott H. L. Sphingomyelin-Cholesterol Domains in Phospholipid Membranes: Atomistic Simulation. Biophys. J. 2004, 87, 1092–1100. 10.1529/biophysj.104.041939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedlovszky P.; Medvedev N. N.; Mezei M. Effect of Cholesterol on the Properties of Phospholipid Membranes. 3. Local Lateral Structure. J. Phys. Chem. B 2004, 108, 465–472. 10.1021/jp0307912. [DOI] [PubMed] [Google Scholar]
- Falck E.; Patra M.; Karttunen M.; Hyvönen M. T.; Vattulainen I. Lessons of Slicing Membranes: Interplay of Packing, Free Area, and Lateral Diffusion in Phospholipid/cholesterol Bilayers. Biophys. J. 2004, 87, 1076–1091. 10.1529/biophysj.104.041368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S. A.; Jakobsson E.; Scott H. L. Simulation of the Early Stages of Nano-Domain Formation in Mixed Bilayers of Sphingomyelin, Cholesterol, and Dioleylphosphatidylcholine. Biophys. J. 2004, 87, 3312–3322. 10.1529/biophysj.104.046078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzzati V.X-Ray Diffration Studies of Lipid-Water Systems. Biological Membranes; Academic Press: London, 1968; pp 71–123. [Google Scholar]
- Nagle J. F.; Tristram-Nagle S. Structure of Lipid Bilayers. Biochim. Biophys. Acta, Rev. Biomembr. 2000, 1469, 159–195. 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.