Abstract
Protein scaffolds have proven useful for co-localization of enzymes, providing control over stoichiometry and leading to higher local enzyme concentrations, which have led to improved product formation. To broaden their usefulness, it is necessary to have a wide choice of building blocks to mix and match for scaffold generation. Ideally, the scaffold building blocks should function at any location within the scaffold and have high affinity interactions with their binding partners. We examined the utility of orthogonal synthetic coiled coils (zippers) as scaffold components. The orthogonal zippers are coiled coil domains that form heterodimers only with their specific partner and not with other zipper domains. Focusing on two orthogonal zipper pairs, we demonstrated that they are able to function on either end or in the middle of a multiblock assembly. Surface plasmon resonance was employed to assess the binding kinetics of zipper pairs placed at the start, middle, or end of a construct. Size-exclusion chromatography was used to demonstrate the ability of a scaffold with two zipper domains to bind their partners simultaneously. We then expanded the study to examine the binding kinetics and cross-reactivities of three additional zipper pairs. By validating the affinities and specificities of synthetic zipper pairs, we demonstrated the potential for zipper domains to provide an expanded library of scaffolding parts for tethering enzymes in complex pathways for synthetic biology applications.
Introduction
The use of scaffolds for pathway organization is a promising synthetic biology approach that co-localizes functional proteins of the same pathway to provide programmable control over parameters such as enzyme stoichiometry.1−5 This approach has the potential to yield improved product production, as it serves to physically enrich the local concentration of enzymes and substrates, which may be especially beneficial for multienzyme cascades. The most successful protein-based scaffolds have been built from modular protein–protein interaction domains connected by peptide linkers. Enzymes were expressed as fusions with interaction partners that self-assembled onto the scaffold. The number of repeats of an interaction domain on the scaffold controls the stoichiometry, and the use of multiple interaction domains facilitates the incorporation of various enzymes. In one example, a 77-fold increase in mevalonate biosynthesis was realized through scaffolding of three enzymes.4 To be scalable, a wide variety of binding domains with tight affinity and high specificity need to be available for scaffold construction. Ideally the building blocks should be able to function on either end or in the middle of a scaffold construct.
To date, most of the work has utilized metazoan signaling proteins (SH3-, PDZ-, and GBD-binding domain) or cohesin domains as scaffold components.4,6,7 Other systems, such as tetratricopeptide repeat affinity protein–peptide interaction domains have been proposed as alternate interaction modules.8 These components are useful; however, the interactions of the aforementioned domains are typically in the micromolar range.8,9 Ideally, researchers would benefit from a wide variety of scaffolding/interaction domains with a range of affinities.
Coiled coil domains (zippers) offer additional scaffold components. These domains offer good affinity, tunable from the micromolar down to sub-nanomolar.10 Until recently, researchers did not have access to a large set of coiled coils with specific and predictable interactions. Now, however, several groups have described sets of designed synthetic orthogonal zippers.11−14 The availability of a variety of orthogonal zippers has wide ranging applications from use in synthetic biology to the design of non-natural protein structures such as tetrahedral protein cages.15,16 The motivation behind our work was to investigate the ability of these synthetic zipper domains to function as building blocks for protein scaffolds.
We initially focused on two pairs of SYNZIP designed coiled coil domains first described by the Keating laboratory (SYZNIPs 1 and 2 and SYNZIPs 3 and 4) known to be orthogonal.12,17 Orthogonal zipper domains form interactions only with their partners; in this case, SYNZIP 1 with 2 and SYNZIP 3 with 4. To gain the most utility, the scaffold building blocks must retain their binding function when expressed at either end or within the middle of the scaffold assembly. Each zipper was constructed as a Bglbrick part18 for facile mixing of parts. The zippers were fused to single-domain antibody (sdAb) domains19,20 to facilitate protein production and characterization. Binding affinities for the zipper pairs were measured using surface plasmon resonance (SPR), and size-exclusion chromatography was utilized to demonstrate the ability of a scaffold with two zippers to bind their partners simultaneously. We expanded our investigation using surface plasmon resonance to examine the interactions of two additional pairs of SYNZIP designed zippers,12,17 as well as a heterodimerizing pair consisting of a basic and an acidic zipper derived from a eukaryotic protein.21
This study demonstrated that the synthetic zippers maintained their function when placed at different locations in a multidomain construct substantiating their promise as domains for use in the scaffolding of enzymes. In addition, this work confirmed the original binding and specificity data reported by the group that produced the SYNZIP proteins. The availability of validated protein building blocks (parts) with well-characterized interactions is beneficial for a variety of synthetic biology applications.22
Results and Discussion
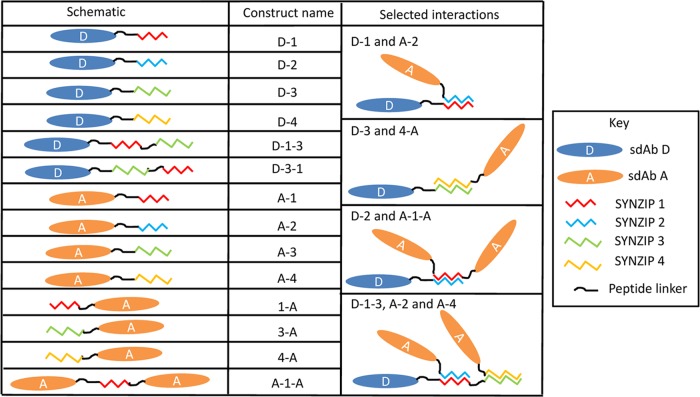
Our work started by evaluating two pairs of synthetic zippers. The constructs were assembled using the Bglbrick cloning strategy,18 where each was a combination of a single-domain antibody (sdAb) domain19,20 and one or two SYNZIP domains.12,17 Each domain was separated by a 12–15 amino acid long linker. Two series of constructs were built. The first included a ricin-binding sdAb, referred to here as D (originally termed D12fneg23), produced as a C-terminal fusion with individual SYNZIPs (1, 2, 3, and 4) and SYNZIP domain pairs (1, 3 and 3, 1). The second set consisted of a staphylococcal enterotoxin B-binding sdAb (ACVE,24 referred here as A) in combinations with each of the SYNZIPs (1, 2, 3, and 4) expressed at the C-terminal, SYNZIPs 1, 3, and 4 at the N-terminal, and SYNZIP 1 sandwiched between two A domains. The constructs were designated by abbreviating the domains in the order they occurred using the letter designation of the sdAb and the number of the SYNZIP. Figure 1 shows a schematic overview of each of the initial constructs. All of the protein sequences are provided in the Supporting Information (Figure S1). Figure 1 also presents the cartoon depictions of several of the interactions expected between the zipper pairs within the constructs.
Figure 1.
List of SYNZIP constructs assembled to examine the ability of the zippers to function at the C-terminus, N-terminus, or interior of a multidomain protein. A schematic is shown for each construct along with cartoons of some of the interactions that we characterized.
SdAb contain a conserved disulfide bond making periplasmic expression preferred, thus all the sdAb-SYNZIP fusions were expressed in the periplasm. Each construct was found to have a protein expression of at least 1 mg/L, with the C-terminal fusions producing ∼9–20 mg/L. Each construct was prepared at least twice; the average protein yields along with the average deviation are summarized in Table 1. Interestingly the C-terminal fusions with A produced at least 5 times better than the N-terminal fusions, and the construct with a SYNZIP sandwiched between two A domains. However, this might be particular to this sdAb and not a general feature of the SYNZIP domains. The scaffolds containing the D sdAb and two SYNZIPs produced ∼10–12 mg/L.
Table 1. Protein Yields.
| construct | average yield, mg/L (average deviation) |
|---|---|
| D-1 | 17 (1) |
| D-2 | 12 (4) |
| D-3 | 14 (2) |
| D-4 | 12 (2) |
| D-1-3 | 12 (3) |
| D-3-1 | 10 (2) |
| A-1 | 18 (1) |
| A-2 | 8 (2) |
| A-3 | 19 (2) |
| A-4 | 18 (4) |
| 1-A | 2 (0.4) |
| 3-A | 1.7 (0.2) |
| 4-A | 1.6 (0.4) |
| A-1-A | 1.4 (0.4) |
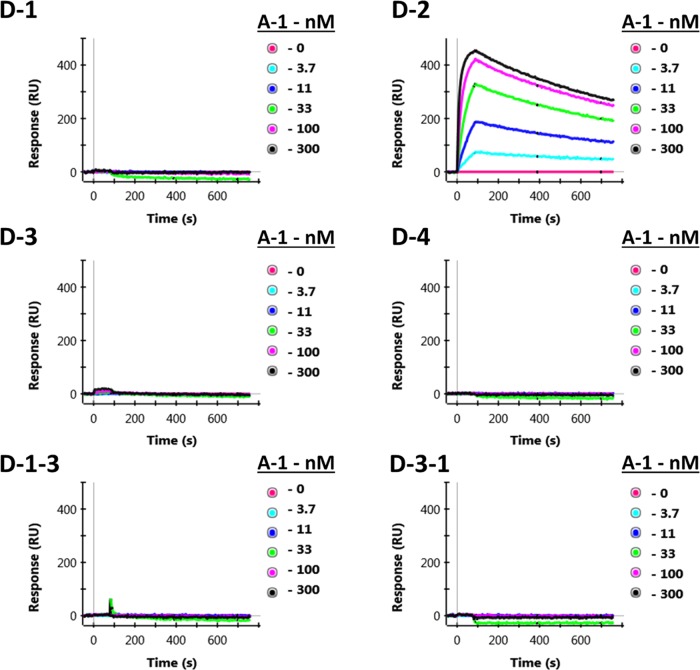
Surface plasmon resonance (SPR) was used to measure the binding kinetics of the SYNZIP interactions and verify their specificity. The BioRad ProteOn XPR36 enabled simultaneous interrogation of six immobilized sdAb-SNYZIP constructs with five concentrations of an sdAb-SYNZIP-based construct in solution. Oriented immobilization of the fused SYNZIP was provided through the interaction of the D sdAb with ricin covalently attached to the chip; a cartoon of the experiment is shown in Figure S2 of the Supporting Information. The D sdAb binds to ricin with a very high affinity (equilibrium dissociation constant (KD) = 0.04 nM),23 which allowed us to evaluate accurately the affinity of the SYNZIP pairs. The SPR results are summarized in Tables 2 and 3. Table 2 provides the KD for each interacting pair, whereas Table 3 includes ka and kd, the measured on and off rate constants. An example set of the SPR data is shown in Figure 2; other data sets are presented in the Supporting Information (Figure S3). Previously, the affinity of the SYNZIP 1–2 and 3–4 pairs had been reported as <10 and <30 nM, respectively.17 Our results measured by SPR are in general agreement with the earlier reports. Critical for their potential use in scaffolding schemes, our work verifies that the SYNZIP 1–2 and 3–4 interactions are specific and demonstrated that the SYNZIP domains are able to specifically recognize their partner when incorporated at the start, middle, or end of a fusion protein.
Table 2. Binding Affinities (KD) of SYNZIP Constructs As Determined by SPR.
| immobilized |
||||||
|---|---|---|---|---|---|---|
| in solution | D-1, nM | D-2, nM | D-3, nM | D-4, nM | D-1-3, nM | D-3-1, nM |
| A-1 | –a | 1.5 | – | – | – | – |
| A-2 | 16 | – | – | – | 26 | 28 |
| A-3 | – | – | – | 34 | – | – |
| A-4 | – | – | 11 | – | 74 | 39 |
| 1-A | – | 5.5 | – | – | – | – |
| 3-A | – | – | – | 17 | – | – |
| 4-A | – | – | 29 | – | 135 | 63 |
| A-1-A | – | 0.9 | – | – | – | – |
“–” indicates no binding observed.
Table 3. On and Off Rate Constants for Interacting SYNZIP Pairs As Determined by SPR.
| in solution | immobilized | ka (1/Ms) | kd (1/s) |
|---|---|---|---|
| A-1 | D-2 | 5.2 × 105 | 7.8 × 10–4 |
| A-2 | D-1 | 1.5 × 105 | 2.5 × 10–3 |
| A-2 | D-1-3 | 1.5 × 105 | 4.0 × 10–3 |
| A-2 | D-3-1 | 1.1 × 105 | 3.0 × 10–3 |
| A-3 | D-4 | 4.0 × 105 | 1.4 × 10–2 |
| A-4 | D-3 | 1.7 × 105 | 1.9 × 10–2 |
| A-4 | D-1-3 | 3.4 × 105 | 2.5 × 10–2 |
| A-4 | D-3-1 | 5.1 × 105 | 2.0 × 10–2 |
| 1-A | D-2 | 2.9 × 105 | 1.6 × 10–3 |
| 3-A | D-4 | 7.5 × 105 | 1.3 × 10–2 |
| 4-A | D-3 | 7.0 × 105 | 2.0 × 10–2 |
| 4-A | D-1-3 | 2.0 × 105 | 2.7 × 10–2 |
| 4-A | D-3-1 | 3.0 × 105 | 1.9 × 10–2 |
| A-1-A | D-2 | 1.9 × 105 | 1.7 × 10–4 |
Figure 2.
Representative SPR data. D-1, D-2, D-3, D-4, D-1-3, and D-3-1 were immobilized on six individual columns of the SPR chip. Dilutions of the A-1 construct (300, 100, 33, 11, 3.7, and 0 nM) were flowed over the chip in rows and are noted to the right of each trace. Data are presented for binding of A-1 to each of the six immobilized D-SYNZIP constructs.
It is imperative that the domains of a scaffold can bind their partners simultaneously. Size-exclusion chromatography was employed to address this question. Results are shown in Figure 3 (also in the Supporting Information, Figure S4). No assembly was observed with the control constructs (A-1 and A-3 with a D-1-3 or D-3-1 scaffold; Figure S4), as judged by the fact that the profile of the combinations was essentially the sum of the profile of the individual components. As shown in Figure 3, when D-3-1 is combined with A-2 or A-4, the combined peak runs at a smaller volume, indicating that D-3-1 is able to bind both the SYNZIP 2- and 4-based partner constructs. The combined D-3-1, A-2, and A-4 sample elutes at a smaller volume than any of the components or partial combinations, clearly implying that both A-2 and A-4 are able to bind simultaneously to the scaffold and maintain that interaction during the course of the gel filtration run. Interestingly, the combined D-1-3, A-2, and A-4 sample runs at only a slightly smaller volume than the combined D-1-3 and A-4 peak (13.2026 mL vs 13.3159 mL; Figure S4). As with the D-3-1 scaffold, however, the three-component mix ran primarily as one peak with ∼3 times the height as the individual components. This indicates that the A-2 and A-4 are assembling simultaneously on the D-1-3 scaffold.
Figure 3.
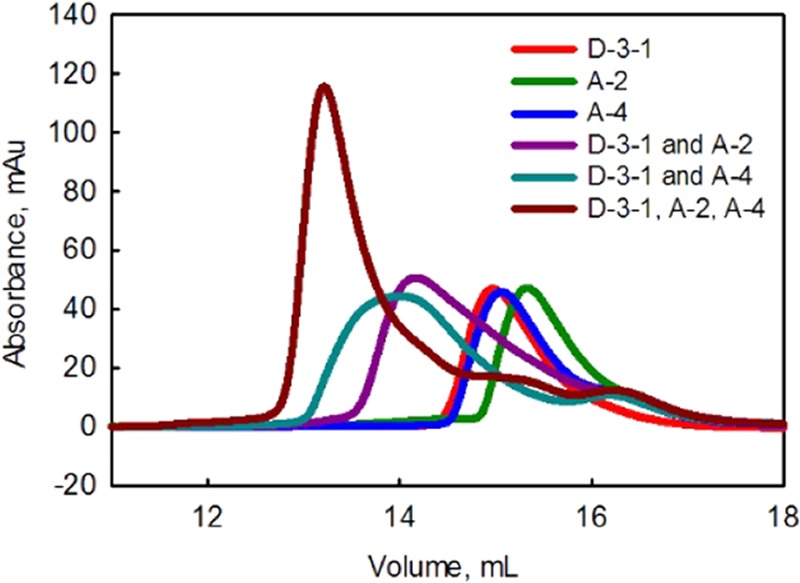
Size-exclusion chromatography. Each component was run separately and then A-2 and A-4 were run in combination with the D-3-1 scaffold. The concentration of each protein was held constant for every experiment.
Next, we produced three additional zipper pairs and measured their binding affinities and cross-reactivities. Four of the zippers were SYNZIPs (5, 6, 17, and 18) that form 2 additional pairs: 5 with 6 and 17 with 18.17 Unlike the other pairs, 17 and 18 were previously shown to interact in an antiparallel orientation. Each new zipper was cloned to the C-terminus of the A or D sdAb. As with the other SYNZIPs, new constructs had excellent protein yields ranging from ∼4 to 25 mg/L (Table S1, Supporting Information). In addition to the SYNZIPs, we produced an acidic and basic zipper pair (R34m and E34m) derived from the B-Zip domain of the chicken transcription factor VBP that has been characterized and utilized in synthetic biology applications as dimer-forming domains.21,22,25 The original acidic and basic zipper sequences each contain a single cysteine. In an effort to avoid the zippers forming disulfide bonds when expressed as sdAb fusions in the periplasm, we mutated the cysteines to serines. Fusions with the E34m zipper produced well (∼13–23 mg/L); however, those with the positively charged R34m zipper had poor protein yields (∼1 mg/L). This is consistent with poor protein expression we had previously obtained for a fusion of an sdAb to a similar positively charged zipper.26
Each of the six additional zippers was immobilized to a chip for SPR analysis. We examined the ability of all 10 zippers to bind, providing information on both affinity and specificity. The binding data are summarized in Table 4, and the kinetics are shown in Table 5. In several cases, weak binding interactions were observed; for example, A-5 has a weak interaction with D-18. In general, the SYNZIP binding affinities observed agreed with what had previously been reported (<15 nM for the 5–6 pair and <10 nM for 17–18 pair).17 The previous characterization of the SYNZIP domains also indicated some cross-reactivity that is consistent with our observations.12,17 Likewise, the E34m–R34m interaction was close to the value of 6 nM that had been previously reported.25
Table 4. Binding Affinities (KD) As Determined by SPR.
| immobilized |
||||||
|---|---|---|---|---|---|---|
| in solution | D-5, nM | D-6, nM | D-17, nM | D-18, nM | D-R34m, nM | D-E34m, nM |
| A-1 | –a | – | – | – | – | – |
| A-2 | – | – | – | – | – | – |
| A-3 | – | – | 15b | – | – | – |
| A-4 | – | 37b | – | – | – | – |
| A-5 | – | 8.4 | – | 327 | – | – |
| A-6 | 36 | – | 34 000b | – | – | – |
| A-17 | – | 33 | – | 1.5 | – | – |
| A-18 | – | – | 12 | – | – | – |
| A-R34m | – | – | – | 268 | – | 10 |
| A-E34m | – | – | – | – | 12 | – |
“–” indicates no binding observed.
Low signal, RU (response units) less than 50.
Table 5. On and Off Rate Constants for Interacting Zipper Pairs As Determined by SPR.
| in solution | immobilized | ka (1/Ms) | kd (1/s) |
|---|---|---|---|
| A-3 | D-17 | 4.4 × 105 | 6.7 × 10–3 |
| A-4 | D-6 | 2.5 × 105 | 9.3 × 10–3 |
| A-5 | D-6 | 3.1 × 105 | 2.6 × 10–3 |
| A-5 | D-18 | 8.15 × 105 | 2.8 × 10–1 |
| A-6 | D-5 | 1.0 × 105 | 3.7 × 10–3 |
| A-6 | D-17 | 2.4 × 103 | 8.2 × 10–2 |
| A-17 | D-6 | 2.0 × 106 | 6.4 × 10–2 |
| A-17 | D-18 | 5.5 × 105 | 8.2 × 10–4 |
| 1-A | D-2 | 2.9 × 105 | 1.6 × 10–3 |
| A-18 | D-17 | 1.9 × 105 | 2.4 × 10–3 |
| A-R34m | D-18 | 6.7 × 107 | 1.8 × 101 |
| A-R34m | D-E34m | 4.6 × 105 | 4.7 × 10–3 |
| A-E34m | D-R34m | 5.5 × 105 | 6.5 × 10–3 |
Both the R34m and SYNZIP 17 zippers were found to be “sticky”, giving a signal on every spot when employing the running buffer used in the binding measurements of the first sets of zippers. Examining the sequences showed that both zippers have five or six more Arg/Lys residues than Glu/Asp. Other zippers had either one additional Arg/Lys or slightly more Glu/Asp (Table S2, Supporting Information). Adding 5 mM MgSO4 to the running buffer eliminated most of the nonspecific interactions while preserving the specific binding of the zipper pairs.
The new zipper constructs were also examined by SPR for their ability to bind to immobilized SYNZIP 1, 2, 3, and 4. The samples were run in buffer with added BSA and MgSO4. No interactions were observed with the exception of SYNZIP 17, which appeared to bind to 2 and 3. This may be due to the sticky nature of the positively charged SYNZIP 17. No binding, however, was observed with the positively charged R34m zipper. The data are reported in Table S3 and Figure S5 in the Supporting Information.
In summation, we demonstrated that the SYNZIPs can be used as effective scaffolding domains, showing that they are able to function at either end or in the middle of a construct. Next, we characterized three other zipper pairs showing the potential for five orthogonal interacting pairs. Our work serves, in part, as an independent verification of the affinity and specificity of the SYNZIP constructs. The methods demonstrated here are of value to evaluate other SYNZIP constructs, or designed orthogonal coiled coil systems to ensure their functionality, and thereby provide a wealth of validated high-affinity interaction domains for synthetic biology and scaffolding applications.
Experimental Section
Construction of Bglbrick Parts and Assembly of Constructs
Cloning enzymes were from New England Biolabs. The BglII site was removed from the pET22b vector using the quick change mutagenesis kit (Agilent), and a Bglbrick compatible version of pET22b termed pET22b-BglII was constructed.27 The DNA coding for each of the SYNZIP sequences12 was synthesized by Eurofins Genomics, with PCR used to introduce linkers and restriction sites for Bglbrick cloning.18 Each sdAb was also cloned into pET22b-BglII. All the Bglbrick parts were constructed with C-terminal Gly Ser linkers. The proteins were produced and purified as described previously,23,24 with the exception that an Enrich SEC 650 column was utilized for size-exclusion purification. Proteins, in PBS containing 0.02% sodium azide, were quickly frozen in dry ice and stored at −80 °C.
Surface Plasmon Resonance
SPR was performed as previously described,23 with the exception that BSA (1 mg/mL) was added to the standard running buffer of PBS with 0.005% Tween-20. Briefly, ricin was immobilized onto every spot on the chip. Next, each of the six D constructs was immobilized along one column. Dilutions of A constructs in the running buffer, including a 0 nM control, were flowed along each of the six rows. The first type of chip contained D-1, D-2, D-3, D-4, D-1-3, and D-3-1, whereas a second type of chip was immobilized with D-5, D-6, D-17, D-18, D-R34m, and D-E34m. The first type of chip was prepared three separate times, whereas the second type was made once. Only limited nonspecific binding interaction was apparent between the initial constructs, it was not until testing the A and D constructs on the second chip that nonspecific binding became more obvious, thus both chips were run an additional time using running buffer containing 1 mg/mL BSA and 5 mM MgSO4 to minimize these undesirable interactions. All of the interactions were measured at least two times, some as many as six times. The data were fit using a global analysis 1:1 Langmuir model with the standard error always less than 10%, typically ∼1%; variation between multiple tests of the same sample was within a factor of 2, and a representative data set was used in the tables and shown in the Supporting Information.
Size-Exclusion Chromatography
The analysis was carried out using a BioLogic DuoFLow chromatography system (Bio-Rad) with an Enrich SEC 650 column. The samples were made up to 250 μL with PBS. The protein concentrations (∼20 μM) were used to generate a signal of around 45–50 mAU. The mixtures (∼20 μM each component) were incubated on the bench top at least 15 min before injecting.
Acknowledgments
This work was supported by NRL base funds. Thanks to Dan Zabetakis for help with the TOC figure.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b00156.
Protein sequences for all constructs; a cartoon of the SPR experiments, SPR data, size-exclusion chromatography (controls and assembly on the D-1-3 scaffold), and tables of yield, affinity, and charge content (PDF)
Author Contributions
G.P.A., L.C.S.-L., J.L.L., and E.R.G. performed the experiments and wrote the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Horn A. H. C.; Sticht H. Synthetic Protein Scaffolds Based on Peptide Motifs and Cognate Adaptor Domains for Improving Metabolic Productivity. Front. Bioeng. Biotechnol. 2015, 3, 191–198. 10.3389/fbioe.2015.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K.-H.; Chen R. P.; Sun Q.; Chen L.; Tsai S.-L.; Chen W. Synthetic scaffolds for pathway enhancement. Curr. Opin. Biotechnol. 2015, 36, 98–106. 10.1016/j.copbio.2015.08.009. [DOI] [PubMed] [Google Scholar]
- Lee H.; DeLoache W. C.; Dueber J. E. Spatial organization of enzymes for metabolic engineering. Metab. Eng. 2012, 14, 242–251. 10.1016/j.ymben.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Dueber J. E.; Wu G. C.; Malmirchegini G. R.; Moon T. S.; Petzold C. J.; Ullal A. V.; Prather K. L.; Keasling J. D. Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 2009, 27, 753–759. 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- Chen R.; Chen Q.; Kim H.; Siu K.-H.; Sun Q.; Tsai S.-L.; Chen W. Biomolecular scaffolds for enhanced signaling and catalytic efficiency. Curr. Opin. Biotechnol. 2014, 28, 59–68. 10.1016/j.copbio.2013.11.007. [DOI] [PubMed] [Google Scholar]
- Moon T. S.; Dueber J. E.; Shiue E.; Prather K. L. J. Use of modular, synthetic scaffolds for improved production of glucaric acid in engineered E. coli. Metab. Eng. 2010, 12, 298–305. 10.1016/j.ymben.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Tsai S.-L.; Oh J.; Singh S.; Chen R.; Chen W. Functional Assembly of Minicellulosomes on the Saccharomyces cerevisiae Cell Surface for Cellulose Hydrolysis and Ethanol Production. Appl. Environ. Microbiol. 2009, 75, 6087–6093. 10.1128/AEM.01538-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speltz E. B.; Nathan A.; Regan L. Design of Protein-Peptide Interaction Modules for Assembling Supramolecular Structures in Vivo and in Vitro. ACS Chem. Biol. 2015, 10, 2108–2115. 10.1021/acschembio.5b00415. [DOI] [PubMed] [Google Scholar]
- Whitaker W. R.; Dueber J. E.. Metabolic Pathway Flux Enhancement by Synthetic Protein Scaffolding. In Method Enzymol; Voigt C., Ed.; Academic Press, 2011; Chapter 19, pp 447–468. [DOI] [PubMed] [Google Scholar]
- Thomas F.; Boyle A. L.; Burton A. J.; Woolfson D. N. A Set of de Novo Designed Parallel Heterodimeric Coiled Coils with Quantified Dissociation Constants in the Micromolar to Sub-nanomolar Regime. J. Am. Chem. Soc. 2013, 135, 5161–5166. 10.1021/ja312310g. [DOI] [PubMed] [Google Scholar]
- Gradišar H.; Jerala R. De novo design of orthogonal peptide pairs forming parallel coiled-coil heterodimers. J. Pept. Sci. 2011, 17, 100–106. 10.1002/psc.1331. [DOI] [PubMed] [Google Scholar]
- Reinke A. W.; Grant R. A.; Keating A. E. A Synthetic Coiled-Coil Interactome Provides Heterospecific Modules for Molecular Engineering. J. Am. Chem. Soc. 2010, 132, 6025–6031. 10.1021/ja907617a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromley E. H. C.; Sessions R. B.; Thomson A. R.; Woolfson D. N. Designed α-Helical Tectons for Constructing Multicomponent Synthetic Biological Systems. J. Am. Chem. Soc. 2009, 131, 928–930. 10.1021/ja804231a. [DOI] [PubMed] [Google Scholar]
- Fletcher J. M.; Boyle A. L.; Bruning M.; Bartlett G. J.; Vincent T. L.; Zaccai N. R.; Armstrong C. T.; Bromley E. H. C.; Booth P. J.; Brady R. L.; Thomson A. R.; Woolfson D. N. A Basis Set of de Novo Coiled-Coil Peptide Oligomers for Rational Protein Design and Synthetic Biology. ACS Synth. Biol. 2012, 1, 240–250. 10.1021/sb300028q. [DOI] [PubMed] [Google Scholar]
- Gradišar H.; Božič S.; Doles T.; Vengust D.; Hafner-Bratkovič I.; Mertelj A.; Webb B.; Šali A.; Klavžar S.; Jerala R. Design of a single-chain polypeptide tetrahedron assembled from coiled-coil segments. Nat. Chem. Biol. 2013, 9, 362–366. 10.1038/nchembio.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badieyan S.; Sciore A.; Eschweiler J. D.; Koldewey P.; Cristie-David A. S.; Ruotolo B. T.; Bardwell J. C. A.; Su M.; Marsh E. N. G. Symmetry-Directed Self-Assembly of a Tetrahedral Protein Cage Mediated by de Novo-Designed Coiled Coils. ChemBioChem 2017, 18, 1888–1892. 10.1002/cbic.201700406. [DOI] [PubMed] [Google Scholar]
- Thompson K. E.; Bashor C. J.; Lim W. A.; Keating A. E. SYNZIP Protein Interaction Toolbox: in Vitro and in Vivo Specifications of Heterospecific Coiled-Coil Interaction Domains. ACS Synth. Biol. 2012, 1, 118–129. 10.1021/sb200015u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. C.; Dueber J. E.; Leguia M.; Wu G. C.; Goler J. A.; Arkin A. P.; Keasling J. D. BglBricks: A flexible standard for biological part assembly. J. Biol. Eng. 2010, 4, 1–12. 10.1186/1754-1611-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyldermans S. Nanobodies: Natural Single-Domain Antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- Goldman E. R.; Liu J. L.; Zabetakis D.; Anderson G. P. Enhancing Stability of Camelid and Shark Single Domain Antibodies: An Overview. Front. Immunol. 2017, 8, 865. 10.3389/fimmu.2017.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya A.; Ruvinov S. B.; Gal J.; Moll J. R.; Vinson C. A Heterodimerizing Leucine Zipper Coiled Coil System for Examining the Specificity of a Position Interactions: Amino Acids I, V, L, N, A, and K. Biochemistry 2002, 41, 14122–14131. 10.1021/bi020486r. [DOI] [PubMed] [Google Scholar]
- Grünberg R.; Ferrar T. S.; van der Sloot A. M.; Constante M.; Serrano L. Building blocks for protein interaction devices. Nucleic Acids Res. 2010, 38, 2645–2662. 10.1093/nar/gkq152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner K. B.; Liu J. L.; Zabetakis D.; Lee A. B.; Anderson G. P.; Goldman E. R. Improving the biophysical properties of anti-ricin single-domain antibodies. Biotechnol. Rep. 2015, 6, 27–35. 10.1016/j.btre.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. L.; Goldman E. R.; Zabetakis D.; Walper S. A.; Turner K. B.; Shriver-Lake L. C.; Anderson G. P. Enhanced production of a single domain antibody with an engineered stabilizing extra disulfide bond. Microb. Cell Fact. 2015, 14, 158–165. 10.1186/s12934-015-0340-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashor C. J.; Helman N. C.; Yan S.; Lim W. A. Using Engineered Scaffold Interactions to Reshape MAP Kinase Pathway Signaling Dynamics. Science 2008, 319, 1539–1543. 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- Goldman E. R.; Anderson G. P.; Brozozog-Lee P. A.; Zabetakis D. SdAb heterodimer formation using leucine zippers. Proc. SPIE 2013, 872313 10.1117/12.2016145. [DOI] [Google Scholar]
- Goldman E. R.; Broussard A.; Anderson G. P.; Liu J. L. Bglbrick strategy for the construction of single domain antibody fusions. Heliyon 2018, 3, e00474 10.1016/j.heliyon.2017.e00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.