Highlights
-
•
The genetic variant rs145204276 is associated with the susceptibility of OS.
-
•
Patients with genotype del/del of rs145204276 had significantly higher expression of GAS5 in the OS tissues.
-
•
SNP rs145204276 could play a functional role in the development and progression of OS by altering the methylation status of GAS5 promoter.
Key words: Osteosarcoma, Long non-coding RNA, Polymorphism, GAS5, Susceptibility
Abstract
Background
Previous studies showed that genetic variant rs145204276 in the promoter region of GAS5 was associated with the development of human cancer including colorectal cancer and hepatocellular cancer. This study aimed to investigate the role of rs145204276 in the development of osteosarcoma (OS).
Methods
132 OS patients and 1270 healthy controls were recruited for the genotyping analysis of rs145204276. Promoter methylation level of GAS5 was determined for all patients. The tumor tissues and the adjacent normal tissue were collected from 42 patients during surgery and the relative expression of GAS5 was then quantified by Real-time PCR. The Chi-square test was used to determine the difference of genotype and allele frequency between the patients and the controls. The gene expression and the percentage of methylation alleles were compared among different genotypes of rs145204276 with One-way ANOVA test.
Results
Compared with the controls, patients were found to have significantly lower rate of genotype del/del (7.6% vs. 8.7%, p = 0.024). The frequency of allele del was significantly lower in the patients than in the controls (23.5% vs. 30.1%, p = 0.021). Compared with than patients with genotype ins/ins, those with genotype del/del had remarkably higher expression of GAS5 (0.0033 ± 0.0019 vs. 0.0018 ± 0.0006, p < 0.001). Patients with genotype del/del were found to have obviously hypermethylation at the 7th CpG site as compared with those with genotype ins/ins (38.7% ± 21.1% vs. 20.5% ± 8.2%, p < 0.001).
Conclusions
The genetic variant rs145204276 is functionally associated with the susceptibility of OS, which can function as a protective factor in the incidence of OS possibly through the regulation of GAS5.
1. Introduction
Osteosarcoma (OS) is the most common pediatric bone cancer with the 5-year survival rate estimated to range from 40% to 75% [1]. Over 30% of OS patients may finally experience distant metastases and a poor prognosis [2]. Although the genetic background of OS has been extensively studied, the molecular mechanisms underlying the initiation and progression of OS remains poorly understood. Therefore, investigations on the molecular etiology of OS are essential to improve the treatment of this devastating disease.
The functional role of long non-coding RNA (lncRNAs) in the biological activity of malignant tumor has been widely investigated [3], [4]. By regulating the expression of target gene through different mechanisms, lncRNAs have been reported to be associated the growth and metastasis of a variety of cancer [5], [6], [7]. Aberrant upregulation of oncogenic lncRNA TUG1 was observed in B-cell malignancies, bladder cancer, hepatocellular carcinoma and OS [8]. The role of lncRNA CCAT1 has been well documented in OS, gastric cancer (GC), colorectal cancer and hepatocellular carcinoma [9]. Recently, the growth arrest specific transcript 5 (GAS5) was identified as a tumor-suppressor, including breast cancer, gastric cancer, colorectal cancer and bladder cancer [10], [11], [12], [13]. Ye et al reported that overexpression of GAS5 may suppress cell growth and EMT of OS through the miR-221/ARHI pathway [14]. However, the regulatory mechanism underlying down-expressed GAS5 in OS tissues remains poorly understood.
The polymorphism rs145204276 in the promoter region of GAS5 may regulate its expression level by influencing the methylation status [15]. Tao et al. [15] reported that the allele del of rs145204276 could regulate the expression of GAS5 and thus increased the risk of hepatocellular carcinoma. Zheng et al.[10] observed that the allele del of rs145204276 was associated with a decreased risk of colorectal cancer. To our knowledge, however, there is a paucity of knowledge concerning the association between rs145204276 and the expression of GAS5 in OS. The objective of the current study was to investigate the role of rs145204276 in the development of OS.
2. Methods
2.1. Subjects
The current study was approved by the local institutional review board. A total of 132 OS patients who came to our clinic center for treatment between 2006 January and 2017 March were included. The diagnosis of OS was validated by a multiple disciplinary team composed of a senior orthopedic surgeon, a radiological specialist and a pathologist. Patients received no previous medical treatment before the surgery. 1270 healthy controls were recruited during physical exanimations. Demographic data of the patients were collected from the medical record, including age, gender, tumor size, histological differentiation, Anneking Stage, and presence of distant metastasis. Written informed consent was obtained from all the subjects for the collection of blood or tissue samples.
2.2. Genotyping assay
The genomic DNA was extracted from peripheral blood of each subject using genomic DNA purification kit (Qiagen, Tokyo). DNA fragments containing rs145204276 were amplified using the following primers: F-TCCCGACTGAGGAGGAAGAGCA; R-AACACCGTCCCGGAAGTGAAA as previously reported [15]. The PCR products were then analyzed by 7% non-denaturing polyacrylamide gel electrophoresis, and the results were visualized by silver staining. The genotype was classified as del/del, del/ins or ins/ins for each patient. 10% samples were randomly sequenced for duplication analysis, which yielded a concordance rate of 100%.
2.3. Expression of GAS in tissues of the patients
The tumor tissues and the adjacent normal tissue were collected from 42 patients during surgery. The tissues were stored at −80 °C, and total RNA was extracted with a commercial kit (CWBio. Co. Ltd). The relative expression of GAS5 was quantified by Real-time PCR (RT-PCR) on Roche Light Cycler 480 system with the following primer 5′- CTTCTGGGCTCAAGTGATCCT-3′ and 5′- TTGTGCCATGAGACTCCATCAG-3′. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal control. The primer for GAPDH designed as 5′-GTCAACGGATTTGGTCTGTATT-3′ and 5′-AGTCTTCTGGGTGGCAGTGAT-3′. The amplification procedures were composed of an initial denaturation step of 95 °C for 10 min, 40 amplification cycles at 95 °C for 10 s, annealing at 60 °C for 20 s and elongation at 72 °C for 10 s. The relative expression of GAS5 was normalized using the ΔΔCt method as previously reported [15].
2.4. DNA methylation analysis
Promoter methylation level of GAS5 was determined using quantitative bisulfite pyrosequencing by the EpigenDx Inc. (Worcester, MA, USA). The target sequence of the 7th CpG site in GAS5 promoter region was listed in Table 1. The bisulfite conversion was carried out with 100 ng extracted DNA using the EpiTect Bisulfite Kit (QIAGEN) according to the manufacturer's protocol. The converted DNA was then amplified using PyroMark PCR Kit (QIAGEN) with primers mentioned above. The methylation status of each CpG site was analyzed as artificial T/C SNP using QCpG software (Qiagen Pyrosequencing). Each pyrosequencing assay was performed a minimum of three times. DNA methylation analysis was subsequently conducted using the Sequenom EpiTYPER system (Sequenom Inc., USA) as described previously.
Table 1.
Primers sequences and selected CpG sites in the promoter region.
| Primer name | Sequences |
|---|---|
| GAS5-p1 | 5′-AATATAGTTTAGGAAGTGAAATTTT-3′ |
| GAS5-p2 | 5′-GTTTTTTTTTTTTTTATTTTTAGA-3′ |
| GAS5-p3 | 5′-TTTTAGTAGGGAGGAG-3′ |
| CpG site | CCACCCCCTCCCACGGAGCGGGCGACGTGCCGGAAGGAAA TCACTCAGCCTTACACCGCCCCCCCTTCCCCCATCCCCA GAGCTTTCCTTGCCTCGCGCCCCCTCCCCTCTCTGCTCTT CCTCCTCAGTCGGGAGGAGGGCGGGAGCACGGCATCACGTGGACGGTCATGTCTCTGCCCACAATGGCG |
2.5. Statistical analyses
SPSS version 19.0 (SPSS Inc., Chicago, USA) was used for the data analysis. The Hardy-Weinberg equilibrium (HWE) test was performed for all the subjects. The Student's t test was used to analyze the continuous date and the Chi-square test was used to analyze the categorical data. Odds ratio (OR) was calculated to evaluate the contribution of rs145204276 to the risk of OS. The effect size (ϕ) was defined as the value of the square root of the Chi square value divided by total number of subjects. Specifically, a ϕ value of 0.1 was defined as small effect, 0.3 as medium effect and 0.5 as large effect. The gene expression and the percentage of methylation alleles were compared among different genotypes of rs145204276 with One-way ANOVA test. The relationship between the gene expression level and the percentage of methylation was determined by Pearson correlation analysis. The statistical significance was set at p < 0.05.
3. Results
3.1. Demographic data
As shown in Table 2, there was no significant difference between the cases and the controls in terms of age and gender. For the subjects included in the genotyping analysis, the mean age was 31.3 ± 18.4 years for the patients and 30.8 ± 12.3 years for the controls, respectively. Clinical features of the patients, including tumor size, histological differentiation, Enneking Stage and presence of distant metastasis, were summarized in Table 2.
Table 2.
Baseline characteristics of the patients and the controls.
| Features | Patients (n = 132) | Controls (n = 1270) | p | |
|---|---|---|---|---|
| Gender | 0.76 | |||
| Male | 76 | 748 | ||
| female | 56 | 522 | ||
| Age (years) | 0.83 | |||
| > 20 | 58 | 546 | ||
| ≤ 20 | 74 | 724 | ||
| Enneking stages | N/A | |||
| I | 19 | – | ||
| IIA | 22 | – | ||
| IIB | 44 | – | ||
| III | 13 | – | ||
| Histologic type | N/A | |||
| Osteoblastic | 75 | – | ||
| Chondroblastic | 37 | – | ||
| Fibroblastic | 5 | |||
| Mixed | 15 | |||
| Tumor size (cm) | N/A | |||
| > 5 | 91 | – | ||
| ≤ 5 | 41 | – | ||
| Tumor metastasis | N/A | |||
| Presence | 45 | – | ||
| Absence | 87 | – | ||
3.2. Association of rs145204276 with the susceptibility of oS
The frequency of rs145204276 of the cases and the controls were summarized in Table 3. HWE test indicated no selection bias for either group (p > 0.05). Compared with the controls, patients were found to have significantly lower rate of genotype del/del (7.6% vs. 8.7%, p = 0.024). The frequency of allele del was significantly lower in the patients than in the controls (23.5% vs. 30.1%, p = 0.021), with an OR of 0.72 (95% CI = 0.54–0.97). The effect size (ϕ) of rs145204276 was 0.07.
Table 3.
Comparison of the genotype and allele frequency of rs145204276 between the patients and controls.
| Genotype |
p | Allele |
p | Odds ratio (95% CI a) | ||||
|---|---|---|---|---|---|---|---|---|
| del/del | del/ins | ins/ins | del | ins | ||||
| Patients (n = 132) | 10 (7.6%) | 42 (31.8%) | 80 (60.6%) | 0.024 | 62 (23.5%) | 204 (76.5%) | 0.021 | 0.72 (0.54–0.97) |
| Controls (n = 1270) | 111 (8.7%) | 543 (42.8%) | 616 (48.5%) | 765 (30.1%) | 1775 (69.9%) | |||
3.3. The relationship between rs145204276 and progression and metastasis of oS
The tumor size was compared among different genotypes to determine the relationship between rs145204276 and the progression of OS. As shown in Table 4, the tumor size of patients with genotype del/del were remarkably smaller than that of patients with genotype ins/ins (4.1 cm ± 1.7 cm vs. 5.9 cm ± 2.4 cm, p = 0.04).
Table 4.
Comparison of tumor size and metastasis among different genotypes of rs145204276.
| Genotype |
p | |||
|---|---|---|---|---|
| del/del | del/ins | ins/ins | ||
| Tumor size (cm) | 4.1 ± 1.7 | 5.6 ± 1.9 | 5.9 ± 2.4 | 0.04 |
| Distant metastasis | 0.19 | |||
| Yes (n = 45) | 3 | 11 | 31 | |
| No (n = 87) | 7 | 31 | 49 | |
The incidence of distant metastasis was also compared among different genotypes to determine whether rs145204276 is associated with the metastasis of OS. As shown in Table 4, patients with genotype del/del had a numerically lower but statistically similar rate of distant metastasis of OS than those with ins/ins (30.0% vs. 38.7% p = 0.19).
3.4. Expression of GAS5 in OS tissues
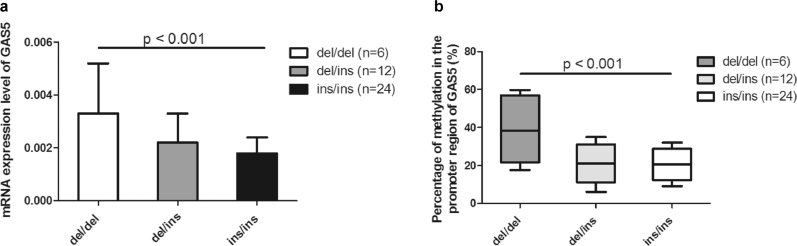
The relative mRNA expression of GAS5 was significantly lower in the tumor tissue than in the adjacent normal tissues (0.0021 ± 0.0012 vs. 0.0039 ± 0.0022, p < 0.001). As shown in Fig. 1a, patients with different genotypes of rs145204276 were found to have significantly different GAS5 expression. Compared with patients with genotype ins/ins, those with genotype del/del had remarkably higher relative expression of GAS5 (0.0033 ± 0.0019 vs. 0.0018 ± 0.0006, p < 0.001).
Fig. 1.
Comparison of the gene expression and the percentage of methylation among different genotypes of rs145204276. (a) Patients with genotype del/del had remarkably higher expression of GAS5 than those with genotype ins/ins (0.0033 ± 0.0019 vs. 0.0018 ± 0.0006, p < 0.001). (b) Patients with genotype del/del had obviously hypermethylation as compared with those with genotype ins/ins (38.7% ± 21.1% vs. 20.5% ± 8.2%, p < 0.001).
3.5. Methylation status of GAS5
As shown in Fig. 1b, there was significant difference of methylation percentage in the 7th CpG site. Patients with genotype del/del were found to have obviously hypermethylation as compared with those with genotype ins/ins (38.7% ± 21.1% vs. 20.5% ± 8.2%, p < 0.001). In addition, the methylation percentage was significantly correlated with the expression of GAS5 (r = 0.364, p = 0.02).
4. Discussion
With the development of sequencing technology, it was revealed that protein-coding sequences only occupy a small proportion of the whole human genome [16]. Increasing evidence showed that lncRNAs could be involved in cellular processes of human cancer [5], [6], [7]. In previous studies, a number of lncRNAs have been found to participate in the development and progression of OS [17], [18], [19]. Zhao et al [20] observed that CCAT1 was upregulated in OS tissues and involved in the proliferation and migration of the cancer cells. As a rising star of tumor-suppressors, the abnormal expression of GAS5 has recently been reported to play a role in a variety of human cancer including OS [12], [14], [15]. For the first time, we systematically investigated the regulatory capacity of the genetic variant on the expression of GAS5 in OS tissues and further evaluated its association with the susceptibility of OS. We found that the allele del of rs145204276 was significantly associated with a decreased risk of OS. Mover, patients with genotype del/del had significantly higher expression of GAS5 in the OS tissues. Apparently, the up-regulation of GAS5 and presence of genotype del/del could play a protective role in the development of OS. It was noteworthy that a ϕ value of 0.07 indicated a small effect of this variant on the development of OS, which was in line with an odds ratio of 0.72. Generally, common variants have a small effect in the development of complex disease, and can only explain a limited proportion of the overall variance. Herein, more factors need to be identified to better clarify the etiology of OS.
In the current study, we investigated the association of rs145204276 with the progression of OS and found it was significantly associated with tumor size. It has been well documented that GAS5 could function as a suppressor of cancer cells and inhibit cell proliferation and migration through different molecular mechanisms [11], [12], [14], [15]. Guo et al [21] reported that GAS5 could suppress the transition of G1/S phase of GC cells by up-regulation of P21 and suppression of CDK6. Liu et al [11] found that GAS5 could act as a ceRNA in GC and suppress the negative regulation of miR-23a. In a recent study, GAS5 was reported to function as a competing endogenous RNA for miR-221 and suppress cell growth of OS by regulating the miR-221/ARHI pathway [14]. To summarize, the definite mechanism underlying the regulatory effect of GAS5 on the development and progression of human cancer remains obscure. Further investigations are warranted to determine whether GAS5 can serve as a potential therapeutic target for OS.
Located in the promoter region of GAS5, allele del of rs145204276 could result in an increased expression of GAS5 in colorectal cancer and hepatocellular carcinoma [10], [15]. Through luciferase activity analysis, Tao et al. [15] found that rs145204276 could affect the transcriptional activity of GAS5. The authors speculated that rs145204276 could possibly regulate GAS5 expression through the methylation of CpG islands in the promoter region [15]. In this study, we confirmed that genotype del/del of rs145204276 was associated with a remarkably higher rate of methylation in the 7th CpG site of GAS5 promoter. Moreover, the methylation percentage was significantly correlated with the mRNA expression level of GAS5 in OS tissues. Therefore, it is most likely that rs145204276 played a functional role in the development and progression of OS by altering the methylation status of GAS5 promoter.
There were two limitations in this study that should be addressed. First, the sample size of the OS patients included in the genotyping analysis was relatively small due to a rare incidence of this type of cancer. In the future multi-center study, more OS patients need to be recruited for a stronger statistical power. Second, the mechanism underlying the influence of rs145204276 on methylation status of the GAS5 promoter remains undetermined, which is worthy of further investigation in the future study.
5. Conclusions
The genetic variant rs145204276 is functionally associated with the susceptibility of OS, which may function as a protective factor in the incidence of OS possibly via regulation of GAS5. The mechanism underlying the influence of rs145204276 on methylation status of the GAS5 promoter needs to be further investigated.
Conflict of interest statement
The authors declare no conflicts of interest.
Funding
This work was supported by the Nanjing Key Program of Medical Science and Technology Development (Grant No. ZKX14021).
Acknowledgement
We wish to thank all the patients who participated in this study.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jbo.2018.03.001.
Appendix. Supplementary materials
References
- 1.Ogura K., Fujiwara T., Yasunaga H., Matsui H., Jeon D.G., Cho W.H., Hiraga H., Ishii T., Yonemoto T., Kamoda H., Ozaki T., Kozawa E., Nishida Y., Morioka H., Hiruma T., Kakunaga S., Ueda T., Tsuda Y., Kawano H., Kawai A. Development and external validation of nomograms predicting distant metastases and overall survival after neoadjuvant chemotherapy and surgery for patients with nonmetastatic osteosarcoma: A multi-institutional study. Cancer. 2015;121:3844–3852. doi: 10.1002/cncr.29575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aljubran A.H., Griffin A., Pintilie M., Blackstein M. Osteosarcoma in adolescents and adults: survival analysis with and without lung metastases. Ann. Oncol. 2009;20:1136–1141. doi: 10.1093/annonc/mdn731. [DOI] [PubMed] [Google Scholar]
- 3.Hu G., Yang T., Zheng J., Dai J., Nan A., Lai Y., Zhang Y., Yang C., Jiang Y. Functional role and mechanism of lncRNA LOC728228 in malignant 16HBE cells transformed by anti-benzopyrene-trans-7,8-dihydrodiol-9,10-epoxide. Molecular carcinogenesis 54 Suppl. 2015;1:E192–E204. doi: 10.1002/mc.22314. [DOI] [PubMed] [Google Scholar]
- 4.Kong X.P., Yao J., Luo W., Feng F.K., Ma J.T., Ren Y.P., Wang D.L., Bu R.F. The expression and functional role of a FOXC1 related mRNA-lncRNA pair in oral squamous cell carcinoma. Mol. Cellular Biochem. 2014;394:177–186. doi: 10.1007/s11010-014-2093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ulger Y., Dadas E., Yalinbas Kaya B., Sumbul A.T., Genc A., Bayram S. The analysis of lncRNA HOTAIR rs12826786 C>T polymorphism and gastric cancer susceptibility in a Turkish population: lack of any association in a hospital-based case-control study. Irish J. Med. Sci. 2017;186:859–865. doi: 10.1007/s11845-017-1596-x. [DOI] [PubMed] [Google Scholar]
- 6.Heilmann K., Toth R., Bossmann C., Klimo K., Plass C., Gerhauser C. Genome-wide screen for differentially methylated long noncoding RNAs identifies Esrp2 and lncRNA Esrp2-as regulated by enhancer DNA methylation with prognostic relevance for human breast cancer. Oncogene. 2017;36:6446–6461. doi: 10.1038/onc.2017.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Botti G., Collina F., Scognamiglio G., Aquino G., Cerrone M., Liguori G., Gigantino V., Malzone M.G., Cantile M. LncRNA HOTAIR polymorphisms association with cancer susceptibility in different tumor types. Current Drug Targets. 2017 doi: 10.2174/1389450118666170622091940. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 8.Li Z., Shen J., Chan M.T., Wu W.K. TUG1: a pivotal oncogenic long non-coding RNA of human cancers. Cell Proliferation. 2016;49:471–475. doi: 10.1111/cpr.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xin Y., Li Z., Shen J., Chan M.T., Wu W.K. CCAT1: a pivotal oncogenic long non-coding RNA in human cancers. Cell Proliferation. 2016;49:255–260. doi: 10.1111/cpr.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng Y., Song D., Xiao K., Yang C., Ding Y., Deng W., Tong S. LncRNA GAS5 contributes to lymphatic metastasis in colorectal cancer. Oncotarget. 2016;7:83727–83734. doi: 10.18632/oncotarget.13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X., Jiao T., Wang Y., Su W., Tang Z., Han C. Long non-coding RNA GAS5 acts as a molecular sponge to regulate miR-23a in gastric cancer. Minerva medica. 2016 [Epub ahead of print] [PubMed] [Google Scholar]
- 12.Li W., Zhai L., Wang H., Liu C., Zhang J., Chen W., Wei Q. Downregulation of LncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget. 2016;7:27778–27786. doi: 10.18632/oncotarget.8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z., Wang W., Jiang J., Bao E., Xu D., Zeng Y., Tao L., Qiu J. Downregulation of GAS5 promotes bladder cancer cell proliferation, partly by regulating CDK6. PloS one. 2013;8:e73991. doi: 10.1371/journal.pone.0073991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye K., Wang S., Zhang H., Han H., Ma B., Nan W. Long noncoding RNA GAS5 suppresses cell growth and epithelial-mesenchymal transition in osteosarcoma by regulating the miR-221/ARHI pathway. J. Cellular Biochem. 2017;118:4772–4781. doi: 10.1002/jcb.26145. [DOI] [PubMed] [Google Scholar]
- 15.Tao R., Hu S., Wang S., Zhou X., Zhang Q., Wang C., Zhao X., Zhou W., Zhang S., Li C., Zhao H., He Y., Zhu S., Xu J., Jiang Y., Li L., Gao Y. Association between indel polymorphism in the promoter region of lncRNA GAS5 and the risk of hepatocellular carcinoma. Carcinogenesis. 2015;36:1136–1143. doi: 10.1093/carcin/bgv099. [DOI] [PubMed] [Google Scholar]
- 16.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nature Rev. Genetics. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 17.Liu K., Hou Y., Liu Y., Zheng J. LncRNA SNHG15 contributes to proliferation, invasion and autophagy in osteosarcoma cells by sponging miR-141. J. Biomedical Sci. 2017;24:46. doi: 10.1186/s12929-017-0353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong D., Wang Y. Knockdown of lncRNA HULC inhibits proliferation, migration, invasion, and promotes apoptosis by sponging miR-122 in osteosarcoma. J. Cellular Biochem. 2017 doi: 10.1002/jcb.26273. [DOI] [PubMed] [Google Scholar]
- 19.Jia D., Niu Y., Li D., Liu Z. LncRNA C2dat1 promotes cell proliferation, migration, and invasion by targeting MiR-34a-5p in Osteosarcoma Cells. Oncol. Res. 2017 doi: 10.3727/096504017X15024946480113. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Zhao J., Cheng L. Long non-coding RNA CCAT1/miR-148a axis promotes osteosarcoma proliferation and migration through regulating PIK3IP1. Acta biochimica et biophysica Sinica. 2017;49:503–512. doi: 10.1093/abbs/gmx041. [DOI] [PubMed] [Google Scholar]
- 21.Guo X., Deng K., Wang H., Xia J., Shan T., Liang Z., Yao L., Jin S. GAS5 inhibits gastric cancer cell proliferation partly by modulating CDK6. Oncol. Res. Treat. 2015;38:362–366. doi: 10.1159/000433499. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.