Abstract
Encysted embryos of Artemia are among the most stress-resistant eukaryotes partly due to the massive amount of a cysteine-rich protein termed artemin. High number of cysteine residues in artemin and their intramolecular spatial positions motivated us to investigate the role of the cysteine residues in the chaperone-like activity of artemin. According to the result of Ellman’s assay, there are nine free thiols (seven buried and two exposed) and one disulfide bond per monomer of artemin. Subsequent theoretical analysis of the predicted 3D structure of artemin confirmed the data obtained by the spectroscopic study. Native and reduced/modified forms of artemin were also compared with respect to their efficiency in chaperoning activity, tertiary structure, and stability. Since the alkylation and reduction of artemin diminished its chaperone activity, it appears that its chaperoning potential depends on the formation of intermolecular disulfide bond and the presence of cysteine residues. Comparative fluorescence studies on the structure and stability of the native and reduced protein revealed some differences between them. Due to the redox-dependent functional switching of artemin from the less to more active form, it can be finally suggested as a redox-dependent chaperone.
Keywords: Artemin, Cysteine-rich protein, Redox-dependent chaperone, Disulfide bond
Introduction
The extremophile Artemia is a crustacean with high stress resistance to harsh environmental conditions (Gajardo and Beardmore 2012). Diapause-destined embryos of Artemia exhibit an exceptional level of tolerance to unfavorable environmental conditions including temperature extremes, organic solvents, anoxia, repeated hydration–dehydration, ultraviolet, ionizing radiations, and desiccation (Liang and MacRae 1999; Clegg and Campagna 2006; Clegg 2011; Clegg 1997). There are several factors contributing to the stress tolerance of this crustacean including the synthesis, composition, and structure of the shell (Robbins et al. 2010; MacRae 2003) and the presence of several well-known intracellular molecular chaperones including p26 (Liang et al. 1997), artemin (MacRae 2003), and ArHsp21 and ArHsp22 (Qiu and MacRae 2008a, b). Artemin is a cysteine and histidine-rich protein representing about 12% of soluble protein in cysts (Chen et al. 2003) while it is almost absent during other developmental stages of the species (Chen et al. 2007). High structural stability, abundant accumulation, chaperone activity, and thermal resistance characteristics of artemin suggest that the protein contributes to cyst stress resistance (King et al. 2014).
Previous research revealed a chaperone-like activity for artemin which is responsible for protecting transfected cells and preventing protein aggregation in vitro (Chen et al. 2007). We also have presented some structural information about artemin from Artemia urmiana (Urmia Lake, Iran) based on its proposed 3D structural model (Rasti et al. 2009). The chaperone-like activity of artemin was investigated, and the results showed that artemin can prevent protein aggregation during refolding and improve the recovery of the lost biological activity of denatured enzymes (Shahangian et al. 2011; Shirzad et al. 2011). According to our recent studies, the chaperone activity of artemin was investigated under different stress conditions in bacterial cells using luciferase as an intracellular reporter (Takalloo et al. 2016, 2017), and the recent study demonstrated that artemin can protect proteins against oxidative and salt stress conditions.
Due to the high content of cysteine residues and their distribution in artemin, they may be regarded as main regulatory agents that play a key role in artemin function. This assumption motivated us to investigate the potential role of cysteine residues on artemin chaperone activity. Furthermore, the ability of cysteines to undergo oxidation/reduction reactions is considered as an important factor contributing to the physiological properties of artemin (Hu et al. 2011). Given the cytoplasmic localization of artemin and the role of glutathione (GSH) in maintaining thiols in the reduced state, this redox system may affect the oxidation–reduction of artemin, thereby mediating the formation of reversible disulfide bonds. Accordingly, it has been suggested that artemin can act as a reducing reservoir in protecting cells against oxidation and prevents modification of other proteins such as tubulins (Landino et al. 2002; Day et al. 2003).
Conformational changes as well as oligomer–dimer transition are required for chaperoning activity of some small heat shock proteins (sHSPs). The dimeric form is predominantly considered as the active form of chaperones for binding substrates under stress conditions (Gu et al. 2002). In some cases, the dimeric active structure of chaperones is formed via disulfide linkage, a mechanism which may also be proposed for artemin which possesses its similar structural properties. There is no strong evidence so far to support the relationship between such conformational changes and the chaperoning activity of artemin. Accordingly, herein, we have investigated whether the cysteine residues, their oxidation–reduction state, and consequent conformational changes can affect the chaperoning activity of artemin.
Materials and methods
Chemicals
5,5′-Dithiobis (2-nitrobenzoic acid) (DTNB), dithiothreitol (DTE), bovine carbonic anhydrase (CA), ρ-nitrophenyl acetate, 4-aminoantipyrine, and horseradish peroxidase (HRP) were purchased from Sigma (St. Louis, MO, USA). Iodoacetamide (IAM), isopropyl-b-D-thiogalactopyranoside (IPTG), and kanamycin were obtained from Invitrogen (Carlsbad, CA, USA). Nickel-nitrilotriacetic acid (Ni-NTA) agarose was provided by Qiagen (Hilden, Germany), and Q-Sepharose column was obtained from Bio-Rad (CA, USA). Phenol, guanidine hydrochloride (GdmCl), hydrogen peroxide (H2O2), β-mercaptoethanol, and all other chemicals were obtained from Merck (Darmstadt, Germany).
Artemin purification from Artemia cysts
Artemin purification was performed as previously described (Rasti et al. 2009) through the following steps: homogenization of hydrated encysted embryos of Artemia urmiana, centrifugation of the homogenate, precipitation of the supernatant with 60% (NH4)2SO4, overnight dialysis, heat treatment at 70 °C for 7 min, and ion exchange chromatography using Q-Sepharose column. Artemin was eluted from the column at 300 mM NaCl in 20 mM Tris-HCl buffer, pH 7.4.
Expression and purification of recombinant artemin
Protein expression was carried out in Escherichia coli BL21 (DE3) cells as described previously (Shahangian et al. 2011). IPTG was added when the culture reached an OD600 of about 0.6 and incubation continued for 6 h. All bacterial cultures were then harvested by centrifugation at 5000×g for 15 min. The pellet was resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole, pH 8.0), disrupted by sonication, clarified by centrifugation (14,000×g, 25 min, 4 °C), and stored at − 70 °C. Purification of the His-tagged protein was carried out using Ni-NTA agarose column. Bound proteins were eluted with a buffer containing 50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole, pH 8.0.
Preparation of reduced artemin
For complete reduction of purified artemin, the protein was incubated in 20 mM Tris-HCl buffer pH 7.4 with [GSH]/[GSSG] ratio of 100:1 ([GSH] = 10 mM and [GSSG] = 0.1 mM) and 30 mM DTE at 37 °C for 30 min. The reduced samples were then used for chaperone activity assay and fluorescence measurements.
Modification of artemin
Recombinant artemin was incubated in denaturing buffer containing 20 mM Tris buffer pH 8.0, 100 mM NaH2PO4, 8 M urea, and 10 mM DTE for 5 h at 4 °C. Denaturation of the protein and reduction of its disulfide bonds were followed by alkylation of the resulting thiol groups with 100 mM IAM (Rudolph and Lilie 1996; Mayer and Buchner 2004; Lin et al. 2007). Refolding of artemin was performed by gradual 10-fold dilution in refolding buffer containing 50 mM Tris-HCl buffer, pH 8.0, 50 mM NaCl, 50 mM glycine, 2 mM MgCl2, 5 mM GSH, and 0.05 mM GSSG with 10% glycerol and 0.005% Tween 20. The refolding reaction was allowed to proceed overnight at 4 °C. Refolded artemin was then precipitated by 80% (NH4)2SO4. The excess DTE was removed by dialysis against 20 mM Tris-HCl buffer, pH 7.4 for 24 h. The chaperone activity of the modified protein was evaluated by recovery of denatured CA, as the protein substrate.
SDS-PAGE analysis
Purified artemin from Artemia embryos and recombinant artemin were analyzed by reducing and non-reducing sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (12%) followed by Coomassie Brilliant Blue staining.
Chaperone-like activity assays
Denatured HRP was prepared by incubating the enzyme (1 mg/ml) in 0.2 mM phosphate buffer, pH 7.0 containing 6 M GdmCl at 25 °C for 2.5 h. Protein denaturation was confirmed by measuring the enzyme activity. Renaturation of the denatured HRP was conducted by rapid 80-fold dilution of the denatured samples into freshly prepared renaturation buffer (30 mM DTE or [GSH] = 10 mM and [GSSG] = 0.1 mM to create the reducing environment), containing different concentrations of reduced and native artemin. The enzyme activity of the samples was measured after 30 min of incubation at room temperature. The molar ratio of chaperone (artemin)/substrate (HRP) was the same for all samples. HRP activity was determined at room temperature by measuring an increase in absorbance at 510 nm. The 1 ml assay mixture containing 50 μl of enzyme solution (0.02 mg/ml), fresh substrate solution of 2.5 mM 4-aminoantipyrine coupled to 170 mM phenol, 1.7 mM H2O2, and total volume was set by adding 200 mM phosphate buffer, pH 7.0 (Trinder 1969).
Native CA (10 mg/ml) was denatured by overnight incubation in denaturating buffer (20 mM Tris-sulfate, pH 7.75, containing 6 M GdmCl) at room temperature. Protein unfolding was confirmed by measuring the enzyme activity. Renaturation of the denatured CA was conducted by rapid 50-fold dilution of the protein mixture (at the final CA concentration of 0.2 mg/ml) into freshly prepared renaturation solution (20 mM Tris-sulfate, pH 7.75) in the presence of various concentrations of native and modified artemin. The enzyme activity of the samples was measured after 2 h of incubation at room temperature. The molar ratio of chaperone (artemin)/substrate (CA) was the same for all subjects we examined. The enzymatic activity of the renatured CA solutions was determined based on ρ-nitrophenyl acetate esterase activity, according to Pocker and Stone (1967). Briefly, 45 μl of 52 mM ρ-nitrophenyl acetate in dry acetonitrile was added to 455 μl of CA solution in order to make a solution of 0.2 mg/ml CA, 12.8 mM Tris/sulfate, pH 7.75, and 4.7 mM ρ-nitrophenyl acetate. After mixing for 5 s, the increasing rate of p-nitrophenolate production was monitored by measuring absorbance at 400 nm as a function of time. The untreated CA samples were also assayed as control sets.
In both experiments, the yield of enzyme refolding was expressed as percentage of activity relative to the untreated enzymes (control samples) under identical conditions. The outcome was expressed as percentage of activities exhibited by control.
Fluorescence study
Intrinsic fluorescence measurements of the native and reduced artemin were performed at room temperature using a Varian Cary Eclipse fluorescence spectrophotometer (Agilent Technologies, Santa Clara, CA) in a quartz cell of 1-cm path length. The concentration of enzymes used for fluorescence studies was 10 μg/ml. The excitation wavelength was set at 280 nm, and the emission spectra were monitored in the wavelength range of 300–400 nm. Excitation and emission bandwidths were 5.5 nm.
Chemical induced denaturation
Chemical denaturation of reduced and native artemin was performed using increasing concentrations of GdmCl at pH 7.4. After overnight incubation at room temperature with different concentrations of the denaturant, unfolding curves of the denatured samples were monitored by fluorescence measurements at 25 °C. The standard free energy of protein unfolding (∆Gc) was calculated as a function of GdmCl concentration by Eq. 1 and was drawn against the denaturant concentrations. To obtain the protein stability parameters in standard conditions, the ∆G curve was extended to 0 M guanidine hydrochloride and ∆G (H2O) was extracted (Monsellier and Bedouelle 2005; Bolen and Santoro 1988; Ahmad et al. 1992).
.
| 1 |
Quantification of thiol and disulfide
The free SH content of artemin was determined using Ellman’s reagent according to the method of Beveridge et al. (1974). Artemin was denatured under non-reducing condition with 8 M urea at pH 8.0. The reagent was prepared by dissolving 4 mg of DTNB reagent in 1 ml of 0.1 M sodium-phosphate buffer containing 0.1 M EDTA (reaction buffer), pH 8.0. Total free thiol content of artemin was determined by mixing 100 μl of artemin solution, 1 ml reaction buffer with 8 M urea, and 20 μl Ellman’s reagent at 25 °C for 15 min, and then, the absorbance at 412 nm was recorded using a UV-3000 spectrophotometer (Pharmacia Biotech). To determine the exposed protein thiol content, the procedure was performed as described above, except that the reaction buffer was denaturant-free. The reaction buffer with and without 8 M urea was included as blank for total and exposed free thiols, respectively. The SH content was calculated by measuring the absorbance at 412 nm using the extinction coefficient of 13,600 M−1 cm−1 for 2-nitro-5-thiobenzoate (NTB) (Eq. 2).
| 2 |
Accessible surface area of the cysteine residues
The structural model of artemin, previously constructed and confirmed by various validation tests, was used (Rasti et al. 2009). The accessible surface area (ASA) of cysteine residues was calculated using What If web server (http://swift.cmbi.ru.nl/whatif/), and their positions were shown in 3D model of artemin.
Results
SDS-PAGE analysis
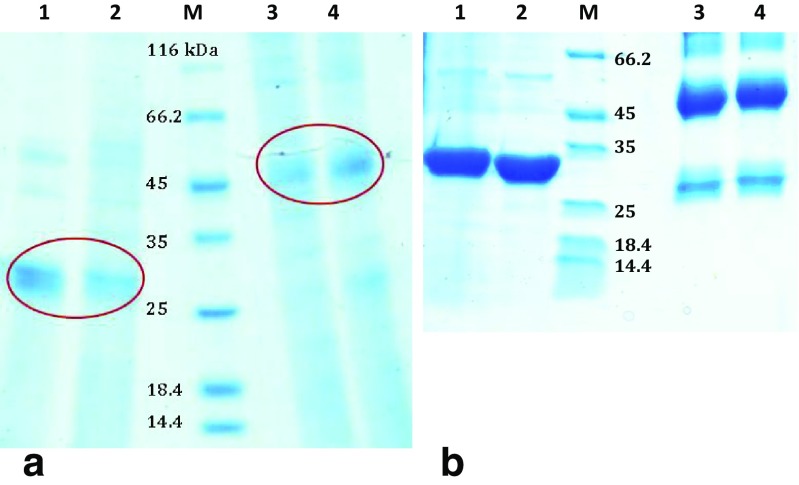
According to Fig. 1a, the apparent molecular weight of the purified artemin from Artemia cysts was estimated to be about 27 kDa using reducing SDS-PAGE (lanes 1 and 2), while under non-reducing condition, this band was appeared about 50 kDa (lanes 3 and 4). Similar results were obtained for recombinant artemin purified from bacterial cells (Fig. 1b).
Fig. 1.
SDS-PAGE analysis of purified wild-type and recombinant artemin under reducing and non-reducing conditions. a Purified artemin from Artemia cysts in a monomeric form on reduced SDS-PAGE with a single band about 27 kDa (lanes 1 and 2) and in a dimeric form on non-reduced SDS-PAGE with a band about 50 kDa (lanes 3 and 4). b Purified recombinant artemin from Escherichia coli cells, under reducing (1 and 2) and non-reducing (3 and 4) conditions
In vitro chaperone activity of reduced and native artemin
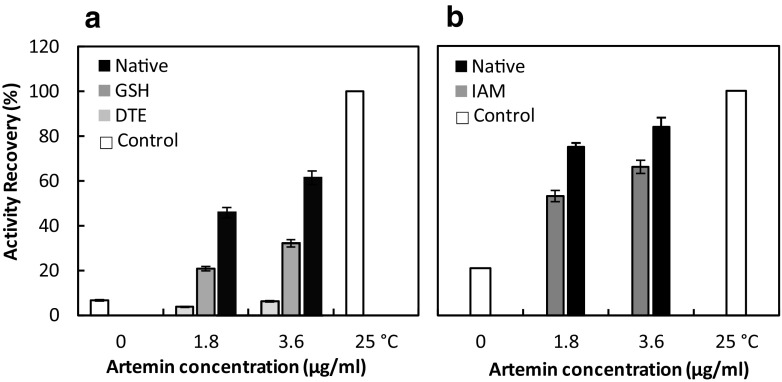
The refolding efficiency of denatured HRP was investigated in the presence of different concentrations of reduced and native artemin when its disulfide bonds were broken by GSH and DTE. The reducing reagents GSH and DTE cleave the disulfide bond of the chaperone, and the alkylation agent IAM blocks all the reduced cysteine residues and prevents formation of the disulfide bond. Reduction of artemin (by GSH) increased the refolding yield of denatured HRP in a concentration-dependent manner, although it was not as effective as native artemin (Fig. 2a). The recovery yield of the denatured HRP was nearly 60% in the presence of native artemin, whereas it was markedly diminished by 10-fold and 3-fold in the presence of artemin reduced by DTE and GSH, respectively. Since the activity recovery of denatured HRP in the presence and absence of reduced artemin (by DTE) was the same, it can be concluded that artemin lost its chaperoning potency after reduction by DTE. Although the denatured enzyme activity was not regained in the presence of reduced artemin (by DTE), the activity recovery was promoted from 6 to 30% at 3.6 μg/ml of reduced artemin (by GSH). Since DTE is a more potent reductant agent of disulfide bonds, it forms a more stable reduced state of artemin as compared to GSH. It indicates that more efficient cleavage of the specific disulfide bond with DTE results in a complete loss of the chaperone activity revealing the importance of the disulfide bond in the function of artemin.
Fig. 2.
Assisted refolding of HRP (a) and CA (b) in the presence of various concentrations of reduced/modified and native artemin. The reducing reagents GSH and DTE cleave the disulfide bond of the chaperone, and the alkylation agent IAM blocks all reduced cysteine residues and prevents formation of the disulfide bond. The reduced and alkylated artemin were used to assess the recovery yield of denatured HRP (a) and CA (b), respectively. a The denatured HRP (1 mg/ml) was diluted to a final concentration of 0.0125 mg/ml using the appropriate refolding buffers containing different amounts of native artemin (black) and reduced artemin in the presence of GSH 10 mM (dark gray) and DTE 30 Mm (light gray) at 25 °C. b The denatured CA (10 mg/ml) was diluted to a final concentration of 0.2 mg/ml using the appropriate refolding buffers containing different amounts of native artemin (black) and modified artemin with IAM 100 mM (dark gray) at 25 °C. The residual activity of denatured HRP and CA was reported as the percentage recovery relative to the untreated enzymes (white) under similar experimental conditions. Results are the means for triplicate measurements
The effect of disulfide bond cleavage on the chaperone activity of artemin was evaluated after chemical modification of artemin using IAM. As a sulfhydryl reactive alkylating agent, IAM blocks reduced cysteines, thereby preventing the formation of disulfide bonds. According to Fig. 2b, alkylation of the thiol groups affected the ability of artemin to refold denatured CA. The refolding yield of the enzyme was almost 50 and 65% at 1.8 and 3.6 μg/ml of the chaperone concentrations, respectively, compared to 75 and 84% for the native one. The yield of activity recovery for the control sample was about 20%.
Structural changes of reduced artemin
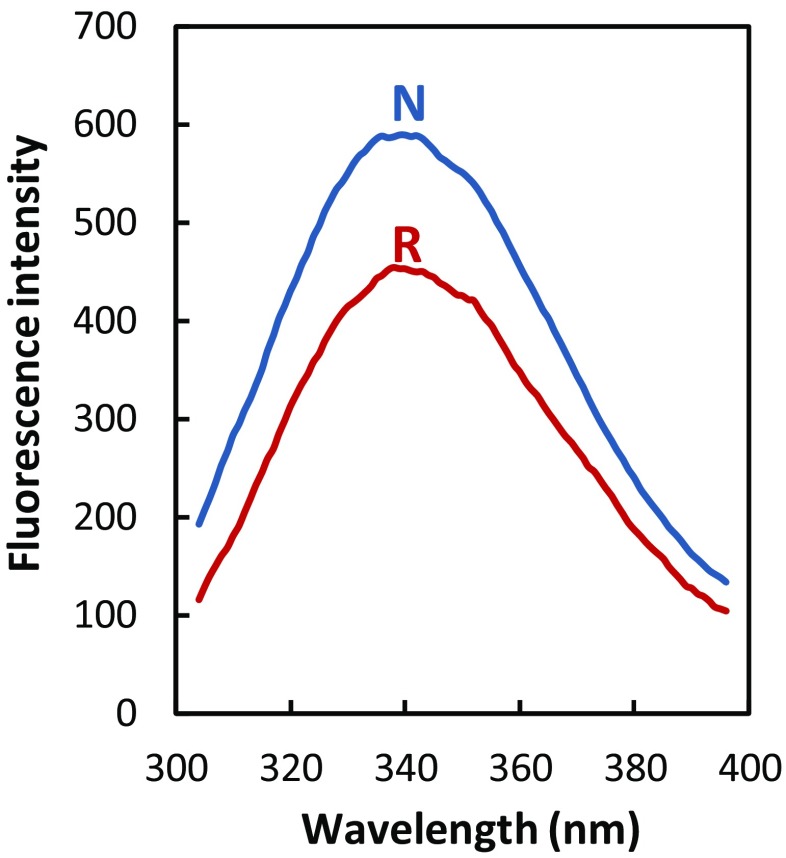
Analysis of intrinsic fluorescence spectra revealed that both reduced and native artemin have a λmax at ∼ 340 nm, implying that there were no significant changes in the tryptophan environment of the reduced sample. A slight decrease in fluorescence intensity of the reduced protein reflects the conformational changes taking place in the artemin structure as a result of the reduction of disulfide bonds (Fig. 3).
Fig. 3.
Intrinsic fluorescence spectra of native (N) and reduced artemin (R). The protein concentration was 10 μg/ml, and the excitation wavelength was set at 280 nm
GdmCl-induced denaturation curve of native and reduced artemin
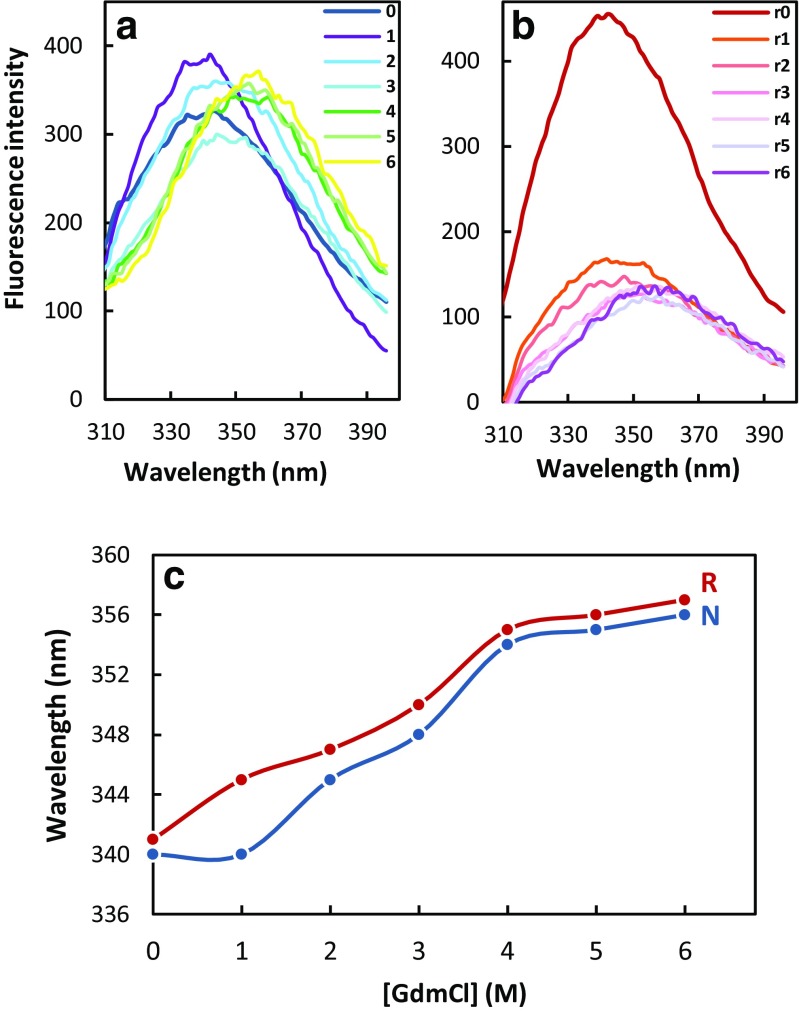
The thermodynamic stability of a protein is the difference of free energy between its native and unfolded states in physiological conditions and can be deduced from equilibrium constant between these two conformational states and thus from the measurement of concentrations. Since the concentrations of unfolded states of a protein in physiological conditions are very low, the values of the protein stability are measured and extrapolated to the standard conditions. Here, the effect of disulfide bond on the thermodynamic stability of artemin was studied by GdmCl-induced unfolding transitions at pH 7.4 via tryptophan fluorescence (Fig. 4).
Fig. 4.
Denaturation of native (a) and reduced (b) artemin induced by GdmCl. The reduced sample was prepared in 20 mM Tris/HCl containing 30 mM DTE, pH 7.4. Curves 0–6 and r0–r6 correspond to fluorescence spectra of native and reduced artemin, respectively, in the presence of 0–6 M GdmCl. c GdmCl-induced unfolding transitions of reduced (R) and native (N) artemin. All protein samples (10 μg/ml) were incubated overnight in the presence of various concentrations of GdmCl. Data correspond to emission intensities measured at an excitation wavelength of 280 nm
As seen in Fig. 4a, b, the reduced protein started to unfold at 1 M GdmCl, while the native form was stable up to 4 M GdmCl. According to Fig. 4b, the disulfide bond cleavage significantly affected the stability of reduced artemin. As depicted in Fig. 4c, fluorescence emission maximum, as well as the fluorescence quantum yield of protein samples, was considerably influenced by increasing the denaturant concentration. Due to the exposure of the chromophores to the solvent, the emission maximum showed a red shift from 340 to 375 nm. The quantum yield was significantly reduced, representing perturbations of the tertiary structure of the protein.
The unfolding midpoint was also higher for the native form. The midpoint of the unfolding transition (Cm) was calculated at 2.419 M for native artemin, whereas it was found to be around 1.896 M for the reduced sample revealing that the reduced protein lost its folded structure under much lower concentrations of denaturant in comparison with native artemin (Table 1). Therefore, it is demonstrated that native artemin is considerably more resistant to GdmCl-induced denaturation than the reduced protein. Thus, the disulfide bond formation in native artemin stabilizes artemin structure. In addition, the free energy at 0 M guanidine hydrochloride (∆G (H2O)) was calculated to be 4.83 and 3.19 kcal/mol for native and reduced artemin, respectively (Table 1). This comparison indicates that native artemin is more stable than the reduced protein.
Table 1.
Thermodynamic parameters associated with GdmCl-induced denaturation of native and reduced artemin
| Cm (M) | ∆G (H2O) | |
|---|---|---|
| Native | 2.419 | 4.83 kJ/mol |
| Reduced | 1.896 | 3.19 kJ/mol |
Free SH and S–S contents
To analyze the cysteine residues with regard to their ability for disulfide bond formation, the number of free exposed and buried thiols was quantified using DTNB reaction under non-denaturing and denaturing conditions. Each subunit contains a high level of free cysteine content, 9 out of 10, of which two thiols are exposed, while the other ones are buried (Table 2). Therefore, the presence of one inter-subunit disulfide bond between two monomers of artemin can be predicted.
Table 2.
Quantification of thiol content of artemin using Ellman’s test
| Number of free -SH | |
|---|---|
| Surface -SH | ~ 2 |
| Total free -SH | ~ 9 |
Accessibility of the cysteine residues
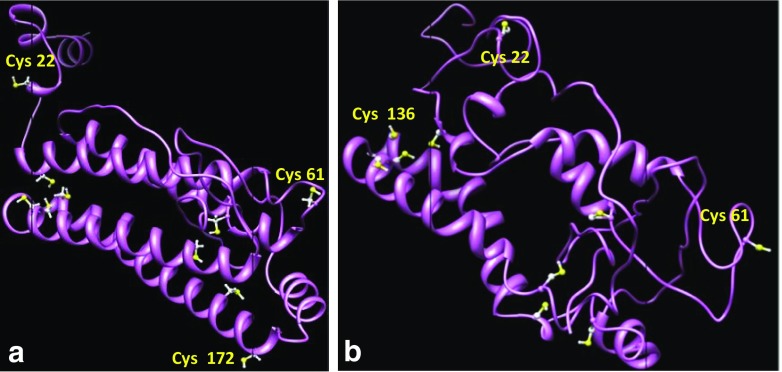
In comparison with the result of Ellman’s method, theoretical studies showed the presence of an additional exposed cysteine per every monomer of artemin, probably responsible for inter-subunit disulfide bond formation. Theoretical data also indicated that at least three cysteines have the most accessible surfaces for inter-subunit disulfide bond formation. According to molecular dynamic (MD) results, Cys22 in the N-terminal area of artemin in both conditions has the most surface accessibility (Table 3). Figure 5 shows the cysteine positions in proposed models before (a) and after (b) MD.
Table 3.
Accessibe surface area of cysteine residues in artemin before (− MD) and after (+ MD) molecular dynamic process. Italic residues represent maximum values of ASA
| Before MD | After MD | ||||||
|---|---|---|---|---|---|---|---|
| Accessible surface area (Å2) | Accessible surface area (Å2) | ||||||
| Residue | Total | Backbone | Side chain | Residue | Total | Backbone | Side chain |
| CYS22 | 34.2701 | 5.1959 | 29.0742 | CYS22 | 32.3792 | 8.2582 | 24.1210 |
| CYS32 | 1.4422 | 0.3117 | 1.1304 | CYS32 | 2.3650 | 1.9335 | 0.4315 |
| CYS61 | 30.1140 | 10.0488 | 20.0652 | CYS61 | 23.2348 | 1.2043 | 22.0305 |
| CYS71 | 0.0000 | 0.0000 | 0.0000 | CYS71 | 6.0804 | 0.4174 | 5.6630 |
| CYS119 | 0.2157 | 0.0000 | 0.2157 | CYS119 | 2.2199 | 1.5727 | 0.6472 |
| CYS136 | 23.2348 | 4.2785 | 18.9563 | CYS136 | 25.4024 | 2.5931 | 22.8093 |
| CYS137 | 0.2157 | 0.0000 | 0.2157 | CYS137 | 1.0787 | 0.0000 | 1.0787 |
| CYS144 | 0.0000 | 0.0000 | 0.0000 | CYS144 | 0.6472 | 0.0000 | 0.6472 |
| CYS166 | 5.1776 | 0.0000 | 5.1776 | CYS166 | 1.9033 | 0.1559 | 1.7474 |
| CYS172 | 34.5257 | 7.4233 | 27.1024 | CYS172 | 8.1978 | 0.0000 | 8.1978 |
Fig. 5.
Predicted three-dimensional structure of artemin before (a) and after (b) molecular dynamic simulation. As shown in Table 2, there are three exposed cysteines in both molecular dynamic processes: Cys22, Cys61, and Cys136 have the maximum ASA
Discussion
Molecular chaperones interact with hydrophobic surfaces of partially folded/unfolded proteins to stabilize non-native proteins, inhibit protein misfolding, and prevent aggregation of protein substrates (Gething and Sambrook 1992). In our previous studies, we found artemin to be a potent molecular chaperone due to its ability to prevent chemical aggregation of protein substrates and conferring stress tolerance to bacterial cells (Shahangian et al. 2011; Takalloo et al. 2016). Artemin is a cysteine-rich protein, and it is suggested that the residues can modulate redox-dependent function of artemin through disulfide bond formation (Hu et al. 2011). Therefore, in the present work, the role of cysteine residues, their oxidation–reduction state, on the chaperone-like activity of artemin has been investigated using theoretical, functional, and structural studies.
The following findings, indicated by our studies, suggest that formation and cleavage of a disulfide bond influence structure and function of artemin, meaning that the activity of artemin must be under redox control. (i) Two SDS-PAGE bands of 27 and 50 kDa correspond to monomeric and dimeric forms of artemin, respectively (Fig. 1). (ii) The chaperone-like activity of artemin has been shown to be affected by its reduction and alkylation (Fig. 2). (iii) Conformational changes exist between reduced and native artemin. Fluorescence measurements have indicated that the tertiary structure of the chaperone was partially affected under reducing conditions, suggesting that artemin is susceptible to shifts in intracellular redox state (Fig. 3). (iv) Structural stability of artemin was declined under reducing conditions, suggesting that the disulfide bond formation stabilizes the structure of native artemin compared to the reduced protein (Fig. 4).
Typically, sHSPs typically act as oligomeric complexes in different sizes. There are several studies showing that subunit association/dissociation in certain sHsps, Hsp18.1 and Hsp27 in particular, can affect their chaperoning activity. α-Crystalline also exhibits the enhanced chaperone-like activity when its oligomerization state changes under heat shock treatment. Also, in wheat Hsp16.9, the oligomer dissociation into dimers is required for substrate binding through exposed hydrophobic surfaces (Gu et al. 2002). Based on the facts mentioned above, it seems that subunit conformational changes are required for chaperoning activity of chaperones. In addition, specific small heat shock proteins such as Hsp25 and Hsp33 can form dimeric structures based on the formation of intermolecular disulfide bonds under oxidizing conditions (Zavialov et al. 1998). Regarding the similar structural properties of artemin and also the obtained experimental results from present study, a similar mechanism can be suggested for artemin as well.
Cysteine residues generally act as the cellular redox sensors through formation of disulfide bonds. The activity of specific molecular chaperones such as HSP33 and 2-Cys peroxiredoxins is directly controlled by oxidation status of their redox-sensitive cysteines (Kumsta and Jakob 2009). In normal physiological conditions, HSP33 is inactive, but upon exposure to oxidative stress, the protein is active (Janda et al. 2004). There is evidence suggesting that activation of the protein is performed through the formation of a reversible disulfide bond. In this case, partial unfolding of the chaperone is promoted by oxidative stress-induced disulfide bond formation that results in exposure of the high-affinity binding sites involved in substrate binding (Ilbert et al. 2007). Another example is human protein disulfide isomerase (hPDI) that undergoes large redox-dependent conformational changes required for regulation of its chaperone activity (Wang et al. 2012). The thiol moiety of cysteine is extremely sensitive to oxidation and can form a disulfide bond with another thiol group (Miki and Funato 2012). Since cysteines are the most redox-sensing residues of proteins and ionizable under pH variations (Wouters et al. 2010), they can rapidly detect subtle changes in redox status and trigger functional and structural changes in proteins (Barford 2004; Niforou et al. 2014). Herein, the functional role of the cysteine residues of artemin has been investigated by a refolding assay in the presence of the reduced/modified and native forms of artemin (Figs. 3 and 4). As seen in the results, lower refolding yield was obtained by using reduced/modified artemin compared to its native form. Our results also indicated that the intermolecular disulfide bond of artemin is functionally important and its cleavage makes a remarkable alteration in the chaperone function. Disulfide bonds are extremely rare in intracellular proteins due to the reductive nature of the cytoplasm (Jacob et al. 2004; Gilbert 1993; Claiborne et al. 1999). Because of the cytoplasmic localization of artemin in Artemia cysts, the physiological role of the cysteine residues may depend on the formation of the reversible disulfide bond under an oxidation/reduction reaction. It is supposed that this reaction likely plays an essential role in protecting other proteins and the cells against oxidation (Chen et al. 2003).
The evaluation of equilibrium unfolding of artemin by the denaturant revealed a high stability of the native protein compared to the reduced one (Fig. 4). Accordingly, it is suggested that denaturation using GdmCl leads to both dissociation of the protein dimers to monomers and unfolding of the monomers. Fluorescence studies indicated that by disruption of the disulfide bond between the cysteine residues in reduced artemin, the sensitivity of the chaperone increased considerably so that at the very low denaturant concentrations, the structure of artemin was remarkably changed. Our results are also consistent with several previous findings indicating that disulfide bonds confer extra stability to proteins (McAuley et al. 2008; Betz 1993).
Ellman’s assay revealed that 9 out of 10 thiols are free in artemin monomers and there is only one cysteine involved in intermolecular disulfide bond formation (Table 2). As indicated by molecular modeling, in each artemin monomer, Cys22, 61, and 136 have maximum values of ASA among 10 cysteines (Table 3), and one of them is probably involved in intermolecular disulfide bond. Cys22 shows the highest accessible surface area, and it is thought to be the responsible residue for disulfide bond formation. The computational analysis showed a good agreement with the experimental results (Fig. 5).
In conclusion, findings of the present study indicate that artemin may act as a redox-sensing protein by forming a reversible disulfide bond to function as a powerful molecular chaperone. Two monomers of artemin form the chaperone dimer through an intramolecular disulfide bond that is connected by the next neighbor cysteine residues. Disulfide bond formation converts artemin into the more active and stable form, whereas cleavage of the disulfide bond converts artemin into the reduced monomeric form exhibiting low chaperone activity. This property may have a predominate role in intracellular defense against ROS-induced oxidative damage.
Funding information
The authors express their gratitude to the research council of Tarbiat Modares University and University of Guilan for financial support during the course of this project.
Footnotes
Bita Mosaddegh and Zeinab Takalloo contributed equally to this work.
References
- Ahmad F, Yadav S, Taneja S. Determining stability of proteins from guanidinium chloride transition curves. Biochem J. 1992;287(2):481–485. doi: 10.1042/bj2870481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barford D. The role of cysteine residues as redox-sensitive regulatory switches. Curr Opin Struc Biol. 2004;14(6):679–686. doi: 10.1016/j.sbi.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Betz SF. Disulfide bonds and the stability of globular proteins. Protein Sci. 1993;2(10):1551–1558. doi: 10.1002/pro.5560021002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge T, Toma SJ, Nakai S. Determination of SH-and SS-groups in some food proteins using Ellman’s reagent. J Food Sci. 1974;39(1):49–51. doi: 10.1111/j.1365-2621.1974.tb00984.x. [DOI] [Google Scholar]
- Bolen DW, Santoro MM. Unfolding free energy changes determined by the linear extrapolation method. 2. Incorporation of. DELTA. G. degree. NU values in a thermodynamic cycle. Biochemistry. 1988;27(21):8069–8074. doi: 10.1021/bi00421a015. [DOI] [PubMed] [Google Scholar]
- Chen T, Amons R, Clegg JS, Warner AH, MacRae TH. Molecular characterization of artemin and ferritin from Artemia franciscana. Eur J Biochem. 2003;270(1):137–145. doi: 10.1046/j.1432-1033.2003.03373.x. [DOI] [PubMed] [Google Scholar]
- Chen T, Villeneuve TS, Garant KA, Amons R, MacRae TH. Functional characterization of artemin, a ferritin homolog synthesized in Artemia embryos during encystment and diapause. FEBS J. 2007;274(4):1093–1101. doi: 10.1111/j.1742-4658.2007.05659.x. [DOI] [PubMed] [Google Scholar]
- Claiborne A, Yeh JI, Mallett TC, Luba J, Crane EJ, Charrier V, Parsonage D. Protein-sulfenic acids: diverse roles for an unlikely player in enzyme catalysis and redox regulation. Biochemistry. 1999;38(47):15407–15416. doi: 10.1021/bi992025k. [DOI] [PubMed] [Google Scholar]
- Clegg J. Embryos of Artemia franciscana survive four years of continuous anoxia: the case for complete metabolic rate depression. J Exp Biol. 1997;200(Pt 3):467–475. doi: 10.1242/jeb.200.3.467. [DOI] [PubMed] [Google Scholar]
- Clegg JS. Stress-related proteins compared in diapause and in activated, anoxic encysted embryos of the animal extremophile, Artemia franciscana. J Insect Physiol. 2011;57(5):660–664. doi: 10.1016/j.jinsphys.2010.11.023. [DOI] [PubMed] [Google Scholar]
- Clegg JS, Campagna V. Comparisons of stress proteins and soluble carbohydrate in encysted embryos of Artemia franciscana and two species of Parartemia. Comp Biochem Physiol B: Biochem Mol Biol. 2006;145(2):119–125. doi: 10.1016/j.cbpb.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Day RM, Gupta JS, MacRae TH. A small heat shock/α-crystallin protein from encysted Artemia embryos suppresses tubulin denaturation. Cell Stress Chaperones. 2003;8(2):183–193. doi: 10.1379/1466-1268(2003)008<0183:ASHCPF>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajardo GM, Beardmore JA. The brine shrimp Artemia: adapted to critical life conditions. Front Physiol. 2012;3:1–8. doi: 10.3389/fphys.2012.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething MJ, Sambrook J. Protein folding in the cell. Nature. 1992;355(6355):33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Gilbert HF. Molecular and cellular aspects of thiol-disulfide exchange. Adv Enzymol Relat Areas Mol Biol. 1993;63:69–172. doi: 10.1002/9780470123096.ch2. [DOI] [PubMed] [Google Scholar]
- Gu L, Abulimiti A, Li W, Chang Z. Monodisperse Hsp16. 3 nonamer exhibits dynamic dissociation and reassociation, with the nonamer dissociation prerequisite for chaperone-like activity. J Mol Biol. 2002;319(2):517–526. doi: 10.1016/S0022-2836(02)00311-X. [DOI] [PubMed] [Google Scholar]
- Hu Y, Bojikova-Fournier S, King AM, MacRae TH. The structural stability and chaperone activity of artemin, a ferritin homologue from diapause-destined Artemia embryos, depend on different cysteine residues. Cell Stress Chaperones. 2011;16(2):133–141. doi: 10.1007/s12192-010-0225-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilbert M, Horst J, Ahrens S, Winter J, Graf PCF, Lilie H, Jakob U. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat Struct Mol Biol. 2007;14(6):556–563. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob C, Holme AL, Fry FH. The sulfinic acid switch in proteins. Org Biomol Chem. 2004;2(14):1953–1956. doi: 10.1039/B406180B. [DOI] [PubMed] [Google Scholar]
- Janda I, Devedjiev Y, Derewenda U, Dauter Z, Bielnicki J, Cooper DR, Graf PC, Joachimiak A, Jakob U, Derewenda ZS. The crystal structure of the reduced, Zn 2+-bound form of the B. subtilis Hsp33 chaperone and its implications for the activation mechanism. Structure. 2004;12(10):1901–1907. doi: 10.1016/j.str.2004.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AM, Toxopeus J, MacRae TH. Artemin, a diapause-specific chaperone, contributes to the stress tolerance of Artemia franciscana cysts and influences their release from females. J Exp Biol. 2014;217(10):1719–1724. doi: 10.1242/jeb.100081. [DOI] [PubMed] [Google Scholar]
- Kumsta C, Jakob U. Redox-regulated chaperones. Biochemistry. 2009;48(22):4666–4676. doi: 10.1021/bi9003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landino LM, Hasan R, McGaw A, Cooley S, Smith AW, Masselam K, Kim G. Peroxynitrite oxidation of tubulin sulfhydryls inhibits microtubule polymerization. Arch Biochem Biophys. 2002;398(2):213–220. doi: 10.1006/abbi.2001.2729. [DOI] [PubMed] [Google Scholar]
- Liang P, MacRae TH. The synthesis of a small heat shock/α-crystallin protein in Artemia and its relationship to stress tolerance during development. Develop Biol. 1999;207(2):445–456. doi: 10.1006/dbio.1998.9138. [DOI] [PubMed] [Google Scholar]
- Liang P, Amons R, MacRae TH, Clegg JS. Purification, structure and in vitro molecular-chaperone activity of Artemia p26, a small heat shock/α-crystallin protein. Eur J Biochem. 1997;243(1-2):225–232. doi: 10.1111/j.1432-1033.1997.0225a.x. [DOI] [PubMed] [Google Scholar]
- Lin Z, Lei H, Cao P. Expression, purification, and in vitro refolding of soluble tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) Protein Expr Purif. 2007;51(2):276–282. doi: 10.1016/j.pep.2006.07.026. [DOI] [PubMed] [Google Scholar]
- MacRae TH. Molecular chaperones, stress resistance and development in Artemia franciscana. Sem Cell Develop Biol. 2003;14:251–258. doi: 10.1016/j.semcdb.2003.09.019. [DOI] [PubMed] [Google Scholar]
- Mayer M, Buchner J. Refolding of inclusion body proteins. In: Decker J, Reischl U, editors. Molecular diagnosis of infectious diseases. Totowa, (NJ): Humana Press Inc; 2004. pp. 239–245. [Google Scholar]
- Mcauley A, Jacob J, Carl GK, Westland K, Lee HJ, Stephen RB, Rehder D, Gerd RK, David NB, Matsumura M. Contributions of a disulfide bond to the structure, stability, and dimerization of human IgG1 antibody CH3 domain. Protein Sci. 2008;17:95–106. doi: 10.1110/ps.073134408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Funato Y. Regulation of intracellular signalling through cysteine oxidation by reactive oxygen species. J Biochem. 2012;151(3):255–261. doi: 10.1093/jb/mvs006. [DOI] [PubMed] [Google Scholar]
- Monsellier E, Bedouelle H. Quantitative measurement of protein stability from unfolding equilibria monitored with the fluorescence maximum wavelength. Protein Eng Des Sel. 2005;18(9):445–456. doi: 10.1093/protein/gzi046. [DOI] [PubMed] [Google Scholar]
- Niforou K, Cheimonidou C, Trougakos IP. Molecular chaperones and proteostasis regulation during redox imbalance. Redox Biol. 2014;2:323–332. doi: 10.1016/j.redox.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocker Y, Stone JT. The catalytic versatility of erythrocyte carbonic anhydrase. III. Kinetic studies of the enzyme-catalyzed hydrolysis of p-nitrophenyl acetate. Biochemistry. 1967;6(3):668–678. doi: 10.1021/bi00855a005. [DOI] [PubMed] [Google Scholar]
- Qiu Z, MacRae TH. ArHsp21, a developmentally regulated small heat-shock protein synthesized in diapausing embryos of Artemia franciscana. Biochem J. 2008;411(3):605–611. doi: 10.1042/BJ20071472. [DOI] [PubMed] [Google Scholar]
- Qiu Z, MacRae TH. ArHsp22, a developmentally regulated small heat shock protein produced in diapause-destined Artemia embryos, is stress inducible in adults. FEBS J. 2008;275(14):3556–3566. doi: 10.1111/j.1742-4658.2008.06501.x. [DOI] [PubMed] [Google Scholar]
- Rasti B, Shahangian SS, Sajedi RH, Taghdir M, Hasannia S, Ranjbar B. Sequence and structural analysis of artemin based on ferritin: a comparative study. Biochim Bipphys Acta. 2009;1794(10):1407–1413. doi: 10.1016/j.bbapap.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Robbins HM, Van Stappen G, Sorgeloos P, Sung YY, MacRae TH, Bossier P. Diapause termination and development of encysted Artemia embryos: roles for nitric oxide and hydrogen peroxide. J Exp Biol. 2010;213(9):1464–1470. doi: 10.1242/jeb.041772. [DOI] [PubMed] [Google Scholar]
- Rudolph R, Lilie H. In vitro folding of inclusion body proteins. FASEB J. 1996;10(1):49–56. doi: 10.1096/fasebj.10.1.8566547. [DOI] [PubMed] [Google Scholar]
- Shahangian SS, Rasti B, Sajedi RH, Khodarahmi R, Taghdir M, Ranjbar B. Artemin as an efficient molecular chaperone. Protein J. 2011;30(8):549–557. doi: 10.1007/s10930-011-9359-4. [DOI] [PubMed] [Google Scholar]
- Shirzad F, Sajedi RH, Shahangian SS, Rasti B, Mosadegh B, Taghdir M, Hosseinkhani S. Deletion of extra C-terminal segment and its effect on the function and structure of artemin. Int J Biol Macromol. 2011;49(3):311–316. doi: 10.1016/j.ijbiomac.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Takalloo Z, Sajedi RH, Hosseinkhani S, Asghari SM. Real-time monitoring of artemin in vivo chaperone activity using luciferase as an intracellular reporter. Arch Biochem Biophys. 2016;610:33–40. doi: 10.1016/j.abb.2016.09.016. [DOI] [PubMed] [Google Scholar]
- Takalloo Z, Sajedi RH, Hosseinkhani S, Moazzenzade T. Artemin protects cells and proteins against oxidative and salt stress. Int J Biol Macromol. 2017;95:618–624. doi: 10.1016/j.ijbiomac.2016.11.088. [DOI] [PubMed] [Google Scholar]
- Trinder P. Determination of glucose in blood using glucose oxidase with an alternative oxygen acceptor. Ann Clin Biochem. 1969;6(1):24–27. doi: 10.1177/000456326900600108. [DOI] [Google Scholar]
- Wang CYJ, Huo L, Wang L, Feng W, Wang CC. Human protein-disulfide isomerase is a redox-regulated chaperone activated by oxidation of domain a′. J Biol Chem. 2012;287(2):1139–1149. doi: 10.1074/jbc.M111.303149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters MA, Fan SW, Haworth NL. Disulfides as redox switches: from molecular mechanisms to functional significance. Antioxid Redox Signal. 2010;12(1):53–91. doi: 10.1089/ars.2009.2510. [DOI] [PubMed] [Google Scholar]
- Zavialov A, Benndorf R, Ehrnsperger M, Zav’yalov V, Dudich I, Buchner J. The effect of the intersubunit disulfide bond on the structural and functional properties of the small heat shock protein Hsp25. Int J Biol Macromol. 1998;22(3-4):163–173. doi: 10.1016/S0141-8130(98)00014-2. [DOI] [PubMed] [Google Scholar]