Abstract
l-Threonine is an important supplement in the food industry. It is currently produced through fermentation of Escherichia coli but requires additional purification steps to remove E. coli endotoxin. To avoid these steps, it is desirable to use Corynebacterium glutamicum, a microorganism generally regarded as safe. Engineering of C. glutamicum to increase production of l-threonine has mainly focused on gene regulation as well as l-threonine export or carbon flux depletion. In this study, we focus on the negative feedback inhibition produced by l-threonine on the enzyme homoserine kinase (ThrB). Although l-threonine binds to allosteric sites of aspartate kinase (LysC) and homoserine dehydrogenase (Hom), serving as a noncompetitive inhibitor, it acts as a competitive inhibitor on ThrB. This is problematic when attempting to engineer enzymes that are nonresponsive to increasing cellular concentrations of l-threonine. Using primary structure alignment as well as analysis of the Methanocaldococcus jannaschii ThrB (MjaThrB) active site in complex with l-threonine (inhibitor of ThrB) and l-homoserine (substrate of ThrB), a conserved active-site alanine residue (A20) in C. glutamicum ThrB (CglThrB) was predicted to be important for differential interactions with l-threonine and l-homoserine. Through site-directed mutagenesis, we show that one variant of C. glutamicum ThrB, CglThrB-A20G, retains wild-type enzymatic activity, with dramatically decreased feedback inhibition by l-threonine. Additionally, by solving the first Corynebacterium X-ray crystal structure of homoserine kinase, we can confirm that the changes in l-threonine affinity to the CglThrB-A20G active site derive from loss of van der Waals interactions.
Introduction
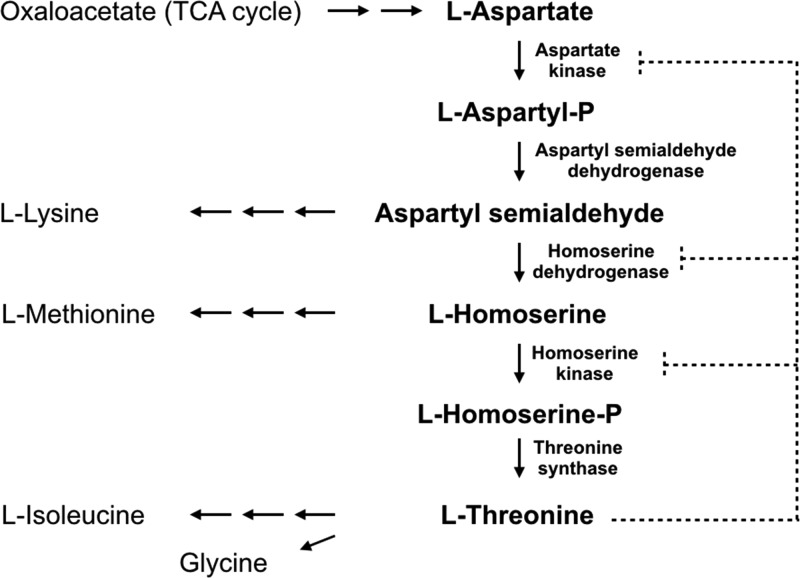
l-Threonine is an essential amino acid for mammals and has a wide spectrum of commercial uses, such as a food additive and agricultural feed supplement. Mainly produced through microbial fermentation of Escherichia coli, considerable efforts have been directed toward more efficient production of l-threonine via metabolic engineering.1 From l-aspartate, the biosynthesis of l-threonine comprises five successive reactions sequentially catalyzed in Corynebacterium glutamicum by aspartate kinase (LysC), aspartyl semialdehyde dehydrogenase (Asd), homoserine dehydrogenase (Hom), homoserine kinase (ThrB), and threonine synthase (ThrC) (Figure 1).
Figure 1.
Biosynthetic pathway of l-threonine and its regulation in C. glutamicum. l-Threonine regulates its own biosynthetic pathway by allosterically inhibiting aspartate kinase (LysC) and homoserine dehydrogenase (Hom) in a noncompetitive manner and homoserine kinase (ThrB) as a competitive inhibitor (shown in broken lines). Solid lines indicate branch pathways related to the l-threonine biosynthetic pathway.
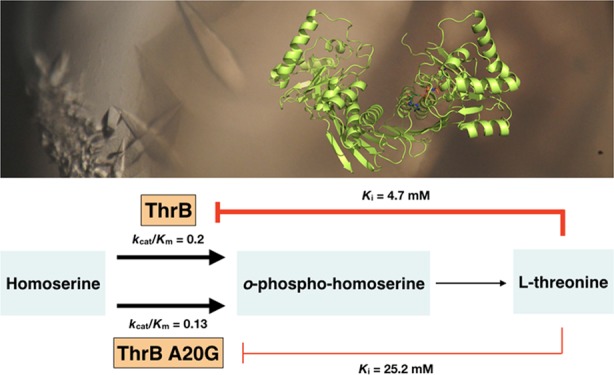
The level of l-threonine production in C. glutamicum is tightly regulated in four different ways (Figure 1). First, available carbon flux is routed from l-threonine biosynthesis mainly toward the competing l-lysine and l-methionine metabolic pathways.2 Second, intracellular l-threonine is depleted through the production of l-isoleucine and glycine.3−6 Third, both the hom and thrB genes in C. glutamicum are clustered in a single operon that is repressed by l-methionine, possibly by a transcriptional attenuation-like regulation mechanism.7,8 Finally, three enzymes in the biosynthetic pathway of l-threonine are regulated by feedback inhibition: homoserine dehydrogenase (Hom) is subject to feedback inhibition by l-threonine and aspartate kinase (LysC) by both l-lysine and l-threonine. These two enzymes are inhibited by l-threonine in an allosteric noncompetitive manner, whereas homoserine kinase (ThrB) is inhibited by l-threonine in a competitive inhibitory mechanism.9−11
To afford higher levels of l-threonine production, several metabolic engineering strategies have been implemented. First, overexpression of the homr-thrB operon that includes a feedback-resistant homoserine dehydrogenase allele is a strategy that has been one of the most successful ones to redirect the carbon flux from the l-lysine branch into the l-threonine branch, leading to the accumulation of both l-threonine and l-homoserine.12,13 The second strategy has been to reduce l-threonine depletion toward l-isoleucine and glycine by mutation of both glyA and ilvA responsible for the conversion of l-threonine into glycine and l-isoleucine, respectively.14 Another strategy was to enhance the efflux of l-threonine, which typically accumulates in the cell and thus downregulates the biosynthetic enzymes in a feedback manner.4,15 Both the introduction of a recombinant plasmid containing the permease thrE gene and the overexpression of the E. coli genes rhtA, rhtC, and yeaS, previously inserted into C. glutamicum, lead to an increased production of l-threonine and a decrease in accumulation of l-lysine, glycine, l-isoleucine, and l-homoserine.4,15−17
As described above, there are three enzymes (LysC, Hom, and ThrB) in the l-threonine biosynthetic pathway that are feedback inhibited by the end-product l-threonine (Figure 1). Among these, feedback-resistant mutations were successfully made in genes encoding for LysC and Hom and overexpression of these mutant alleles resulted in a 3.5-fold higher production of l-threonine in C. glutamicum.12,13 However, it has been difficult to produce feedback-resistant homoserine kinase because l-threonine inhibits the enzyme in a competitive manner with the substrate l-homoserine.11 Therefore, mutations that block the binding of l-threonine at the active site would also inhibit the binding of l-homoserine and thus severely diminish the enzymatic activity of the enzyme.
In this report, an attempt was made to render the active site of C. glutamicum ThrB (CglThrB) more selective toward l-homoserine than l-threonine. First, we successfully crystallized CglThrB-WT with a resolution of 2.2 Å. Aligning this structure with the archaea Methanocaldococcus jannaschii ThrB (MjaThrB) X-ray crystal structure that was solved with both homoserine and threonine, we hypothesized that alanine 12 in MjaThrB (A20 in CglThrB) may support the binding of l-threonine and that the alteration of the residue might affect the binding of l-threonine without affecting that of l-homoserine. To test this possibility, we mutated A20 in CglThrB to various amino acid residues; we tested the enzymatic activity of these variants. The C. glutamicum ThrB-A20G (CglThrB-A20G) showed a similar activity as wild-type ThrB (CglThrB-WT), whereas other mutations abolished or greatly diminished the activity. Subsequent inhibition assays showed that CglThrB-A20G was less inhibited by l-threonine compared with CglThrB-WT. Taken together, we have successfully released competitive feedback inhibition of CglThrB without losing much of the enzymatic activity via structural analysis and site-specific mutation.
Results
Identification and Role of the Conserved ThrB Active-Site Residues
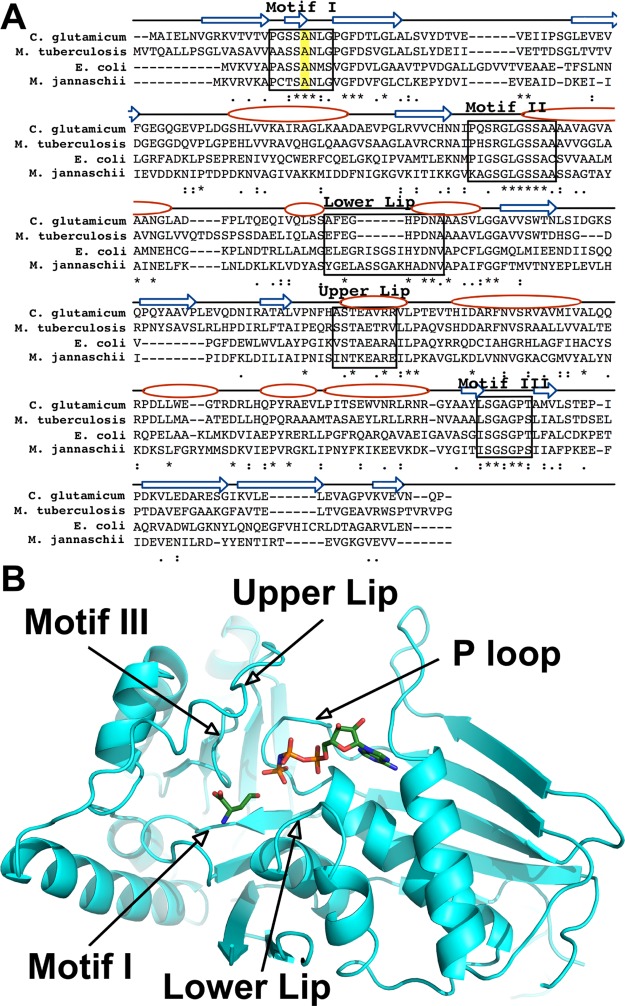
The enzymes of the galactokinase, homoserine kinase, mevalonate kinase, and phosphomevalonate kinase (GHMP) family possess three conserved structural motifs, motif I, II, and III (Figure 2).18 Two additional motifs are known to be important for the binding amino acid substrate or inhibitor in ThrB, the lower and upper lips.19,20 Of these motifs, motif I is the only motif directly interacting with the side chains of the l-homoserine substrate or l-threonine inhibitor, thereby allowing potential discrimination between the branched inhibitor, l-threonine, and the linear substrate, l-homoserine. To determine the exact position of that conserved motif in C. glutamicum ThrB, we performed a pairwise protein sequence alignment using the ThrB sequences encoded by E. coli and Mycobacterium tuberculosis. Because X-ray crystal structures of the M. jannaschii-encoded ThrB in complex with l-threonine and l-homoserine have been solved (PDB codes: 1H73 and 1H72, respectively), the M. jannaschii ThrB sequence was also used in the alignment (Figure 2).19 A conserved amino acid sequence, −SSAN–, in motif I was identified in this alignment.
Figure 2.
(A) Alignment of C. glutamicum homoserine kinase (CglThrB) sequence with those of E. coli, M. jannaschii, and M. tuberculosis using CLUSTAL O (1.2.4). Conserved residues are shown with asterisk (*), residues with strongly similar properties are shown with a colon (:), and residues with weakly similar properties are shown with a period (.). The conserved Ala residues in motif I are highlighted in yellow. The arrows represent β-strands, whereas the ovals represent α-helices. (B) Structure of C. glutamicum ThrB (CglThrB-WT) with modeled l-homoserine and AMP-PNP based on the structure of M. jannaschii ThrB, with these compounds bound in the active site.
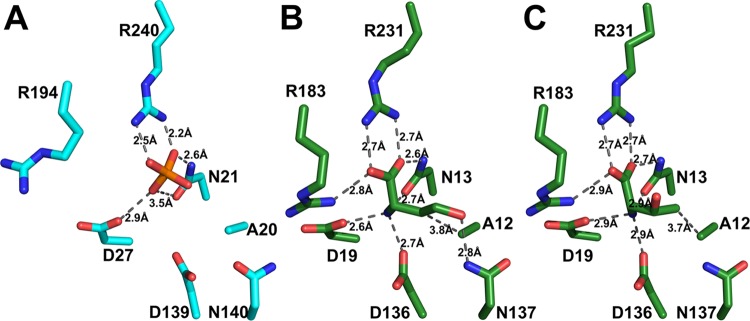
A full-length structure of CglThrB-WT was solved at 2.2 Å resolution. Aligning that structure with the X-ray crystal structure of M. jannaschii in complex with l-threonine and l-homoserine (Figure 3B,C, respectively), it was confirmed that the active sites of the two proteins were structurally similar and that the alanine (A12) of that motif was the only residue directly interacting with the side-chain methyl moiety of l-threonine. Thus, the conserved alanine residue (A20) in motif I of C. glutamicum ThrB offered the highest potential for producing ThrB variants exhibiting enhanced selectivity of l-threonine over l-homoserine (Figure 3A).
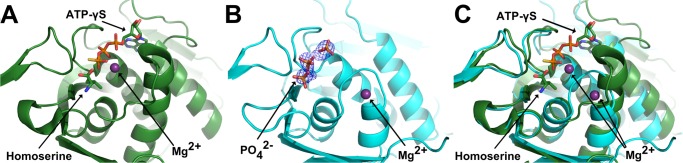
Figure 3.
Interactions of C. glutamicum ThrB (A in cyan) or M. jannaschii ThrB (B and C in green) between binding site amino acids and substrates or ligands. (A) A phosphate anion is bound in the active site of CglThrB-WT, preventing binding of substrates or inhibitors within the amino acid binding site. (B) l-homoserine bound into the active site of M. jannaschii ThrB (PDB: 1H72). (C) l-threonine bound into the active site of M. jannaschii ThrB (PDB: 1H73).
There are multiple strategies to retain wild-type ThrB activity while decreasing affinity for the competitive inhibitor, l-threonine. One strategy is to engineer new interactions that support substrate binding but weaken inhibitor binding. For example, one can retain hydrophobicity of the active site responsible for binding the ethyl moiety of the l-homoserine side chain but add steric bulk at position A20 in C. glutamicum (corresponding to A12 in MjaThrB), promoting steric hindrance with the methyl branch of the l-threonine side chain. To test this strategy, A20 of CglThrB was mutated into either a valine or a leucine residue. A second strategy to test the importance of A20 and its role in substrate/inhibitor binding is to change the polarity by creation of a serine mutation. It is the least disrupting variant because it minimizes steric effects. A third strategy is to decrease the strength of the van der Waals interaction between the A20 and the hydrophobic portion of the corresponding substrate/inhibitor side chain. In this case, a glycine mutation that likely weakens van der Waals interactions between the methyl group of l-threonine and A20 was made to evaluate its importance in selectivity between l-threonine and l-homoserine.
CglThrB-A20G Mutant Enzyme: As Active As CglThrB-WT
Activity of each purified enzyme was measured in an end-point assay using the ADP Colorimetric/Fluorometric Assay Kit (Sigma-Aldrich). CglThrB-A20V and CglThrB-A20L lost their activity, but CglThrB-A20S retained 44% of the wild-type activity (Figure 4). The last mutant, CglThrB-A20G showed a similar level of enzymatic activity to CglThrB-WT (Figure 4).
Figure 4.
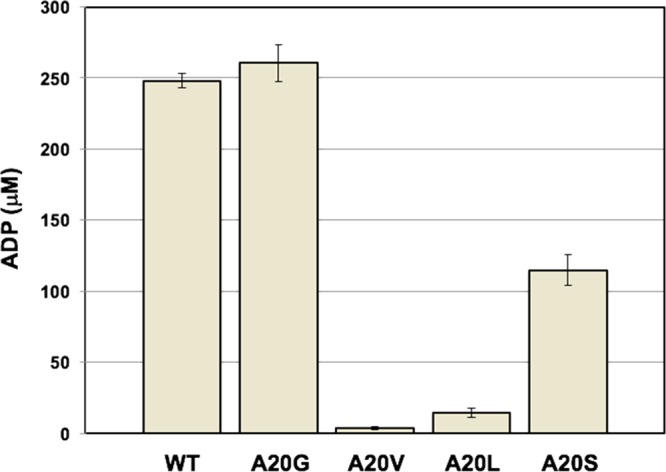
Enzymatic activity of WT and four variants of CglThrB. CglThrB-A20G is as active as CglThrB-WT. The concentration of ADP (Y axis) was determined after incubating each enzyme with l-homoserine and ATP at 27 °C for 30 min. WT: CglThrB-WT, A20G: CglThrB-A20G, A20V: CglThrB-A20V, A20L: CglThrB-A20L, and A20S: CglThrB-A20S.
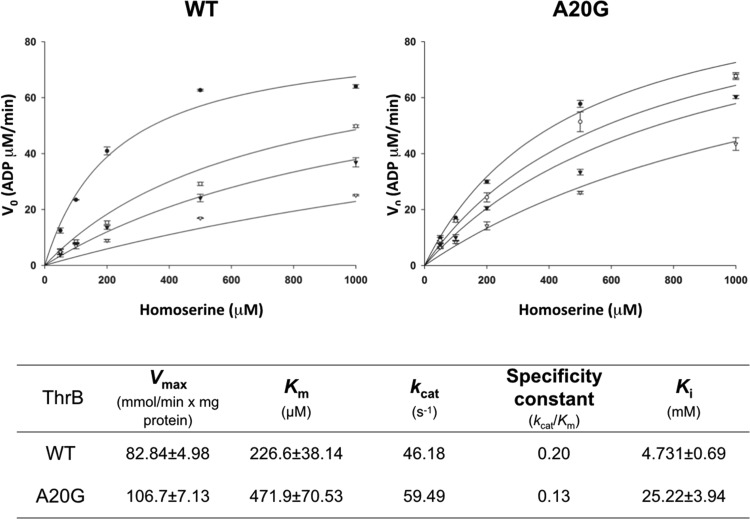
To further assess the enzymatic activity of CglThrB-WT and the CglThrB-A20G variant and the effect of changing l-homoserine concentrations, steady-state kinetic experiments were performed with saturating concentrations of ATP. Under these pseudo-first-order conditions, the Km for l-homoserine in CglThrB-A20G was determined to be only 2-fold higher than the Km of CglThrB-WT and resulting in only a 35% decrease in the specificity constant (Figure 5).
Figure 5.
Kinetic reaction rates in the presence of 0, 10, 20, and 50 mM l-threonine of CglThrB-WT and CglThrB-A20G variant. Each datum represents the value of three determinations (n = 3). l-Threonine concentration: ●: 0 mM, ○: 10 mM, ▼: 20 mM, and ∇: 50 mM.
CglThrB-A20G Exhibits Reduced Feedback Inhibition by l-Threonine
Because CglThrB-A20G was the only mutant retaining wild-type activity, that variant was tested for changes in negative feedback inhibition by l-threonine in an end-point assay (Figure 6) and steady-state enzyme kinetics (Figure 5). CglThrB-WT exhibits a Ki of 4.73 mM, whereas CglThrB-A20G showed an approximately 5.4-fold higher Ki for l-threonine, with a final value of 25.22 mM (Figure 5). For example, the 5 mM l-Threonine inhibited wild-type ThrB activity by 40% but inhibited CglThrB-A20G by only 16% (Figure 6). l-Threonine (10 mM), which is unlikely to occur in vivo, inhibits 65% of CglThrB-WT activity but it only inhibits 38% of CglThrB-A20G activity.
Figure 6.
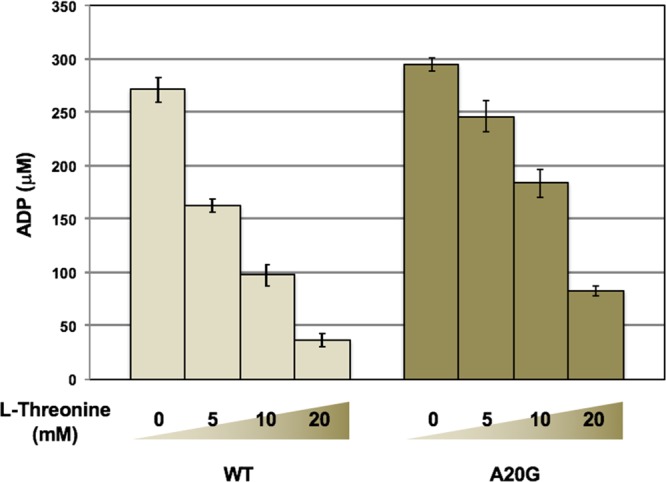
CglThrB-A20G has eliminated much of the feedback inhibition while retaining wild-type enzymatic activity. The reaction conditions were the same as those in Figure 4 except that different concentrations of l-threonine (0, 5, 10, and 20 mM) were used.
Structural Analysis of CglThrB-WT
As mentioned previously (Figure 2), homoserine kinases have five important regions interacting with their substrate and inhibitor. The first structure of CglThrB-WT solved at 1.8 Å resolution displays hydrolysis of the upper lip as well as the helix stabilizing it, suggesting it could have an important role in binding the substrate/inhibitor (Figure 7).
Figure 7.
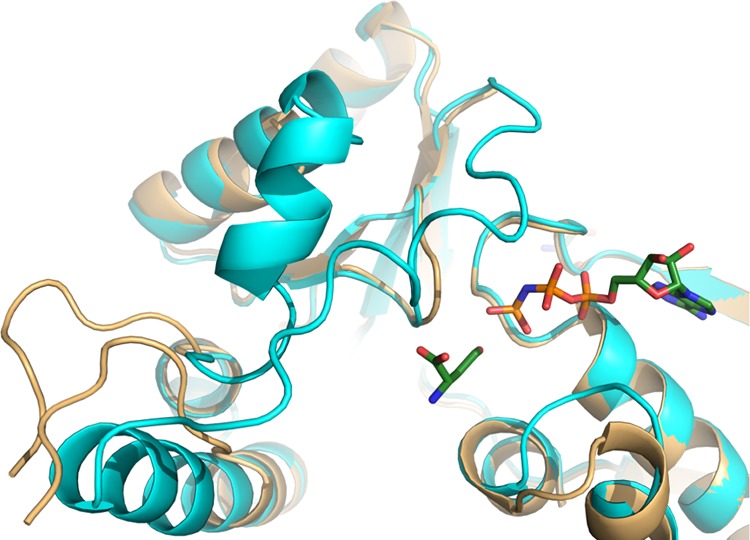
Hydrolyzed structure of CglThrB-WT (gold) superimposed with the full-length CglThrB-WT (cyan) containing modeled l-homoserine and AMP-PNP from 1H72.
The full-length structure was solved at 2.2 Å resolution and exhibited high B-factors for residues of both the upper and lower lips. This fully intact structure of CglThrB-WT was obtained with three phosphate or sulfate ions in the active site. One anion is bound in the amino acid binding site interacting with R240, N21, and D27, which are conserved residues required for coordinating both the α-carboxylate moiety and the α-amino moiety of amino acid substrate or inhibitors (Figure 3). The other two ions directly interact with the P-loop.21 Additionally, a magnesium ion is present in the active site, resulting in the displacement of the lower lip (Figure 8).
Figure 8.
Significant shift of the lower lip that interacts with both magnesium and the amino acid present in the active site. (A) M. jannaschii ThrB-WT with l-isoleucine, magnesium (purple sphere) and ATP-γS bound into its active site (PDB: 1H74). (B) CglThrB-WT with magnesium (purple sphere) and three phosphate/sulfate bound in the active site. The omit map of all three phosphates/sulfates is contoured to 3σ. (C) Superposition of both M. jannaschii and CglThrB-WT with both their magnesium (purple spheres) and l-isoleucine and ATP-γS from 1H74.
Discussion
To promote higher levels of l-threonine production by fermentation of C. glutamicum, the negative feedback inhibition of ThrB by l-threonine needs to be suppressed. After analyses of the available structures from M. jannaschii in complex with both the end-product inhibitor, l-threonine, and the natural substrate, l-homoserine, the alanine 12 residue, was identified as being the only amino acid present in the active site that would allow for discrimination of the substrate and competitive inhibitor by the enzyme. To determine the impact of varying the conserved alanine (A20) in C. glutamicum ThrB, the A20 residue was mutated into a leucine and a valine. These mutations were meant to introduce a steric hindrance, preventing binding of l-threonine while still affording the binding of the l-homoserine substrate. Another mutation to a serine residue was also performed to introduce polarity into the active site, creating an unfavorable environment that rendered the active site less compatible to binding of the hydrophobic methyl moiety of the l-threonine side chain. A final mutation into a glycine was also introduced with the rationale that this mutation may suppress a low-energy van der Waals interaction between the methyl group of l-threonine and the alanine side chain, which is an interaction that does not occur when l-homoserine binds within the active site.
It was shown in the end-point enzyme assay that while mutations to either leucine or valine resulted in complete loss of activity, the serine variant lost only 47% of WT activity. This suggests that the steric hindrance between the amino acid ligand and the side chain at position 20 of CglThrB has a more significant detrimental impact on enzymatic activity than a change in polarity at this position of the active site. This is likely due to weakening of binding of the l-homoserine amino acid in the A20L and A20V variants due to steric hindrance that does not occur in the A20S variant. Alternatively, these variants may still bind the substrate but the side-chain hydroxyl moiety of l-homoserine is then positioned in a location or an orientation that disallows attack on the γ-phosphate of ATP. The last variant, CglThrB-A20G, retained complete WT activity. The subsequent assays with l-threonine showed a dramatic loss of negative feedback inhibition in CglThrB-A20G, with a Ki that is 5.3-fold lower than that of CglThrB-WT. Because we determined the activity of CglThrB-A20G by measuring ADP production, not o-phosphohomoserine production, we cannot rule out the possibility that A20G mutation simply affords the hydrolysis of ATP instead of phosphoryl transfer to l-homoserine.
To investigate further the interaction of l-threonine and l-homoserine with the active site of CglThrB, the protein was crystallized. Interestingly, the first apo structure that was solved at 1.8 Å, showed hydrolysis of the upper lip as well as the α-helix directly adjacent due to structural dynamics in that region, as later evidenced by elevated B-factors in the analogous region of the full-length protein structure (Figure 7). A full-length structure solved at 2.2 Å also shows high dynamicity of the upper and lower lip with high B-factors in these regions. Additionally, crystallization of both CglThrB-A20G and CglThrB-A20V has been attempted. However, the mutations seem to either destabilize the active site or increase flexibility in the active site, thereby suppressing crystallization.
Because only apo structures of CglThrB-WT (PDB: 5WAT) were obtained, interactions between the enzymes and the amino acid substrates or inhibitors can only be inferred from interactions observed in the M. jannaschii ThrB structures. However, three sulfate/phosphate molecules are present in the active site and exhibit interactions with the CglThrB enzyme that are similar to the interactions of amino acids with the M. jannaschii active site. For instance, both N13 and R231 form identical interactions with the sulfate present in the CglThrB-WT structure that are analogous to interactions formed by these conserved active site residues with the carboxylic acid moiety of the amino acid present in M. jannaschii. Similarly, N21 and D27 form interactions with the sulfate present in the CglThrB-WT that are comparable to interactions between MjaThrB-WT and the amino moiety of the amino acid substrate or inhibitor. Differences were, however, observed. For example, a 1.0 to 1.7 Å shift was observed in the position of the D139 side chain, resulting from the absence of interactions with the amino moiety of the substrate (Figure 3). In a similar manner, the lack of interaction between R194 and the carboxylic moiety of the amino acid results in the R194 side chain interacting with solvent (Figure 3). Additionally, although the magnesium in MjaThrB-WT is interacting with the phosphate of APTγS and the lower lip, it is solvent-exposed in CglThrB-WT and is coordinated by the lower lip, resulting in interactions with the two sulfates present in the ATP binding site. This further supports the high dynamicity of the active site of actinobacterial ThrB.
In conclusion, we have demonstrated that a single mutation at a conserved residue at the active site in homoserine kinase decreases the feedback inhibition by l-threonine while maintaining the enzymatic activity. To our knowledge, this was the first case where competitive feedback inhibition was successfully released via structural analysis and site-specific mutation. A similar approach may be used to create feedback-insensitive mutants in other biosynthetic enzymes. Additionally, competitive inhibitors that bind to ThrB homolog of medically important bacterial pathogens may be designed on the basis of these determined structures.
Materials and Methods
Primary Structure Alignment of Homoserine Kinase from E. coli, C. glutamicum, M. tuberculosis, and M. jannaschii
The alignment used to determine conserved motifs and amino acids of interest to our study was performed using Clustal Omega.22,23 The Uniprot codes for the respective proteins are P00547, I6Y665, A0A072ZHQ8, and Q58504.
Bacterial Strains and Growth Conditions
E. coli Top 10 (Invitrogen) (Table 1) was used as a cloning host and E. coli BL21 (DE3) for expression of recombinant proteins and for the determination of l-threonine production. Cultures were grown at 37 °C in Luria-Bertani (LB) broth or solid medium with chloramphenicol (34 μg/mL).
Table 1. Strains and Plasmids Used in This Work.
| description | reference or source | |||
|---|---|---|---|---|
| strain | E. coli | Top 10 | E. coli host for molecular cloning | Invitrogen |
| BL21 (DE3) | E. coli host for protein expression | Invitrogen | ||
| C. glutamicum | ATCC 13032 | genomic DNA as a template for polymerase chain reaction (PCR) | ATCC | |
| plasmid | pACYCDuet-1 | duel expression vector (Cmr) | Invitrogen | |
| pCK448 | pACYCDuet-1 carrying C. glutamicumthrB-6his under PT7 | in house | ||
| pCK449 | pACYCDuet-1 carrying C. glutamicumthrBA16G-6his under PT7 | in house | ||
| pCK450 | pACYCDuet-1 carrying C. glutamicumthrBA16V-6his, this study under PT7 | in house | ||
| pCK453 | pACYCDuet-1 carrying C. glutamicumthrBA16L-6his, this study under PT7 | in house | ||
| pCK454 | pACYCDuet-1 carrying C. glutamicumthrBA16S-6his, this study under PT7 | in house | ||
Plasmid Construction
The thrB-his genes were amplified by PCR using ThrBFor and ThrBRev primers (Table 2) and the C. glutamicum ATCC 13032 genomic DNA as a template. PCR product was cloned into the pACYCDuet-1 plasmid (Novagen) behind a T7 promoter. A20 of CglthrB was mutated to glycine, valine, serine, or leucine using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). The CglthrB-A20G allele was PCR-amplified using primers c59g and c59g_antisense and pACYCDuet-1-PT7-thrB-6his as template. The thrB-A20V allele was amplified with c59t and c59t_antisense primers, the thrB-A20S allele with g58t and g58t_antisense, and the thrB-A20L allele with g58c_c59t and g58c_c59t_antisense primers. All plasmid and primers are listed in Tables 1 and 2, respectively.
Table 2. Primers Used in This Work.
| primer name | sequence |
|---|---|
| ThrBFor | 5′-CCT CAT ATG GCA ATT GAA CTG AAC GTC-3′ |
| ThrBRev | 5′-GGA CTC GAG CTA GTG GTG ATG ATG GTG GTG AGG TTG GTT AAC TTC AAC CTT G-3′ |
| c59g | 5′-TAC CTG GAT CTT CTG GAA ACC TCG GAC CTG G-3′ |
| c59g_antisense | 5′-CCA GGT CCG AGG TCC AGA AGA TCC AGG TA-3′ |
| c59t | 5′-GTA CCT GGA TCT TCT GTA AAC CTC GGA CCT GGC-3′ |
| c59t_antisense | 5′-GCC AGG TCC GAG GTT TAC AGA AGA TCC AGG TAC-3′ |
| g58t | 5′-GGT ACC TGG ATC TTC TTC AAA CCT CGG ACC TGG-3′ |
| g58t_antisense | 5′-CCA GGT CCG AGG TTT GAA GAA GAT CCA GGT ACC-3′ |
| g58c_c59t | 5′-GGT ACC TGG ATC TTC TCT AAA CCT CGG ACC TGG C-3′ |
| g58c_c59t_antisense | 5′-GCC AGG TCC GAG GTT TAG AGA AGA TCC AGG TAC C-3′ |
Expression and Purification of the ThrB Proteins
All constructs were used to transform E. coli BL21 (DE3), and transformants were grown in LB broth with 34 μg/mL chloramphenicol. Gene expression was induced with 1 mM isopropyl -d-1-thiogalactopyranoside at mid-log phase for 3 h at 37 °C. Cell pellets were stored at −80 °C until cells were lysed in a 1× PBS buffer containing protease inhibitors (Roche) by using a French Press (Thermo Scientific). Proteins were purified using Bio-Scale Mini Profinity IMAC Cartridges and the Profinia Protein Purification System (Bio-Rad). The proteins were then dialyzed overnight in 25 mM Tris pH 7.4. The purified proteins were then run on a sodium dodecyl sulfate polyacrylamide gel electrophoresis, stained with GelCode Blue (Pierce Biotechnology Inc.), and visualized on ChemiDoc XRS (Bio-Rad).
In Vitro Enzyme Assay and Kinetic Analysis of ThrB
Enzyme activity of ThrB (phosphorylation of l-homoserine) was determined in end-point assays by measuring a reaction product ADP via the ADP Colorimetric/Fluorometric Assay Kit (Sigma-Aldrich). Ultrapure ATP (Sigma-Aldrich) was diluted in 10 mM Tris-Cl, pH 7.0. Enzymes (CglThrB-WT, CglThrB-A20G, CglThrB-A20V, CglThrB-A20L, and CglThrB-A20S) and l-homoserine were diluted in an assay buffer provided by the manufacturer. Each enzyme was assayed for 5 min at 27 °C using 40 nM enzyme, 1 mM l-homoserine, 2 mM ATP, 10 mM MgSO4, and 0.5 M KCl in 250 mM Tris pH 7.8 in a 96-well microtiter plate. The temperature was then shifted to 99 °C for 2 min to terminate the reaction. ADP assay buffer (Sigma-Aldrich) was added and further incubated at 27 °C for 30 min. The standard curve for ADP was obtained by incubating 0, 10, 20, 50, 100, and 200 μM of ADP in the ADP assay buffer. ADP concentrations were monitored by the MultiskanGO microplate spectrophotometer (Thermo Fisher Scientific) at 620 nm. Assays for the feedback inhibition were performed similarly using 5, 10, or 20 mM l-threonine.
Kinetic analysis of CglThrB-WT and CglThrB-A20G was conducted in similar reaction conditions, with addition of l-threonine at 0, 10, 20, or 50 mM concentrations. These reactions were assayed with 50, 100, 200, 500, and 1000 μM homoserine. Reads were taken at 30 s intervals. After incubating for 5 min (this condition was determined in a separate control experiment where the product homoserine-phosphate formation was still linearly proportional to incubation time), ThrB was inactivated by heating at 99 °C for 2 min. The concentration of ADP from each reaction was determined as described above, and Km and Vmax values were determined by using the SigmaPlot 12.5 (Systat software).
Crystallization of the CglThrB-WT
The CglThrB protein concentrated to 16.2 mg/mL in 20 mM Tris pH 7.5, 150 mM NaCl, 50 mM KCl, and 50 mM MgCl2 was used for crystallization experiments. Using the hanging-drop vapor diffusion method, CglThrB crystals were grown with 2 μL of protein solution and 2 μL of well solution. Drops were equilibrated against 100 μL of well solution containing 0.25 M ammonium sulfate, 25% poly(ethylene glycol) 3350, and 0.1 M N-(2-hydroxyethyl)piperazine-N′-ethanesulfonic acid pH 7.5. Protein was co-crystallized with either methionine or ATPγS and cysteine for the 1.8 and 2.2 Å structures, respectively. No cryoprotectant was used for flash-freezing the crystals.
Diffraction Experiments and Structural Determination of CglThrB-WT
X-ray diffraction experiments were carried out at the LS-CAT beamline at the Advanced Photon Source of Argonne National Labs, Argonne, IL. The CglThrB-WT structure was solved using diffraction data collected at a wavelength of 1.07819 Å. Diffraction data were integrated and scaled using HKL300024 (Table 3).
Table 3. Data Collection and Refinement Statisticsa.
| CglThrB-hydrolyzed | CglThrB full length | |
|---|---|---|
| data collection | ||
| space group | P41212 | I222 |
| cell dimensions | ||
| a, b, c (Å) | 46.23, 46.23, 267.30 | 68.08, 130.08, 195.95 |
| α, β, γ (deg) | 90, 90, 90 | 90, 90, 90 |
| resolution (Å) | 50.0–1.8 | 50.0–2.2 |
| Rmerge | 0.052 | 0.030 |
| I/σ(I) | 28.77 (6.40) | 16.10 (3.25) |
| completeness (%) | 97.5 (95.2) | 99.8 (95.4) |
| redundancy | 23.4 (19.5) | 4.7 (4.7) |
| refinement | ||
| resolution (Å) | 34.97–1.80 | 42.34–2.14 |
| no. of unique reflections | 27 709 (2 537) | 48 115 (4 566) |
| R work/Rfree (%) | 24.14/28.71 (31.24/37.21) | 19.84/23.66(20.75/25.62) |
| no. of atoms | ||
| protein | 2058 | 4553 |
| ligand/ion | 5 | 65 |
| water | 61 | 185 |
| B-factors (Å2) | ||
| protein | 49.50 | 51.00 |
| ligand/ion | 47.00 | 53.30 |
| water | 43.30 | 44.50 |
| rms deviations | ||
| bond lengths (Å) | 0.006 | 0.009 |
| bond angles (deg) | 1.040 | 1.080 |
| Ramachandran | ||
| favored (%) | 96.22 | 96.73 |
| outliers (%) | 0.00 | 0.00 |
The necessary data were obtained from one crystal. Values in parentheses are for the highest shells.
A search model for CglThrB-WT was made on the basis of the 4rpf structure and used for molecular replacement using Phaser in PHENIX. Rigid body refinement, simulated annealing, positional, and B-factor refinements were performed using PHENIX.25 Manual refinement of both structures was performed using COOT.26
Glossary
Abbreviations
- Mja
Methanocaldococcus jannaschii
- Mtb
Mycobacterium tuberculosis
- Cgl
Corynebacterium glutamicum
Author Contributions
⊥ C.P. and Y.K. contributed equally to the work.
Author Contributions
C.P. and C.-M.K. wrote the original draft, reviewed, and edited the manuscript. D.R.R. and Y.K. reviewed and edited the manuscript. D.R.R. and C.P. did the sequence alignment and analysis of the structures. Y.K., S.-K.L., J.-W.S., and C.-M.K. designed the experiments, and Y.K., S.-K.L., J.B., and E.L. constructed vectors for expression of wild-type and mutant proteins. C.-M.K., Y.K., and S.-K.L. characterized ThrB-WT and mutants activity in vitro. C.P. purified and crystallized ThrB-WT and determined its X-ray structure. All authors analyzed the results and approved the final version of the manuscript.
This work was supported by the financial support from California State University Stanislaus C.-M.K. and University of Toledo to D.R.R. This work was also supported by a grant from the Next-Generation BioGreen 21 Program (No. PJ00900705), Korea Rural Development Administration, Republic of Korea to C.-M.K.
The authors declare no competing financial interest.
References
- Becker J.; Wittmann C. Systems and synthetic metabolic engineering for amino acid production-the heartbeat of industrial strain development. Curr. Opin. Biotechnol. 2012, 23, 718–726. 10.1016/j.copbio.2011.12.025. [DOI] [PubMed] [Google Scholar]
- Schrumpf B.; Schwarzer A.; Kalinowski J.; Puhler A.; Eggeling L.; Sahm H. A functionally split pathway for lysine synthesis in Corynebacterium glutamicium. J. Bacteriol. 1991, 173, 4510–4516. 10.1128/jb.173.14.4510-4516.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möckel B.; Eggeling L.; Sahm H. Functional and structural analyses of threonine dehydratase from Corynebacterium glutamicum. J. Bacteriol. 1992, 174, 8065–8072. 10.1128/jb.174.24.8065-8072.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic P.; Willuhn J.; Sahm H.; Eggeling L. Identification of glyA (encoding serine hydroxymethyltransferase) and its use together with the exporter ThrE to increase L-threonine accumulation by Corynebacterium glutamicum. Appl. Environ. Microbiol. 2002, 68, 3321–3327. 10.1128/AEM.68.7.3321-3327.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rückert C.; Pühler A.; Kalinowski J. Genome-wide analysis of the L-methionine biosynthetic pathway in Corynebacterium glutamicum by targeted gene deletion and homologous complementation. J. Biotechnol. 2003, 104, 213–228. 10.1016/S0168-1656(03)00158-5. [DOI] [PubMed] [Google Scholar]
- Kalinowski J.; Bathe B.; Bartels D.; Bischoff N.; Bott M.; Burkovski A.; Dusch N.; Eggeling L.; Eikmanns B. J.; Gaigalat L.; Goesmann A.; Hartmann M.; Huthmacher K.; Kramer R.; Linke B.; McHardy A. C.; Meyer F.; Mockel B.; Pfefferle W.; Puhler A.; Rey D. A.; Ruckert C.; Rupp O.; Sahm H.; Wendisch V. F.; Wiegrabe I.; Tauch A. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J. Biotechnol. 2003, 104, 5–25. 10.1016/S0168-1656(03)00154-8. [DOI] [PubMed] [Google Scholar]
- Henkin T. M.; Yanofsky C. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. Bioessays 2002, 24, 700–707. 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- Mateos L. M.; Pisabarro A.; Patek M.; Malumbres M.; Guerrero C.; Eikmanns B. J.; Sahm H.; Martin J. F. Transcriptional analysis and regulatory signals of the hom-thrB cluster of Brevibacterium lactofermentum. J. Bacteriol. 1994, 176, 7362–7371. 10.1128/jb.176.23.7362-7371.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A.; Tomita T.; Kuzuyama T.; Nishiyama M. Mechanism of concerted inhibition of alpha2beta2-type hetero-oligomeric aspartate kinase from Corynebacterium glutamicum. J. Biol. Chem. 2010, 285, 27477–27486. 10.1074/jbc.M110.111153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer J. A.; Solow-Cordero D. E.; Sinskey A. J. A C-terminal deletion in Corynebacterium glutamicum homoserine dehydrogenase abolishes allosteric inhibition by L-threonine. Gene 1991, 107, 53–59. 10.1016/0378-1119(91)90296-N. [DOI] [PubMed] [Google Scholar]
- Miyajima R.; Otsuka S.-I.; Shiio I. Regulation of aspartate family amino acid biosynthesis in Brevibacterium flavum. I. Inhibition by amino acids of the enzymes in threonine biosynthesis. J. Biochem. 1968, 63, 139–148. 10.1093/oxfordjournals.jbchem.a128754. [DOI] [PubMed] [Google Scholar]
- Colón G. E.; Jetten M. S.; Nguyen T. T.; Gubler M. E.; Follettie M. T.; Sinskey A. J.; Stephanopoulos G. Effect of inducible thrB expression on amino acid production in Corynebacterium lactofermentum ATCC 21799. Appl. Environ. Microbiol. 1995, 61, 74–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinscheid D. J.; Kronemeyer W.; Eggeling L.; Eikmanns B. J.; Sahm H. Stable Expression of hom-1-thrB in Corynebacterium glutamicum and Its Effect on the Carbon Flux to Threonine and Related Amino Acids. Appl. Environ. Microbiol. 1994, 60, 126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debabov V. G. In The Threonine Story; Advances in Biochemical Engineering/Biotechnology; Springer, 2003; Vol. 79, pp 113–136. [DOI] [PubMed] [Google Scholar]
- Diesveld R.; Tietze N.; Furst O.; Reth A.; Bathe B.; Sahm H.; Eggeling L. Activity of exporters of Escherichia coli in Corynebacterium glutamicum, and their use to increase L-threonine production. J. Mol. Microbiol. Biotechnol. 2009, 16, 198–207. 10.1159/000142530. [DOI] [PubMed] [Google Scholar]
- Kruse D.; Kramer R.; Eggeling L.; Rieping M.; Pfefferle W.; Tchieu J. H.; Chung Y. J. Jr.; Saier M. H.; Burkovski A. Influence of threonine exporters on threonine production in Escherichia coli. Appl. Microbiol. Biotechnol. 2002, 59, 205–210. 10.1007/s00253-002-0987-7. [DOI] [PubMed] [Google Scholar]
- Livshits V. A.; Zakataeva N. P.; Aleshin V. V.; Vitushkina M. V. Identification and characterization of the new gene rhtA involved in threonine and homoserine efflux in Escherichia coli. Res. Microbiol. 2003, 154, 123–135. 10.1016/S0923-2508(03)00036-6. [DOI] [PubMed] [Google Scholar]
- Zhou T.; Daugherty M.; Grishin N. V.; Osterman A. L.; Zhang H. Structure and mechanism of homoserine kinase: prototype for the GHMP kinase superfamily. Structure 2000, 8, 1247–1257. 10.1016/S0969-2126(00)00533-5. [DOI] [PubMed] [Google Scholar]
- Krishna S. S.; Zhou T.; Daugherty M.; Osterman A.; Zhang H. Structural basis for the catalysis and substrate specificity of homoserine kinase. Biochemistry 2001, 40, 10810–10818. 10.1021/bi010851z. [DOI] [PubMed] [Google Scholar]
- Andreassi J. L. 2nd; Leyh T. S. Molecular functions of conserved aspects of the GHMP kinase family. Biochemistry 2004, 43, 14594–14601. 10.1021/bi048963o. [DOI] [PubMed] [Google Scholar]
- Singh S. K.; Yang K.; Karthikeyan S.; Huynh T.; Zhang X.; Phillips M. A.; Zhang H. The thrH gene product of Pseudomonas aeruginosa is a dual activity enzyme with a novel phosphoserine:homoserine phosphotransferase activity. J. Biol. Chem. 2004, 279, 13166–13173. 10.1074/jbc.M311393200. [DOI] [PubMed] [Google Scholar]
- Sievers F.; Wilm A.; Dineen D.; Gibson T. J.; Karplus K.; Li W.; Lopez R.; McWilliam H.; Remmert M.; Soding J.; Thompson J. D.; Higgins D. G. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon M.; McWilliam H.; Li W.; Valentin F.; Squizzato S.; Paern J.; Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010, 38, W695–W699. 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W.; Cymborowski M.; Otwinowski Z.; Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2006, 62, 859–866. 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkoczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L. W.; Kapral G. J.; Grosse-Kunstleve R. W.; McCoy A. J.; Moriarty N. W.; Oeffner R.; Read R. J.; Richardson D. C.; Richardson J. S.; Terwilliger T. C.; Zwart P. H. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]