Abstract
Viral infections of the epididymis are associated with epididymitis, which damages the epithelium and impairs fertility. We showed previously that innate immune response genes were differentially expressed in the corpus and cauda region of the human epididymis in comparison to the caput. Here we investigate the antiviral defense response mechanisms of human epididymis epithelial (HEE) cells. Toll-like receptor (TLR) 3 and retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) are enriched in HEE cells from the corpus and cauda region. These HEE cells show an enhanced response to antiviral ligands (poly(I:C) and HSV-60), as shown by increased IFN-β mRNA expression and IFN-β secretion. Nuclear translocation of phosphorylated p65 occurs after poly(I:C) exposure. In addition, paired box 2 (PAX2), which was implicated in regulating antiviral response pathways, is required for basal expression of the DNA sensor, Z-DNA binding protein (ZBP1) and type I interferon, in caput but not in cauda cells.
Introduction
The epithelial lining of the human epididymis has a critical role in maintaining the health of the male genital ducts and hence preserving fertility. Bacterial, yeast and viral infections of the ducts may damage the epithelium, in turn impairing the maturation and movement of spermatozoa and compromising male fertility (Schuppe et al., 2017). Though these infections are more commonly caused by bacteria than viruses, rubulavirus, Coxsackie-B, Herpesvirus (HSV) and Human papilloma viruses (HPV) are likely viral pathogens in the epididymis (Dejucq and Jegou, 2001, Kapranos et al., 2003, Emerson et al., 2007, Vuorinen et al., 2014). Antiviral innate immunity is mediated by two families of pattern recognition receptors (PRRs), which identify viral pathogens through pathogen-associated molecular patterns (PAMPs). One family includes Toll-like receptor (TLR)-3, -7, -8, and -9 that recognize double-stranded RNA (dsRNA) or single-stranded RNA (ssRNA) at the membrane of endosomes. The second group is the cytoplasmic retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), including three viral dsRNA sensors: retinoic acid-inducible gene I (RIG-I) (encoded by the DDX58 gene), melanoma differentiation-associated gene 5 (MDA-5) (encoded by IFIH1) and their negative regulator, LGP-2 (laboratory of genetics and physiology-2)/DexH-Box Helicase 58 (encoded by the DHX58 locus) (Loo and Gale, 2011). Recognition of the invading virus by these PRR molecules initiates signaling pathways that result in the production of type I interferons (IFNs) and proinflammatory cytokines (Schroder and Bowie, 2005, Chiang et al., 2014). IFNs bind to specific cellular receptors, to induce the synthesis of several proteins with antiviral activity: Ubiquitin-like protein ISG15 (ISG15), 2′,5′-oligoadenylate synthetase (2′,5′-AS or OAS), double-stranded RNA-activated protein kinase (PKR), and Mx protein and (Schoggins and Rice, 2011). In addition to RLRs, other DNA sensors are known, including Z-DNA Binding Protein 1 (ZBP1 or DAI), Absent In Melanoma 2 (AIM2), Interferon Gamma Inducible Protein 16 (IFI16 or p204), RNA Polymerase III (reviewed in Paludan and Bowie (2013)).
TLR3/RLR signaling has been characterized in human and mouse testis-derived cells (Le Tortorec et al., 2008, Zhu et al., 2013, Zhao et al., 2014) and in the mouse epididymis (Zhu et al., 2015) but to date, not in the human epididymis. The human epididymis is anatomically and functionally distinct from the mouse organ and has 3 functionally distinct regions: the head (caput), body (corpus) and tail (cauda). We previously reported a robust protocol for culturing primary epithelial cells from each of these regions (Leir et al., 2015). Gene expression analysis by RNA-seq revealed differentially expressed genes (DEG) in caput, corpus and cauda epithelial cells (Browne et al., 2016). Among notable DEGs were proteins involved in the defense response of the epididymis which were substantially enriched in the corpus and cauda regions. To further understand the significance of these gene expression patterns, we investigated the antiviral responses to PRR ligands in the different regions of the epididymis.
Materials and Methods
Cell Culture
De-identified human epididymis tissue was obtained with Institutional Review Board approval. HEE cells were isolated and cultured as described previously (Leir et al., 2015). Sub-confluent/confluent 24-h serum-starved cells were treated with polyinosinic-polycytidylic acid (poly(I:C), 2 μg/ml) or transfected (using Lipofectamine® 2000) with HSV-60 (0.6 μg/ml), 2′3′-cGAMP (cyclic GMP-AMP)) or their appropriate negative controls (all from InvivoGen, San Diego, CA) for the indicated times. Cells were washed in cold sterile PBS and immediately harvested for RNA extraction using TRIzol® (Thermo Scientific (Waltham, MA, USA)) or for whole cell lysate using NET Buffer (Leir and Harris, 2011). For cellular fractionation experiments the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Scientific) was used to isolate cytoplasmic and nuclear fractions from HEE cells.
SiRNA-mediated depletion of PAX2
Cells at 40–50% confluence were transfected with non-targeting control- or PAX2-targeting siRNA (sc-37007 and sc-38745, Santa Cruz) using RNAiMAX Lipofectamine® transfection reagent (Life Technologies, Grand Island, NY) at a final concentration of 20 nM. At 72 h post transfection, cells were harvested for RNA extraction.
RNA-seq
RNA libraries were prepared from 2 μg of total RNA from three replicates (of one donor) from control- and PAX2-siRNA transfected cells. RNA quality was confirmed by NanoDrop measurement of OD 260/280 and 260/230 ratios. RNA-seq libraries were prepared using the TruSeq RNA Sample Preparation Kit v2 per the manufacturer’s Low-Throughput protocol (Illumina). The libraries were sequenced on an Illumina HiSeq2500 machine. Data were analyzed using TopHat and Cufflinks (Trapnell et al., 2012). All data are deposited at GEO:GSE104944. Biological processes associated with differentially expressed genes were identified using DAVID (Huang da et al., 2009a, Huang da et al., 2009b).
Reverse transcription and quantitative PCR (RT-qPCR)
Reverse transcription and quantitative PCR (RT–qPCR) was performed by standard protocols. cDNA was synthesized using a TaqMan Reverse Transcription kit (Applied Biosystems) with random hexamers, and qPCR experiments used SYBR Green. The primers specific for each target gene are shown in Suppl. Table SI and expression was normalized to β2-microglobulin (B2M). For RNA-seq validation experiments RNA from 3 different donor HEE cultures was used.
Western blotting
Cells were lysed in buffer containing 1% (vol/vol) protease inhibitor cocktail (Sigma) and western blots performed as described previously (Leir and Harris, 2011). Antibodies used were: TLR3 (#62566, Abcam); phospho-p65 (#3033), IRF3 (#11904), MDA5 (#5321), RIG-1 (#3743) and ISG15 (#2758) all from Cell Signaling Technology; p65 (sc-372 and sc-8008) and Lamin A/C (sc-7292) from Santa Cruz; and β-tubulin (T4026, Sigma).
IFN-β ELISA
Cell culture supernatant from vehicle and poly(I:C)-treated cells was collected, cleared by centrifugation at 280 × g and stored at −80°C. Interferon-β (IFN-β) protein was quantified using a bioluminescent ELISA kit (LumiKine, InvivoGen) according to the manufacturer’s protocol.
Statistical analysis
All data are shown as mean ± SD. Statistical tests were performed with GraphPad Prism 6 software using one-way ANOVA and Bonferroni post-hoc test and values of P < 0.05 were considered significant.
Results
Viral sensor genes are differentially expressed in the corpus and cauda regions of the human epididymis
To determine the functional signature of the different regions of the human epididymis, we earlier performed RNA-seq analysis of caput, corpus and cauda tissue and epithelial cells cultured from each section at passage 2 or 3 from three donors (Browne et al., 2016). Inspection of antiviral sensor expression in these data shows a greater abundance of transcripts from viral sensor genes (TLR3, DDX58 and IFIH1) in the corpus and cauda compared to the caput (Figure 1A and Suppl. Table II). The RNA-seq data were validated at the mRNA level by RT-qPCR (Figure 1B) and increased expression of TLR3, DDX58 and IFIH1 protein in cauda compared to caput cells was confirmed by western blotting (Figure 1C). Also evident in Suppl. Table I is the greater expression of genes encoding other important components of antiviral signaling in corpus and cauda cells in comparison to caput. These include the signaling protein interleukin-1 receptor-associated kinase 2 (IRAK2), the transcription factor interferon regulatory factor 7 (IRF7), the antiviral proteins myxovirus resistance 1 and 2 (MX1 and MX2) and 2′-5′-oligoadenylate synthetase 1, 2 and 3 (OAS1, OAS2 and OAS3).
Figure 1. Differential distribution of viral sensors in HEE cells from caput, corpus and cauda epididymis.
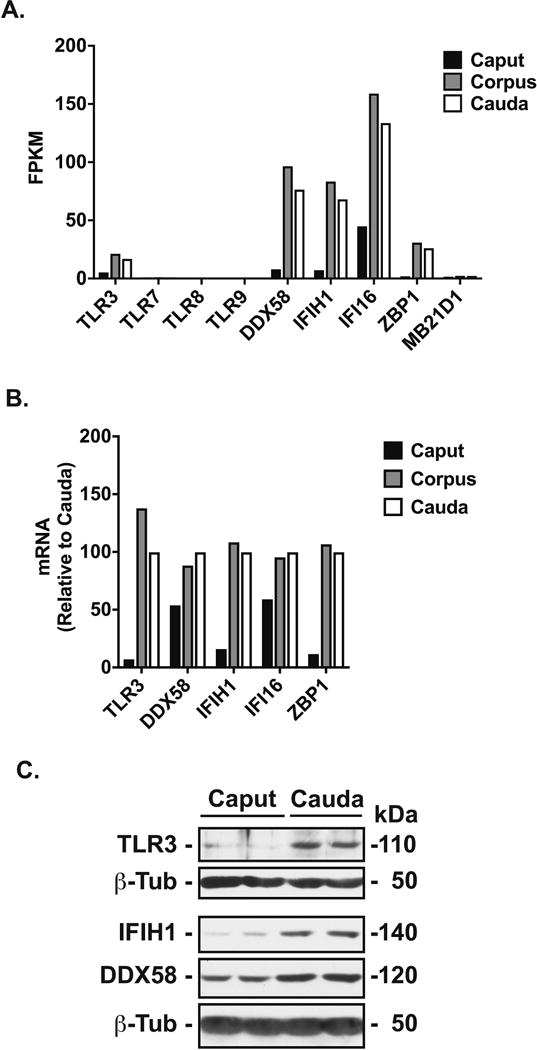
A, Region-specific gene expression in caput (black bar), corpus (gray bar) and cauda (white bar) cells, is shown as FPKM (average of 3 donors) (Browne et al., 2016). B, RT-qPCR analysis of the mRNA of viral sensors in caput, corpus and cauda cells from one of the donors shown in A; data are normalized to β-2-microglobulin and expressed relative to cauda. C, Western blots probed with antibodies specific for TLR3, DDX58, IFIH1 and β-tubulin (β-Tub) loading control.
Cauda HEE cells show greater antiviral responses to TLR3 and RLR ligands than do caput cells
To characterize antiviral responses in the human epididymis epithelium, we exposed HEE cells to TLR3 and RLR ligands (polyinosinic-polycytidylic acid, poly(I:C) and HSV-60). Poly(I:C) is a synthetic analog of double-stranded RNA, that contains a molecular pattern associated with viral infection. It serves as a ligand for both TLRs and RLRs. HSV-60 is a 60 bp oligonucleotide containing viral (herpes simplex virus 1 genome) DNA motifs, which only activates RLRs (Unterholzner et al., 2010). Since TLR3 and RLRs are both cytosolic rather than integral to the cell membrane, we first tested if doses of poly(I:C) (2 μg/ml) and HSV-60 (0.6 μg/ml) reported previously (Zhu et al., 2015) were directly taken up by HEE cells from the culture medium, or required lipid-mediated transfection for effective uptake. Activation of expression of the antiviral ISG15 ubiquitin-like modifier (ISG15) mRNA measured by RT-qPCR was used as a marker of ligand: receptor binding and hence uptake (Suppl. Figure SI). A comparable increase in ISG15 mRNA was seen in poly(I:C)-treated corpus cells with or without Lipofectamine® 2000 demonstrating that this ligand was freely taken up into HEE cells. In contrast, lipid-mediated transfection of HSV-60 was required for HSV-60-induced up-regulation of ISG15 mRNA.
Next, to compare antiviral responses of the human caput and cauda epididymis, we performed RT-qPCR analysis of interferon beta (IFN-β) mRNA in vehicle or poly(I:C)-treated caput and cauda cells (Figure 2A). IFN-β mRNA in poly(I:C)-treated cauda cells was substantially higher than in poly(I:C)-treated caput cells at all time points tested (2, 6 and 24h) and with maximum activation at 6h (P<0.0001, n=3, Figure 2A). Consistent with the IFN-β mRNA results, cauda cells treated with poly(I:C) for 48h secreted significantly more IFN-β protein into their growth media than did caput cells, as measured by IFN-β ELISA (P<0.01, n=3, Figure 2B). This differential effect was also seen at an earlier time point (24h) (Suppl. Figure SII). Moreover, abundance of the antiviral protein, ISG15 and associated p65 phosphorylation in 24h and 48h poly(I:C)-treated cauda cells was correspondingly elevated compared to the same treatment of caput cells (Figure 2C). Subcellular fractionation of cauda cells treated with vehicle or Poly(I:C) revealed that both total and phosphorylated forms of p65 accumulated in the nucleus 2h after Poly(I:C) stimulation and were still evident there 15 h later (Figure 2D).
Figure 2. Effect of poly(I:C) on antiviral responses in HEE cells.
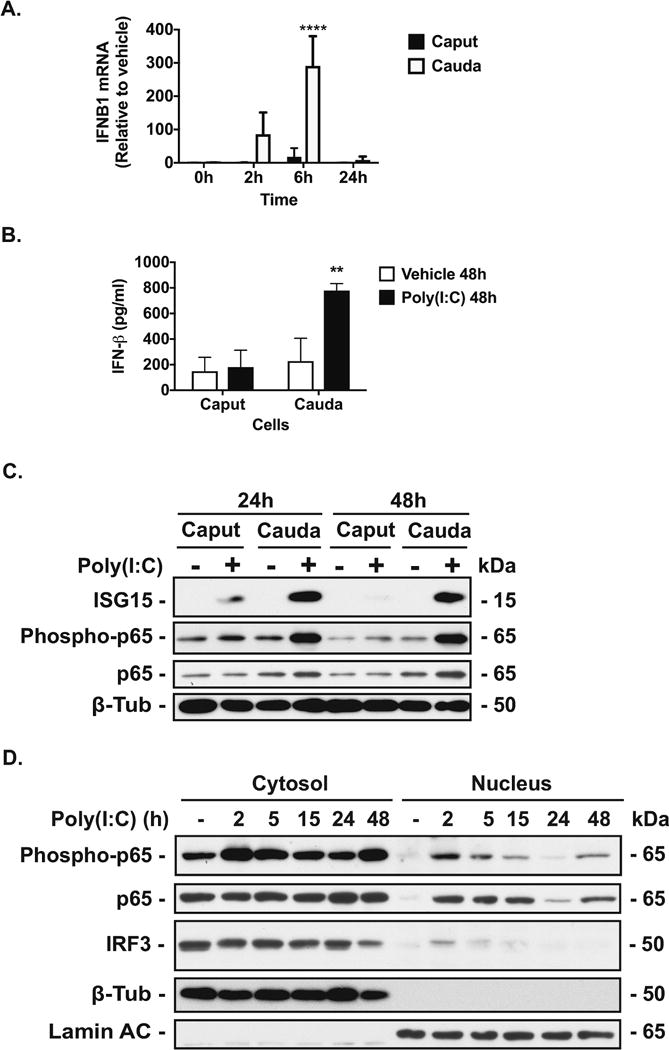
A, RT-qPCR analysis of IFNB1 mRNA in caput and cauda cells treated with vehicle or poly(I:C) (2 μg/ml) for 0, 2, 6 and 24h. Data are normalized to β-2-microglobulin and expressed relative to vehicle. B, IFN-β protein secretion (determined by ELISA) from caput and cauda cells treated with vehicle or poly(I:C) (2 μg/ml) for 48h. **P < 0.001 and ****P < 0.0001 versus vehicle (n=3). C. Western blots probed with antibodies specific for ISG15 and phosphorylated p65 in caput and cauda cells treated with vehicle- or poly(I:C) for 24h and 48h. D. Western blots probed with antibodies specific for p65 (phosphorylated and total forms) and IRF3 in cytosolic and nuclear fractions of cauda cells treated with vehicle- or poly(I:C) for 2, 5, 15, 24 and 48h. β-tubulin (β-Tub) and Lamin A/C provided cytosolic and nuclear loading controls, respectively. For all experiments, cells were serum-starved for 24h prior to vehicle/poly(I:C) treatment.
RT-qPCR analysis demonstrated greater IFN-β mRNA in HSV-60-transfected cauda cells than in similarly treated caput cells at 6h (P<0.05, n=3, Figure 3A). The IFNB1 mRNA response was also greater in cauda cells than in caput cells following exposure to 2′3′-cGAMP (an agonist of stimulator of interferon genes, STING) (Zhang et al., 2013) (Suppl. Figure SIII). Consistent with these data, greater levels of the antiviral protein, ISG15 were detected in western blots of cell lysates from HSV-60-transfected cauda cells (24 and 48h) compared to similarly -treated caput cells. Also, p65 was not phosphorylated in response to HSV-60 in caput cells (Figure 3B).
Figure 3. Effect of HSV60 on antiviral responses in HEE cells.
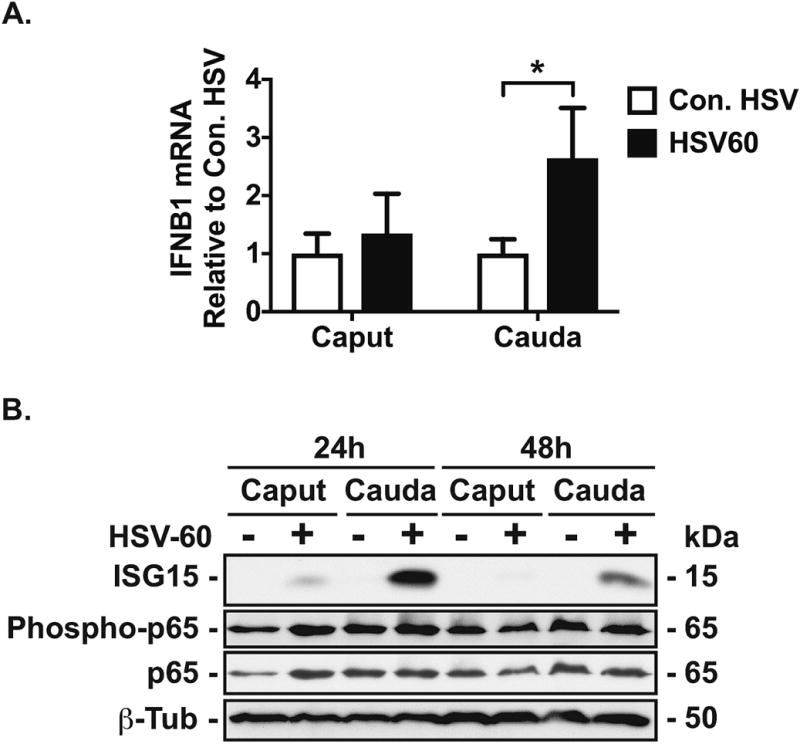
A, RT-qPCR analysis of IFNB1 mRNA in caput and cauda HEE cells transfected with HSV-60 or control-HSV (0.6 μg/ml) for 6h. Data are normalized to β-2 microglobulin (mean ± SD, n = 3, *P < 0.05 (con. HSV, clear bars; HSV-60 black bar). B, Western blot with antibodies specific to ISG15 and phosphorylated p65 in HSV-60-transfected caput and cauda cells. β-tubulin and p65 provided loading controls. For all experiments, cells were serum-starved for 24h prior to vehicle/HSV-60 transfection.
Transcriptional Regulation of Antiviral Responses of HEE Cells
We showed previously that the developmental transcription factor (TF) PAX2 has a pivotal role in the biology of human epididymis epithelial cell lines (Browne et al 2014). To determine whether PAX2 is also involved in the differentiated functions of primary HEE cells, including the innate immune response, we performed siRNA-mediated depletion of PAX2 in HEE caput cells, followed by RNA-seq analysis. Three replicas of caput cells were transfected with the specific siRNAs or with a non-targeting control siRNA. Efficacy of the siRNA-mediated reduction in PAX2 protein is shown by western blot in Figure 4A. RNA-seq libraries were generated for each replica and six libraries sequenced together (Suppl. Table SIII). A Multi-Dimensional Scaling plot shows that the control- and PAX2-siRNA-treated samples clustered together in two separate groups (Suppl. Figure SIV). RNA-seq data were analyzed by TopHat and Cufflinks (Trapnell et al., 2012) to obtain estimates of the expression levels of transcripts. PAX2-depletion in caput HEE cells differentially regulated the expression of 2131 transcripts of which 1006 were repressed and 1125 were activated, by at least 1.3-fold (FPKM ≥ 0.3) (Suppl. Table SIV). A gene ontology process enrichment analysis by DAVID (Huang da et al., 2009a, Huang da et al., 2009b) was then applied to the list of genes that were down-regulated following PAX2-depletion. The top 3 most significant processes (“type-I interferon signaling”, “negative regulation of viral genome replication” and “defense response to virus”) were all related to the innate antiviral immune response of the epididymis (Figure 4B). The full list of genes in the top 6 processes is provided in Suppl. Table SV. Next, to validate the RNA-seq data, RT-qPCR was used to measure transcript levels of a sub-set of genes (Figure 4C) in RNA samples from three independent HEE donors of PAX2 or negative control siRNA-transfected caput cells (Figure 4D). We confirmed the repression after PAX2 depletion of genes encoding ZBP1, a DNA sensor involved in the antiviral response (P < 0.05, n=3) and IFNB1 (P < 0.01, n=3). PAX2-depletion did not significantly change DDX58, IFIH1, ISG15 and OAS1 mRNAs (Figure 4D). Moreover, when peaks of open chromatin (DHS) in caput HEE cells (Yang et al., 2016) were examined, a site with a PAX2 binding motif was found in the first intron of the ZP1 locus. (Figure 4E).
Figure 4.
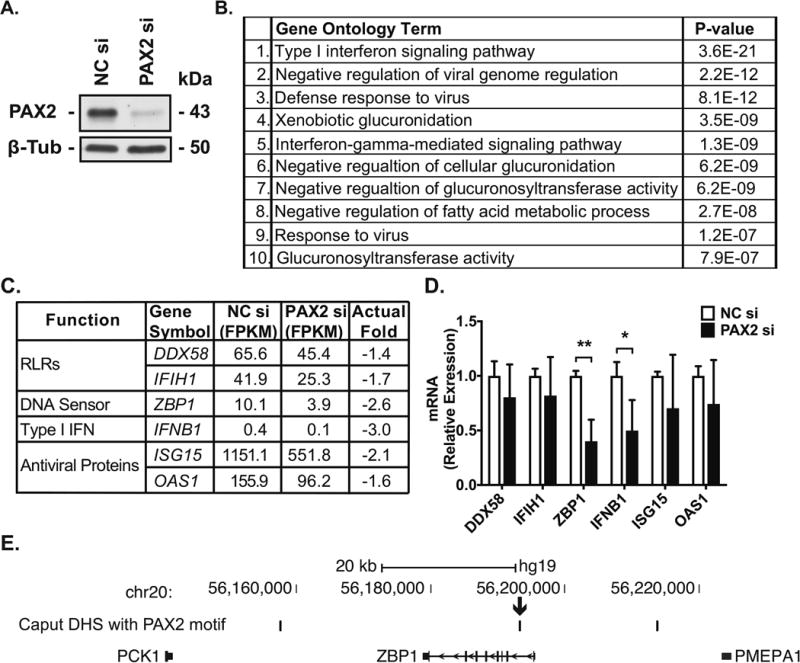
PAX2-depletion reveals its role in regulating the expression of antiviral-related genes in HEE cells. A, Efficacy of siRNA-mediated depletion of PAX2 (PAX2si) in caput cells shown by western blot of cell lysates probed with an antibody specific for PAX2 (43 kDa). β-tubulin (β-Tub) as the loading control. Negative control siRNA (NC). B, Gene ontology analysis by DAVID on the genes repressed in PAX2-depletion compared to control caput HEE cells. C, Antiviral-related genes repressed in siRNA mediated PAX2-depleted caput HEE cells compared to control. D, RT-qPCR validation of differentially expressed genes in PAX2-siRNA-depleted and control siRNA-treated cells. Data are normalized to β-2 microglobulin (mean ± SD, n = 3, *P < 0.05 and **P < 0.01 compared to negative control siRNA (NC, clear bars, PAX2 siRNA black bar). E, Identification of novel candidate cis-regulatory elements in PAX2-regulated genes in caput cells. Peaks of caput-selective open chromatin containing PAX2-binding motifs are shown above the ZBP1 gene, arrow denotes a DHS in the first intron.
Next, we examined the contribution of PAX2 to antiviral-related gene expression changes upon poly(I:C)-stimulation of caput cells. Efficacy of the siRNA-mediated reduction in PAX2 protein is shown by western blot in Suppl. Figure V. RT-qPCR was used to measure transcript levels in independent samples (from 3 HEE cell donors) of control- or PAX2-depleted caput cells treated with vehicle or Poly(I:C). PAX2-depletion had no effect on poly(I:C)-stimulated ZBP1, DDX58, IFIH1, ISG15 mRNA levels but enhanced IFNB1 mRNA abundance (Suppl. Figure V). Though the antiviral responses of cauda cells were the focus of the earlier experiments described here, the differentiated properties of cauda cells make them problematic to use for siRNA-mediated depletion experiments (Leir et al., 2015) and unpublished. Instead we tested corpus cells, which express similar amounts of PAX2 as cauda cells and also show a robust antiviral response. The expression of ZBP1, DDX58, IFIH1, ISG15 and IFNB1 were all increased in control corpus cells following Poly(I:C) treatment. Knockdown of PAX2 did not change this activation (Suppl. Figure SVI). These data suggest the RNA-seq data are robust and confirm the important role of PAX2 in regulating the basal expression of genes involved in the antiviral response only in the caput epididymis epithelium.
Discussion
Here we show that both the expression and functional activity of viral sensors in the human epididymis epithelium (HEE) is region-specific. We also show a role for the PAX2 transcription factor in coordinating the expression of specific antiviral-related genes. Viral sensor expression and activity were characterized previously in the mouse epididymis (Zhu et al., 2015) and human testis-derived cells (Le Tortorec et al., 2008). Hence, the data presented here enables both species and cell-type specific comparisons with the human epididymis.
Our previous RNA-seq analysis of HEE cells derived from caput, corpus and cauda regions of the epididymis showed an abundance of antiviral-related genes in the corpus/cauda compared to the caput (Browne et al., 2016). Here we confirm that the HEE cells express the double-stranded viral RNA sensors; TLR3, DDX58, IFIH1 and the DNA sensors, IFI16 and ZBP1 with greatest abundance in the corpus/cauda. In contrast, the single-stranded viral RNA sensors, TLR7/8 (Heil et al., 2004) and TLR9, which detects unmethylated CpG dinucleotides (Hemmi et al., 2000) were poorly expressed in all HEE cells.
Recognition of invading virus by pattern recognition receptors initiates signaling pathways resulting in the production of type I interferons (e.g. interferon-β, IFN-β) and pro-inflammatory cytokines. So, we next compared cellular and secreted IFN-β and ISG15 levels produced in caput and cauda cells exposed to TLR3/RLR ligands. A greater increase in cellular IFNB1 mRNA was seen in response to all ligands, in the cauda than in the caput. Consistently, cauda cells secreted more IFN-β protein than caput cells, in response to poly(I:C) treatment. Secreted IFN-β binds to its receptor on cells to initiate JAK-STAT signaling that produces the interferon-stimulated gene factor 3 (ISGF3) complex. In turn this complex enters the nucleus, binds to IFN stimulated response elements (ISREs) and induces interferon-stimulated genes (ISGs), such as ISG15 (Schindler and Darnell, 1995). In response to either poly(I:C) or HSV-60, cellular ISG15 antiviral protein levels were greater in cauda than in caput cells. However, increased p65 phosphorylation was seen only in poly(I:C)-treated cauda cells and there was little response to HSV-60. This is consistent with observations in the mouse, where p65 phosphorylation was evident in Poly(I:C)- but not in HSV-60-stimulated epithelial cells isolated from the entire length of epididymis (Zhu et al., 2015). The greater and more consistent antiviral responses evoked by poly(I:C) than by either HSV-60 or 2′3′-cGAMP correlates with the ability of poly(I:C) to activate both TLR3 and RLRs (Karpus et al., 2012), whereas HSV-60 or 2′3′-cGAMP only activate RLRs (Unterholzner et al., 2010, Gao et al., 2015, Ren et al., 2015).
Increased expression and activity of viral sensors in the distal epididymis compared to the proximal regions may reflect the ascending nature of epididymitis. Caput cells may also be enriched for negative regulators of antiviral signaling. Several RLR repressors are reported in other cells (Quicke et al., 2017), of which, Ring Finger Protein 122 (RNF122) and SEC14 Like Lipid Binding 1 (SEC14L) are more abundant in caput compared to cauda cells (Suppl. Table SII of Browne et al., (2016)). RNF122 binds to RIG-I (DDX58) to promote its degradation (Yang et al., 2011) and SEC14L encodes a protein that prevents DDX58 interacting with MAVS/IPS1 (Li et al., 2013). With respect potential inhibition of TLR3 signaling in caput cells, these are enriched for the TF, Interferon Regulatory Factor 8 (IRF8) (Browne et al., 2016) which negatively regulates poly(I:C)-stimulated TLR3 expression in human monocyte-derived dendritic cells (Fragale et al., 2011).
Gene expression patterns along the epididymis epithelium are established and maintained by specific TF networks that coordinate region-specific functions. Previous work identified PAX2 binding sites to be overrepresented in open chromatin (the location of regulatory elements) in both immortalized epididymis cells (Browne et al., 2014) and primary HEE cells (Yang et al., 2016). The contribution of PAX2 to the transcriptome of HEE cells was determined by RNA-seq, following siRNA-mediated depletion of PAX2. A gene ontology process enrichment analysis on DEGs after PAX2 depletion found genes associated with defense response to virus, type I interferon signaling and negative regulation of viral genome replication to be among the most significantly down-regulated. Of particular were genes encoding viral sensors and proteins with antiviral activity. Our data suggest that in caput cells, PAX2 is required for basal- but not Poly(I:C)- stimulated expression of ZBP1 and IFNB1, which encode a viral DNA sensor (ZBP1) and type I interferon (IFN-β), respectively. PAX2 is differentially expressed along the epididymis epithelium with greater abundance in the caput (Browne et al., 2016). In corpus cells, PAX2 was not required for basal ZBP1 or IFNB1 expression, nor for Poly(I:C)-stimulated RLR or DNA sensor expression. Cauda levels of PAX2 were too low to assess its contribution to basal- or ligand-induced sensor expression or activity. Hence, we have not determined which TFs mediate basal and TLR3/RLR-mediated antiviral responses of corpus and cauda cells, though IRF7 and IRF8 which are both more abundant here than in the caput are potential candidates (Browne et al., 2016).
Supplementary Material
-
·
Antiviral gene transcripts in human epididymis are more abundant in the corpus and cauda than in the caput.
-
·
These antiviral genes include the viral sensors TLR3 and RIG-I-like receptors (RLRs).
-
·
Corpus and cauda epididymis epithelial cells are most responsive to viral ligands.
-
·
PAX2 enhances basal ZBP1 and IFNβ expression in caput but not cauda cells.
Acknowledgments
We are grateful to the anonymous men with testicular cancer who consented to provide epididymis tissue for these studies. We thank Dr. Rui Yang for bioinformatic analysis. We acknowledge use of the Leica DM6000 widefield microscope in the Light Microscopy Imaging Facility at Case Western Reserve University made available through the Office of Research Infrastructure (NIH-ORIP) Shared Instrumentation Grant (S10RR021228).
Funding
This work was supported by NIH grant R01HD068901 (PI: A.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
J.A.B., S.-H.L., S.E.E. and A.H. acquired, analyzed and interpreted data. J.A.B. and A.H. drafted the article. All authors revised and approved the article.
Conflict of Interest
None declared.
References
- Schuppe HC, Pilatz A, Hossain H, Diemer T, Wagenlehner F, Weidner W. Urogenital Infection as a Risk Factor for Male Infertility. Dtsch Arztebl Int. 2017;114:339–346. doi: 10.3238/arztebl.2017.0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejucq N, Jegou B. Viruses in the mammalian male genital tract and their effects on the reproductive system. Microbiol Mol Biol Rev. 2001;65:208–31. doi: 10.1128/MMBR.65.2.208-231.2001. first and second pages, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranos N, Petrakou E, Anastasiadou C, Kotronias D. Detection of herpes simplex virus, cytomegalovirus, and Epstein-Barr virus in the semen of men attending an infertility clinic. Fertil Steril. 2003;79(Suppl 3):1566–70. doi: 10.1016/s0015-0282(03)00370-4. [DOI] [PubMed] [Google Scholar]
- Emerson C, Dinsmore WW, Quah SP. Are we missing mumps epididymo-orchitis? Int J STD AIDS. 2007;18:341–2. doi: 10.1258/095646207780749754. [DOI] [PubMed] [Google Scholar]
- Vuorinen T, Osterback R, Kuisma J, Ylipalosaari P. Epididymitis caused by coxsackievirus A6 in association with hand, foot, and mouth disease. J Clin Microbiol. 2014;52:4412–3. doi: 10.1128/JCM.02441-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–92. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M, Bowie AG. TLR3 in antiviral immunity: key player or bystander? Trends Immunol. 2005;26:462–8. doi: 10.1016/j.it.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Chiang JJ, Davis ME, Gack MU. Regulation of RIG-I-like receptor signaling by host and viral proteins. Cytokine Growth Factor Rev. 2014;25:491–505. doi: 10.1016/j.cytogfr.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1:519–25. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38:870–80. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Tortorec A, Denis H, Satie AP, Patard JJ, Ruffault A, Jegou B, Dejucq-Rainsford N. Antiviral responses of human Leydig cells to mumps virus infection or poly I:C stimulation. Hum Reprod. 2008;23:2095–103. doi: 10.1093/humrep/den207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Chen Q, Yan K, Liu Z, Li N, Zhang X, Yu L, Chen Y, Han D. RIG-I-like receptors mediate innate antiviral response in mouse testis. Mol Endocrinol. 2013;27:1455–67. doi: 10.1210/me.2013-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Zhu W, Xue S, Han D. Testicular defense systems: immune privilege and innate immunity. Cell Mol Immunol. 2014;11:428–37. doi: 10.1038/cmi.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Zhao S, Liu Z, Cheng L, Wang Q, Yan K, Chen Q, Wu H, Han D. Pattern recognition receptor-initiated innate antiviral responses in mouse epididymal epithelial cells. J Immunol. 2015;194:4825–35. doi: 10.4049/jimmunol.1402706. [DOI] [PubMed] [Google Scholar]
- Leir SH, Browne JA, Eggener SE, Harris A. Characterization of primary cultures of adult human epididymis epithelial cells. Fertil Steril. 2015;103:647–54 e1. doi: 10.1016/j.fertnstert.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne JA, Yang R, Leir SH, Eggener SE, Harris A. Expression profiles of human epididymis epithelial cells reveal the functional diversity of caput, corpus and cauda regions. Mol Hum Reprod. 2016;22:69–82. doi: 10.1093/molehr/gav066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leir SH, Harris A. MUC6 mucin expression inhibits tumor cell invasion. Exp Cell Res. 2011;317:2408–19. doi: 10.1016/j.yexcr.2011.07.021. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangda W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51:226–35. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Kerschner JL, Gosalia N, Neems D, Gorsic LK, Safi A, Crawford GE, Kosak ST, Leir SH, Harris A. Differential contribution of cis-regulatory elements to higher order chromatin structure and expression of the CFTR locus. Nucleic Acids Res. 2016;44:3082–94. doi: 10.1093/nar/gkv1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–9. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–5. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- Schindler C, Darnell JE., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–51. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- Karpus ON, Heutinck KM, Wijnker PJ, Tak PP, Hamann J. Triggering of the dsRNA sensors TLR3, MDA5, and RIG-I induces CD55 expression in synovial fibroblasts. PLoS One. 2012;7:e35606. doi: 10.1371/journal.pone.0035606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Tao J, Liang W, Zhao M, Du X, Cui S, Duan H, Kan B, Su X, Jiang Z. Identification and characterization of phosphodiesterases that specifically degrade 3′3′-cyclic GMP-AMP. Cell Res. 2015;25:539–50. doi: 10.1038/cr.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren A, Wang XC, Kellenberger CA, Rajashankar KR, Jones RA, Hammond MC, Patel DJ. Structural basis for molecular discrimination by a 3′,3′-cGAMP sensing riboswitch. Cell Rep. 2015;11:1–12. doi: 10.1016/j.celrep.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quicke KM, Diamond MS, Suthar MS. Negative regulators of the RIG-I-like receptor signaling pathway. Eur J Immunol. 2017;47:615–628. doi: 10.1002/eji.201646484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YK, Qu H, Gao D, Di W, Chen HW, Guo X, Zhai ZH, Chen DY. ARF-like protein 16 (ARL16) inhibits RIG-I by binding with its C-terminal domain in a GTP-dependent manner. J Biol Chem. 2011;286:10568–80. doi: 10.1074/jbc.M110.206896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MT, Di W, Xu H, Yang YK, Chen HW, Zhang FX, Zhai ZH, Chen DY. Negative regulation of RIG-I-mediated innate antiviral signaling by SEC14L1. J Virol. 2013;87:10037–46. doi: 10.1128/JVI.01073-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragale A, Stellacci E, Ilari R, Remoli AL, Lanciotti A, Perrotti E, Shytaj I, Orsatti R, Lawrence HR, Lawrence NJ, Wu J, Rehli M, Ozato K, Battistini A. Critical role of IRF-8 in negative regulation of TLR3 expression by Src homology 2 domain-containing protein tyrosine phosphatase-2 activity in human myeloid dendritic cells. J Immunol. 2011;186:1951–62. doi: 10.4049/jimmunol.1000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne JA, Yang R, Song L, Crawford GE, Leir SH, Harris A. Open chromatin mapping identifies transcriptional networks regulating human epididymis epithelial function. Mol Hum Reprod. 2014;20:1198–207. doi: 10.1093/molehr/gau075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


