Abstract
In animals, commensal microbes modulate various physiological functions, including behavior. While microbiota exposure is required for normal behavior in mammals, it is not known how widely this dependency is present in other animal species. We proposed the hypothesis that the microbiome has a major influence on the behavior of the vinegar fly (Drosophila melanogaster), a major invertebrate model organism. Several assays were used to test the contribution of the microbiome on some well-characterized behaviors: defensive behavior, sleep, locomotion, and courtship in microbe-bearing, control flies and two generations of germ-free animals. None of the behaviors were largely influenced by the absence of a microbiome, and the small or moderate effects were not generalizable between replicates and/or generations. These results refute the hypothesis, indicating that the Drosophila microbiome does not have a major influence over several behaviors fundamental to the animal’s survival and reproduction. The impact of commensal microbes on animal behaviour may not be broadly conserved.
Introduction
During birth, a human newborn is colonized by diverse microbial species, initiating a complex and poorly understood molecular dialogue between the host and symbiotic microbes. Perturbation of this microbial community during early life is believed to disrupt a range of core physiological processes1–10. A key physiological system found to be critically dependent on the early life microbiome is the brain; evidence shows that the microbiome affects brain function by modulating early brain development11–16. For example, germ-free (GF) mice have abnormal function of their hypothalamic-pituitary-adrenal (HPA) axis, an important neurohormonal system; exposing GF mice to microbes during early life is sufficient to normalize HPA axis function15,17. While microbial exposure in later life is insufficient to normalize the behavior of GF mice12,18, normal function is restored by exposing either postnatal pups15,19 or their mothers to microbial colonization16,20. Thus, normal mammalian brain development and function rely on the microbiome. An intact mammalian gut microbiota is also required for normal anxiety-like behaviour and locomotion7,16,21. However, it is unclear whether this requirement is unique to mammals, or is a principle of brain development that applies broadly to other animal clades.
To investigate the microbe-brain relationship in other clades, we examined the role of the microbiome in the vinegar fly (Drosophila melanogaster), a major model organism for the neurobiology of behavior. Drosophila has several experimental advantages as a model for analyzing the microbiome-physiology interaction: it has a simple microbiome of typically only 5–30 taxa22,23; is highly tractable for a range of experimental manipulations; and offers large sample sizes that render high statistical precision. In addition, between mammals and flies, many of the fundamental mechanisms underlying host-microbial dynamics are conserved. Conserved microbe-interacting systems in the two clades include Toll-like receptor-based immune responses24, metabolism25, and insulin-like peptide signaling26,27. Moreover, as with mammals, the Drosophila microbiome influences fly development across generations28,29. While the microbiome clearly modulates Drosophila immunity and development27, much less is known about commensal microbial effects on behavior.
We examined the role of the Drosophila microbiome in four well-characterized behaviors: anxiety-related wall following, locomotion, sleep, and courtship. In each of these behavioral paradigms, we found no substantial differences between GF and control flies. As the first analysis of cross-generational microbe-regulated Drosophila behaviour, this study indicates that the strong microbiome-brain interactions seen in mammals are not generalizable to all behaviors in all animal clades.
Results
Generation of germ-free adults
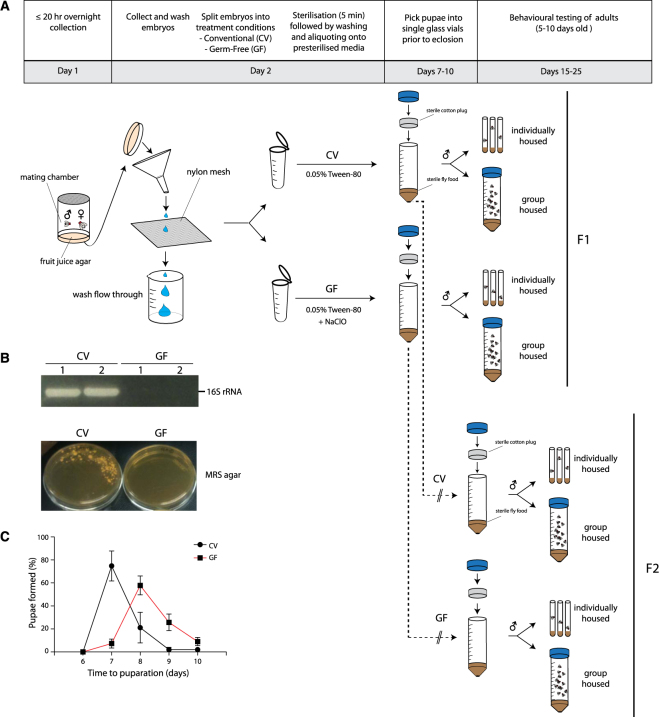
To test whether removal of the parental microbiome is capable of modulating behavioural outcomes in their offspring, we used Wolbachia-free Canton-S flies to generate first generation (F1) GF, second generation (F2) GF, and conventional (CV) flies for behavioural profiling. We first homogenized and plated individual CV flies on MRS agar to characterize bacterial components in the wild-type Drosophila gut. PCR amplification and sequencing of 16 S rRNA gene regions from individual colonies (N = 6) confirmed that previously identified bacterial genera Lactobacillus and Acetobacter were present in CV flies23,30 (Table S1). To generate GF flies, fertilized Canton-S embryos were treated with bleach (Fig. 1A). F1 flies were deprived of microbes during embryogenesis whereas F2 flies were germ-free prior to fertilization. A nutrient-rich medium was chosen to reduce the magnitude of developmental delays as a potential confounding factor in observed phenotypes27. The effectiveness of bacterial removal was confirmed by a negative PCR result for the bacterial 16S rRNA gene and the absence of colonies upon plating of fly homogenates onto agar permissive to the growth of gut bacteria (Fig. 1B). Consistent with previous reports in nutrient rich media, developmental delays (−1.33 days [95CI −1.47, −1.19]) were observed in GF flies, providing additional confirmation that the flies were indeed GF (Fig. 1C)27.
Figure 1.
Drosophila embryo bleaching yields GF flies. (A) Preparation of GF and CV flies prior to behavioural phenotyping. Prior to eclosion, pupae were picked into fresh vials and housed individually or in groups of 20. A full methodological description is provided in Methods. (B) Validation of adult GF fly production by PCR of the 16S rRNA gene on fly homogenate (upper panel) in duplicate and culturing fly homogenate on MRS media (lower panel). A negative PCR result for the 16S rRNA gene and negative growth on MRS agar were used to confirm the absence of microbes. (C) F1 GF flies were monitored for developmental delays by measuring the number of days it took for GF flies to form pupae. Error bars represent average % pupae formation +/− S.E.M. Bleaching of Drosophila embryos took 19% longer to form pupae post bleach treatment (−1.33 days [CI95 −1.47, −1.19], P = 1.0 × 10−4, N = 112, 213].
Individually housed GF and CV flies display similar wall-following behavior
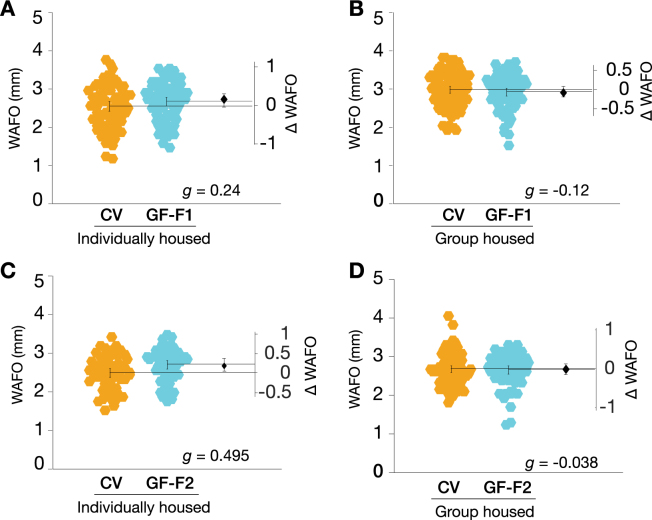
The microbiome has been shown to influence rodent behaviour, including locomotion and anxiety7,12,14–16. Drosophila wall following (WAFO) is governed by similar genetic mechanisms as rodent defense behaviours, and is an anxiety-related behaviour31. We hypothesized that flies without a microbiota, like rodents, would display decreased defense behaviour; we also proposed that this low-anxiety phenotype would be counteracted by social isolation31. We tested WAFO in two generations of GF and CV flies, raised in groups or isolation. Comparison of GF flies with control animals refuted this hypothesis: in three of four conditions, there were only trivial differences in WAFO between GF and CV flies (Fig. 2A,B,D). In the fourth condition, individually housed F2 GF flies exhibited a slight increase in WAFO relative to the CV controls (Fig. 2C), though the effect size was small (g = 0.495). Thus, the Drosophila microbiota does not have a major influence on anxiety-like behaviour.
Figure 2.
GF flies exhibit mostly trivial changes in wall following relative to CV controls. (A) F1 individually housed GF flies did not exhibit substantially altered WAFO activity relative to CV counterparts (g = 0.236, P = 0.152, N = 75, 74). (B) F1 group housed GF flies had largely unaltered WAFO activity (g = −0.118, P = 0.521, N = 96, 80). (C) F2 individually housed GF flies exhibited a small elevation in WAFO activity (g = 0.495, P = 8.829e-03, N = 57, 56), whereas (D) F2 group housed GF flies exhibited essentially no difference in WAFO activity from CV controls (g = –0.038, P = 5.125e-01, N = 89, 66).
Locomotion is mildly elevated in second-generation GF flies
Tracking data from the WAFO assay was also analyzed to determine walking speed over a 10 min interval. In F1 adults, the removal of microbes had only trivial effects on locomotor activity; this was true for both socially naive and group housed GF flies (Fig. 3A,B). However, the F2 GF flies exhibited moderately elevated locomotor activity relative to CV controls (Fig. 3C,D); this was the case for both single- and grouped-housed animals. These data indicated that the microbiota plays no detectable role in modulating Drosophila brief-interval locomotor activity in the first generation, but suggested that the second generation was moderately hyperactive.
Figure 3.
Removal of Drosophila microbiota mildly increases locomotion in the F2 generation. (A) The average locomotion speed of F1 germ-free (GF) singly housed flies was elevated relative to conventional (CV) control animals (+0.09 mm/s [95CI −0.03, +0.21], g = 0.237, P = 1.002e-01, N = 75, 74). (B) F1 group housed GF flies displayed an increase in locomotion relative to CV controls (+0.07 mm/s [95CI −0.05, +0.18], g = 0.17, P = 3.357e-01, N = 96, 80). (C) Individually housed F2 GF flies walked 24.5% faster than controls (+0.31 mm/s [95CI + 0.15, +0.47], g = 0.608, P = 1.08 × 10−3, N = 57, 56). (D) Group housed F2 GF flies walked 36.5% faster than controls (+0.30 mm/s [95CI + 0.18, +0.41], g = 0.683, P = 8.5 × 10−5, N = 89, 66).
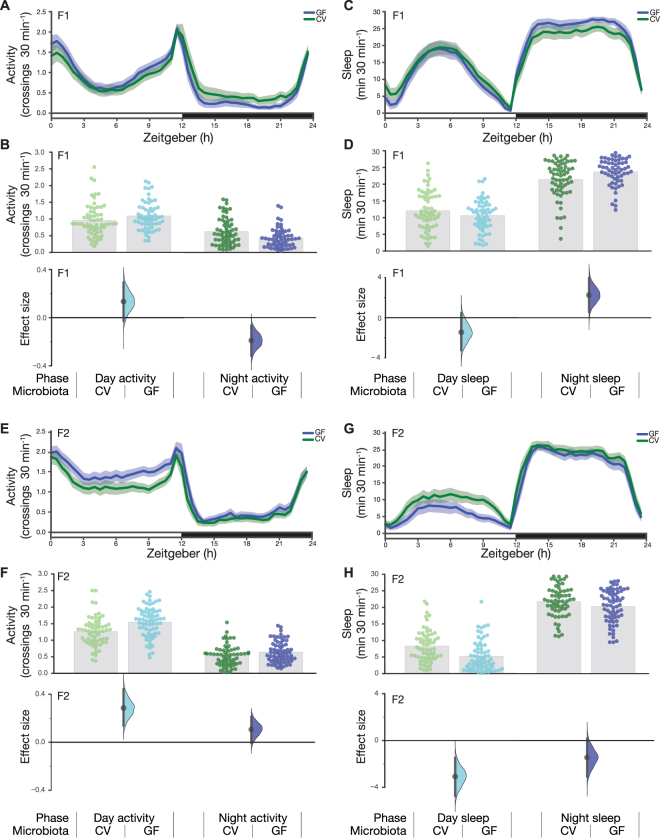
Microbe removal has little impact on activity or sleep
The increased activity observed in the 10 min WAFO assay led us to investigate hyperactivity in F2 GF flies. To examine this phenotype, we recorded activity over six days in both generations. Congruent with the WAFO speed data, the GF F1 flies had only a modest elevation in daytime activity (Fig. 4A,B); during the night, GF F1 flies were moderately less active than CV flies and slept slightly more (Fig. 4C,D). Concordant with the WAFO assay, F2 GF flies displayed moderately elevated daytime activity, along with a small increase in night time activity relative to controls (Fig. 4E, F); similarly, F2 GF flies slept less than controls (Fig. 4G,H).
Figure 4.
Germ-free flies do not have activity or sleep defects. (A) Activity measured as photobeam crossings using the Drosophila Activity Monitoring System (DAMS) assay for group housed F1 CV and GF flies. (B) In the day, GF flies made an average of 1.09 crossings/min (N = 59), while CV flies made an average of 0.96 crossings/min (N = 63), an increase of 0.13 [95CI −0.03, 0.29] (P = 0.11). At night, GF flies showed a slight decrease of −0.19 crossings/min [95CI −0.32, −0.07] (P = 4.16 × 10−3) (C) Sleep in group housed F1 CV and GF flies. (D) In the day, GF flies slept for 10.61 minutes per 30 min (N = 59), while CV flies slept an average of 12.06/30 min (N = 63), a mean sleep decrease of −1.44 minutes [95CI −3.25, 0.46] (P = 0.14). At night, GF flies showed a mild increase in sleep, with a mean difference of +2.26/30 min [95CI 0.56, 3.98] (P = 0.01). (E) Activity measured as beam crossings in the DAM assay for group housed F2 CV and GF flies. (F) GF flies are slightly more active than CV flies. During the day, CV flies made an average of 1.26 crossings/min (N = 70), while GF flies made an average of 1.55 crossings/min (N = 67), an increase of +0.29 [95CI 0.14, 0.44] (P = 3.81 × 10−4). At night, GF flies showed an minor increase of +0.11 crossings/min [95CI −0.007, 0.21] (P = 0.059). (G) Sleep in group housed F2 CV and GF flies. (H) During the day, GF flies slept for 5.23/30 min (N = 70), while CV flies slept an average of 8.30 min (N = 67), a mean sleep decrease of −3.07 minutes [95CI −4.62, −1.39] (P = 3.08 × 10−4). At night, GF flies showed a mild decrease in sleep, with a mean difference of −1.45 min per 30 min [95CI −3.09, 0.18] (P = 0.089).
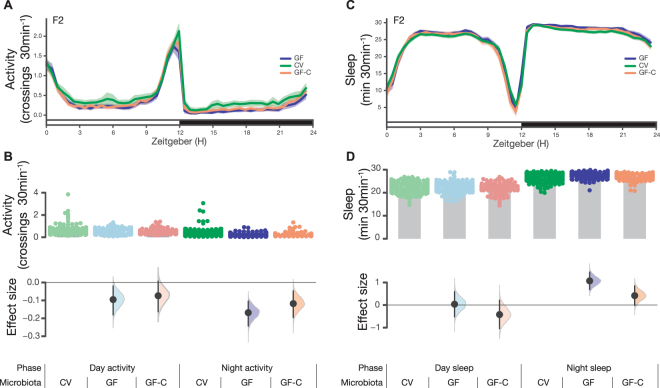
We aimed to generalize these findings to a second Drosophila stock, and to determine if the hyperactivity was reversible with microbiota reintroduction. However, in the replicate experiment, the elevated-activity phenotype was not reproduced. Indeed, relative to controls, the second batch of F2 GF flies were slightly less active (Fig. 5A,B). This difference between batches was also reflected in the sleep estimates (Figs 4G,H, 5C,D). Additionally, microbiota reintroduction had almost no effect (Fig. 5). To our knowledge, neither batch of experiments was flawed: sampling error and heterogeneity are typical causes of such differences between replicates32–34. Thus, taken together, these results refute the hypothesis that the fly microbiota has a major influence on either Drosophila locomotor activity or sleep.
Figure 5.
Conventional, germ-free, and colonized germ-free flies display similar sleep. (A) Activity measured as photobeam crossings using the Drosophila Activity Monitoring System (DAMS) assay for group housed F2 CV, GF, and GF-C flies. (B) In the day, CV, GF, and GF-C flies made an average of 0.60 crossings/min (N = 159), 0.51 crossings/min (N = 115), and 0.53 crossings/min (N = 90) respectively. Relative to CV, GF flies showed a decrease of −0.09 [95CI −0.18, −0.02] (P = 0.04). Relative to CV, GF-C flies showed a decrease of 0.07 [95CI −0.16, 0.01] (P = 0.15). At night, CV, GF, and GF-C flies made an average of 0.38 crossings/min (N = 159), 0.21 crossings/min (N = 115) and 0.27 crossings/min (N = 90) respectively. Relative to CV, GF flies showed a decrease of −0.17 [95CI −0.24, −0.10] (P = 0.00002). Relative to CV, GF-C flies showed a decrease of −0.12 [95CI −0.20, −0.05] (P = 0.009). (C) Sleep estimates of group-housed F2 CV, GF, and GF-C flies. (D) In the day, CV, GF, and GF-C flies slept for 22.30 minutes per 30 min (N = 159), 22.34 minutes per 30 min (N = 115), and 21.88 minutes per 30 min (N = 90) respectively. Relative to CV, GF flies showed a mean sleep increase of 0.04 minutes [95CI −0.52, 0.59] (P = 0.9). Relative to CV, GF-C flies showed a mean sleep decrease of −0.42 minutes [95CI −1.05, 0.21] (P = 0.17). At night, CV, GF, and GF-C flies slept for 26.54 minutes per 30 min (N = 159), 27.61 minutes per 30 min (N = 115) and 26.95 minutes per 30 min (N = 90) respectively. Relative to CV, GF flies showed a mean sleep increase of 1.07 minutes [95CI 0.69, 1.46] (P = 0.0000004). Relative to CV, GF-C flies showed a mean sleep increase of 0.42 minutes [95CI −0.01, 0.85] (P = 0.07).
Conventional females are slightly more attractive than germ-free females
Previous reports have described microbiota-dependent mating preferences35,36, though this is controversial37. In these studies, dietary shifts were used to perturb microbiota compositions over multiple generations. In contrast, a recent report failed to detect a role for the Drosophila microbiota in mating preferences after dietary shifts and antibiotic exposure37. To test whether direct microbe removal impacts microbiota-dependent attractiveness, conventional male flies were placed in a chamber together with decapitated GF and CV female bodies, and their courtship was monitored for 60 minutes (Fig. 6A). Courting wildtype males had a modest preference for CV females over GF females; this was true for both F1 (median proportion = 0.714 [95%CI 0.583; 0.855], P = 0.211) and F2 females (median proportion = 0.699 [95% CI 0.387; 0.945], P = 0.073). These results demonstrate that the Drosophila microbiota has just a minor effect on the attractiveness of either F1 or F2 females.
Figure 6.
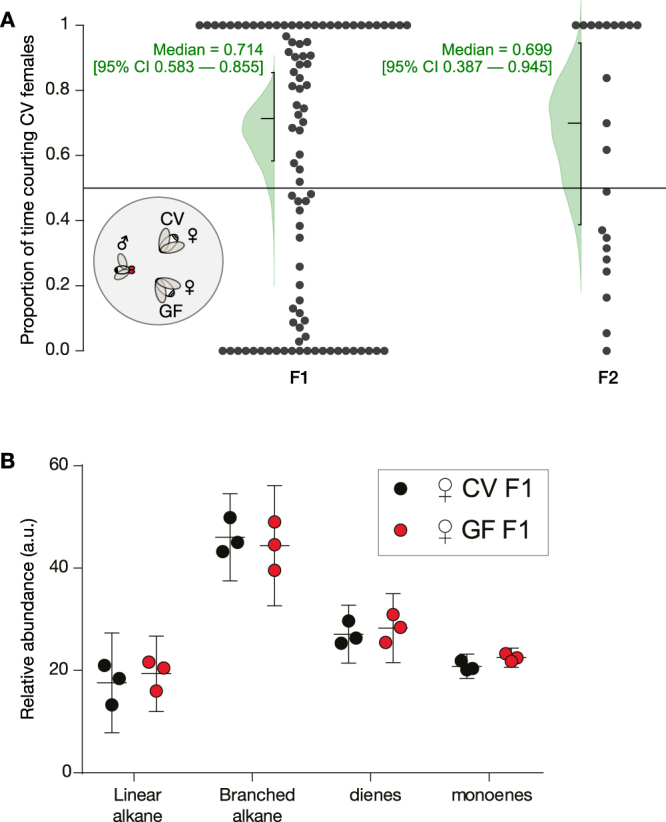
Germ-free female bodies are mildly more attractive than conventional female bodies. (A) Inset: A two-way choice courtship assay where conventionally raised (CR) males from fly stocks were aspirated into a chamber containing CV and GF female bodies. Main axes: 2-choice courtship assay of CV vs. GF female over a 60 min period. Only trials displaying courtship ≥ 1 min were analysed. Data was normalized by removing all non-courtship related behaviour throughout the 60 minute period. F1 (n = 92) and F2 (n = 21) courtships were analysed separately. The medians and their confidence intervals are given in green text; the green curves are bootstrap-estimated distributions of the medians. (B) Relative abundance of CHC chemical classes determined by GC/MS. Data represent the average relative abundance (middle line) and confidence intervals (top and bottom lines) of 3 replicate sets per condition, with each set consisting of 8 females. Relative abundance (arbitrary units, a.u.) is calculated by dividing the area under each CHC peak by the area under the hexacosane internal standard peak. Mean differences between CV and GF relative abundance: linear alkane −1.797 (95CI −8.681, +5.087, P = 0.9239), branched alkane +1.651 (95CI −5.232, +8.535, P = 0.9427), diene −1.174 (95CI −8.058, +5.71, P = 0.9830), monoene −1.71 (95CI −8.594, +5.173, P = 0.9355).
GF females have a normal cuticular hydrocarbon profile
Female Drosophila attractiveness depends on the types of lipids on the cuticle38. To see if there were differences that could explain the mild preference for CV females, we examined F1 female cuticular hydrocarbon (CHC) production with gas chromatography/mass spectrometry (GC/MS). We detected only trivial differences in the overall amount of CHCs between CV and GF flies (Fig. 6B). Relative abundances of individual CHCs are provided in Supplementary Figure S1 and Supplementary Table S2.
Discussion
Here, we report the first examination of microbiota dependant behaviours in Drosophila across generations. We observed that microbiota removal had minor (and/or inconsistent) effects on anxiety-related behaviour7,16,20,39, locomotion16,21, sleep, and sexual attractiveness (Table 1). In summary, these findings suggest that the microbiota-gut-brain axis does not have a strong, consistent influence over several important Drosophila behaviours: defense, motor activity, and reproduction.
Table 1.
Summary of effect sizes for all behavior assays.
| Figure | Assay | F1 GF | F2 GF | F2 GF-C |
|---|---|---|---|---|
| 2 | WAFO - IH | 0.24 | 0.495 | ND |
| 2 | WAFO - GH | −0.12 | −0.04 | ND |
| 3 | Locomotion - IH | 0.24 | 0.61 | ND |
| 3 | Locomotion - GH | 0.17 | 0.68 | ND |
| 4 | Activity - Day | 0.29 | 0.64 | ND |
| 4 | Activity - Night | −0.53 | 0.33 | ND |
| 4 | Sleep - Day | −0.27 | −0.65 | ND |
| 4 | Sleep -Night | 0.46 | −0.30 | ND |
| 5 | Activity - Day | ND | −0.26 | −0.19 |
| 5 | Activity - Night | ND | −0.53 | −0.35 |
| 5 | Sleep - Day | ND | 0.02 | −0.18 |
| 5 | Sleep - Night | ND | 0.64 | 0.23 |
| 6 | Courtship | 0.71 md | 0.70 md | ND |
Summary of effect sizes for germ-free flies assayed for 13 behavioral metrics in 26 experiments. All effect sizes are Hedges’ g, except for courtship preference, which is a median proportion (md). Of the 24 Hedges’ g values, seven are moderate effect sizes; none are reproduced between replicate (Figs 5 vs. 4) or generation (F2 GF vs. F1 GF). IH = individually housed; GH = group-housed; GF-C = colonized germ-free flies.
A range of studies have shown that germ-free rodents display abnormal defense behaviours, establishing that the indigenous mammalian microbiota is required for normal anxiety7,8,15,16,39,40. Surprisingly, experiments in two generations of germ-free flies indicate that this interaction is either minimal or absent in lab-raised Drosophila. Similarly, while there is work showing that the microbiota affects rodent activity levels16, the present study shows that this phenotype is not generalizable to flies. All the present experiments used sample sizes chosen to yield precise estimates of moderate effect sizes or larger (N = ≥56). If the microbiota influence on activity was moderate or large, it would have been measurable across generations and generalizable across conditions in a reproducible manner; this was not the case. Different stocks of the same laboratory strains can have differing microbiomes41,42; while this could be used to explain the between-laboratories differences, it cannot explain the between-assays and between-generations discrepancies. Moreover, independent experiments examining sleep in germfree flies also found only trivial effects43. Together, the varying, moderate-to-trivial effect sizes in several locomotion assays best support a parsimonious interpretation: that the microbiota’s influence over fly activity levels is relatively minor.
Previous work relating the microbiota to Drosophila courtship used antibiotics or diet-induced shifts to assess its role in mating preferences35,36. Our bleach-sterilization protocol removed the microbiota directly, allowing us to quantify the influence of the microbiota on female attractiveness. Our findings show that males court CV females with only a mild preference over GF females. This outcome contrasts with earlier reports demonstrating a major role for the microbiota in fly mate choice35,36. While it remains plausible that the use of CR males directly from laboratory stocks (instead of CV males) could have masked mating preferences between animals with a more similar microbiota complement, the current observations are in line with recent work finding that microbial changes leave mate choice unaltered37.
Although many processes are conserved between mammals and flies, the present findings suggest the microbiota-gut-brain axis is not. There are at least three possible explanations for this difference. First, as rodent neuroscience uses small sample sizes and is affected by publication bias, these statistical factors may have amplified the rodent microbe–brain effect sizes that have been reported34,44,45. Second, there are important differences between mammals and flies, for example, the brain’s relative size: it is plausible that the increased mammalian brains’ higher nutritional demand could mean that mammals rely more critically on microbial-derived nutrients46. Third, it is worth considering that common lab-type fly strains have lost much of their original microbial diversity30; it may be worthwhile examining how wild-type microbial strains influence (recently captive) wild-type fly behaviour42.
In conclusion, although the microbiota impacts diverse aspects of Drosophila biology including immunity, metabolism, and development27,47–49, the present evidence indicates that the microbiota influence on Drosophila behaviour is minor17,50.
Materials and Methods
Drosophila husbandry
An isogenic Wolbachia-free Drosophila melanogaster Canton-S strain was used in all experiments. The absence of Wolbachia spp. was verified by PCR using wsp 81 F (5′-TGGTCCAATAAGTGATGAAGAAAC-3′) and wsp 691 R (5′-AAAAATTAAACGCTACTCCA-3′)51 on crude DNA extract of pooled homogenate from five flies. Flies were raised on autoclaved cornmeal-dextrose-yeast agar food (IMCB52 and EMBL53 recipe available at Zenodo) at 25 °C under a 12:12 hr day/night cycle.
Materials and media preparation
Bacteria isolated from D. melanogaster were cultivated on DeMan, Rogosa, and Sharpe (MRS) medium (Sigma cat. # 69966) containing 1.5% bacteriological agar. Embryos were sterilized using a 1:1 diluted 5% sodium hypochlorite (NaClO) bleach solution (FairPrice) and 0.05% Tween-80 (Sigma cat. # P8074). All solutions used were sterilized by autoclaving at 121 °C, 100 kPa for 15 minutes or passing through a 0.22 μm filter, where indicated, prior to use.
Characterization of bacterial taxa in Drosophila
Bacterial colonies were isolated from the guts of Drosophila Canton-S on MRS agar and grown at 30 °C. Isolates were identified by sequencing the 16S rRNA PCR product generated using 8FE (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492 R (5′-GGMTACCTTGTTACGACTT-3′) primers. PCR products were sequenced using the 8FE primer and were blasted against the National Center for Biotechnology Information database to assign bacterial identity, results are summarised in Table S1.
Generation of germ-free and colonized Drosophila
F1 generation germ-free flies were prepared similarly to a previously described method54. Briefly, eggs deposited overnight onto fruit juice agar were pooled then split into low protein binding 1.5 mL microfuge tubes with a ~100 μL final egg bed volume. Subsequent steps were performed aseptically in a Class II biological safety cabinet. Eggs were washed twice with 1 mL of 0.05% Tween-80. Chorions were removed by exposing to 2.5% hypochlorite and 0.05% Tween-80 with gentle inverting of tubes for 5 min. Control conventional (CV) flies were prepared in parallel by exposure to 0.05% Tween-80 only. Eggs were washed twice with 1 mL of 0.05% Tween-80 solution followed by resuspension in 1 mL of 0.05% Tween-80 solution. 100 μL aliquots of treated eggs were then dispensed onto 10 mL of autoclaved fly food in 50 mL Falcon™ tubes. Pupae were isolated into autoclaved glass vials containing 2 mL fly food and capped with sterile cotton buds. Sexes were determined at eclosion and male experimental flies were either housed individually or in groups of 20. F2 generation GF and CV flies were prepared by transferring F1 adults to autoclaved food aseptically and F2 pupae were collected in the same way as F1. Quality control of GF fly preparation was performed at the end of each generation cycle with 5 adult flies. The absence of bacteria was verified by PCR for the 16S rRNA gene using 8FE (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492 R (5′-GGMTACCTTGTTACGACTT-3′) and plating fly homogenates onto MRS agar48,54.
For re-colonization of germ-free flies, monocultures of Acetobacter pomorum and Lactobacillus sp. were grown overnight at 30 °C with rotation in 2 mL of MRS broth. 50 μL aliquots of overnight culture from each strain was used to directly inoculate the surface of pre-autoclaved fly food inside 50 mL Falcon™ tubes. Dechorionated embryos were then deposited on top of the inoculated fly food and capped with sterilized cotton buds.
Gas chromatography/mass spectrometry analysis
5 day old female D. melanogaster were anaesthetized on ice then collected in 1.8 mL glass vials (Wheaton, New Jersey, USA). 120 μL of hexane containing 10 μg/mL hexacosane (Sigma-Aldrich #241687) as an internal standard was added to triplicate vials (Nflies = 8 per vial) and incubated at room temperature for 20 minutes. 100 μL aliquots were transferred to fresh glass vials and were evaporated using a gentle stream of nitrogen. Samples were stored at −20 °C until analysis.
Analysis by gas chromatography/mass spectrometry (GC/MS) was performed with a QP2010 system (Shimadzu, Kyoto, Japan) equipped with a DB-5 column (5% Phenyl-methylpolysiloxane column; 30 m length, 0.25 mm ID, 0.25 μm film thickness; Agilent Technologies, CA, USA). Ionization was achieved by electron ionization (EI) at 70 eV. One microliter of the sample was injected using a splitless injector. The helium flow was set at 1.9 mL/min. The column temperature program began at 50 °C, increased to 210 °C at a rate of 35 °C/min, then increased to 280 °C at a rate of 3 °C/min. A mass spectrometer was set to unit mass resolution and 3 scans/sec, from m/z 37 to 700. Chromatograms and mass spectra were analysed using GCMSsolution software (Shimadzu).
The relative abundance of each CHC species was calculated by dividing the area under the peak by the area of the hexacosane internal standard peak. The total area for each CHC chemical class (linear alkane, branched alkane, diene, or monoene) was calculated and the relative abundance from GF flies was then compared with that of the control CV flies using two-way ANOVA (GraphPad Prism 5, GraphPad Software Inc., CA, USA).
Courtship choice
Courtship assays were performed using adult flies aged 7–10 days old, corresponding to sexual maturity. A socially naïve conventionally raised (CR) male was aspirated into a courtship chamber (35 × 10-mm) at 23.3 °C and 60% humidity containing one of each freshly decapitated GF female and a CV female 10–15 mm apart. Courtship behaviour (orienting, wing extension, wing vibration, or attempted copulation towards either female) was observed and recorded for 60 min. Courtship choice was calculated by taking a ratio of the amount of time spent by the male displaying courtship behaviours toward one female target to the total time the male spent courting either target. Trials with courtship activity lasting less than 1 min were omitted from analysis.
Wall-following behaviour
The wall-following assay was performed as previously described31. Briefly, for each behavioural assay, male flies were anesthetized in an ice-chilled vial for 30 seconds before being placed individually into an arena with forceps. The arena array was covered with an anti-reflection glass sheet (Edmund Optics, NJ, USA) and placed in an incubator. The array was lit from the sides by white LED microscope lamps (Falcon Illumination, Malaysia). To image the flies, an AVT Guppy F-046B CCD camera (Stemmer Imaging, Germany) equipped with a 12 mm CCTV-type lens was positioned above the behavioural arenas and connected to a computer via an IEEE 1394 cable. Flies were allowed to freely explore the arena during a 10-min test session. Flies’ positions were determined from a live video feed with CRITTA, custom software written in LabVIEW (National Instruments, U.S.A.), which extracts centroid x–y position data and records it to a binary file for offline analysis in MATLAB. Wall following (mean distance from the center of the chamber in mm) and mean speed (mm/sec) were calculated with custom scripts in MATLAB. The differences between control and experimental animal behaviors were reported as effect sizes (standardized mean differences, Hedges’ g). Hedges’ g and its bootstrapped 95% confidence interval were calculated with the Measurements of Effect Size toolbox (MES) in MATLAB55. Hedges’ g is a variant of Cohen’s d standardized effect size that uses pooled SD and adjusts for bias56.
Activity and sleep assays
Group-housed male flies were placed in sterilized 65 mm glass tubes containing sterilized fly food on one end. The experiments were conducted at 12:12 hour light-dark cycle for a period of 6 days in an environment-controlled incubator (25 °C, 60% humidity). The first round of experiments were conducted at IMCB, the second at the Theodor-Boveri Institute. Fly activity was measured by recording the number of photobeam crossings in 1-minute bins using the Drosophila Activity Monitoring System (DAMS, Trikinetics, MA, USA). To generate the activity plots, six days of beam-crossing data were averaged into one 24-h interval and binned by 30 min. Then the binned activity data for the control and experimental flies were averaged separately, and represented as a line plot with the 95% confidence intervals. Sleep analysis was performed in Python using the pandas (McKinney 2011) and NumPy packages (Van Der Walt et al. 2011). Fly beam crossing data from the DAMS experiments were imported to the software and sleep events were identified as any period where the flies were inactive for at least 5 consecutive minutes (Shaw et al. 2000). Flies that did not move for 24 consecutive hours or more were considered dead and removed from the dataset. Then, the six-day sleep data was averaged into one day and binned by 30 minutes. The results were represented as a line plot along with the 95% confidence intervals. Mean sleep duration for the individual flies were represented with the bootstrapped 95% confidence intervals57.
Behavioral statistics and data analysis
For behavior data, estimation methods were used exclusively33: no significance testing was conducted, and P values were reported pro forma only. Confidence intervals (95%) were calculated via bootstrap methods (10,000 samples taken)57, and adjusted with bias correction and acceleration57. A smoothed bootstrap was used for the median bootstrap calculations. The analysis was performed and visualized either in Python using the seaborn58,59 and scikits-bootstrap packages60. Several descriptors were used to describe effect size ranges: trivial (0 < g < 0.2); small (0.2 < g < 0.5); moderate (0.5 < g < 0.8); and large (g > 0.8)32. Sample sizes are reported as the N of animals.
Data availability
All data generated or analyzed during this study are included in this published article.
Electronic supplementary material
Acknowledgements
We thank Lisa Maier, Victoria Hewitt and Florian Szardenings for constructive criticism of this manuscript. We thank Shimadzu Asia Pacific for generously providing access to the GC-MS instrument. FM, JYC and ACC were supported by grants MOE-2013-T2-2-054 from the Ministry of Education; JH was supported by the A*STAR Scientific Scholars Fund. TT was supported by a Singapore International Graduate Award from the A*STAR Graduate Academy. ACC was supported by grants 1231AFG030 and 1431AFG120 from the A*STAR Joint Council Office. FM, JYC, TT and ACC were supported by a Biomedical Research Council block grant to the Institute of Molecular and Cell Biology and by a Ministry of Health block grant to Duke-NUS Medical School. JYY was supported by the Singapore National Research Foundation (grant NRF-RF2010-06). JS and SP were supported by grants from LKC School of Medicine SUG, Swedish Grant VR and SCELSE.
Author Contributions
Conceptualization: J.S. and J.Y.Y.; Methodology: J.S. (germ-removal methods); Software: J.H. and T.T. (Python) and F.M. (Matlab); Investigation: J.S. and S.H.N. (germ removal, courtship), D.R. (sleep), F.M. (locomotion, wall following), J.Y.C. (germ removal, sleep), and Y.N.C. (GC-MS analysis); Data Analysis: J.H. (courtship, sleep), T.T. (sleep), J.Y.C. (sleep) and F.M. (locomotion, WAFO); Writing – Original Draft: J.S.; Writing – Revision: J.S., J.Y.Y., A.C.C., F.M., S.P., C.F.H.; Visualization: J.S., F.M., J.H., T.T. and J.Y.Y.; Supervision: A.C.C. and J.Y.Y.; Project Administration: A.C.C. and J.Y.Y.; Funding Acquisition: A.C.C., S.P., C.F.H. and J.Y.Y.
Competing Interests
The authors declare no competing interests.
Footnotes
Joel Selkrig and Farhan Mohammad contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-28764-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Joanne Y. Yew, Email: jyew@hawaii.edu
Adam Claridge-Chang, Email: claridge-chang.adam@duke-nus.edu.sg.
References
- 1.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016;16:341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrieta M-C, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015;7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 3.Russell SL, et al. Perinatal antibiotic-induced shifts in gut microbiota have differential effects on inflammatory lung diseases. J. Allergy Clin. Immunol. 2015;135:100–109. doi: 10.1016/j.jaci.2014.06.027. [DOI] [PubMed] [Google Scholar]
- 4.Sonnenburg JL, Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsiao EY, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stilling RM, Bordenstein SR, Dinan TG, Cryan JF. Friends with social benefits: host-microbe interactions as a driver of brain evolution and development? Front. Cell. Infect. Microbiol. 2014;4:147. doi: 10.3389/fcimb.2014.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sampson TR, Mazmanian SK. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe. 2015;17:565–576. doi: 10.1016/j.chom.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selkrig J, Wong P, Zhang X, Pettersson S. Metabolic tinkering by the gut microbiome: Implications for brain development and function. Gut Microbes. 2014;5:369–380. doi: 10.4161/gmic.28681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharon G, Sampson TR, Geschwind DH, Mazmanian SK. The Central Nervous System and the Gut Microbiome. Cell. 2016;167:915–932. doi: 10.1016/j.cell.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fung TC, Olson CA, Hsiao EY. Interactions between the microbiota, immune and nervous systems in health and disease. Nat. Neurosci. 2017;20:145–155. doi: 10.1038/nn.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goyal MS, Venkatesh S, Milbrandt J, Gordon JI, Raichle ME. Feeding the brain and nurturing the mind: Linking nutrition and the gut microbiota to brain development. Proc. Natl. Acad. Sci. USA. 2015;112:14105–14112. doi: 10.1073/pnas.1511465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster, J. A. & McVey Neufeld, K.-A. Gut–brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 36, 305–312 (2013/5). [DOI] [PubMed]
- 13.Ogbonnaya ES, et al. Adult Hippocampal Neurogenesis Is Regulated by the Microbiome. Biol. Psychiatry. 2015;78:e7–9. doi: 10.1016/j.biopsych.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 14.Borre YE, et al. Microbiota and neurodevelopmental windows: implications for brain disorders. Trends Mol. Med. 2014;20:509–518. doi: 10.1016/j.molmed.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Clarke G, et al. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol. Psychiatry. 2013;18:666–673. doi: 10.1038/mp.2012.77. [DOI] [PubMed] [Google Scholar]
- 16.Diaz Heijtz R, et al. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. USA. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sudo N, et al. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J. Physiol. 2004;558:263–275. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neufeld K-AM, Kang N, Bienenstock J, Foster JA. Effects of intestinal microbiota on anxiety-like behavior. Commun. Integr. Biol. 2011;4:492–494. doi: 10.4161/cib.15702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desbonnet L, Clarke G, Shanahan F, Dinan TG, Cryan JF. Microbiota is essential for social development in the mouse. Mol. Psychiatry. 2014;19:146–148. doi: 10.1038/mp.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishino R, et al. Commensal microbiota modulate murine behaviors in a strictly contamination-free environment confirmed by culture-based methods. Neurogastroenterol. Motil. 2013;25:521–528. doi: 10.1111/nmo.12110. [DOI] [PubMed] [Google Scholar]
- 21.Montagner A, et al. Hepatic circadian clock oscillators and nuclear receptors integrate microbiome-derived signals. Sci. Rep. 2016;6:20127. doi: 10.1038/srep20127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broderick NA, Lemaitre B. Gut-associated microbes of Drosophila melanogaster. Gut Microbes. 2012;3:307–321. doi: 10.4161/gmic.19896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong CNA, Ng P, Douglas AE. Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ. Microbiol. 2011;13:1889–1900. doi: 10.1111/j.1462-2920.2011.02511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akira S, Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 25.Wong ACN, Vanhove AS, Watnick PI. The interplay between intestinal bacteria and host metabolism in health and disease: lessons from Drosophila melanogaster. Dis. Model. Mech. 2016;9:271–281. doi: 10.1242/dmm.023408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vrieze A, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913–6.e7. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 27.Shin SC, et al. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science. 2011;334:670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- 28.Elgart M, et al. Impact of gut microbiota on the fly’s germ line. Nat. Commun. 2016;7:11280. doi: 10.1038/ncomms11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fridmann-Sirkis Y, et al. Delayed development induced by toxicity to the host can be inherited by a bacterial-dependent, transgenerational effect. Front. Genet. 2014;5:27. doi: 10.3389/fgene.2014.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong AC-N, Chaston JM, Douglas AE. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J. 2013;7:1922–1932. doi: 10.1038/ismej.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohammad F, et al. Ancient Anxiety Pathways Influence Drosophila Defense Behaviors. Curr. Biol. 2016;26:981–986. doi: 10.1016/j.cub.2016.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cumming, G. & Calin-Jageman, R. Introduction to the New Statistics: Estimation, Open Science, and Beyond (Routledge, 2016).
- 33.Claridge-Chang A, Assam PN. Estimation statistics should replace significance testing. Nat. Methods. 2016;13:108–109. doi: 10.1038/nmeth.3729. [DOI] [PubMed] [Google Scholar]
- 34.Mohammad F, et al. Concordance and incongruence in preclinical anxiety models: Systematic review and meta-analyses. Neurosci. Biobehav. Rev. 2016 doi: 10.1016/j.neubiorev.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 35.Sharon G, et al. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2010;107:20051–20056. doi: 10.1073/pnas.1009906107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Najarro MA, Sumethasorn M, Lamoureux A, Turner TL. Choosing mates based on the diet of your ancestors: replication of non-genetic assortative mating in Drosophila melanogaster. PeerJ. 2015;3:e1173. doi: 10.7717/peerj.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leftwich PT, Clarke NVE, Hutchings MI, Chapman T. Gut microbiomes and reproductive isolation in Drosophila. Proc. Natl. Acad. Sci. USA. 2017;114:12767–12772. doi: 10.1073/pnas.1708345114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernández MP, Kravitz EA. Aggression and courtship in Drosophila: pheromonal communication and sex recognition. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2013;199:1065–1076. doi: 10.1007/s00359-013-0851-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neufeld, K. M., Kang, N., Bienenstock, J. & Foster, J. A. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol. Motil. 23, 255–64, e119 (2011). [DOI] [PubMed]
- 40.Bercik, P. et al. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology141, 599–609, 609.e1–3 (2011). [DOI] [PubMed]
- 41.Ren C, Webster P, Finkel SE, Tower J. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 2007;6:144–152. doi: 10.1016/j.cmet.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Chandler JA, Lang JM, Bhatnagar S, Eisen JA, Kopp A. Bacterial communities of diverse Drosophila species: ecological context of a host-microbe model system. PLoS Genet. 2011;7:e1002272. doi: 10.1371/journal.pgen.1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ertekin, D. & Shaw, P. J. Influence of young age microbiome on adult sleep behavior in D. Melanogaster. 10.5281/zenodo.1211175 (2018).
- 44.Sena ES, van der Worp HB, Bath PMW, Howells DW, Macleod MR. Publication bias in reports of animal stroke studies leads to major overstatement of efficacy. PLoS Biol. 2010;8:e1000344. doi: 10.1371/journal.pbio.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Button KS, et al. Power failure: why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci. 2013;14:365–376. doi: 10.1038/nrn3475. [DOI] [PubMed] [Google Scholar]
- 46.Leitão-Gonçalves R, et al. Commensal bacteria and essential amino acids control food choice behavior and reproduction. PLoS Biol. 2017;15:e2000862. doi: 10.1371/journal.pbio.2000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Storelli G, et al. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 2011;14:403–414. doi: 10.1016/j.cmet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 48.Ryu J-H, et al. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- 49.Lee W-J, Brey PT. How microbiomes influence metazoan development: insights from history and Drosophila modeling of gut-microbe interactions. Annu. Rev. Cell Dev. Biol. 2013;29:571–592. doi: 10.1146/annurev-cellbio-101512-122333. [DOI] [PubMed] [Google Scholar]
- 50.Borrelli L, et al. Probiotic modulation of the microbiota-gut-brain axis and behaviour in zebrafish. Sci. Rep. 2016;6:30046. doi: 10.1038/srep30046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou W, Rousset F, O’Neil S. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. Biol. Sci. 1998;265:509–515. doi: 10.1098/rspb.1998.0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laboratories, T. L. S. Recipe for Drosophila media. 10.5281/zenodo.1185451 (2018).
- 53.Lab, C. N.-V. Recipe for EMBL Heidelberg Drosophila media. 10.5281/zenodo.1196651 (2018).
- 54.Ridley EV, Wong AC-N, Westmiller S, Douglas AE. Impact of the resident microbiota on the nutritional phenotype of Drosophila melanogaster. PLoS One. 2012;7:e36765. doi: 10.1371/journal.pone.0036765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hentschke H, Stüttgen MC. Computation of measures of effect size for neuroscience data sets. Eur. J. Neurosci. 2011;34:1887–1894. doi: 10.1111/j.1460-9568.2011.07902.x. [DOI] [PubMed] [Google Scholar]
- 56.Borenstein, M., Hedges, L. V., Higgins, J. P. T. & Rothstein, H. R. Introduction to Meta-Analysis. (John Wiley & Sons, 2011).
- 57.DiCiccio TJ, Efron B. Bootstrap Confidence Intervals. Stat. Sci. 1996;11:189–228. doi: 10.1214/ss/1032280214. [DOI] [Google Scholar]
- 58.Waskom, M. L., Frank, M. C. & Wagner, A. D. Adaptive Engagement of Cognitive Control in Context-Dependent Decision Making. Cereb. Cortex10.1093/cercor/bhv333 (2016). [DOI] [PubMed]
- 59.Waskom, M. et al. seaborn: v0.7. 0 (January 2016). 10.5281/zenodo.45133 (Zenodo, 2016).
- 60.Evans, C. scikits-bootstrap. GitHub (2014). Available at: https://github.com/cgevans/scikits-bootstrap. (Accessed: 4th July 2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.