Abstract
Cellular membranes must undergo remodeling to facilitate critical functions including membrane trafficking, organelle biogenesis, and cell division. An essential step in membrane remodeling is membrane fission, in which an initially continuous membrane surface is divided into multiple, separate compartments. The established view has been that membrane fission requires proteins with conserved structural features such as helical scaffolds, hydrophobic insertions, and polymerized assemblies. In this review we discuss these structure-based fission mechanisms and highlight recent findings from several groups that support an alternative, structure-independent mechanism of membrane fission. This mechanism relies on lateral collisions among crowded, membrane-bound proteins to generate sufficient steric pressure to drive membrane vesiculation. As a stochastic process, this mechanism contrasts with the paradigm that deterministic protein structures are required to drive fission, raising the prospect that many more proteins may participate in fission than previously thought. Paradoxically, our recent work suggests that intrinsically disordered domains may be among the most potent drivers of membrane fission, owing to their large hydrodynamic radii and substantial chain entropy. This stochastic view of fission also suggests new roles for the structure-based fission proteins. Specifically, we hypothesize that in addition to driving fission directly, the canonical fission machines may facilitate the enrichment and organization of bulky disordered protein domains in order to promote membrane fission by locally amplifying protein crowding.
Graphical Abstract
Introduction
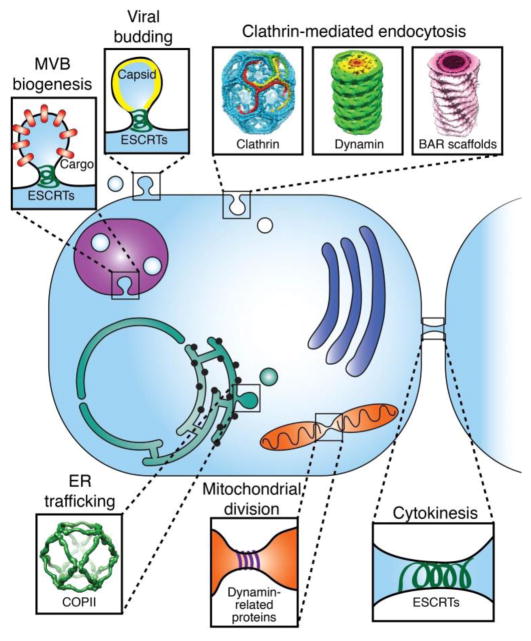
Membrane fission, the process of separating an initially continuous membrane into two or more distinct compartments, is essential for cellular life. Basic cellular functions that require membrane fission include endocytosis [1], assembly of intracellular trafficking vesicles [2], division of mitochondria and other organelles [3], viral budding [4], cytokinesis [5], and biogenesis of multivesicular bodies [6], among others (Fig. 1).
Figure 1.
Membrane fission throughout the cell. Each fission event requires significant energetic contributions from diverse protein machines. Conserved structural features and assembly properties are found in all of the fission proteins highlighted here, including hydrophobic insertions, scaffolding, and polymerization. Clathrin cage structure from Fotin et al, Nature 2004 [13]. Super-constricted dynamin helix: EMDB 2701 [14]. Endophilin N-BAR scaffold on a 28 nm-diameter membrane tube: EMDB 2007 [15]. COPII cage structure: EMDB 1232 [16].
Although membrane fission is required for many essential cellular activities, the lipid bilayer is resistant to the deformations and remodeling events required for fission [7]. The elastic theory of membranes predicts that the energetic barrier to membrane fission is in the range of 400–800 kBT [8]. Therefore, the proteins that orchestrate membrane fission must provide significant energetic contributions in order to overcome this barrier. While a variety of proteins participate in fission, the established view has been that specific, conserved structural features are required to drive the process efficiently [9]. These structural features can be grouped into three major categories: hydrophobic insertions, cylindrical scaffolds, and cytoskeletal assemblies [10].
In this review, we discuss these structure-based mechanisms of membrane fission and explore an emerging, stochastic mechanism of membrane fission that is independent of protein structure. Specifically, this stochastic perspective on fission suggests that when proteins densely cover a membrane surface, lateral collisions among the proteins generate significant steric pressure that can bend membranes [11] and ultimately overcome the energetic barriers to fission [12]. This inherently non-specific mechanism can utilize both folded and intrinsically disordered protein domains. We conclude by exploring the potential interplay between structure-based and stochastic fission mechanisms and suggest ways in which the two mechanisms may work together to drive fission robustly in cells.
Established fission-driving proteins share conserved structural features
Hydrophobic insertions and membrane fission
Hydrophobic insertions are portions of proteins which insert directly into the hydrophobic lipid tail region of the membrane. Insertions have been proposed to drive membrane curvature by expanding the area of one lipid monolayer leaflet relative to the opposite leaflet, forcing the bilayer to take on curvature [17]. An important class of hydrophobic insertions is the amphipathic helix, which consists of both hydrophobic and hydrophilic residues separated onto distinct faces of the helix [18]. Amphipathic helices insert shallowly into the membrane, parallel to the plane of the bilayer [19]. One of the first amphipathic helices implicated in membrane bending and fission is at the N-terminus of the epsin N-terminal homology (ENTH) domain, a 20 kDa domain of the clathrin accessory protein epsin [20, 21]. In vitro work from the McMahon group demonstrated that residues 3–15 of the ENTH domain fold into an amphipathic helix, called helix 0, upon binding to the lipid phosphatidylinositol-4,5-bisphosphate [PtdIns(4,5)P2] [20]. The authors suggested that once folded, membrane insertion of helix 0 drives membrane curvature directly [20]. Evidence for this conclusion came from mutating a single, hydrophobic leucine of helix 0 to a series of more hydrophilic residues (L6H, L6Q, and L6E ENTH). While wild-type ENTH was able to transform initially spherical vesicles into highly curved cylindrical tubules, the more hydrophilic mutants showed a progressive loss of this capacity. More recent work from the McMahon group found that wild-type ENTH also drove membrane fission in vitro and in cells, while hydrophilic mutants displayed a defect in fission [21]. These findings demonstrate that the hydrophobicity of the amphipathic helix is essential to membrane remodeling by ENTH.
Amphipathic helix insertion has been implicated in other contexts of membrane bending and fission as well. For example, Sar1, a core component of the COPII pathway with similar molecular weight to the ENTH domain, exposes a 23-amino acid amphipathic helix upon binding to GTP. Insertion of this amphipathic helix into the membrane is thought to facilitate fission of COPII-coated vesicles, based on studies in cells and in vitro from the Schekman group [22]. The GTPase Arf1 is thought to drive fission of COPI-coated vesicles by a similar mechanism, based on in vitro and cell-based studies [23]. Arf1 is involved in recruitment of the coatomer complex to the membrane, and inserts a 17-amino acid amphipathic helix into the membrane after binding GTP [23]. Arf1’s amphipathic helix is also modified at its N-terminus with a myristoyl group, which further strengthens association of the helix with the membrane [24]. Amphipathic helices are also implicated in viral budding. The M2 protein of influenza, a homotetrameric, transmembrane ion channel, contains a 17-amino acid N-terminal amphipathic helix on its cytoplasmic domain [25]. Work from the Lamb group found that influenza M2 localizes preferentially to the necks of budding virions [26], suggesting that the amphipathic helices of the M2 channel promote the curvature of the neck and facilitate fission [26, 27]. Amphipathic helices have been implicated in a variety of additional contexts, including shaping of photoreceptor organelles [28], fission of caveolae [29], and Golgi membrane fission [30].
Protein scaffolds and membrane fission
Cylindrical protein scaffolds are another important category of protein structures involved in membrane bending and fission [9]. Protein scaffolds drive membrane curvature by forcing membranes to adopt the curved geometry of the scaffold. One of the most studied scaffold-forming proteins is the canonical fission machine dynamin. Dynamin is best known for driving fission of the necks of clathrin-coated membrane buds, though it is also involved in a variety of other membrane shaping processes, including mitochondrial division and endosome fission [31] (Fig. 1). Dynamin is a 100 kDa protein composed of a GTPase domain, a bundle signaling element, a stalk domain, and a pleckstrin homology (PH) domain. Dynamin self-assembles into a helical collar around curved membrane tubes [32], and inserts the variable loop 1 of the PH domain into the membrane after binding PtdIns(4,5)P2 [33]. Binding and hydrolyzing GTP causes the dynamin helical assembly to undergo conformational changes that constrict its diameter, forcing the underlying membrane tube into a highly curved state [34]. In order for the tube to undergo spontaneous fission, the inner diameter of the tube must constrict to approximately 4 nm, as suggested in theoretical studies [8]. Because this inner diameter is similar to the thickness of the membrane bilayer, the lipids of the inner membrane leaflet become closely apposed and are thought to proceed spontaneously through the hemi-fission step, where the inner leaflets fuse resulting in a single-layer, micellar tubule. This hemi-fission intermediate is unstable, and progresses spontaneously to full fission, yielding two separate membrane bilayers [8]. Recent work from the Hinshaw group revealed the structure of a super-constricted, pre-fission dynamin helix with a 3.7 nm lumenal diameter [14] (Fig. 1), demonstrating that dynamin assemblies constrict membrane tubes to a fission-competent diameter.
Another essential scaffold-forming protein involved in membrane bending is the bin-amphiphysin-rvs (BAR) domain, a conserved crescent-shaped structure found in many endocytic adaptor proteins [35, 36]. Domains of the BAR family display a range of intrinsic curvatures [37]. The N-terminal, amphipathic helix-containing BAR (N-BAR) domains are the most highly curved, and are found in several late-stage endocytic adaptors including amphiphysin and endophilin [38] (Fig. 1). Membrane insertion of the amphipathic helices is thought to strengthen binding of N-BAR domains to membrane surfaces, as well as to assist in generation of membrane curvature in collaboration with scaffolding [36]. However, this contribution has been challenged by biophysical data from the Baumgart group [39]. Specifically, this work found that mutations or truncations of the helices of an N-BAR domain did not significantly alter the density of protein required to induce membrane remodeling [39]. Extended-FCH-BAR (F-BAR) domains are more gently curved in comparison to N-BAR domains, and therefore generate shallower membrane curvature [40, 41]. For example, the F-BAR domain-containing proteins FCHo1/2, early-arriving accessory proteins of clathrin-mediated endocytosis, help initiate clathrin-coated pits [42, 43]. Inverse BAR (I-BAR) domains are curved in the opposite direction compared to N-BAR and F-BAR, and are therefore implicated in generating membrane protrusions such as filopodia [44]. Finally, Pinkbar is an approximately flat BAR domain, which promotes the formation of planar, plasma membrane sheets at cell-cell junctions in intestinal cells [45].
BAR domain-containing proteins assist in driving membrane fission primarily by recruiting dynamin [46]. However, emerging evidence suggests that N-BAR domain-containing proteins such as endophilin can drive membrane fission directly. Specifically, in vitro work from the Johannes and Bassereau groups found that endophilin-coated membrane tubes undergo fission when a rapid pulling force is applied [47]. The authors hypothesized that the endophilin scaffold limits the flow of lipids during pulling, leading to spontaneous reduction in the diameter of the tube until fission occurs. More recent in vitro and cell-based studies from the Bassereau and Callan-Jones groups provided further experimental and theoretical evidence for this tube pulling-based fission mechanism by BAR scaffolds, which they call friction-driven scission (FDS) [48].
A final group of essential scaffolding proteins involved in membrane bending and fission is the endosomal sorting complex required for transport (ESCRT) machinery [49]. ESCRTs are best known for sorting and encapsulating protein cargos into intralumenal vesicles of multivesicular bodies (MVBs), but ESCRTs are also involved in other membrane shaping processes including cytokinetic abscission and viral budding [50] (Fig. 1). ESCRTs are unique from dynamin and BAR domains as they drive “reverse-topology” membrane budding and fission, in which the membrane bends away from the cytoplasm. The ESCRT machinery consists of complexes 0, I, II, and III. ESCRT-0, I, and II are involved in binding and sorting cargo and driving the initial curvature of the bud, while the ESCRT-III complexes drive membrane fission [51, 52].
The main assembly subunit of ESCRT-III, Snf7 in yeast, forms spiral assemblies on membranes with a preferred radius of curvature of 25–30 nm, as shown in in vitro work from the Roux and Scheuring groups [53]. This study found that as Snf7 spirals grow, the inner rings become over-bent relative to this preferred radius of curvature, while the outer rings are under-bent. Snf7 spirals thereby act as mechanical “springs,” storing energy that can be released when the spiral buckles out of the plane of the membrane, driving membrane curvature [53]. The direction of buckling by ESCRT-III polymers depends on the specific ESCRT-III subunit. While Snf7 scaffolds coat the inner surface of the membrane tube, other ESCRT-III subunits prefer to coat the external surface of the tube, as shown in studies performed in vitro and in cells by the Frost and Hanson groups [54]. The ability of ESCRT-III polymers to adapt to a variety of membrane curvatures suggests how ESCRT proteins are capable of driving diverse membrane shaping processes throughout the cell. Finally, Snf7 also contains an N-terminal, 11-amino acid amphipathic helix which inserts into the membrane. Live cell studies from the Emr group showed that this amphipathic helix is essential for Snf7 membrane binding, but likely does not contribute to membrane curvature generation, as helix insertion would oppose the reverse-topology curvature created by the Snf7 scaffold [55]. While membrane curvature generation by ESCRT-III is becoming well-understood, the exact mechanisms of membrane fission by ESCRT-III remain debated [50].
Cytoskeletal assemblies and membrane fission
Polymerization of the actin cytoskeleton at membrane surfaces is another important, structure-based driver of membrane fission [56–58]. During endocytosis, actin polymerization imparts a force along the membrane necks of clathrin-coated pits, driving neck constriction and promoting membrane fission [59, 60]. While actin polymerization is required for endocytosis in yeast, it is less clearly essential in mammalian cells [61]. Nonetheless, actin is known to play a role in endocytosis in mammalian cells, for example in counteracting high membrane tension to ensure robust fission, as shown in work from the Kirchhausen group [62].
It is unlikely that any one of the mechanisms outlined above is alone responsible for fission. In particular, clathrin-mediated endocytosis is carried out by a complex network of interacting proteins [63], suggesting that several mechanisms likely work together to ensure robust fission. For example, work from the de Camilli group showed that BAR proteins and actin polymerization work upstream of dynamin to drive membrane curvature prior to dynamin-mediated fission, and even drive fission when dynamin is knocked out [64]. Another study from the same group showed that epsin helps drive curvature of clathrin-coated endocytic structures by recruiting the actin cytoskeleton [65]. Clathrin-independent endocytic pathways also likely rely on multiple fission mechanisms. A report from the Johannes group demonstrated that several fission proteins, including endophilin, actin, and dynamin, work in an additive manner to drive fission in cells [47]. All of these proteins likely facilitate fission in a recently characterized, clathrin-independent pathway, fast endophilin-mediated endocytosis (FEME) [66].
The fission proteins discussed above all rely on specific structural or assembly features to drive fission. However, another unifying property of these protein structures is that they are inherently localized on one side of the membrane, ensuring that they act asymmetrically to induce curvature from an initially flat membrane surface. Whether these proteins insert hydrophobic regions, assemble into scaffolds, or physically push on membranes, each protein acts preferentially on one side of the membrane, and not on the other, in order to drive bending and fission. From this perspective, we next examine how nonspecific physical effects, which can also act asymmetrically on membranes, can contribute to membrane bending and fission by increasing membrane spontaneous curvature [67].
Nonspecific physical mechanisms can raise membrane spontaneous curvature
Osmotic gradients can increase membrane spontaneous curvature
Osmotic imbalance across the membrane is an important example of a nonspecific physical stimulus that can raise membrane spontaneous curvature. Early in vitro work from the Helfrich group reported tubular protrusions from vesicles exposed to a hypertonic external solution. The authors hypothesized that osmotically-driven water efflux from these vesicles induced a flow of lipids from the inner to outer membrane leaflets, leading to an asymmetry in leaflet area which raised membrane spontaneous curvature [68]. Later in vitro work from the Parikh group showed that osmotically-induced spontaneous curvature is sufficient to induce a pearling instability, in which the membrane is transformed into a set of curved vesicles, or “pearls,” connected by narrow membrane tethers. When the diameter of these tethers became sufficiently narrow, the tethers underwent spontaneous fission leading to the separation of the pearled vesicles [69]. Similarly, in vitro work from the Frolov group showed that osmotically-induced water efflux was capable of driving membrane fission. In these experiments, exposure of membrane nanotubes to a hypertonic external solution drove squeezing of the tube diameter until spontaneous fission occurred [70].
Membrane-anchored polymers generate steric pressure that increases membrane spontaneous curvature
The addition of high concentrations of membrane-anchored polymers has also been demonstrated to raise membrane spontaneous curvature. Specifically, when polymers densely cover a membrane surface, the lateral confinement of the polymers creates steric pressure. This pressure acts to reduce polymer entropy and extend the polymer from an initially mushroom-like configuration into a more straightened, brush-like configuration [71]. Theoretical work from Lipowsky suggested that when polymers asymmetrically crowd one membrane leaflet, the polymers will raise membrane spontaneous curvature in order to expand the available surface area, thereby reducing the loss in polymer entropy due to lateral steric pressure [72]. Polymer crowding at membrane surfaces has also been demonstrated to promote micelle formation, in experimental work from the Sportelli group [73]. This work showed that inclusion of high surface densities of polyethylene glycol (PEG) conjugated to membrane lipids promoted lateral expansion, or stretching, of the bilayer. When steric pressure among crowded PEG chains became comparable to the tensile strength of the bilayer, the bilayer was destabilized and transformed into lipid micelles, the high curvature of which provides a larger volume per PEG chain [73]. Interestingly, lipid micelles resemble the hemi-fission intermediate, in which the membrane consists of a single lipid monolayer prior to complete membrane fission, suggesting that lateral pressure exerted by polymer crowding can overcome one of the key energetic barriers to fission.
Steric pressure from protein crowding provides a structure-independent mechanism of membrane bending
The ability of steric pressure from crowding among membrane-anchored polymers to raise spontaneous curvature suggests that any membrane-bound molecule, including proteins, should be able to drive membrane curvature and fission through a crowding mechanism. Indeed, a growing body of work is revealing that asymmetric crowding among proteins at membrane surfaces generates significant steric pressure that can drive membrane bending and even membrane fission. In 2010 our group reported that when green fluorescent protein (GFP) bound to giant unilamellar vesicles at high density, the proteins drove the formation of long, tubular protrusions from the vesicles [74]. The length of these tubules increased with the concentration of GFP, suggesting that the driving force for tubulation was dependent on the surface density of membrane-bound protein. The ability of GFP to drive membrane bending in vitro, confirmed in recent work from the Baumgart group [75], was surprising because GFP plays no physiological role in membrane curvature in the cell and contains none of the structural features thought to be required for membrane bending. How can a simple globular protein such as GFP drive membrane curvature when bound to membranes at high surface coverage?
We proposed that crowded membrane-bound proteins generate steric pressure, analogous to a compressed gas. Specifically, the translational entropy of the proteins on the membrane surface results in lateral protein-protein collisions, which raise steric pressure nonlinearly with increasing coverage of the membrane surface by proteins. When the difference in steric pressure between the two membrane leaflet surfaces becomes sufficiently high, membrane curvature should increase to relieve the pressure and expand the area per protein molecule at the mid-plane of the attached protein layer (Fig. 2A). Notably, this view of steric pressure from protein crowding only accounts for lateral, repulsive collisions. Other physical effects may also influence steric pressure, including electrostatic repulsions and changes in rotational entropy. Future studies are needed to examine these diverse contributions in greater detail.
Figure 2.
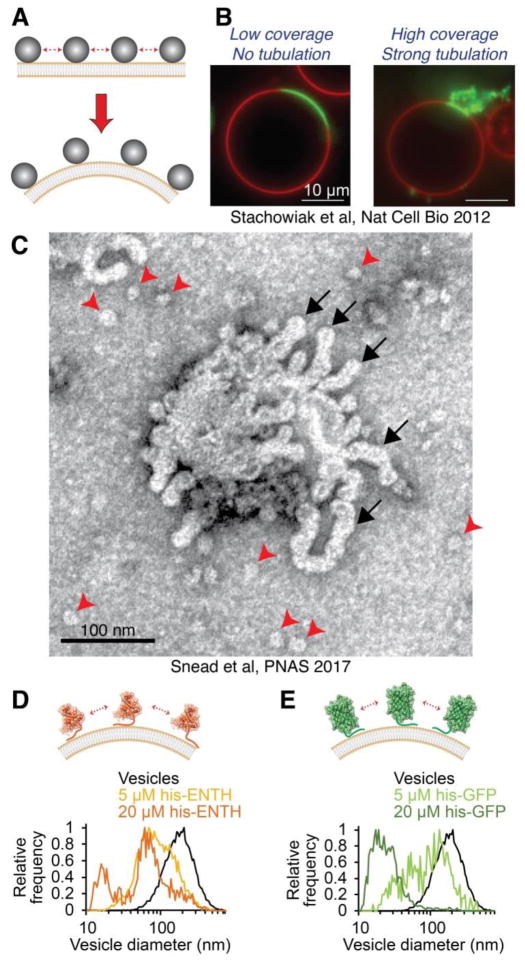
Protein crowding provides a stochastic driving force for membrane bending and fission. (A) Collisions among membrane-bound proteins densely covering the membrane surface generate steric pressure. The membrane takes on curvature to relieve steric pressure until crowding energy is balanced by the bending energy of the membrane. (B) When wild-type ENTH domain is bound to giant unilamellar vesicles at sufficient coverage, the resulting steric pressure drove membrane tubulation. Image from Stachowiak et al, Nature Cell Biology 2012 [76]. (C) Wild-type ENTH drove membrane fission in addition to membrane curvature, as evidenced by the presence of highly curved membrane tubules (black arrows) and fission vesicles (red arrowheads) after exposure of initially low-curvature vesicles to protein. (D) A version of the ENTH domain lacking the membrane-inserting amphipathic helix (his-ENTH) and recruited to membranes by a histidine-NTA interaction formed highly curved fission vesicles when crowded at membrane surfaces. (E) Histidine-tagged green fluorescent protein (his-GFP) also drove potent membrane fission by protein crowding, despite lacking any structural feature traditionally associated with fission. Data in (C – E) from Snead et al, PNAS 2017 [12].
A 2012 in vitro study from our group demonstrated that endocytic adaptor proteins can also drive membrane curvature using a crowding mechanism [76]. Specifically, this work examined membrane curvature by the ENTH domain of epsin, which, as discussed above, had been thought to drive curvature through insertion of an amphipathic helix [20, 21]. This work revealed that ENTH generated highly curved tubules from vesicles in a manner that was dependent on the surface coverage of membrane-bound protein (Fig. 2B). In particular, when the amphipathic helix of ENTH was mutated to reduce its hydrophobicity, or completely replaced with a non-inserting, synthetic membrane binding motif, the protein drove membrane curvature with equal efficiency in comparison to wild-type ENTH, provided all proteins covered equivalent fractions of the membrane surface. Specifically, when protein covered 20–30% of the membrane surface, ENTH consistently drove potent membrane tubulation, in agreement with the protein crowding mechanism described above. This study demonstrated that protein crowding is the underlying mechanism of membrane curvature generation by the ENTH domain, and that hydrophobic insertions promote crowding by attaching proteins tightly to the membrane surface, rather than directly bending membranes [76].
Protein crowding and membrane fission
In principle, any of the non-specific drivers of membrane bending described above also has the potential to fully vesiculate membranes, provided the associated change in membrane spontaneous curvature is sufficient to overcome the energetic barriers to membrane fission [67]. Indeed, a recent in vitro study from our group has demonstrated that protein crowding can also drive membrane fission [12] (Fig. 2C). In particular, previous work from the McMahon group showed that a series of ENTH mutants with decreasing hydrophobicity of the membrane-inserting helix drove substantially reduced membrane fission compared to wild-type ENTH [21]. In contrast, our study showed that the membrane binding affinity of L6E ENTH, the most hydrophilic of the mutant family, was significantly weaker in comparison to wild-type ENTH. Consequently, comparing wild-type ENTH and L6E ENTH at equal membrane coverage by proteins revealed that the two proteins drove equivalent membrane fission. Moreover, ENTH consistently formed fission products of similar curvature at a given membrane coverage by proteins, regardless of the helix hydrophobicity, and even for ENTH domains recruited to membranes by a histidine-NTA interaction rather than a helix insertion (Fig. 2D). Similarly, histidine-tagged GFP drove the formation of highly curved fission vesicles when bound to membranes at high coverage (Fig. 2E) [12]. Collectively, these findings revealed that protein crowding provides a potent, entropically-driven mechanism of membrane fission. Notably, protein crowding in the cell is likely facilitated by the collective contributions of many integral and membrane-bound proteins, rather than the individual protein constituents examined during in vitro experiments.
Protein crowding and membrane shaping throughout the cell
A significant body of work from several groups has implicated protein crowding in shaping membranes in a variety of cellular contexts. In vitro studies from the Baumgart group showed that membrane bending by the N-BAR domain of endophilin depends on the coverage of the membrane surface by proteins [77], and is independent of amphipathic helix insertion [39]. Additionally, a recent study performed in cells and in vitro from the Walther group revealed that protein crowding plays an important role in dictating the protein content of lipid droplets [78]. Specifically, proteins must compete for limited space on the surfaces of densely crowded lipid droplets, and certain proteins become displaced when lipid droplets shrink during lipolysis.
Work in yeast cells from the Miller group showed that the packaging of bulkier cargo molecules requires a more rigid COPII coat to accommodate the more highly crowded lumen [79]. Similarly, earlier work in mammalian cells from the Silvius group demonstrated that the targeting of lipid-anchored protein cargo to clathrin-independent endocytic pathways is determined by the physical bulk of the protein, regardless of the biochemical properties of the protein or its lipid anchor [80]. Moreover, recent mammalian cell experiments from our group showed that bulky, intrinsically disordered cargoes are excluded from clathrin-coated pits, owing to the greater steric pressure exerted in comparison to an equal number of smaller cargo proteins [81]. Collectively, these results imply that steric pressure among endocytic cargo proteins, which must be overcome during membrane bending and vesiculation, displaces bulky receptors that lack sufficient biochemical affinity for endocytic coats [82].
In contrast, steric pressure among transmembrane cargo proteins may actually assist in driving vesicle formation in the case of reverse-topology membrane bending and fission by the ESCRT machinery. Recent papers from the Roux and Scheuring groups and Kirchhausen and Teis groups hypothesized that asymmetric crowding among cargo proteins on the opposite membrane leaflet of ESCRT assemblies may help form intralumenal vesicles during MVB biogenesis [53, 83]. Similarly, budding of enveloped viruses, another context of reverse-topology membrane bending, may also be assisted by protein crowding. Specifically, early work from Vennema et al. showed that lateral interactions among viral envelope proteins alone, in the absence of the nucleocapsid, drive membrane budding and fission, suggesting that asymmetric crowding among viral surface proteins on the extracellular leaflet may provide a potent force for viral budding [84]. Interestingly, the influenza surface proteins hemagglutinin and neuraminidase have large ectodomains [85], consistent with this hypothesis. The potential collaborations between protein crowding and the mechanisms of reverse-topology membrane fission are fascinating but remain to be explored in depth.
Membrane curvature by protein crowding has also been implicated in human diseases. An in vitro and cell-based study from the Baumgart group found that previously-identified disease mutations affecting T-tubule formation by the protein BIN1 caused decreased membrane binding, thereby inhibiting BIN1 from driving membrane curvature partially via a crowding mechanism [86]. In another example, an in vitro study from the Lee group found that α-synuclein, an intrinsically disordered, amyloid-forming protein involved in Parkinson’s disease [87], is capable of driving curvature of membranes with no net charge, despite interacting with such membranes very weakly [88]. The lack of a requirement for specific binding lipids or protein structural features led the authors to hypothesize that α-synuclein may drive curvature through a crowding mechanism. Collectively, results from several groups in diverse systems have revealed that protein crowding plays an essential role in driving membrane curvature in multiple cellular contexts.
Intrinsically disordered domains as drivers of membrane bending and fission
Disordered domains of endocytic proteins drive membrane bending and fission
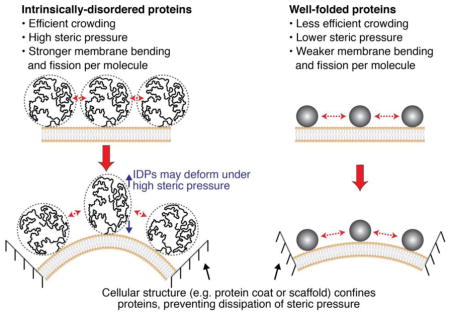
An important prediction of the protein crowding mechanism for membrane bending and fission is that larger proteins should drive fission more efficiently than smaller proteins, as fewer copies of larger proteins are required to reach a crowded membrane coverage. In line with this reasoning, intrinsically-disordered protein (IDP) domains lack a defined structure and occupy significantly larger volumes than folded proteins of equal molecular weight [89] (Fig. 3). IDPs have previously been demonstrated to generate entropic pressure that facilitates several cellular processes. For example, cytoskeletal components of neuronal axons called neurofilaments contain large IDP domains of 300–600 amino acids [90]. Analogous to a bottlebrush or pipe cleaner, neurofilament IDP domains radiate outward along the length of the filaments, creating a volume that excludes neighboring IDPs. Neurofilament IDP domains thereby control the spacing between filaments and the overall diameter of the axon [90, 91] (Fig. 3A). Similarly, the titin protein in muscle contains very large IDP domains of up to a few thousand amino acids, which act as entropic springs that stretch and compress under applied forces in order to provide the elasticity of skeletal muscle [92, 93].
Figure 3.
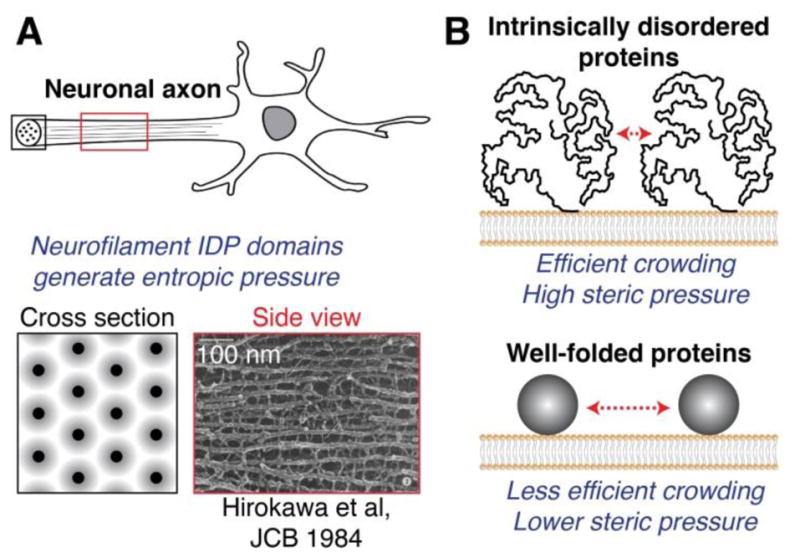
Intrinsically disordered proteins generate entropic pressure and are efficient drivers of membrane bending and fission. (A) Axonal neurofilaments are an important cellular example of entropic pressure generated by IDP domains. Neurofilament IDPs radiate outward along filaments, creating zones of exclusion which repel neighboring filaments and control filament spacing. Neurofilament IDP domains thereby determine the overall diameter of the axon. Electron micrograph from Hirokawa et al, Journal of Cell Biology 1984 [96]. (B) IDP domains crowd membrane surfaces more efficiently than well-folded proteins of equal molecular weight. IDP domains therefore require fewer protein copies to drive membrane bending and fission through a crowding mechanism, in comparison to well-folded domains of equal molecular weight.
Inspired by the potential of IDP domains to generate steric pressure, an in vitro study from our group demonstrated that membrane-bound IDP domains are potent drivers of membrane curvature [81]. This study utilized the clathrin adaptor proteins epsin and AP180, which both contain C-terminal IDP domains of greater than 400 amino acids [94, 95]. Our experiments demonstrated that the large hydrodynamic radii of these domains facilitate efficient crowding of the membrane surface, with fewer membrane-bound protein copies required to reach a crowded coverage in comparison to smaller, well-folded proteins like the ENTH domain of epsin (Fig. 3B). As expected, the IDP domains drove membrane curvature more efficiently than the ENTH domain, demonstrating that steric pressure among IDP domains provides a potent force for membrane shaping despite a lack of well-defined protein structure [81].
More recent work from our group also demonstrated that IDPs are potent drivers of membrane fission [12]. Specifically, we found that full-length epsin, which contains both the ENTH domain and the large, C-terminal IDP domain, drove fission at a substantially lower number density of membrane-bound proteins than the ENTH domain alone. This concept of increasing the steric bulk of a protein to enhance its fission activity applied to other fission proteins as well. In particular, we created a version of Sar1, a small, globular protein involved in fission of COPII-coated vesicles [22], fused to the large, C-terminal IDP domain of AP180. Experiments revealed that this bulky Sar1-AP180 fusion protein drove fission at a lower number density of membrane-bound proteins in comparison to Sar1 alone. Collectively these findings demonstrated that the physical bulk of IDP domains can substantially enhance the efficiency of fission-driving proteins.
Physical basis of membrane shaping by disordered domains
As described previously, the lateral collisions among crowded membrane-bound proteins generate steric pressure, analogous to a compressed gas. This steric pressure can be estimated using a non-ideal equation of state, for example the Carnahan-Starling equation, which approximates the proteins as non-interacting, membrane-bound discs [97]. This model predicts that steric pressure will increase nonlinearly with increasing coverage of the membrane surface by proteins [76].
Although non-ideal equations of state like Carnahan-Starling provide a useful estimate of steric pressure from crowding of well-folded proteins [76, 98], such models are less appropriate for inherently flexible disordered protein domains. Specifically, IDPs are deformable polymers, capable of compressing under applied pressure and interdigitating with neighboring IDP chains [99, 100]. Therefore, other IDP models are needed in order to better account for IDP deformation and chain interdigitating under crowded conditions. A recent report from the Schuler group explored the ability of several models to predict IDP behavior under crowding by molecules of varying molecular weight [99]. For example, the “Gaussian cloud” model estimates the mass distribution of an IDP as a Gaussian function centered around the center of mass of the protein [101]. This model allows for rigid-sphere crowding molecules that are much smaller than the size of the IDP chain itself to penetrate the IDP. However, when the size of the crowder became close to the size of the IDP, the crowder no longer penetrated the chain, and the model predictions became similar to those of a non-ideal equation of state. Therefore, more complex polymer theories are needed to predict IDP compaction under crowding by polymer-like crowders of similar size to the IDP. Indeed, the authors found that a renormalized form of Flory-Huggins theory provided reasonable predictions of IDP compaction over a wide range of crowder sizes, in good agreement with the experimental measurements [99].
In addition to deforming under crowding conditions, IDPs also have complex, sequence-dependent charge profiles and pH sensitivity [102]. Changes in salt concentration or pH can therefore strongly influence the mechanical properties and hydrodynamic radii of IDPs. For example, a study from the Kumar group found that ionic strength and pH dramatically affected the thickness of a brush layer of IDPs assembled on a rigid substrate [103]. Specifically, when pH was altered, the IDP behavior shifted from that of a polyelectrolyte, with residues of mostly like charge, to a polyampholyte, with residues of opposite charge. This transition caused the IDPs to compress from an initially expanded state, caused by repulsive forces among like charges, to a more compact state, caused by electrostatic interactions and salt bridges formed between oppositely charged residues [103]. Based on these findings, it is expected that the size and mechanical properties of the IDP domains involved in membrane shaping should also be strongly influenced by changes in pH and ionic strength. Such changes could alter the ability of the IDPs to generate steric pressure via protein crowding. Future theoretical models should account for the polyelectrolyte or polyampholyte behavior of IDPs in order to better understand how crowded IDPs generate steric pressure.
Cellular contexts in which IDP domains may assist in membrane bending and fission
Intriguingly, structural disorder is found in many proteins involved in vesicle trafficking [104] (Table 1), suggesting that bulky, disordered proteins may play an important yet poorly understood role in remodeling cellular membranes. Clathrin-mediated endocytosis employs many proteins with large regions of structural disorder [104, 105], which could assist in shaping clathrin-coated structures and facilitating membrane fission. As previously discussed, the clathrin adaptors epsin and AP180 (Table 1) both contain large IDP domains [94, 95] that can drive membrane bending and fission [12, 81], suggesting that these proteins help shape clathrin-coated structures and sever them from the membrane surface. Additionally, intersectin and FCHo1 (Table 1), involved in the early stages of clathrin-coated pit initiation, also have large IDP domains of greater than 400 amino acids [104]. Crowding by these IDP domains could potentially assist in generating the initial curvature of a pit. The N-BAR domain-containing protein amphiphysin (Table 1), which is recruited to the curved necks of late stage clathrin-coated pits via cooperative interactions with dynamin [106, 107], contains a large IDP domain of greater than 300 amino acids [108]. Similar to amphiphysin, sorting nexin 9 (SNX9, Table 1) is also recruited synergistically with dynamin to membrane necks [107, 109], and contains both a BAR domain and an IDP domain of greater than 100 amino acids [110]. The ability of amphiphysin and SNX9 to cooperatively assemble with dynamin at the necks of clathrin-coated pits, thereby locally concentrating large disordered domains, could provide an intriguing potential fission mechanism that remains to be investigated (Fig. 4). Finally, the clathrin-uncoating protein, auxilin (Table 1), contains a large IDP domain of greater than 400 amino acids [111]. Recent in vitro work from the Lafer and Sousa groups showed that entropic pressure created by auxilin provides a potent driving force for disassembly of the clathrin coat [112], suggesting that IDP domains play an important role in vesicle uncoating.
Table 1.
Intrinsically-disordered protein domains potentially involved in membrane shaping. Values of predicted % disorder (human sequences) come from Pietrosemoli et al., PLOS Computational Biology 2013 [104]. CME: clathrin-mediated endocytosis.
| Protein | Function | Membrane interaction | % Disorder (location) | Major binding partners |
|---|---|---|---|---|
| Epsin1 | Cargo adaptor in CME [20, 94] | ENTH domain [20] | 78 (C-term.) | AP2, clathrin, cargo [63] |
| AP180 | Cargo adaptor in CME [94, 95] | ANTH domain [115] | 28 (C-term.) | AP2, clathrin, cargo [63] |
| Intersectin1 | CME initiation [42] | PH, C2 domains [116] | 28 (middle) | AP2, clathrin, Eps15, FCHo1/2 [63, 116] |
| FCHo1 | CME initiation [42] | F-BAR domain [117] | 48 (middle) | Eps15, intersectin [42] |
| Amphiphysin | Dynamin recruitment [106] | N-BAR domain [38] | 61 (middle) | Dynamin, AP2, clathrin [63] |
| SNX9 | Dynamin recruitment [109] | PX-BAR domain [110] | 27 (middle) | Dynamin, AP2, clathrin [63] |
| Auxilin | Clathrin uncoating [118] | n/a | 45 (middle) | HSC70, clathrin, dynamin [63] |
| Sec16A | COPII accessory [113] | n/a | 71 (N and C-term.) | Sec13/31, Sec23/24, Sar1 [113, 119] |
| Sec31A | COPII coat component [114] | n/a | 34 (middle) | Sec13, Sec23/24, Sec16 [114, 119] |
Figure 4.
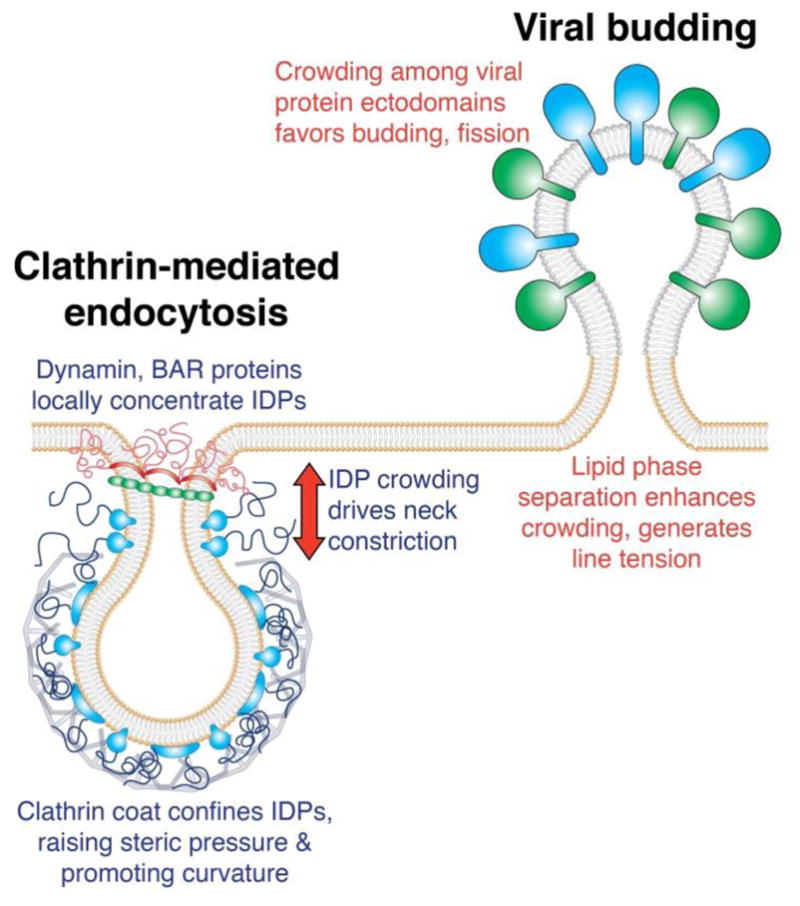
Potential mechanisms of organizing and controlling protein crowding to drive membrane bending and fission.
Large IDP domains are present in intracellular trafficking proteins as well. Sec16 (Table 1), an essential COPII accessory protein involved in cargo capture and coat assembly, contains a disordered region of greater than 1000 amino acids [104, 113], which is thought to be important for spanning long distances to capture the COPII coat components [104]. The potential of this IDP domain to generate steric pressure that could drive budding and fission of COPII-coated vesicles remains to be explored. The COPII coat component Sec31 (Table 1) also has a disordered region of greater than 400 amino acids, thought to be important for binding the Sec23/24 complex [114]. Like Sec16, Sec31 may help promote steric pressure and thereby aid in budding and fission of COPII vesicles.
Notably, attractive interactions among IDPs have the potential to significantly influence steric pressure in a crowded environment. In particular, lateral interactions among neighboring IDPs would tend to reduce the conformational entropy of the IDP, lowering steric pressure. Therefore, many of the IDP domains involved in membrane fission could behave quite differently in a crowded membrane environment compared to the model IDPs examined in previous in vitro studies. Future work is needed to better understand how the physical properties and binding interactions of diverse IDPs influence the steric pressure generated by crowded IDP domains.
Mechanisms of organizing and controlling crowding at presumptive fission sites
The abundance of bulky proteins, especially IDP domains, near sites of membrane fission suggests that such proteins make important contributions toward meeting the energetic demands of fission. However, crowding is inherently stochastic, as it relies on random collisions among proteins attached to membrane surfaces. How might the cell organize protein crowding to ensure fission occurs at desired locations?
Protein coats may provide an efficient means of concentrating and confining proteins within a defined space. As discussed above, the clathrin coat interacts with a complex network of accessory proteins [63], many of which contain large IDP domains [104]. The ability of the clathrin coat to locally confine bulky molecules, increasing steric pressure, may facilitate bending and fission of clathrin-coated pits (Fig. 4). Similarly, assembly of the COPII coat could also enable the concentration of bulky IDP domains on membrane surfaces, including those of Sec16 and Sec31, to drive membrane budding and fission. However, it is important to note that even though the IDPs involved in membrane traffic often do not fold upon encountering their binding partners [43, 120], the clathrin and COPII coats may still limit the mobility and conformational entropy of the proteins they confine, lowering steric pressure. Therefore, continuous and dynamic remodeling of the coat proteins may be necessary to provide the confined IDPs and other accessory proteins with flexibility and mobility in order to build steric pressure. In support of this hypothesis, studies in cells from the Briggs and Kaksonen groups have shown that the clathrin coat is highly dynamic and continuously remodeled throughout maturation [121]. Understanding how crowded adaptor proteins interact with coat scaffolds is a complex biophysical problem that will require detailed measurements and modeling studies.
In addition to constricting membrane tubes, helical protein scaffolds such as dynamin may also help to generate steric pressure by recruiting bulky proteins, such as amphiphysin and SNX9, at curved membrane necks. High local density of the bulky IDP domains of amphiphysin and SNX9 may help to constrict the membrane tube down to a fission-competent diameter. This crowding-mediated tube constriction may then collaborate with the structure-mediated fission activity of dynamin by reducing the energetic barrier to fission that dynamin must overcome (Fig. 4).
Cells may also utilize cytoskeletal assemblies to organize steric factors. For example, the septin family of cytoskeletal proteins acts as a barrier to diffusion of proteins over membrane surfaces, partitioning and confining proteins within defined membrane regions [122]. One such region is the primary cilium, where septins were shown to help maintain the enrichment of signaling proteins, such as the Sonic hedgehog pathway proteins, by preventing diffusion out of the cilium into the surrounding membrane [123]. Sustaining a crowded protein environment for the purpose of driving membrane bending and fission could be an important function of cytoskeletal diffusion barriers.
The separation of membrane lipids into distinct phases could provide another means of concentrating proteins and acutely raising steric pressure. Work from our group [74, 98, 124] and others [125] has shown that distinct lipid phases on synthetic membranes provide a platform for concentrating proteins and bulky molecules in vitro, suggesting that cells may utilize lipid phase separation to locally increase protein concentration. One such cellular context is the assembly of influenza viral buds, during which lipid rafts coalesce into a solid-like lipid phase enriched in cholesterol and sphingolipids [85] (Fig. 4). Raft coalescence is thought to occur as the viral surface proteins hemagglutinin and neuraminidase, both of which are raft-localized proteins, assemble at the bud region. Crowding among these concentrated viral surface proteins may then favor budding and fission of the virus particle (Fig. 4). Further, line tension at the lipid phase boundary, which tends to minimize the length of the boundary interface, has also been shown to promote membrane constriction and fission [126–128]. In particular, line tension is thought to serve as a driving force for fission of endocytic vesicles in yeast [57, 129]. These findings suggest that line tension may work cooperatively with protein crowding to drive fission of phase-separated membrane regions. However, molecular crowding has been shown to drive the dissolution of lipid membrane domains [98, 124], suggesting that steric pressure can actually disrupt the very domain which confines the proteins. Therefore, other cellular structures and mechanisms may be needed to help maintain the enrichment of proteins at sites of lipid phase separation.
Perspectives
The mechanisms described above for organizing protein crowding suggest new roles for the canonical, structure-based fission machinery. By enriching and confining bulky proteins within defined membrane spaces, these fission machines may facilitate a sharp increase in steric pressure, driving fission via a crowding mechanism. While this possibility is intriguing, future work remains to improve understanding of how protein coats and scaffolds concentrate steric factors, and how protein crowding can collaborate with structure-based fission mechanisms. Specifically, while it is increasingly clear that protein crowding provides a potent force for driving membrane bending and fission, much work remains in order to understand how protein crowding might help to drive membrane fission in diverse contexts throughout the cell. In particular, much of our understanding of the protein crowding mechanism has come from studies on endocytic proteins, but crowding likely contributes to a broader class of cellular processes. Further, it is becoming clear that intrinsically disordered domains are important drivers of membrane remodeling, in contrast with the long-standing view that the capacity to shape membranes arises from specific, structured domains. However, the role of structural disorder in membrane shaping and fission is only beginning to be understood. Future work should take a closer look at disordered regions near sites of membrane fission, and examine the collective contributions from both structured and unstructured domains.
As discussed above, the development of appropriate theoretical and computational models will be essential toward improving our understanding of the biophysics of membrane fission. Simple analytical models based on non-ideal equations of state provide useful estimates of the contribution of protein crowding to membrane curvature, but the interplay between crowding and structure-based mechanisms requires more sophisticated modeling approaches [60]. Further, development of appropriate theories is needed to identify and predict regions of protein structural disorder near sites of membrane fission. For example, the Pappu group has developed computational tools which predict the physical and chemical properties of IDPs based on amino acid sequence [130]. Such tools will aid in understanding how IDPs generate steric pressure and ultimately participate in membrane fission in diverse cellular contexts.
Finally, the majority of our mechanistic understanding of protein crowding and membrane bending and fission has come from in vitro work using purified membrane-protein systems. However, these questions must be examined in the context of live cells in order to understand whether protein crowding can provide physiologically relevant forces for bending and driving fission of cellular membranes. In such studies, it is essential to quantify the molecular stoichiometry at fission sites, in order to estimate the magnitudes of crowding and steric pressure. Fortunately, calibrated single-molecule studies in live cells [131, 132] and even living tissues [133] provide a powerful means of quantitatively monitoring protein recruitment to sites of membrane fission. Advanced imaging approaches and single-molecule techniques like these will be essential for improving our understanding of how diverse mechanisms contribute to shaping the membranes of living cells.
A significant body of work from multiple laboratories has collectively demonstrated the importance of protein crowding in shaping and driving fission of cellular membranes. These findings suggest rich opportunities for future research that will ultimately elucidate the synergistic contributions from both stochastic and structure-based biophysical mechanisms to membrane remodeling in the cell.
Highlights.
Membrane fission is essential for basic cellular functions.
Structured proteins are viewed as the traditional drivers of membrane fission.
Steric pressure from protein crowding is a structure-independent fission mechanism.
IDPs may be among the most potent fission drivers, owing to their large size.
Future work is needed to understand how IDPs shape membranes throughout the cell.
Acknowledgments
J.C.S. acknowledges funding from the National Institutes of Health (R01GM112065). W.T.S. acknowledges the support of a Ruth L. Kirschstein NRSA Predoctoral Fellowship from the National Institutes of Health (F31GM121013), as well as a fellowship from the Graduate School at UT Austin. The authors declare no conflict of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Conner S, Schmid S. Regulated portals of entry into the cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 2.Corda D, Carcedo C, Bonazzi M, Luini A, Spano S. Molecular aspects of membrane fission in the secretory pathway. Cellular and Molecular Life Sciences. 2002;59:1819–32. doi: 10.1007/PL00012508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoppins S, Lackner L, Nunnari J. The machines that divide and fuse mitochondria. Annual Review of Biochemistry. 2007;76:751–80. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- 4.Votteler J, Sundquist WI. Virus budding and the ESCRT pathway. Cell Host Microbe. 2013;14:232–41. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlton J, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: A role for the ESCRT machinery. Science. 2007;316:1908–12. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 6.Hurley J, Boura E, Carlson L, Rozycki B. Membrane Budding. Cell. 2010;143:875–87. doi: 10.1016/j.cell.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helfrich W. Elastic Properties of Lipid Bilayers - Theory and Possible Experiments. Zeitschrift Fur Naturforschung C-a Journal of Biosciences. 1973;C 28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 8.Kozlovsky Y, Kozlov M. Membrane fission: Model for intermediate structures. Biophysical Journal. 2003;85:85–96. doi: 10.1016/S0006-3495(03)74457-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmerberg J, Kozlov M. How proteins produce cellular membrane curvature. Nature Reviews Molecular Cell Biology. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 10.McMahon H, Gallop J. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–6. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 11.Stachowiak J, Brodsky F, Miller E. A cost-benefit analysis of the physical mechanisms of membrane curvature. Nature Cell Biology. 2013;15:1019–27. doi: 10.1038/ncb2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snead W, Hayden C, Gadok A, Zhao C, Lafer E, Rangamani P, et al. Membrane fission by protein crowding. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E3258–E67. doi: 10.1073/pnas.1616199114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fotin A, Cheng Y, Sliz P, Grigorieff N, Harrison S, Kirchhausen T, et al. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–9. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- 14.Sundborger A, Fang S, Heymann J, Ray P, Chappie J, Hinshaw J. A Dynamin Mutant Defines a Superconstricted Prefission State. Cell Reports. 2014;8:734–42. doi: 10.1016/j.celrep.2014.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mim C, Cui H, Gawronski-Salerno J, Frost A, Lyman E, Voth G, et al. Structural Basis of Membrane Bending by the N-BAR Protein Endophilin. Cell. 2012;149:137–45. doi: 10.1016/j.cell.2012.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stagg S, Gurkan C, Fowler D, LaPointe P, Foss T, Potter C, et al. Structure of the Sec13/31 COPII coat cage. Nature. 2006;439:234–8. doi: 10.1038/nature04339. [DOI] [PubMed] [Google Scholar]
- 17.Campelo F, McMahon H, Kozlov M. The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophysical Journal. 2008;95:2325–39. doi: 10.1529/biophysj.108.133173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drin G, Antonny B. Amphipathic helices and membrane curvature. Febs Letters. 2010;584:1840–7. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 19.Hristova K, Wimley W, Mishra V, Anantharamiah G, Segrest J, White S. An amphipathic alpha-helix at a membrane interface: A structural study using a novel X-ray diffraction method. Journal of Molecular Biology. 1999;290:99–117. doi: 10.1006/jmbi.1999.2840. [DOI] [PubMed] [Google Scholar]
- 20.Ford M, Mills I, Peter B, Vallis Y, Praefcke G, Evans P, et al. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–6. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 21.Boucrot E, Pick A, Camdere G, Liska N, Evergren E, McMahon H, et al. Membrane Fission Is Promoted by Insertion of Amphipathic Helices and Is Restricted by Crescent BAR Domains. Cell. 2012;149:124–36. doi: 10.1016/j.cell.2012.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee M, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell. 2005;122:605–17. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 23.Beck R, Prinz S, Diestelkotter-Bachert P, Rohling S, Adolf F, Hoehner K, et al. Coatomer and dimeric ADP ribosylation factor 1 promote distinct steps in membrane scission. Journal of Cell Biology. 2011;194:765–77. doi: 10.1083/jcb.201011027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antonny B, BeraudDufour S, Chardin P, Chabre M. N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry. 1997;36:4675–84. doi: 10.1021/bi962252b. [DOI] [PubMed] [Google Scholar]
- 25.Tian C, Gao P, Pinto L, Lamb R, Cross T. Initial structural and dynamic characterization of the M2 protein transmembrane and amphipathic helices in lipid bilayers. Protein Science. 2003;12:2597–605. doi: 10.1110/ps.03168503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossman J, Jing X, Leser G, Lamb R. Influenza Virus M2 Protein Mediates ESCRT-Independent Membrane Scission. Cell. 2010;142:902–13. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt N, Mishra A, Wang J, DeGrado W, Wong G. Influenza Virus A M2 Protein Generates Negative Gaussian Membrane Curvature Necessary for Budding and Scission. Journal of the American Chemical Society. 2013;135:13710–9. doi: 10.1021/ja400146z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khattree N, Ritter L, Goldberg A. Membrane curvature generation by a C-terminal amphipathic helix in peripherin-2/rds, a tetraspanin required for photoreceptor sensory cilium morphogenesis. Journal of Cell Science. 2013;126:4659–70. doi: 10.1242/jcs.126888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walser P, Ariotti N, Howes M, Ferguson C, Webb R, Schwudke D, et al. Constitutive Formation of Caveolae in a Bacterium. Cell. 2012;150:752–63. doi: 10.1016/j.cell.2012.06.042. [DOI] [PubMed] [Google Scholar]
- 30.Pagliuso A, Valente C, Giordano L, Filograna A, Li G, Circolo D, et al. Golgi membrane fission requires the CtBP1-S/BARS-induced activation of lysophosphatidic acid acyltransferase delta. Nature Communications. 2016:7. doi: 10.1038/ncomms12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antonny B, Burd C, De Camilli P, Chen E, Daumke O, Faelber K, et al. Membrane fission by dynamin: what we know and what we need to know. Embo Journal. 2016;35:2270–84. doi: 10.15252/embj.201694613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hinshaw J, Schmid S. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–2. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- 33.Ramachandran R, Pucadyil T, Liu Y, Acharya S, Leonard M, Lukiyanchuk V, et al. Membrane Insertion of the Pleckstrin Homology Domain Variable Loop 1 Is Critical for Dynamin-catalyzed Vesicle Scission. Molecular Biology of the Cell. 2009;20:4630–9. doi: 10.1091/mbc.E09-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferguson S, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nature Reviews Molecular Cell Biology. 2012;13:75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frost A, Unger V, De Camilli P. The BAR Domain Superfamily: Membrane-Molding Macromolecules. Cell. 2009;137:191–6. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mim C, Unger V. Membrane curvature and its generation by BAR proteins. Trends in Biochemical Sciences. 2012;37:526–33. doi: 10.1016/j.tibs.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simunovic M, Voth G, Callan-Jones A, Bassereau P. When Physics Takes Over: BAR Proteins and Membrane Curvature. Trends in Cell Biology. 2015;25:780–92. doi: 10.1016/j.tcb.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peter B, Kent H, Mills I, Vallis Y, Butler P, Evans P, et al. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science. 2004;303:495–9. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 39.Chen Z, Zhu C, Kuo CJ, Robustelli J, Baumgart T. The N-Terminal Amphipathic Helix of Endophilin Does Not Contribute to Its Molecular Curvature Generation Capacity. J Am Chem Soc. 2016;138:14616–22. doi: 10.1021/jacs.6b06820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman E, et al. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–17. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDonald NA, Gould KL. Linking up at the BAR: Oligomerization and F-BAR protein function. Cell Cycle. 2016;15:1977–85. doi: 10.1080/15384101.2016.1190893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henne W, Boucrot E, Meinecke M, Evergren E, Vallis Y, Mittal R, et al. FCHo Proteins Are Nucleators of Clathrin-Mediated Endocytosis. Science. 2010;328:1281–4. doi: 10.1126/science.1188462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma L, Umasankar P, Wrobel A, Lymar A, McCoy A, Holkar S, et al. Transient Fcho1/2.Eps15/R.AP-2 Nanoclusters Prime the AP-2 Clathrin Adaptor for Cargo Binding. Developmental Cell. 2016;37:428–43. doi: 10.1016/j.devcel.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Millard T, Bompard G, Heung M, Dafforn T, Scott D, Machesky L, et al. Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. Embo Journal. 2005;24:240–50. doi: 10.1038/sj.emboj.7600535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pykalainen A, Boczkowska M, Zhao H, Saarikangas J, Rebowski G, Jansen M, et al. Pinkbar is an epithelial-specific BAR domain protein that generates planar membrane structures. Nature Structural & Molecular Biology. 2011;18:902–U59. doi: 10.1038/nsmb.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daumke O, Roux A, Haucke V. BAR Domain Scaffolds in Dynamin-Mediated Membrane Fission. Cell. 2014;156:882–92. doi: 10.1016/j.cell.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 47.Renard H, Simunovic M, Lemiere J, Boucrot E, Garcia-Castillo M, Arumugam S, et al. Endophilin-A2 functions in membrane scission in clathrin-independent endocytosis. Nature. 2015;517:493. doi: 10.1038/nature14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simunovic M, Manneville J, Renard H, Evergren E, Raghunathan K, Bhatia D, et al. Friction Mediates Scission of Tubular Membranes Scaffolded by BAR Proteins. Cell. 2017:170. doi: 10.1016/j.cell.2017.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hurley J, Hanson P. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nature Reviews Molecular Cell Biology. 2010;11:556–66. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schoneberg J, Lee I, Iwasa J, Hurley J. Reverse-topology membrane scission by the ESCRT proteins. Nature Reviews Molecular Cell Biology. 2017;18:5–17. doi: 10.1038/nrm.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley J. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–U2. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wollert T, Hurley J. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–U73. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chiaruttini N, Redondo-Morata L, Colom A, Humbert F, Lenz M, Scheuring S, et al. Relaxation of Loaded ESCRT-III Spiral Springs Drives Membrane Deformation. Cell. 2015;163:866–79. doi: 10.1016/j.cell.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCullough J, Clippinger A, Talledge N, Skowyra M, Saunders M, Naismith T, et al. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science. 2015;350:1548–51. doi: 10.1126/science.aad8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buchkovich N, Henne W, Tang S, Emr S. Essential N-Terminal Insertion Motif Anchors the ESCRT-III Filament during MVB Vesicle Formation. Developmental Cell. 2013;27:201–14. doi: 10.1016/j.devcel.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 56.Romer W, Pontani L, Sorre B, Rentero C, Berland L, Chambon V, et al. Actin Dynamics Drive Membrane Reorganization and Scission in Clathrin-Independent Endocytosis. Cell. 2010;140:540–53. doi: 10.1016/j.cell.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 57.Liu J, Kaksonen M, Drubin D, Oster G. Endocytic vesicle scission by lipid phase boundary forces. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10277–82. doi: 10.1073/pnas.0601045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jarsch I, Daste F, Gallop J. Membrane curvature in cell biology: An integration of molecular mechanisms. Journal of Cell Biology. 2016;214:375–87. doi: 10.1083/jcb.201604003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Collins A, Warrington A, Taylor K, Svitkina T. Structural Organization of the Actin Cytoskeleton at Sites of Clathrin-Mediated Endocytosis. Current Biology. 2011;21:1167–75. doi: 10.1016/j.cub.2011.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hassinger J, Oster G, Drubin D, Rangamani P. Design principles for robust vesiculation in clathrin-mediated endocytosis. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E1118–E27. doi: 10.1073/pnas.1617705114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Galletta B, Mooren O, Cooper J. Actin dynamics and endocytosis in yeast and mammals. Current Opinion in Biotechnology. 2010;21:604–10. doi: 10.1016/j.copbio.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boulant S, Kural C, Zeeh J, Ubelmann F, Kirchhausen T. Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nature Cell Biology. 2011;13:1124–U158. doi: 10.1038/ncb2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmid E, McMahon H. Integrating molecular and network biology to decode endocytosis. Nature. 2007;448:883–8. doi: 10.1038/nature06031. [DOI] [PubMed] [Google Scholar]
- 64.Ferguson S, Raimondi A, Paradise S, Shen H, Mesaki K, Ferguson A, et al. Coordinated Actions of Actin and BAR Proteins Upstream of Dynamin at Endocytic Clathrin-Coated Pits. Developmental Cell. 2009;17:811–22. doi: 10.1016/j.devcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Messa M, Fernandez-Busnadiego R, Sun E, Chen H, Czapla H, Wrasman K, et al. Epsin deficiency impairs endocytosis by stalling the actin-dependent invagination of endocytic clathrin-coated pits. Elife. 2014:3. doi: 10.7554/eLife.03311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boucrot E, Ferreira A, Almeida-Souza L, Debard S, Vallis Y, Howard G, et al. Endophilin marks and controls a clathrin-independent endocytic pathway. Nature. 2015;517:460. doi: 10.1038/nature14067. [DOI] [PubMed] [Google Scholar]
- 67.Frolov V, Escalada A, Akimov S, Shnyrova A. Geometry of membrane fission. Chemistry and Physics of Lipids. 2015;185:129–40. doi: 10.1016/j.chemphyslip.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 68.Boroske E, Elwenspoek M, Helfrich W. Osmotic Shrinkage of Giant Egg-Lecithin Vesicles. Biophysical Journal. 1981;34:95–109. doi: 10.1016/S0006-3495(81)84839-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanborn J, Oglecka K, Kraut R, Parikh A. Transient pearling and vesiculation of membrane tubes under osmotic gradients. Faraday Discussions. 2013;161:167–76. doi: 10.1039/c2fd20116j. [DOI] [PubMed] [Google Scholar]
- 70.Bashkirov P, Akimov S, Evseev A, Schmid S, Zimmerberg J, Frolov V. GTPase Cycle of Dynamin Is Coupled to Membrane Squeeze and Release, Leading to Spontaneous Fission. Cell. 2008;135:1276–86. doi: 10.1016/j.cell.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaufmann S, Borisov O, Textor M, Reimhult E. Mechanical properties of mushroom and brush poly(ethylene glycol)-phospholipid membranes. Soft Matter. 2011;7:9267–75. [Google Scholar]
- 72.Lipowsky R. Bending of membranes by anchored polymers. Europhysics Letters. 1995;30:197–202. [Google Scholar]
- 73.Montesano G, Bartucci R, Belsito S, Marsh D, Sportelli L. Lipid membrane expansion and micelle formation by polymer-grafted lipids: Scaling with polymer length studied by spin-label electron spin resonance. Biophysical Journal. 2001;80:1372–83. doi: 10.1016/S0006-3495(01)76110-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stachowiak J, Hayden C, Sasaki D. Steric confinement of proteins on lipid membranes can drive curvature and tubulation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7781–6. doi: 10.1073/pnas.0913306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen Z, Atefi E, Baumgart T. Membrane Shape Instability Induced by Protein Crowding. Biophysical Journal. 2016;111:1823–6. doi: 10.1016/j.bpj.2016.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stachowiak J, Schmid E, Ryan C, Ann H, Sasaki D, Sherman M, et al. Membrane bending by protein-protein crowding. Nature Cell Biology. 2012;14:944–9. doi: 10.1038/ncb2561. [DOI] [PubMed] [Google Scholar]
- 77.Shi Z, Baumgart T. Membrane tension and peripheral protein density mediate membrane shape transitions. Nature Communications. 2015:6. doi: 10.1038/ncomms6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kory N, Thiam A, Farese R, Walther T. Protein Crowding Is a Determinant of Lipid Droplet Protein Composition. Developmental Cell. 2015;34:351–63. doi: 10.1016/j.devcel.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Copic A, Latham C, Horlbeck M, D’Arcangelo J, Miller E. ER Cargo Properties Specify a Requirement for COPII Coat Rigidity Mediated by Sec13p. Science. 2012;335:1359–62. doi: 10.1126/science.1215909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bhagatji P, Leventis R, Comeau J, Refaei M, Silvius J. Steric and not structure-specific factors dictate the endocytic mechanism of glycosylphosphatidylinositol-anchored proteins. Journal of Cell Biology. 2009;186:615–28. doi: 10.1083/jcb.200903102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Busch D, Houser J, Hayden C, Sherman M, Lafer E, Stachowiak J. Intrinsically disordered proteins drive membrane curvature. Nature Communications. 2015:6. doi: 10.1038/ncomms8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Silvius J. Membrane Bending Tug of War. Science. 2012;335:1308–9. doi: 10.1126/science.1220221. [DOI] [PubMed] [Google Scholar]
- 83.Adell M, Migliano S, Upadhyayula S, Bykov Y, Sprenger S, Pakdel M, et al. Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding. Elife. 2017:6. doi: 10.7554/eLife.31652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vennema H, Godeke G, Rossen J, Voorhout W, Horzinek M, Opstelten D, et al. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. Embo Journal. 1996;15:2020–8. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Veit M, Thaa B. Association of influenza virus proteins with membrane rafts. Adv Virol. 2011;2011:370606. doi: 10.1155/2011/370606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu T, Shi Z, Baumgart T. Mutations in BIN1 Associated with Centronuclear Myopathy Disrupt Membrane Remodeling by Affecting Protein Density and Oligomerization. Plos One. 2014:9. doi: 10.1371/journal.pone.0093060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Trexler A, Rhoades E. Function and Dysfunction of alpha-Synuclein: Probing Conformational Changes and Aggregation by Single Molecule Fluorescence. Molecular Neurobiology. 2013;47:622–31. doi: 10.1007/s12035-012-8338-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jiang Z, de Messieres M, Lee J. Membrane Remodeling by alpha-Synuclein and Effects on Amyloid Formation. Journal of the American Chemical Society. 2013;135:15970–3. doi: 10.1021/ja405993r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hofmann H, Soranno A, Borgia A, Gast K, Nettels D, Schuler B. Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:16155–60. doi: 10.1073/pnas.1207719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kumar S, Yin X, Trapp B, Hoh J, Paulaitis M. Relating interactions between neurofilaments to the structure of axonal neurofilament distributions through polymer brush models. Biophysical Journal. 2002;82:2360–72. doi: 10.1016/S0006-3495(02)75581-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brown H, Hoh J. Entropic exclusion by neurofilament sidearms: A mechanism for maintaining interfilament spacing. Biochemistry. 1997;36:15035–40. doi: 10.1021/bi9721748. [DOI] [PubMed] [Google Scholar]
- 92.Kellermayer M, Smith S, Granzier H, Bustamante C. Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science. 1997;276:1112–6. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 93.Linke W, Kulke M, Li H, Fujita-Becker S, Neagoe C, Manstein D, et al. PEVK domain of titin: An entropic spring with actin-binding properties. Journal of Structural Biology. 2002;137:194–205. doi: 10.1006/jsbi.2002.4468. [DOI] [PubMed] [Google Scholar]
- 94.Kalthoff C, Alves J, Urbanke C, Knorr R, Ungewickell E. Unusual structural organization of the endocytic proteins AP180 and epsin 1. Journal of Biological Chemistry. 2002;277:8209–16. doi: 10.1074/jbc.M111587200. [DOI] [PubMed] [Google Scholar]
- 95.Lafer E. Clathrin-protein interactions. Traffic. 2002;3:513–20. doi: 10.1034/j.1600-0854.2002.30801.x. [DOI] [PubMed] [Google Scholar]
- 96.Hirokawa N, Glicksman M, Willard M. Organization of mammalian neurofilament polypeptides within the neuronal cytoskeleton. Journal of Cell Biology. 1984;98:1523–36. doi: 10.1083/jcb.98.4.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Carnahan N, Starling K. Equation of state for nonattracting rigid spheres. Journal of Chemical Physics. 1969;51:635. [Google Scholar]
- 98.Scheve C, Gonzales P, Momin N, Stachowiak J. Steric Pressure between Membrane-Bound Proteins Opposes Lipid Phase Separation. Journal of the American Chemical Society. 2013;135:1185–8. doi: 10.1021/ja3099867. [DOI] [PubMed] [Google Scholar]
- 99.Soranno A, Koenig I, Borgia M, Hofmann H, Zosel F, Nettels D, et al. Single-molecule spectroscopy reveals polymer effects of disordered proteins in crowded environments. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:4874–9. doi: 10.1073/pnas.1322611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hong J, Gierasch L. Macromolecular Crowding Remodels the Energy Landscape of a Protein by Favoring a More Compact Unfolded State. Journal of the American Chemical Society. 2010;132:10445–52. doi: 10.1021/ja103166y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Minton A. Models for excluded volume interaction between an unfolded protein and rigid macromolecular cosolutes: Macromolecular crowding and protein stability revisited. Biophysical Journal. 2005;88:971–85. doi: 10.1529/biophysj.104.050351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Uversky V. Intrinsically Disordered Proteins and Their Environment: Effects of Strong Denaturants, Temperature, pH, Counter Ions, Membranes, Binding Partners, Osmolytes, and Macromolecular Crowding. Protein Journal. 2009;28:305–25. doi: 10.1007/s10930-009-9201-4. [DOI] [PubMed] [Google Scholar]
- 103.Srinivasan N, Bhagawati M, Ananthanarayanan B, Kumar S. Stimuli-sensitive intrinsically disordered protein brushes. Nature Communications. 2014:5. doi: 10.1038/ncomms6145. [DOI] [PubMed] [Google Scholar]
- 104.Pietrosemoli N, Pancsa R, Tompa P. Structural Disorder Provides Increased Adaptability for Vesicle Trafficking Pathways. Plos Computational Biology. 2013:9. doi: 10.1371/journal.pcbi.1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Owen D, Collins B, Evans P. Adaptors for clathrin coats: Structure and function. Annual Review of Cell and Developmental Biology. 2004;20:153–91. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- 106.Takei K, Slepnev V, Haucke V, De Camilli P. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nature Cell Biology. 1999;1:33–9. doi: 10.1038/9004. [DOI] [PubMed] [Google Scholar]
- 107.Meinecke M, Boucrot E, Camdere G, Hon W, Mittal R, McMahon H. Cooperative Recruitment of Dynamin and BIN/Amphiphysin/Rvs (BAR) Domain-containing Proteins Leads to GTP-dependent Membrane Scission. Journal of Biological Chemistry. 2013;288:6651–61. doi: 10.1074/jbc.M112.444869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Miele A, Watson P, Evans P, Traub L, Owen D. Two distinct interaction motifs in amphiphysin bind two independent sites on the clathrin terminal domain beta-propeller. Nature Structural & Molecular Biology. 2004;11:242–8. doi: 10.1038/nsmb736. [DOI] [PubMed] [Google Scholar]
- 109.Soulet F, Yarar D, Leonard M, Schmid S. SNX9 regulates dynamin assembly and is required for efficient clathrin-mediated endocytosis. Molecular Biology of the Cell. 2005;16:2058–67. doi: 10.1091/mbc.E04-11-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lundmark R, Carlsson S. SNX9-a prelude to vesicle release. Journal of Cell Science. 2009;122:5–11. doi: 10.1242/jcs.037135. [DOI] [PubMed] [Google Scholar]
- 111.Scheele U, Alves R, Frank R, Duwel M, Kalthoff C, Ungewickell E. Molecular and functional characterization of clathrin- and AP-2-binding determinants within a disordered domain of auxilin. Journal of Biological Chemistry. 2003;278:25357–68. doi: 10.1074/jbc.M303738200. [DOI] [PubMed] [Google Scholar]
- 112.Sousa R, Liao H, Cuellar J, Jin S, Valpuesta J, Jin A, et al. Clathrin-coat disassembly illuminates the mechanisms of Hsp70 force generation. Nature Structural & Molecular Biology. 2016;23:821–9. doi: 10.1038/nsmb.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Whittle J, Schwartz T. Structure of the Sec13-Sec16 edge element, a template for assembly of the COPII vesicle coat. Journal of Cell Biology. 2010;190:347–61. doi: 10.1083/jcb.201003092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fath S, Mancias J, Bi X, Goldberg J. Structure and organization of coat proteins in the COPII cage. Cell. 2007;129:1325–36. doi: 10.1016/j.cell.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 115.Miller S, Mathiasen S, Bright N, Pierre F, Kelly B, Kladt N, et al. CALM Regulates Clathrin-Coated Vesicle Size and Maturation by Directly Sensing and Driving Membrane Curvature. Developmental Cell. 2015;33:163–75. doi: 10.1016/j.devcel.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McMahon H, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nature Reviews Molecular Cell Biology. 2011;12:517–33. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 117.Henne W, Kent H, Ford M, Hegde B, Daumke O, Butler P, et al. Structure and analysis of FCHo2 F-BAR domain: A dimerizing and membrane recruitment module that effects membrane curvature. Structure. 2007;15:839–52. doi: 10.1016/j.str.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 118.Fotin A, Cheng Y, Grigorieff N, Walz T, Harrison S, Kirchhausen T. Structure of an auxilin-bound clathrin coat and its implications for the mechanism of uncoating. Nature. 2004;432:649–53. doi: 10.1038/nature03078. [DOI] [PubMed] [Google Scholar]
- 119.Jensen D, Schekman R. COPII-mediated vesicle formation at a glance. Journal of Cell Science. 2011;124:1–4. doi: 10.1242/jcs.069773. [DOI] [PubMed] [Google Scholar]
- 120.Zhuo Y, Ilangovan U, Schirf V, Demeler B, Sousa R, Hinck A, et al. Dynamic Interactions between Clathrin and Locally Structured Elements in a Disordered Protein Mediate Clathrin Lattice Assembly. Journal of Molecular Biology. 2010;404:274–90. doi: 10.1016/j.jmb.2010.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Avinoam O, Schorb M, Beese C, Briggs J, Kaksonen M. Endocytic sites mature by continuous bending and remodeling of the clathrin coat. Science. 2015;348:1369–72. doi: 10.1126/science.aaa9555. [DOI] [PubMed] [Google Scholar]
- 122.Caudron F, Barral Y. Septins and the Lateral Compartmentalization of Eukaryotic Membranes. Developmental Cell. 2009;16:493–506. doi: 10.1016/j.devcel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 123.Hu Q, Milenkovic L, Jin H, Scott M, Nachury M, Spiliotis E, et al. A Septin Diffusion Barrier at the Base of the Primary Cilium Maintains Ciliary Membrane Protein Distribution. Science. 2010;329:436–9. doi: 10.1126/science.1191054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Imam Z, Kenyon L, Carrillo A, Espinoza I, Nagib F, Stachowiak J. Steric Pressure among Membrane-Bound Polymers Opposes Lipid Phase Separation. Langmuir. 2016;32:3774–84. doi: 10.1021/acs.langmuir.6b00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zeno W, Rystov A, Sasaki D, Rishud S, Longo M. Crowding-Induced Mixing Behavior of Lipid Bilayers: Examination of Mixing Energy, Phase, Packing Geometry, and Reversibility. Langmuir. 2016;32:4688–97. doi: 10.1021/acs.langmuir.6b00831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Roux A, Cuvelier D, Nassoy P, Prost J, Bassereau P, Goud B. Role of curvature and phase transition in lipid sorting and fission of membrane tubules. Embo Journal. 2005;24:1537–45. doi: 10.1038/sj.emboj.7600631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lipowsky R. Budding of Membranes Induced by Intramembrane Domains. Journal De Physique II. 1992;2:1825–40. [Google Scholar]
- 128.Allain J, Storm C, Roux A, Amar M, Joanny J. Fission of a multiphase membrane tube. Physical Review Letters. 2004:93. doi: 10.1103/PhysRevLett.93.158104. [DOI] [PubMed] [Google Scholar]
- 129.Liu J, Sun YD, Drubin DG, Oster GF. The Mechanochemistry of Endocytosis. Plos Biology. 2009:7. doi: 10.1371/journal.pbio.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Holehouse A, Das R, Ahad J, Richardson M, Pappu R. CIDER: Resources to Analyze Sequence-Ensemble Relationships of Intrinsically Disordered Proteins. Biophysical Journal. 2017;112:16–21. doi: 10.1016/j.bpj.2016.11.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Aguet F, Antonescu C, Mettlen M, Schmid S, Danuser G. Advances in Analysis of Low Signal-to-Noise Images Link Dynamin and AP2 to the Functions of an Endocytic Checkpoint. Developmental Cell. 2013;26:279–91. doi: 10.1016/j.devcel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cocucci E, Gaudin R, Kirchhausen T. Dynamin recruitment and membrane scission at the neck of a clathrin-coated pit. Mol Biol Cell. 2014;25:3595–609. doi: 10.1091/mbc.E14-07-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Aguet F, Upadhyayula S, Gaudin R, Chou YY, Cocucci E, He K, et al. Membrane dynamics of dividing cells imaged by lattice light-sheet microscopy. Mol Biol Cell. 2016;27:3418–35. doi: 10.1091/mbc.E16-03-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]