Abstract
Layered double hydroxide (LDH) nanoparticles (NPs) are safe and effective vectors for small interfering RNA (siRNA) delivery. However, it is unclear whether there are optimal parameters for the efficient delivery of functional siRNA using LDH NPs. In this research, we comprehensively examined the effect of parameters, such as the mixing method and LDH/siRNA mass ratio on siRNA silencing capability. We first noted that the best way for loading gene segments (25 bp dsDNA and siRNA) is to add gene molecules to 100 nm LDH and then diluting in Dulbecco’s modified Eagle’s medium. Very interestingly, the optimal LDH/gene mass ratio is around 20:1 in terms of cellular uptake amount of gene segments, whereas this ratio is shifted to around 5:1 in terms of target gene silencing efficacy, which has been reasonably explained. The optimization for LDH NP-based gene delivery system may provide the guidance for more efficient in vitro and in vivo siRNA delivery using the optimal parameters.
1. Introduction
Layered double hydroxide (LDH) is a group of anionic clay materials that have attracted increasing attention in recent years for biomedical applications, such as gene delivery, vaccine delivery, and drug delivery.1−3 LDH materials have several unique features, including anionic exchange capacity,4 and the ability to capture organic and inorganic anions.5−7 MgAl-LDH nanoparticles (NPs) have been extensively investigated as vehicles for delivery of genes and drugs to cells,8−11 which are biocompatible, have a high loading capacity, facilitate cellular uptake, and release target biomolecules in a pH-dependent manner.12 Moreover, the inherent surface positive charge and anion exchange property enable MgAl-LDH NPs to carry and deliver anionic biomolecules and pharmaceutical drugs such as oligonucleotides,13 DNAs,14,15 RNAs,10,14 methotrexate,16 5-fluorouracil,13 and anti-inflammatory drugs (diclofenac, gemfibrozil, ibuprofen, and naproxen).11 Recent studies have demonstrated the capacity of LDH NPs to target specific cells15 or subcellular compartments.1 More advantages of LDH NPs as delivery vehicles include the low toxicity, protection of payloads, and high cellular delivery efficacy.17,18 These properties indicate that LDH NPs are a good cellular delivery system for biomolecules such as DNAs or RNAs.
RNA interference (RNAi) is a biological mechanism, in which small interfering RNA (siRNA) or microRNA destroys targeted messenger RNA (mRNA) to suppress specific gene expression.19−21 Because of its preciseness and effectiveness, RNAi has proven to be a promising measure for the treatment of cancer.22−25 However, some impediments hinder its further clinical use, such as low cellular uptake and instability of RNA molecules, under physiological conditions. Thus, it is necessary to devise an efficient gene delivery system for RNAi molecules to enter tumor cells to elicit gene modification effects.26−29
Mg2Al–Cl-LDH nanomaterials have shown great potential to be an efficient delivery system for RNA molecules in RNAi-based treatment of cancers, as reported elsewhere.13,14,30,31 However, it is not clear whether the parameters applied to formulate the LDH/siRNA hybrid system are optimal as there is no such report about the parameter optimization for this system.
Therefore, the objectives of this research were to: (1) examine the effect of LDH NP/siRNA mixing method on the siRNA uptake by MCF-7 cells; (2) investigate the effects of the LDH NP/siRNA mass ratio on the cellular uptake and the target gene knockdown efficacy; and (3) confirm the delivery efficacy of siRNA in terms of target mRNA silence and target protein expression reduction using the optimized parameters. Our findings in this research suggest a set of optimal parameters for efficient siRNA delivery using LDH NPs.
2. Results and Discussion
2.1. Physicochemical Features of LDH NPs
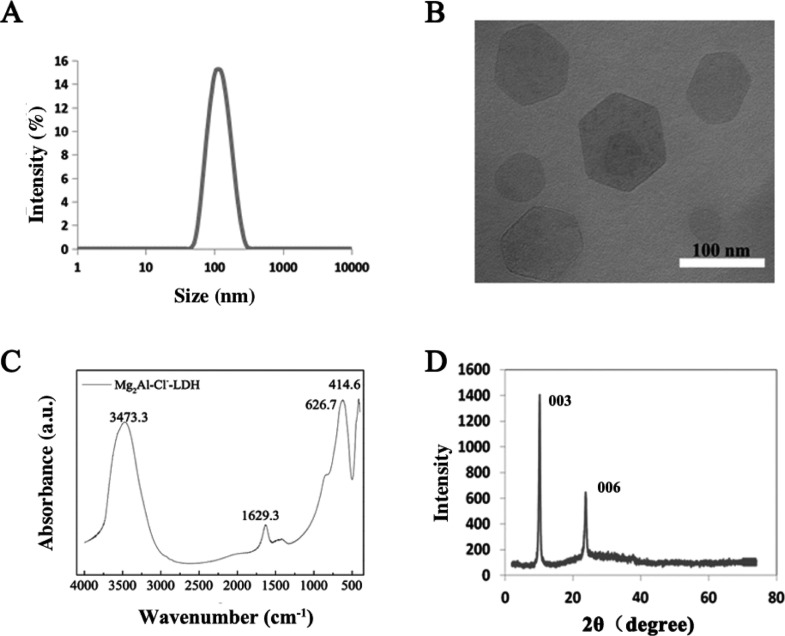
Analysis of homogeneously dispersed Mg2Al–Cl-LDH suspension gave a narrow particle size distribution (Figure 1A). The equivalent mean hydrodynamic diameter was 110 nm with the polydispersity index of 0.099; most particles were distributed within a range of 50–200 nm, and the suspension was transparent. Consistent with previous reports,32−34 Mg2Al–Cl-LDH NPs had an average zeta potential of ∼40 mV in suspension. The transmission electron microscopy (TEM) image (Figure 1B) shows that the LDH crystallites were well-crystallized with a typical hexagonally shaped morphology, as reported previously.32,34 The lateral length of the crystallites was in the range of 60–160 nm, corresponding to the hydrodynamic diameter of most LDH NPs measured by photon correlation spectroscopy (PCS), indicating that LDH NPs were individually dispersed in aqueous solution.
Figure 1.
Characterization of LDH NPs: (A) particle size distribution of Mg2Al–Cl-LDH NPs in suspension; (B) TEM image; (C) FTIR spectrum; and (D) XRD pattern of Mg2Al–Cl-LDH NPs.
The layered structure of as-prepared LDH NPs is confirmed by the X-ray diffraction (XRD) pattern (Figure 1C) and Fourier transform infrared (FTIR) spectrum (Figure 1D). The FTIR spectrum is typical of Mg2Al–Cl-LDH material, as featured by a broad band at 3473 cm–1 (νOH), a peak at 1629 cm–1 (δH2O), and a broad band at 627 and 415 cm–1 (due to M–O vibrations and M–O–H bending, respectively). According to the XRD pattern of Mg2Al–Cl-LDH NPs, this LDH had a basal spacing of 0.781 nm. Taking into account that the thickness of the LDH hydroxide layer is 0.48 nm, the interlayer spacing is 0.31 nm, similar with the size of the chloride ion.32 These data indicate that LDH NPs prepared in this study possess the typical physicochemical properties, as reported previously.8,35,36
2.2. Optimization of the Mixing Method and the Culture Time
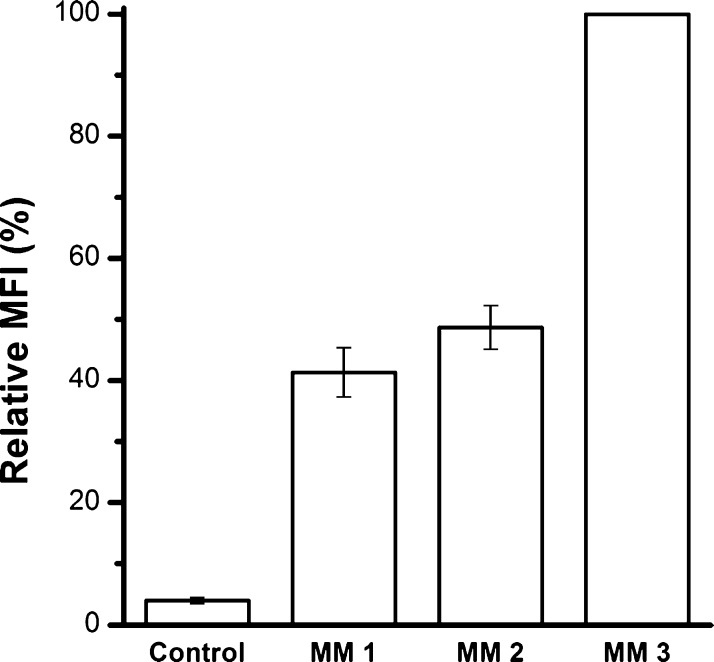
Three mixing methods were designed to load siRNA/double-stranded DNA (dsDNA) onto LDH NPs. siRNA was mimicked by dsDNA-Cy3, and the loading efficiency of each mixing method was quantified by the cellular uptake amount of dsDNA-Cy3-loaded LDH NPs, which was measured in terms of the average cell fluorescence intensity by fluorescence-activated cell sorting (FACS). After LDH/dsDNA hybrids were obtained in one mixing method, they were added in the culture medium for MCF-7 cells to take up for 4 h. Cells were then harvested and fixed for FACS analysis. In each test, the fluorescence intensity of cells from group mixing method 3 (MM 3) was adjusted as 100% and compared with that from the other two groups, as shown in Figure 2. The conclusion is that MM3 exhibits the highest uptake efficiency for the siRNA/dsDNA delivery to MCF-7 cells. This optimal mixing method was employed in the following experiments.
Figure 2.
Optimization of the mixing method. dsDNA was loaded to LDH NPs in MM 1, 2, and 3 at LDH/dsDNA mass ratio = 20:1. Then, the mixture was added into the medium for MCF-7 uptake. The uptake amount was measured in terms of the mean fluorescence intensity (MFI) by FACS. Data presented as the mean ± SE from three independent experiments.
The mixing method affects the LDH/dsDNA delivery efficacy probably through changing the aggregation of LDH NPs when mixing with cell culture medium. As reported previously, the aggregation is concentration- and size-dependent,35 happening through shortening the van del Walls thickness of electron double layers on the LDH NP surface and serum protein bridging effect.37 When LDH is premixed with the medium [mixing method 1 (MM1) and mixing method 2 (MM2)], the surface of LDH NPs is partly occupied with serum proteins and the electron double-layer thickness on the LDH NP surface is decreased by the salt ions, which may induce aggregation of LDH NPs.37 Further, phosphate anions in the medium are also adsorbed on the LDH NP surface, which may significantly reduce the dsDNA loading onto LDH NPs via anion exchange. Both aggregation and less exchange liability may result in some free dsDNA and thus less cellular uptake. When dsDNA is first loaded onto LDH NPs via surface adsorption and interlayer intercalation of dsDNA,38 dsDNA is all adsorbed onto the LDH particle surface. The surface loading normally results in LDH/dsDNA aggregates,37 which may be then better dispersed in culture medium, leading to an increased cellular uptake.
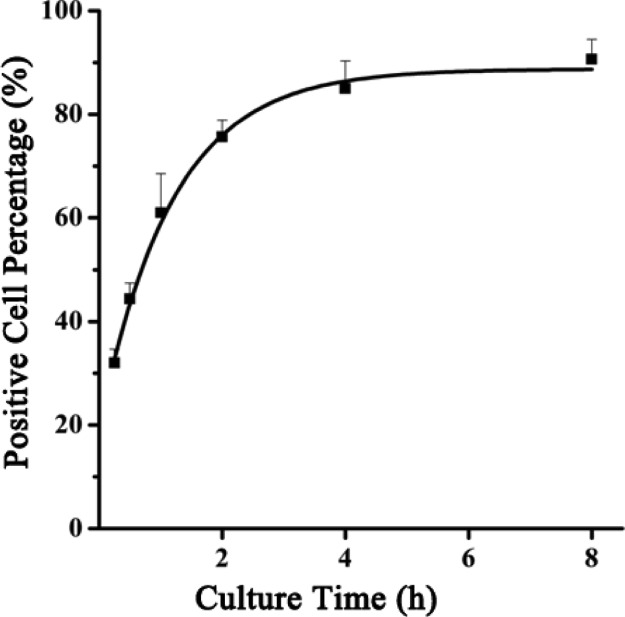
To optimize the cellular uptake time, MCF-7 cells were treated with LDH/dsDNA-Cy3 (MM3, LDH/dsDNA = 20:1) and harvested for FACS analysis at different time points to assess the Cy3-positive cell percentage. As shown in Figure 3, a robust uptake of LDH/dsDNA hybrids was observed during the first hour. After LDH/dsDNA treatment for 4 h, vast majority of MCF-7 cells (>80%) were positive. Extending the culture time to 8 h seemed not to obviously increase the positive cell percentage, as reported previously.35 Thus, 4 h appears to be the optimal culture time.
Figure 3.
Cellular uptake profile of LDH/dsDNA hybrids represented by the positive cell percentage vs culture time. LDH/dsDNA = 20:1, [LDH] = 20.8 μg/mL, and [dsDNA-Cy3] = 80 nM.
2.3. Optimization of the LDH/dsDNA Mass Ratio for Cellular Uptake
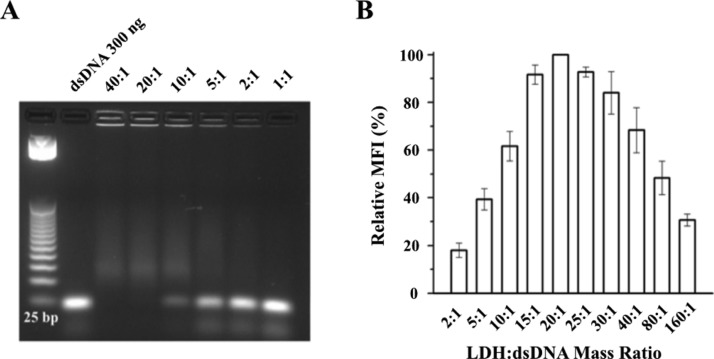
For optimization of the LDH/dsDNA mass ratio for gene delivery, dsDNA (25 bp) was immobilized with LDH NPs at different mass ratios. From the imaging, dsDNA was completely immobilized by LDH NPs when LDH/dsDNA was 20:1 or 40:1 (Figure 4A). At 10:1, most dsDNA was loaded onto LDH, whereas considerable dsDNA was free at 5:1 to 1:1. For comparison, the MFI of cells treated with LDH/dsDNA hybrids at the mass ratio of 20:1 was set at 100%. The MFI from other groups was then compared to give a relative MFI. Clearly, MCF-7 cells took up most of the dsDNA via LDH NPs at the mass ratio between 15:1 and 25:1 (Figure 4B).
Figure 4.
(A) Agarose gel electrophoresis for examining the immobilization of dsDNA by LDH NPs at the mass ratio from 40:1 to 1:1. (B) Relative cellular uptake amount of dsDNA represented by the relative MFI of MCF-7 cells vs the LDH/dsDNA mass ratio at [dsRNA-Cy3] = 80 nM = 1.04 μg/mL.
The relationship between the mass ratio and cellular uptake rate probably results from the trade-off between the dsDNA amount per LDH NP and the concentration of LDH NP number. Because almost all dsDNA-Cy3 is loaded onto LDH NPs at the mass ratio of 15:1 and above (Figure 4A), dsDNA-Cy3 is all adsorbed onto LDH NPs in the mass ratio range indicated in Figure 5B,C, and the dsDNA amount per LDH NP is linearly dependent on the LDH/dsDNA mass ratio. In the mass ratio range in Figure 5A, the maximum dsDNA amount per LDH NP may be achieved at the mass ratio of 10:1 to 2:1 and may be nearly constant, so there is an essential amount of free dsDNA-Cy3 at the mass ratio of 5:1 to 2:1. As also pointed out in previous studies,35,39,40 the cellular uptake efficiency of LDH NPs is almost linearly dependent on the LDH concentration under 20 μg/mL (∼1.0 × 1010 number/mL) and reached a plateau beyond this critical concentration. In the current research, the critical concentration occurs at the LDH/dsDNA mass ratio of around 20:1. Therefore, the dsDNA cellular uptake is monotonically reduced with the mass ratio from 20:1 to 80:1 (Figure 4B) as the LDH NP number taken up by the cells is similar but the dsDNA amount per LDH NP is decreased with the mass ratio from 20:1 to 80:1. With the mass ratio changing from 20:1 to 10:1, the LDH NP number taken up by the cells is decreased but the dsDNA amount per LDH NP is increased. It seems that the former is predominant over the latter so that the dsDNA uptake is decreased overall (Figure 4B). Obviously, the dsDNA amount per LDH NP is maximized in the mass ratio of 10:1 to 2:1, whereas the LDH NP number concentration is decreased linearly; thus, the cellular uptake of dsDNA is reduced accordingly (Figure 4B). It should be mentioned that the aggregation state of LDH/dsDNA hybrids may be slightly varied at different mass ratios as the particle size of these aggregates was mainly distributed in the range of 100–1000 nm in the culture medium.37 We believe that the aggregation state does not significantly affect the dsDNA transfection to MCF-7 cells.
Figure 5.
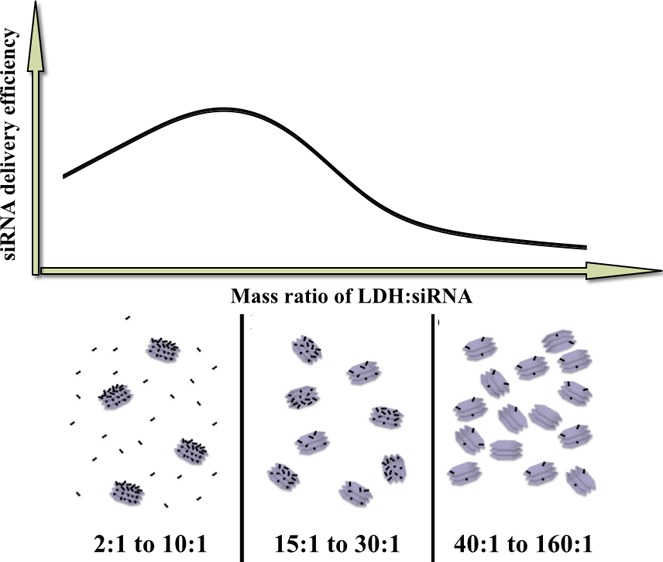
Scheme illustration of the dsRNA loading amount per LDH NPs and the relative LDH NP number at varied LDH/dsDNA mass ratios. (A) At 2:1 to 10:1, LDH NPs have the maximum dsDNA loading with some free dsDNA; (B) at 15:1 to 30:1, LDH NPs have the maximum dsDNA loading without free dsDNA; and (C) at 40:1 to 160:1, each LDH NP loads fewer dsDNA molecules without free dsDNA.
Therefore, it appears that the optimal LDH/dsDNA mass ratio is around 20:1 in terms of the cellular uptake amount of dsDNA-Cy3 at the dsDNA concentration of 80 nM in the culture medium.
2.4. Optimized Functional siRNA Delivery to MCF-7
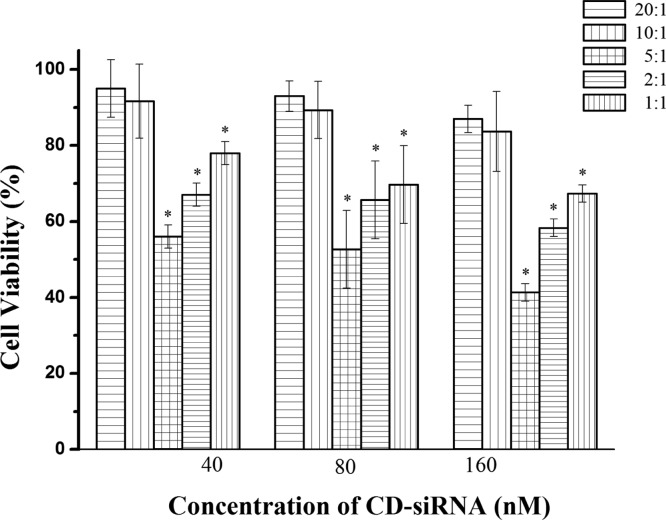
To verify the feasibility of optimized siRNA delivery system, MCF-7 cells were transfected with cell death siRNA (CD-siRNA) at different LDH/siRNA mass ratios ranging from 1:1 to 40:1. MCF-7 cells were incubated with LDH/CD-siRNA for 4 h and followed by incubation for 72 h in standard culture medium. As shown in Figure 6, the MCF-7 cell viability was decreased by approximately 45–60% at a mass ratio of 5:1 with the CD-siRNA concentration from 40 to 160 nM. This decrease of cell viability is much more than that at the mass ratio of 20:1 (5–12%), 10:1 (8–17%), 2:1 (32–42%), and 1:1 (22–33%) under the same conditions, respectively. This observation indicates that the optimal LDH/siRNA mass ratio is around 5:1 in terms of CD-siRNA inhibition efficacy of cancer cells, which is much different from that (20:1) in terms of just cellular uptake by or delivery to MCF-7 cells (Figure 4B).
Figure 6.
Cell viability of MCF-7 transfected with CD-siRNA delivered by LDH NPs under various conditions. Data presented as the mean ± SE from three independent experiments.
The shift of the optimal mass ratio from 20:1 for just cellular uptake to 5:1 for functional siRNA to take effect may be further related to the siRNA release from LDH/siRNA hybrids to the cytosol so as to target and dice specific mRNA. The siRNA release from LDH/siRNA hybrids may occur via two processes, that is, dissolution of LDH NPs and anion exchange. As revealed previously,35,41 cellular internalization of LDH NPs is basically driven by clathrin-mediated endocytosis. During endocytosis, a certain amount of LDH NPs is acidified in the endosome (pH 5–6) to be dissolved and release siRNA molecules, which may also occur in the cytosol (pH 7.4) at a much slower pace after LDH/siRNA hybrids escape from the endosome. As the dissolved LDH NP amount would be very similar for each cell in a similar time scale, thus LDH NP with the maximum siRNA loading would free most siRNA molecules for complexing with RNA-induced silencing complex and dicing the target mRNA. In comparison, double amount of LDH NPs would be taken up by MCF-7 cells at 10:1 as at 5:1; thus, only half amount of free siRNA would be released to the MCF-7 cytosol at 10:1 as at 5:1 and would result in lower inhibition on cell viability. At the mass ratio of 20:1 to 80:1, the release of siRNA is even more difficult, leading to much lower inhibition of tumor cell growth (Figure 6). In contrast, less than half amount of LDH NPs would be taken up by MCF-7 cells at 2:1 as at 5:1 (LDH NP number at 2:1 was about 40% at 5:1 when siRNA was fixed at 40, 80, or 160 nM); thus, less than half amount of siRNA may be released to MCF-7 cytosol at 2:1 as at 5:1 (supposed the siRNA loading per LDH NP is maximum and the same), resulting in much lower cell growth inhibition. In addition, few siRNA molecules on the LDH surface would be exchanged with cytosol anions (Cl– and phosphates) and become free, but this would contribute limitedly to the cell viability.
Therefore, the optimal LDH/siRNA mass ratio seems to be 5:1 for functional siRNA delivery as the result of trading-off of these several processes, that is, the loading amount of dsDNA/siRNA per LDH particle, the number of LDH particles, and the suitable release rate of dsDNA/siRNA. Comparatively, we used an LDH/siRNA mass ratio of 1:1 for siRNA delivery to neuron and NIH3T3 cells in our previous research studies,10,42 which may underlie the delivery capacity of LDH NPs. On the other hand, using the mass ratio of 40:1 for cellular uptake of siRNA35 is not optimal either. Similarly, varied mass ratios (10:1 to 40:1) were used to deliver CD-siRNA to kill cancer cells in combination with 5-FU,13 which may not maximize the biological function of CD-siRNA in killing cancer cells.
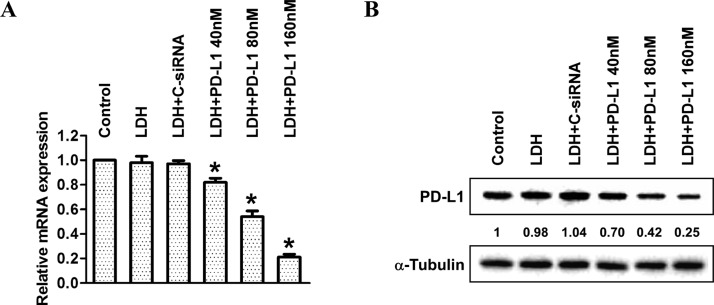
As a feasibility test, the optimized parameters for functional siRNA delivery were used to formulate LDH/PD-L1-siRNA at the concentration of 40, 80, and 160 nM with the LDH/siRNA mass ratio of 5:1, which was used to treat MCF-7 for target gene silence. As shown in Figure 7, the delivered PD-L1 siRNA efficiently silenced PD-L1 mRNA expression (20–80%) and PD-L1 expression (30–75%).
Figure 7.
Downregulation of PD-L1 in MCF-7. (A) Real-time PCR data for the knockdown of PD-L1 mRNAs and (B) western blot showing reduction of PD-L1 expression in MCF-7 treated with LDH/PD-L1-siRNA at the concentration from 40 to 160 nM.
Therefore, this research has optimized and confirmed the LDH NP system for efficient in vitro delivery of genes to cancer cells, which is believed to be also applicable for the in vivo delivery for cancer gene therapy.43,44
3. Conclusions
In this work, the mixing method, cellular uptake time, and LDH/siRNA mass ratio were optimized for functional siRNA delivery to cancer cells. We found that the optimal mixing method was to directly add siRNA into the LDH suspension, followed by a dilution with culture medium, and the optimal cellular uptake time was 4 h, with >80% cells being transfected. In the mass ratio optimization, MCF-7 cells seem to take up the greatest number of dsDNA or siRNA at the LDH/gene mass ratio of 20:1. However, most functional CD-siRNA and PD-L1 siRNA seem to avail and work the best at the mass ratio of around 5:1. Therefore, this research has thus optimized the LDH-based platform for gene delivery in vitro as well as in vivo.
4. Materials and Methods
4.1. Chemicals and Reagents
All samples were prepared under sterile conditions. Sodium hydroxide pellets, magnesium chloride hexahydrate (MgCl2·6H2O), and aluminum chloride hexahydrated (AlCl3·6H2O) were purchased from Ajax Finechem, and Sigma-Aldrich Pty Ltd, respectively. dsDNA-Cy3 was purchased from GeneWorks. CD-siRNA was purchased from QIAGEN Pty. Ltd. PD-L1 siRNA (sense: 5′-AGAcGuAAGcAGuGuuGAAdTsdT-3′ and antisense: 5′-UUcAAcACUGCUuACGUCUdTsdT-3′) and other chemicals and reagents were purchased from Sigma-Aldrich if not illustrated specifically. Water used in experiments was deionized Milli-Q water.
4.2. LDH Preparation
Mg2Al–Cl-LDH NPs were synthesized by coprecipitation-hydrothermal method, which was well established in our group.8 Briefly, a mixture of MgCl2 (0.70 M) and AlCl3 (0.30 M) with a total volume of 10 mL was quickly added to 40 mL of NaOH solution (0.45 M) within 5 s, under vigorous stirring. After 10 min of stirring, the slurry was separated via centrifugation and redispersed in 40 mL of deionized water. The resultant suspension was moved into a stainless steel autoclave with a Teflon lining and heated at 100 °C for 16 h. The final mass concentration of LDH in the obtained transparent suspensions was approximately 10 mg/mL, with the yield of ∼60%.
4.3. Characterization of LDH
The particle size distribution of as-prepared LDH NPs was determined by PCS (Nanosizer Nano ZS, Malvern Instruments) using noninvasive backscatter optics. For TEM imaging, LDH solution was air-dried on a copper grid. The images were obtained on a JEOL 1010A transmission electron microscope at an acceleration voltage of 200 kV. FTIR (PerkinElmer 1760X FTIR) spectrum and XRD (Siemens F-series diffractometer with Cu Kα radiation, λ = 0.15418 nm) pattern were collected to confirm the layered structure of LDH NPs.
4.4. Cell Culture
MCF-7, a widely studied epithelial cancer cell line derived from breast adenocarcinoma, was used as the cancer cell model in this study. Normally, MCF-7 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented by 10% (v/v) fetal bovine serum (FBS) at 37 °C in 5% CO2 atmosphere.
4.5. Optimization of the siRNA Delivery with LDH NPs
LDH NPs were used to load siRNA-mimicking dsDNA-Cy3 using different parameters. The delivery efficiency was evaluated by FACS, which was used to decide the optimal parameters.
For mixing method optimization, we designed three different methods. In MM 1, both dsDNA and LDH suspension (LDH/dsDNA mass ratio was 20:1) were diluted with DMEM containing 10% FBS. After 15 min incubation at room temperature, diluted dsDNA was added to the diluted LDH suspension, and the mixture was left at room temperature for another 15 min. In MM 2, only the LDH suspension was diluted in the medium before mixing with dsDNA, whereas in MM 3, dsDNA was directly added into the LDH suspension, and then the mixture was diluted to the designed volume. The obtained mixture (at the nominal dsDNA-Cy3 concentration of 80 nM) was added to MCF-7 cells seeded in 6-well plates for 24 h (1 × 105 cells per well) and then kept for 4 h. The cells were harvested and fixed for FACS analysis afterwards.
To define the optimal LDH/siRNA mass ratio for transfection, an appropriate dose of dsDNA was mixed with different amounts of LDH in suspension with LDH/dsDNA mass ratios ranging from 2:1 to 160:1, using MM 3 as mentioned above. After adding the mixture, the cells were incubated at 37 °C with 5% CO2 for 4 h and then collected for FACS analysis.
To determine the proper culture time, MCF-7 cells treated with LDH/dsDNA-Cy3 were placed in an atmosphere of 37 °C with 5% CO2. At different time points (0.25, 0.5, 1, 2, 4, and 8 h), cells were collected for FACS analysis.
4.6. FACS Analysis
FACS analysis was performed in a BD Accuri C6 (San Jose, CA, USA) flow cytometer with CFlow Sampler software (Becton Dickinson, Mountain View, USA).
4.7. Cell Viability Assay
Cell viability was assessed by a colorimetric assay using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT). Briefly, MCF-7 cells were cultured in 96-well plates (2.5 × 103 cells per well) and treated with LDH/CD-siRNA at the concentration ranging from 40 to 160 nM. After 24 h, DMEM was supplemented with 20 μL of MTT reagent (0.5 mg/mL) to each well and incubated for 3 h at 37 °C. Thereafter, MTT solution was removed. After addition of 100 μL of dimethyl sulfoxide (DMSO), the plate was incubated for 10 min at 37 °C to dissolve the formazan crystals. Absorbance readings of DMSO extracts were performed at 570 nm with reference to those at 650 nm using the plate reader.
4.8. Knockdown of PD-L1 Gene in MCF-7 with siRNA
PD-L1 siRNA (sense: 5′-AGAcGuAAGcAGuGuuGAAdTsdT-3′ and antisense: 5′-UUcAAcACUGCUuACGUCUdTsdT-3′) was used in this study. For MCF-7 cells, 1 × 105 cells/well were seeded in a 6-well plate and cultured at 37 °C for 4 h in the presence of LDH/PD-L1-siRNA that was formulated using optimal parameters. The culture medium was then replaced with the fresh medium, and transfected MCF-7 cells were cultured for another 72 h at 37 °C, 5% CO2. PD-L1 mRNA and protein level were examined by real-time polymerase chain reaction (PCR) and western blotting, respectively.
4.9. Real-Time PCR
Real-time PCR assays were carried out according to the manufacturer instructions. Briefly, 1.2 mL of trizol/chloroform (1:5, v/v) was mixed with lyse cells and the supernatant was collected by centrifugation (12 500 rpm, 15 min). Subsequently, 2.4 mL of isopropanol was added, followed by centrifugating the supernatant for another 15 min at 12 500 rpm. The pellet was collected and washed by 70% ethanol once. The pellet was resuspended in 50 μL of H2O after drying and diluted with 80 μL of H2O. For each well, 3.5 μL of pellet solution was mixed with 8.5 μL of PCR Master Mixture solution. Real-time PCR was conducted after centrifugation.
4.10. Western Blotting
The relative PD-L1 concentration in cell lysates was estimated using the Pierce BCA Protein Assay Kit (Thermo Scientific). The samples (roughly 10 μg total protein per well) were mixed with protein-loading buffer (Bio-Rad) containing 2-mercaptoethanol. After denaturation by 5 min boiling, the samples were loaded on 4–15% Mini-PROTEAN TGX precast gels sodium dodecyl sulfate polyacrylamide gel (Bio-Rad). The gels were blotted onto poly(vinylidene difluoride) (PVDF) membranes for 90 min @ 80 V; proteins were transferred onto PVDF membranes and blocked for nonspecific binding with Tris-buffered saline with Tween-20 (TBST) [0.05% (v/v) Tween-20 in TBS pH 7.4] plus 5% bovine serum albumin for 1 h at room temperature. PD-L1 RabMab antibodies (ab205921) (Abcam, USA; at dilution of 1:800–1000) were applied overnight at 4 °C. The membrane was washed with TBST and then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (goat antirabbit IgG H&L (HRP), ab97051) (Abcam, USA; at dilution 1:5000) for 2 h. After washing, protein bands were visualized using Clarity western ECL substrate (Bio-Rad) and analyzed using ImageJ v1.40 software.
4.11. Statistical Analysis
All experiments were repeated for three or more times. All data are represented as means ± SEM. Data were analyzed by one-way analysis of variance, followed by multiple comparisons using Tukey’s test within GraphPad Prism 6 software. A p value < 0.05 was considered significant.
Acknowledgments
The authors wish to thank the Australian Research Council (ARC) Discovery Project grant (DP170104643) and AIBN Group Leader PhD living Allowance Scholarship for financial supports. The authors also acknowledge the Queensland Node Fabrication Facility (ANFF-Q) and The University of Queensland for the assistance and facilities.
Author Contributions
Y.W., W.G., and Z.P.X. conceived and designed the experiments. Y.W. and S.T.D. performed the experiments. Y.W., W.G., and Z.P.X. analyzed the data. All authors contributed to the writing of the manuscript. All authors have approved the final version of the manuscript.
The authors declare no competing financial interest.
References
- Xu Z. P.; Niebert M.; Porazik K.; Walker T. L.; Cooper H. M.; Middelberg A. P. J.; Gray P. P.; Bartlett P. F.; Lu G. Q. Subcellular compartment targeting of layered double hydroxide nanoparticles. J. Controlled Release 2008, 130, 86–94. 10.1016/j.jconrel.2008.05.021. [DOI] [PubMed] [Google Scholar]
- Choy J.-H.; Jung J.-S.; Oh J.-M.; Park M.; Jeong J.; Kang Y.-K.; Han O.-J. Layered double hydroxide as an efficient drug reservoir for folate derivatives. Biomaterials 2004, 25, 3059–3064. 10.1016/j.biomaterials.2003.09.083. [DOI] [PubMed] [Google Scholar]
- Choi S.-J.; Choy J.-H. Layered double hydroxide nanoparticles as target-specific delivery carriers: uptake mechanism and toxicity. (Report). Nanomedicine 2011, 6, 803. 10.2217/nnm.11.86. [DOI] [PubMed] [Google Scholar]
- Wang S.; Li Z.; Lu C. Polyethyleneimine as a novel desorbent for anionic organic dyes on layered double hydroxide surface. J. Colloid Interface Sci. 2015, 458, 315–322. 10.1016/j.jcis.2015.07.056. [DOI] [PubMed] [Google Scholar]
- Ma S.; Huang L.; Ma L.; Shim Y.; Islam S. M.; Wang P.; Zhao L.-D.; Wang S.; Sun G.; Yang X.; Kanatzidis M. G. Efficient uranium capture by polysulfide/layered double hydroxide composites. J. Am. Chem. Soc. 2015, 137, 3670. 10.1021/jacs.5b00762. [DOI] [PubMed] [Google Scholar]
- Ma S.; Islam S. M.; Shim Y.; Gu Q.; Wang P.; Li H.; Sun G.; Yang X.; Kanatzidis M. G. Highly efficient iodine capture by layered double hydroxides intercalated with polysulfides. Chem. Mater. 2014, 26, 7114–7123. 10.1021/cm5036997. [DOI] [Google Scholar]
- Yu S.; Wang X.; Chen Z.; Wang J.; Wang S.; Hayat T.; Wang X. Layered double hydroxide intercalated with aromatic acid anions for the efficient capture of aniline from aqueous solution. J. Hazard. Mater. 2017, 321, 111–120. 10.1016/j.jhazmat.2016.09.009. [DOI] [PubMed] [Google Scholar]
- Yan S.; Rolfe B. E.; Zhang B.; Mohammed Y. H.; Gu W.; Xu Z. P. Polarized immune responses modulated by layered double hydroxides nanoparticle conjugated with CpG. Biomaterials 2014, 35, 9508–9516. 10.1016/j.biomaterials.2014.07.055. [DOI] [PubMed] [Google Scholar]
- Barahuie F.; Hussein M.; Fakurazi S.; Zainal Z.. Development of Drug Delivery Systems Based on Layered Hydroxides for Nanomedicine; MDPI AG: Basel, 2014; Vol. 15, pp 7750–7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Y.; Cooper H. M.; Zhang K.; Chen M.; Bartlett P.; Xu Z. P. Efficiency of layered double hydroxide nanoparticle-mediated delivery of siRNA is determined by nucleotide sequence. J. Colloid Interface Sci. 2012, 369, 453–459. 10.1016/j.jcis.2011.12.046. [DOI] [PubMed] [Google Scholar]
- Khan A. I.; Lei L.; Norquist A. J.; O’Hare D. Intercalation and controlled release of pharmaceutically active compounds from a layered double hydroxide. Chem. Commun. 2001, 2342. 10.1039/b106465g. [DOI] [PubMed] [Google Scholar]
- Li X.-S.; Ke M.-R.; Huang W.; Ye C.-H.; Huang J.-D. A pH-Responsive Layered Double Hydroxide (LDH)–Phthalocyanine Nanohybrid for Efficient Photodynamic Therapy. Chem.—Eur. J. 2015, 21, 3310–3317. 10.1002/chem.201404514. [DOI] [PubMed] [Google Scholar]
- Li L.; Gu W.; Chen J.; Chen W.; Xu Z. P. Co-delivery of siRNAs and anti-cancer drugs using layered double hydroxide nanoparticles. Biomaterials 2014, 35, 3331–3339. 10.1016/j.biomaterials.2013.12.095. [DOI] [PubMed] [Google Scholar]
- Chen M.; Cooper H. M.; Zhou J. Z.; Bartlett P. F.; Xu Z. P. Reduction in the size of layered double hydroxide nanoparticles enhances the efficiency of siRNA delivery. J. Colloid Interface Sci. 2013, 390, 275–281. 10.1016/j.jcis.2012.09.033. [DOI] [PubMed] [Google Scholar]
- Desigaux L.; Belkacem M. B.; Richard P.; Cellier J.; Léone P.; Cario L.; Leroux F.; Taviot-Guého C.; Pitard B. Self-assembly and characterization of layered double hydroxide/DNA hybrids. Nano Lett. 2006, 6, 199–204. 10.1021/nl052020a. [DOI] [PubMed] [Google Scholar]
- Choi G.; Huiyan P.; Alothman Z.; Vinu A.; Yun C.-O.; Choy J.-H. Anionic clay as the drug delivery vehicle: tumor targeting function of layered double hydroxide-methotrexate nanohybrid in C33A orthotopic cervical cancer model. (ORIGINAL RESEARCH). Int. J. Nanomed. 2016, 11, 337. 10.2147/ijn.s95611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H.; Xiu K. M.; Xu S. L.; Yang W. T.; Xu F. J. Functionalized Layered Double Hydroxide Nanoparticles Conjugated with Disulfide-Linked Polycation Brushes for Advanced Gene Delivery. Bioconjugate Chem. 2013, 24, 968. 10.1021/bc300683y. [DOI] [PubMed] [Google Scholar]
- Xu F.-J. Versatile types of hydroxyl-rich polycationic systems via O-heterocyclic ring-opening reactions: From strategic design to nucleic acid delivery applications. Prog. Polym. Sci. 2018, 78, 56–91. 10.1016/j.progpolymsci.2017.09.003. [DOI] [Google Scholar]
- Izquierdo M. Short interfering RNAs as a tool for cancer gene therapy. Cancer Gene Ther. 2005, 12, 217–227. 10.1038/sj.cgt.770079. [DOI] [PubMed] [Google Scholar]
- Weinstein S.; Peer D. Rnai Nanomedicines: Challenges and Opportunities within the Immune System. Nanotechnology 2010, 21, 232001. 10.1088/0957-4484/21/23/232001. [DOI] [PubMed] [Google Scholar]
- Huang L.; Liu Y. In Vivo Delivery of RNAi with Lipid-Based Nanoparticles. Annu. Rev. Biomed. Eng. 2011, 13, 507–530. 10.1146/annurev-bioeng-071910-124709. [DOI] [PubMed] [Google Scholar]
- Tsang J.; Lee V. H. F.; Kwong D. L. W. Novel therapy for nasopharyngeal carcinoma—Where are we. Oral Oncol. 2014, 50, 798. 10.1016/j.oraloncology.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Dassie J. P.; Liu X.-y.; Thomas G. S.; Whitaker R. M.; Thiel K. W.; Stockdale K. R.; Meyerholz D. K.; McCaffrey A. P.; McNamara J. O.; Giangrande P. H. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 2009, 27, 839. 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Bathula S. R.; Li J.; Huang L. Multifunctional nanoparticles delivering small interfering RNA and doxorubicin overcome drug resistance in cancer. J. Biol. Chem. 2010, 285, 22639. 10.1074/jbc.m110.125906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Huang L. Targeted delivery of RNAi therapeutics for cancer therapy. Nanomedicine 2010, 5, 1483–1486. 10.2217/nnm.10.124. [DOI] [PubMed] [Google Scholar]
- Song Y.; Dong M.-M.; Yang H.-F. Effects of RNA interference targeting four different genes on the growth and proliferation of nasopharyngeal carcinoma CNE-2Z cells. Cancer Gene Ther. 2011, 18, 297–304. 10.1038/cgt.2010.80. [DOI] [PubMed] [Google Scholar]
- Lu M.; Shan Z.; Andrea K.; Macdonald B.; Beale S.; Curry D. E.; Wang L.; Wang S.; Oakes K. D.; Bennett C.; Wu W.; Zhang X. Chemisorption Mechanism of DNA on Mg/Fe Layered Double Hydroxide Nanoparticles: Insights into Engineering Effective SiRNA Delivery Systems. Langmuir 2016, 32, 2659. 10.1021/acs.langmuir.5b04643. [DOI] [PubMed] [Google Scholar]
- Stein C. A.; Hansen J. B.; Lai J.; Wu S.; Voskresenskiy A.; Høg A.; Worm J.; Hedtjärn M.; Souleimanian N.; Miller P.; Soifer H. S.; Castanotto D.; Benimetskaya L.; Ørum H.; Koch T. Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res. 2010, 38, e3 10.1093/nar/gkp841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K.; Huang L. Achieving efficient RNAi therapy: progress and challenges. Acta Pharm. Sin. B 2013, 3, 213–225. 10.1016/j.apsb.2013.06.005. [DOI] [Google Scholar]
- Li L.; Zhang R.; Gu W.; Xu Z. P. Mannose-conjugated layered double hydroxide nanocomposite for targeted siRNA delivery to enhance cancer therapy. Nanomed. Nanotechnol. Biol. Med. 2017, 10.1016/j.nano.2017.06.006. [DOI] [PubMed] [Google Scholar]
- Li A.; Qin L.; Wang W.; Zhu R.; Yu Y.; Liu H.; Wang S. The use of layered double hydroxides as DNA vaccine delivery vector for enhancement of anti-melanoma immune response. Biomaterials 2011, 32, 469–477. 10.1016/j.biomaterials.2010.08.107. [DOI] [PubMed] [Google Scholar]
- Xu Z. P.; Stevenson G.; Lu C.-Q.; Lu M.; Bartlett P. F.; Gray P. P. Stable Suspension of Layered Double Hydroxide Nanoparticles in Aqueous Solution. J. Am. Chem. Soc. 2006, 128, 36–37. 10.1021/ja056652a. [DOI] [PubMed] [Google Scholar]
- Xu Z. P.; Stevenson G.; Lu C.-Q.; Lu M. Dispersion and Size Control of Layered Double Hydroxide Nanoparticles in Aqueous Solutions. J. Phys. Chem. B 2006, 110, 16923–16929. 10.1021/jp062281o. [DOI] [PubMed] [Google Scholar]
- Xu Z. P.; Jin Y.; Liu S.; Hao Z. P.; Lu G. Q. Surface charging of layered double hydroxides during dynamic interactions of anions at the interfaces. J. Colloid Interface Sci. 2008, 326, 522–529. 10.1016/j.jcis.2008.06.062. [DOI] [PubMed] [Google Scholar]
- Dong H.; Parekh H. S.; Xu Z. P. Particle size- and number-dependent delivery to cells by layered double hydroxide nanoparticles. J. Colloid Interface Sci. 2015, 437, 10–16. 10.1016/j.jcis.2014.09.010. [DOI] [PubMed] [Google Scholar]
- Braterman P.; Xu Z.; Yarberry F.. Layered Double Hydroxides. In Handbook of Layered Materials; Auerbach S., Carrado K., Dutta P., Eds.; Marcel Dekker: New York, 2004; pp 373–474. [Google Scholar]
- Gu Z.; Zuo H.; Li L.; Wu A.; Xu Z. P. Pre-coating layered double hydroxide nanoparticles with albumin to improve colloidal stability and cellular uptake. J. Mater. Chem. B 2015, 3, 3331–3339. 10.1039/c5tb00248f. [DOI] [PubMed] [Google Scholar]
- Andrea K. A.; Wang L.; Carrier A. J.; Campbell M.; Buhariwalla M.; Mutch M.; Macquarrie S. L.; Bennett C.; Mkandawire M.; Oakes K.; Lu M.; Zhang X. Adsorption of Oligo-DNA on Magnesium Aluminum-Layered Double-Hydroxide Nanoparticle Surfaces: Mechanistic Implication in Gene Delivery. Langmuir 2017, 33, 3926. 10.1021/acs.langmuir.6b04172. [DOI] [PubMed] [Google Scholar]
- Gu Z.; Thomas A. C.; Xu Z. P.; Campbell J. H.; Lu G. Q. In vitro sustained release of LMWH from MgAl-layered double hydroxide nanohybrids. Chem. Mater. 2008, 20, 3715–3722. 10.1021/cm703602t. [DOI] [Google Scholar]
- Choy J.-h.; Kwak S.-y.; Park J.-s.; Jeong Y.-j. Cellular uptake behavior of [γ-32P] labeled ATP–LDH nanohybrids. J. Mater. Chem. 2001, 11, 1671–1674. 10.1039/b008680k. [DOI] [Google Scholar]
- Oh J.-m.; Choi S.-J.; Kim S.-T.; Choy J.-H. Cellular Uptake Mechanism of an Inorganic Nanovehicle and Its Drug Conjugates: Enhanced Efficacy Due To Clathrin-Mediated Endocytosis. Bioconjugate Chem. 2006, 17, 1411. 10.1021/bc0601323. [DOI] [PubMed] [Google Scholar]
- Wong Y.; Markham K.; Xu Z. P.; Chen M.; Lu G. Q.; Bartlett P. F.; Cooper H. M. Efficient delivery of siRNA to cortical neurons using layered double hydroxide nanoparticles. Biomaterials 2010, 31, 8770–8779. 10.1016/j.biomaterials.2010.07.077. [DOI] [PubMed] [Google Scholar]
- Li B.; Gu Z.; Kurniawan N.; Chen W.; Xu Z. P. Manganese-Based Layered Double Hydroxide Nanoparticles as a T1-MRI Contrast Agent with Ultrasensitive pH Response and High Relaxivity. Adv. Mater. 2017, 29, 1700373. 10.1002/adma.201700373. [DOI] [PubMed] [Google Scholar]
- Gu W.; Zou H.; Li L.; Carcedo I. G.; Xu Z. P.; Monteiro M. Synergistic inhibition of colon cancer cell growth with nanoemulsion-loaded paclitaxel and PI3K/mTOR dual inhibitor BEZ235 through apoptosis. (ORIGINAL RESEARCH)(Report). Int. J. Nanomed. 2016, 11, 1947. 10.2147/ijn.s100744. [DOI] [PMC free article] [PubMed] [Google Scholar]