SUMMARY
Direct reprogramming of fibroblasts into cardiomyocyte-like cells (iCM) holds great potential for heart regeneration and disease modeling and may lead to future therapeutic applications. Currently, application of this technology is limited by our lack of understanding of the molecular mechanisms that drive direct iCM reprogramming. Using a quantitative mass spectrometry-based proteomic approach, we identified the temporal global changes in protein abundance that occur during initial phases of iCM reprogramming. Collectively, our results show systematic and temporally distinct alterations in levels of specific functional classes of proteins during the initiating steps of reprogramming including extracellular matrix proteins, translation factors, and chromatin-binding proteins. We have constructed protein relational networks associated with the initial transition of a fibroblast into an iCM. These findings demonstrate the presence of an orchestrated series of temporal steps associated with dynamic changes in protein abundance in a defined group of protein pathways during the initiating events of direct reprogramming.
Graphical abstract
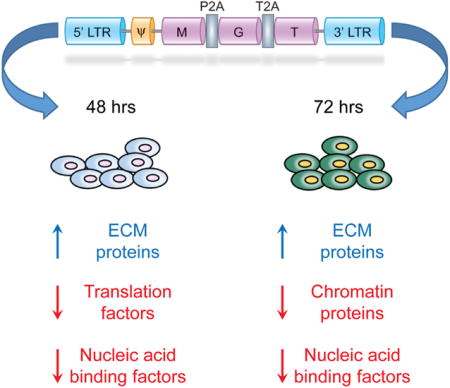
INTRODUCTION
Direct reprogramming of fibroblasts into cardiomyocyte-like cells (iCM) technology holds great promise as an alternative strategy for cardiac regeneration, targeting the abundant population of resident scar-forming fibroblasts in vivo. Alternatively, in vitro conversion of isolated patient cells may provide a personalized screening method for drug efficacy. Currently, the translational application of this technology is limited by our lack of understanding of the molecular mechanisms that drive iCM reprogramming by these transcription factors.
We and others have successfully bypassed the stem/progenitor state to achieve direct conversion of mouse fibroblasts into iCMs in vitro and in vivo, by retroviral overexpression of three cardiac lineage-specific transcription factors, namely Mef2C (M), Gata4 (G), and Tbx5 (T) (Addis et al., 2013; Chen et al., 2012; Christoforou et al., 2013; Ieda et al., 2010; Inagawa et al., 2012; Jayawardena et al., 2012; Muraoka et al., 2014; Nam et al., 2013; Protze et al., 2012; Qian et al., 2012, 2013; Song et al., 2012; Wada et al., 2013). iCMs were observed as early as day 3 based on marker expression and epigenetic landscape re-patterning,(Liu et al., 2016; Wang et al., 2015a) while the electrically coupled, functionally matured iCMs were present ~ 3–4 weeks post-transduction (Qian et al., 2013).
In order to overcome the barrier of low reprogramming efficiency, we have utilized a single-promoter polycistronic construct to induce expression of all three factors in a single cell, allowing for a precise dosage of expression. Additionally, we have shown the stoichiometry of these three lineage-specific transcription factors within this construct, with high expression of Mef2C and low expression of Gata4 and Tbx5 (MGT), leads to increased efficiency and quality of reprogramming (Ma et al., 2015; Mathison et al., 2014; Wang et al., 2015a, 2015b). The underlying mechanisms by which this construct (MGT) governs the direct reprogramming of mouse fibroblasts into iCMs are relatively unknown. Furthermore, we have previously shown transduced fibroblasts underwent a drastic histone mark re-patterning at 72 hr post-transduction of MGT-mediated cardiac reprogramming. This differential re-patterning is correlated with a rapid early activation of the cardiac program and by contrast, a progressive suppression of fibroblast marker genes at later time points (Liu et al., 2016). Therefore, to determine how overexpression of these transcription factors directs the initial phases of cardiac reprogramming, we leveraged our established MGT system and knowledge that the cell fate conversion is likely controlled by molecular events occurring within the first 3 days, to profile proteome alterations that occur 48 and 72 hr post-transduction of MGT. Identification and quantification of the proteome abundance changes that occur 2 and 3 days post-transduction was performed using a multiplexed quantitative mass spectrometry approach. Gene set enrichment analysis of quantified proteins, as a function of protein annotation through evolutionary relationship (PANTHER) protein class, showed systematic and temporally distinct protein abundance alterations in specific functional classes, encompassing extracellular matrix proteins, translation factors, and chromatin-binding proteins. For these selected protein classes, we went on to construct protein relational networks and identify functional networks that are associated with the initial transition of a fibroblast into an iCM. Interestingly, one of the most upregulated proteins was Agrin, a protein recently shown to promote cardiac proliferation and repair (Bassat et al., 2017). Together, our study establishes that early reprogramming of a mouse fibroblast into an iCM occurs through an orchestrated series of temporal steps, and these early temporal events are associated with the differential regulation of protein abundance in a defined set of pathways.
RESULTS AND DISCUSSION
Initiating Events in Re-programming Occur at 48 hr Post-infection
Past studies have suggested that the first events associated with direct reprogramming occur before or at 72 hr post-transduction when cardiac promoters show a reduction of the repressive indicator of transcription trimethylation of histone 3 lysine 27 (H3K27me3) and concomitantly, an enrichment of the activation indicator of transcription, trimethylation of histone 3 lysine 4 (H3K4me3). These events are associated with the expression of the cardiac reporter gene α-MHC-GFP, the subsequent increase in cardiac gene expression and followed shortly thereafter, by the complementary decrease in fibroblast marker expression (Liu et al., 2016). However, no morphological alterations in the phenotype of the fibroblast have been observed over these stages of direct reprogramming. To determine the relative changes in gene expression during the initial stages of reprogramming we conducted qRT-PCR of direct reprogrammed mouse embryonic fibroblasts (MEFs) retrovirally transduced with either the pMX-MGT construct and as a control either pMX-DsRed or pMX-GFP as previously described (Wang et al., 2015a) (Figure S1). Viral infection efficiency was shown to be 81.4% at 48 hr, 78.0% at 72 hr, and 89.2% at 10 days post-infection (Figures S2A–S2G), and the reprogrammed cells were shown to be 13.9% MHC-GFP-positive at 48 hr, 21.9% MHC-GFP-positive at 72 hr, and 31.6% MHC-GFP-positive by day 10 (Figures S3A–S3G and S4A–S4I). Analysis of cardiomyocyte markers showed no change in expression at 24 hr but an increase at 48 hr and 72 hr with a concomitant decrease in fibroblast-associated markers (Figures S5A–S5H). Taken together, these studies would imply that the initiating events in direct re-programming occur at 48 hr post-infection.
Identification and Quantification of MGT Post-transduction Proteomic Profiles
Because only ~ 10% of RNAs that show 2-fold or more change in transcriptional profiling or micro-arrays lead to alterations in protein levels (Bonaldi et al., 2008; Chick et al., 2016; de Sousa Abreu et al., 2009; Nie et al., 2007), we sought to directly measure the quantitative changes of proteins during these initiating events in direct reprogramming using multiplexed tandem mass tag (TMT 6-plex) mass spectrometry-based proteomics. This quantitative multiplexing approach increases throughput and minimizes variability because relative quantification is performed for up to six combined samples in a single analysis (Thompson et al., 2003). Transduced MEFs were harvested at 48 and 72 hr post-transduction. Nuclei were isolated for proteomic analysis (Figure 1A). Proteins from each biological replicate (N = 3) and time point (48 and 72 hr) were individually digested with trypsin, labeled with TMT reagents, and pooled within time points (Figure 1A). Using offline two-dimensional peptide separation paired with online reverse-phase nanoliquid chromatography tandem mass spectrometry, the relative abundance of 3,463 proteins were quantified (minimum of 2 quantified unique spectra). As expected, the distribution of relative protein abundances (MGT/DsRed) was centered at 1:1, and most proteins were quantified with TMT abundance ratios < ±1.3-fold (Figures S6A and S6B). This result confirmed that the between-samples TMT abundance normalization was effective and also that the majority of proteins (~90%) have small to no abundance differences upon MGT expression at these time points. The majority of proteins (3,088) were quantified at both time points (Figure 1B). A positive correlation between protein abundances at 48 and 72 hr was observed, although discordant time-dependent abundance changes were evident (Figure S6C). To compare these MGT-induced, time-dependent proteome changes, we first performed hierarchical clustering of the 3,463 relative protein abundances as a function of time post-transduction (Figure 1C; Table S1). As expected from analysis of the relative abundance distributions (Table S2), the largest cluster of the heatmap also reflected proteins with low to no abundance change (Figure 1C, cluster 7). Several small clusters showed the highest degree of up (#1–#3) and down (#10 and #11) regulation, occurring similarly at both 48- and 72-hr time points. We also observed distinct clusters of proteins that reflected proteins with stronger changes in abundance at 72 hr compared to 48 hr (#6 and #8) and vice versa (#4 and #9), while the remaining cluster (#5) reflected proteins with opposing abundance changes between time points. Overall, these data indicate that the effects of MGT expression are associated with similar degrees of up and downregulation of protein abundance but also that selective modulation of protein abundances often depended on the duration of MGT expression.
Figure 1. Induced Cardiomyocyte Proteomic Analysis.
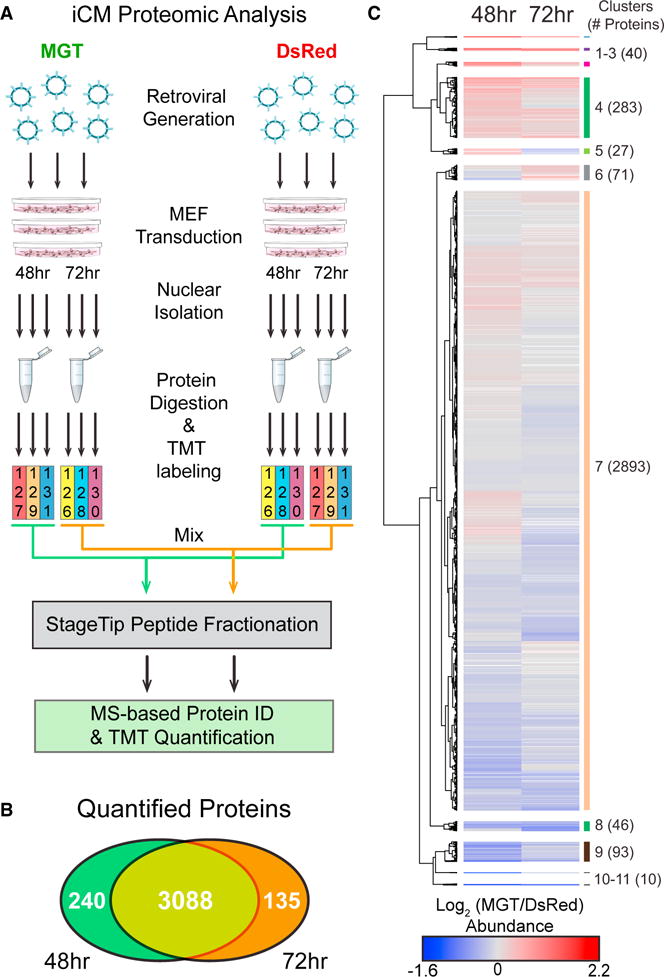
(A) First, retroviruses packaged with either a pMXs-DsRed (Red) construct or a pMXs-MGT (green) construct, were generated. Freshly generated virus was used to transduce MEFs (<p5). Nuclei were isolated from transduced MEFs at 48 and 72 hr post-transduction and then subjected to quantitative TMT MS/MS analysis to identify and quantify proteomic changes.
(B) Comparison of proteins quantified by TMT-based proteomics at 48 and 72 hr post-transduction.
(C) Hierarchical clustering of log2 TMT abundance ratios (MGT/DsRed) visualized as a heatmap showing increased (red), decreased (blue), and unchanged (gray) TMT ratios. Missing values were indicated in white. Heatmap clusters were assigned from a specific dendrogram depth.
Differential Protein Regulation in Response to MGT
Given that these time-dependent proteome changes have a range of magnitudes, particularly more subtle shifts in abundance (Figure 1C), we explored whether analyzing these abundance changes by protein function could identify key proteome signatures associated with MGT expression. Toward this goal, we performed gene set enrichment analysis (GSEA) to identify protein functional classes that may be regulated during reprogramming. This analysis maps protein accessions to genes and assigns the genes to their respective functional ontologies. Then a statistical enrichment test is performed with Bonferroni correction to determine whether the TMT abundance ratios within each ontology classification are statistically up or down-regulated compared to the total distribution of abundance ratios within that same classification. GSEA analysis using the PANTHER protein class ontology (Mi et al., 2016) found 23 protein classes were significantly altered (adjusted p-value <0.05) for at least one of the time points (Table 1). Notably, the number of proteins assigned to each class was similar, supporting that the functional enrichment analysis detected systematic alterations in protein class abundances. The greatest number of proteins were assigned into the nucleic acid binding class (n = 556 at 48 hr and n = 529 at 72 hr post- transduction) and the RNA binding protein class (n = 384 at 48 hr and n = 375 at 72 hr post-transduction), both of which were downregulated (Table 1). Moreover, the most statistically significant protein changes occurred in the protein classes extracellular matrix protein, translation factor, and chromatin/chromatin-binding proteins (Table 1).
Table 1.
PANTHER Protein Classes that Were Differentially Regulated (Corrected p Value <0.05) during Reprogramming
| PANTHER Protein Class | PANTHER ID | Post-reprogramming | |||||
|---|---|---|---|---|---|---|---|
| Annotated Proteins (n) | U/D | p value | |||||
| 48 | 72 | 48 | 72 | 48 | 72 | ||
| Extracellular matrix protein | PC00102 | 70 | 70 | U | U | 8.43E-06 | 6.33E-04 |
| Translation factor | PC00223 | 46 | 46 | D | 1.79E-08 | NS | |
| Chromatin/chromatin-binding protein | PC00077 | 36 | 38 | D | NS | 4.49E-10 | |
| Nucleic acid binding | PC00171 | 556 | 529 | D | D | 1.25E-03 | 0 |
| RNA binding protein | PC00031 | 384 | 375 | D | D | 1.67E-02 | 2.46E-08 |
| Extracellular matrix glycoprotein | PC00100 | 31 | 31 | U | U | 1.07E-02 | 5.60E-03 |
| Translation initiation factor | PC00224 | 34 | 35 | D | 3.24E-06 | NS | |
| Aminoacyl-tRNA synthetase | PC00047 | 18 | 18 | D | 4.68E-03 | NS | |
| Chaperonin | PC00073 | 11 | 11 | D | 1.88E-02 | NS | |
| Ligase | PC00142 | 92 | 88 | D | 3.84E-02 | NS | |
| Receptor | PC00197 | 139 | 137 | U | 8.92E-08 | NS | |
| Transporter | PC00227 | 175 | 177 | U | 4.72E-05 | NS | |
| Extracellular matrix linker protein | PC00101 | 6 | 5 | U | 1.45E-02 | NS | |
| Serine protease | PC00203 | 30 | 31 | U | 2.41E-02 | NS | |
| DNA binding protein | PC00009 | 113 | 105 | D | NS | 1.47E-13 | |
| Histone | PC00118 | 16 | 15 | D | NS | 1.16E-03 | |
| Ribosomal protein | PC00202 | 126 | 125 | D | NS | 9.37E-03 | |
| Transcription factor | PC00218 | 120 | 104 | D | NS | 2.18E-02 | |
| Actin family cytoskeletal protein | PC00041 | 125 | 129 | U | NS | 4.85E-09 | |
| Cytoskeletal protein | PC00085 | 201 | 208 | U | NS | 9.27E-07 | |
| Actin binding motor protein | PC00040 | 20 | 21 | U | NS | 1.55E-02 | |
| Non-motor actin binding protein | PC00165 | 60 | 61 | U | NS | 1.88E-02 | |
| Membrane traffic protein | PC00150 | 141 | 135 | U | NS | 2.56E-02 | |
NS, not significant; U/D, up- or downregulation.
Extracellular Matrix Protein Networks/Agrin Are Predominately Upregulated in Response to MGT
To place the regulated protein classes identified by GSEA into the context of cellular pathways and protein signaling networks, we investigated their functional relationships, including direct and indirect associations, using the STRING database (Szklarczyk et al., 2015). One PANTHER protein class of interest was “extracellular matrix proteins” (n = 70), which had on average a significant upregulation of TMT abundance ratios at both 48 and 72 hr post-transduction (Figure 2A). The functional interactions among these proteins were visualized as STRING interaction networks and included a color-scaled overlay of each protein’s TMT abundance ratio, separately for 48 and 72 hr. Surprisingly, the most upregulated of all ECM proteins was Agrin, a protein that acts to inhibit the Hippo signaling pathway (Chakraborty et al., 2017) (Figures 2B and 2C) and has recently been demonstrated to stimulate cardiac repair and proliferation (Bassat et al., 2017). Taken together, this implies that Agrin has a conserved role in re-programming as well as repair, and further suggests that inhibition of the Hippo pathway could promote cardiac reprogramming. From these networks, we further found that many of the proteins upregulated are those involved in the integrin signaling pathway (Figures 2B and 2C). These included the integrins themselves (Itgb3, Itgb1) and their ECM binding partners, the collagens (e.g., Col10a1, Col10a1, Col15a1, Col1a2, Col4a1) and laminins (Lama5, Lama4, Lamb2, etc.). Integrin signaling is known to play a role in cardiomyogenesis and is essential for hypertrophic growth and proliferation in cardiomyocytes (Hornberger et al., 2000; Ogawa et al., 2000). Thus, these findings would imply that one of the initiating events in direct re-programming is an upregulation in proteins required for scaffolding and those involved in sensing signals for proliferation and differentiation. In support of our data, an overall increase in ECM proteins was also observed in GHMT-infected (Gata4, Hand2, Mef2C, and Tbx5) cells during the first week of reprogramming (Zhao et al., 2015). While we observed that MGT expression leads to an overall increase in the average abundance of ECM proteins at 48 and 72 hr (Figure 2A), the relative abundances for many proteins were less strongly upregulated at 72 versus 48 hr (compare Figure 2B versus 2C). For example, we observed a slight decrease of Col1a1 and Col1a2 from 48 to 72 hr, consistent with our previous study demonstrating a gradual suppression of fibroblast-specific genes in response to MGT (Liu et al., 2016). Inhibition of pro-fibrotic signaling using small molecules that target transforming growth factor-β or Rho-associated kinase pathways converted embryonic fibroblasts into iCMs with greater efficiency, suggesting pro-fibrotic signaling antagonizes cardiac reprogramming (Zhao et al., 2015). Fibroblast signatures can also be suppressed through expression of microRNA mir-133, which acts to inhibit Snai1, a marker for endothelial-to-mesenchymal transition. Mir-133 expression in combination with GMT (Gata4, Mef2C, and Tbx5) in human and mouse fibroblast cultures, increased reprogramming efficiency by 7-fold and shortened the time frame by which cells begin beating (Muraoka et al., 2014). Hypertrophic growth has been specifically linked to the α1 and α3 integrin subunits, both of which were identified as upregulated in our study during iCM reprogramming (Johnston et al., 2009; Ross et al., 1998). Integrin signaling can also activate many signaling pathways in a bidirectional manner (i.e., inside and outside the cell) but requires structural, chemical, and mechanical cues to initiate these pathways (Gjorevski and Nelson, 2009; Sheehy et al., 2012). Because mechanical forces are not present in a culture environment, this may result in altered signaling within these MEFs/iCMs and could provide an explanation for the decreased reprogramming efficiency observed in vitro compared to in vivo (Happe and Engler, 2016; Qian et al., 2012, 2013; Ross and Borg, 2001).
Figure 2. Extracellular Matrix Protein Networks Are Predominantly Upregulated at Both 48 and 72 hr Post-reprogramming.
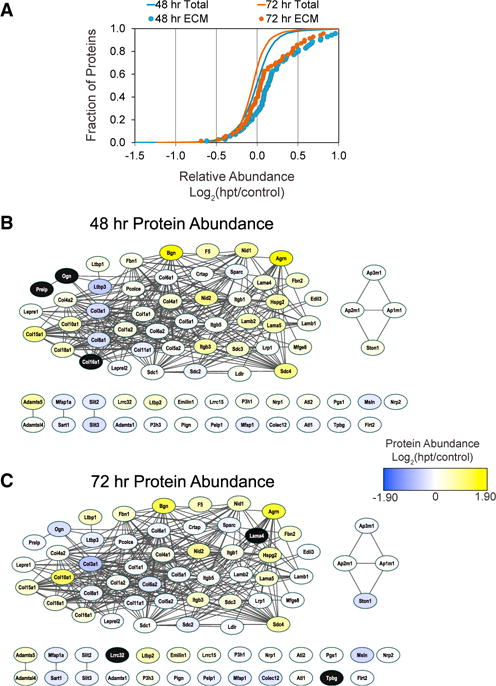
(A) Gene set enrichment analysis of TMT protein ratios found that proteins annotated to the PANTHER protein class extracellular matrix protein (n = 70) were on average significantly upregulated at both 48 (blue circles, p < 8.43E-6) and 72 hr (orange circles, p < 6.3E-4) compared to the distribution of all proteins (orange and blue lines).
(B and C) The functional connectivity and relative abundance of the proteins annotated to extracellular matrix protein networks was visualized by STRING analysis. Node color indicates average relative increase (yellow) or decrease (blue) in protein abundance at 48 and 72 hr post-transduction. Black node color indicates the protein was not detected at that time point. Extracellular matrix (ECM) protein abundance was similarly increased at both time points, although on average the magnitude was greater at 48 hr.
Translation Factor Protein Networks Are Downregulated at 48 hr but Not at 72 hr in Response to MGT
In addition to the upregulation of ECM proteins at 48 and 72 hr post-transduction, we also identified the “translation factor” class (n = 46) that was on average strongly downregulated compared to the distribution of all proteins at 48 hr although not at 72 hr (Figure 3A). Of those proteins involved in translation, the eukaryotic translation initiation factors (Eif) and eukaryotic translation elongation factors (Eef) were the majority of those identified, including Eif1a, Eif4b, Eif5a, Eef1g, and Eef1b2 (Figures 3B and 3C). The role of these factors in iCM reprogramming could be linked to the need for an accelerated rate of protein synthesis associated with cellular hypertrophy and sarcomere formation as these cells transition into cardiomyocytes (Favier et al., 2008). Because protein synthesis consumes large amounts of energy, this cellular mechanism is often “turned off” in response to energy depletion within cells. A specific example of this is the downregulation of elongation factor Eef2 by phosphorylation in cardiomyocytes, in response to energy depletion (McLeod and Proud, 2002). Because we see significant decreases in eukaryotic translation initiation factors (Eif) and eukaryotic translation elongation factors (Eef), including Eef2, at 48 and 72 hr post-transduction, these data are suggestive of an initial transitory burst in cellular response to energy depletion at the early stages of reprogramming.
Figure 3. Translation Factor Networks Are Strongly Downregulated at 48 hr Post-reprogramming but Then Rebound at 72 hr.
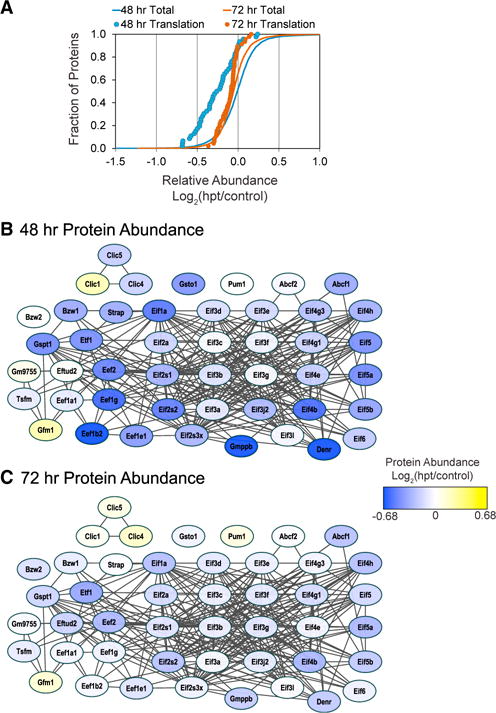
(A) Gene set enrichment analysis of TMT protein ratios found proteins annotated to the PANTHER protein translation factors (n = 46) were on average significantly downregulated at 48 hr (blue circles, p < 1.79E-08) but not at 72 hr (orange circles) compared to the distribution of all proteins (orange and blue lines).
(B and C) The functional connectivity and relative abundance of the proteins annotated to translational protein networks was visualized by STRING analysis. Node color indicates average relative increase (yellow) or decrease (blue) in protein abundance at 48 and 72 hr post-transduction. Individual protein abundances were strongly decreased at 48 hr, but less so at 72 hr when all proteins showed <50% change.
Chromatin Binding Protein Networks Are Downregulated at 72 hr in Response to MGT
By 72 hr, we observed the “chromatin/chromatin binding protein” class of proteins were on average significantly down-regulated in abundance (n = 38) (Figures 4A–4C). While this effect was not detected at 48 hr, individual proteins linked to the Wnt signaling pathway such as Crebbp, ep300, Smarcc1, and Smarce1 (Teo and Kahn, 2010) showed a unique pattern of expression; being upregulated at 48 hr and, as with other chromatin/chromatin binding proteins, downregulated at 72 hr (Figures 4B and 4C). Wnt signaling is primarily involved in cardiogenesis, initiating cardiac specification during early heart formation, progenitor proliferation, and differentiation of cardiomyocytes (Klaus et al., 2007). In the adult heart, Wnt signaling is rarely reported, but has been shown to increase following ischemic myocardial damage and activate fetal gene programs (Duan et al., 2012; Oerlemans et al., 2010). Many of these Wnt-associated proteins were upregulated at 48 hr post-transduction, compared to other proteins in this class, but were down-regulated at 72 hr post-transduction. Akt-interacting proteins, Ep300 and Acin1, followed a similar expression profile to those linked to Wnt signaling, being upregulated at 48 hr post-transduction and downregulated at 72 hr. Recently, an unbiased screen of kinases in cardiac reprogramming identified Akt/ protein kinase B as an effective enhancer and accelerator of cardiac reprogramming when expressed in addition to Gata4, Hand2, Mef2C, and Tbx5 (Zhou et al., 2015).
Figure 4. Chromatin-Associated Protein Networks Are Predominantly Downregulated at 72 hr Post-reprogramming.
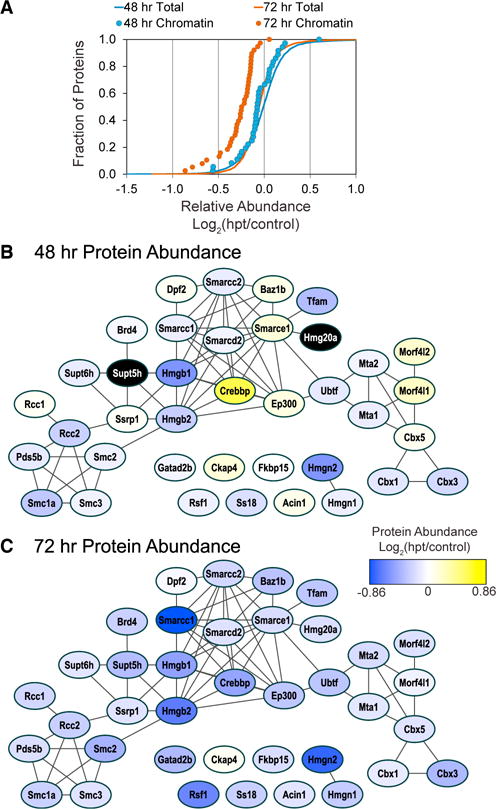
(A)Gene set enrichment analysis of TMT protein ratios found proteins annotated to the PANTHER protein class chromatin/chromatin-binding proteins (n = 38) were on average significantly downregulated at 72 hr (orange circles, p < 4.5E-10) but not at 48 hr (blue circles) compared to the distribution of all proteins (orange and blue lines).
(B and C) The functional connectivity and relative abundance of the proteins annotated to chromatin protein networks was visualized by STRING analysis. Node color indicates average relative increase (yellow) or decrease (blue) in protein abundance at 48 and 72 hr post-transduction. Black node color indicates the protein was not detected at that time point.
Consistently, it has also been reported that treatment with FGF2, FGF10, and VEGF (FFV) and GMT in serum-free conditions enabled reprogramming of fibroblasts into functional iCMs with an increased efficiency, and even eliminated the need for Gata4 during reprogramming. FFV was shown to act directly through P38/MAPK and PI3K/Akt pathways to activate cardiac transcription programs (Yamakawa et al., 2015). Overall, we observe a significant downregulation of chromatin and chromatin binding proteins at 72 hr but not 48 hr post-transduction. This observation would imply that the initiating events that trigger the changes in the chromatin state associated with the conversion of fibroblast to iCMs are completed by this stage. Our results further suggest that the chromatin and chromatin binding proteins not downregulated by 72 hr function to maintain or reinforce this chromatin landscape. These conclusions are consistent with the observation that at 72 hr post-transduction cardiac promoters show a curtailment of repressive indicators of transcription, an enhancement of the activation indicator of transcription and accordantly, the onset of expression of the cardiac reporter gene α-MHC-GFP.
Temporal Changes in ECM, Translation, and Chromatin Binding Protein Networks during Reprogramming
To independently validate our results from the TMT proteomic-based approach, we conducted western blot analysis on proteins found at critical nodes of the ECM and chromatin protein networks. Each western blot was conducted a minimum of three times and quantified by densitometry. Consistent with our findings from TMT, the ECM protein NID2 was upregulated at 72 hr (Figure 5A). We also observed by western blot analysis that the ECM-associated protein Agrin was dramatically upregulated at 72 hr (Figure 5B), while the chromatin-associated proteins, SMARCC1 and HMGB1, showed a marked reduction at 48 hr (Figures 5C and 5D), all of which were consistent with the quantitative TMT proteomics data. Thus, these results further verify that components of distinct protein pathways aretemporally regulated during the initiating steps of direct fibroblast reprogramming to iCMs.
Figure 5. Temporal Changes in ECM, Translation, and Chromatin Binding Protein Networks.
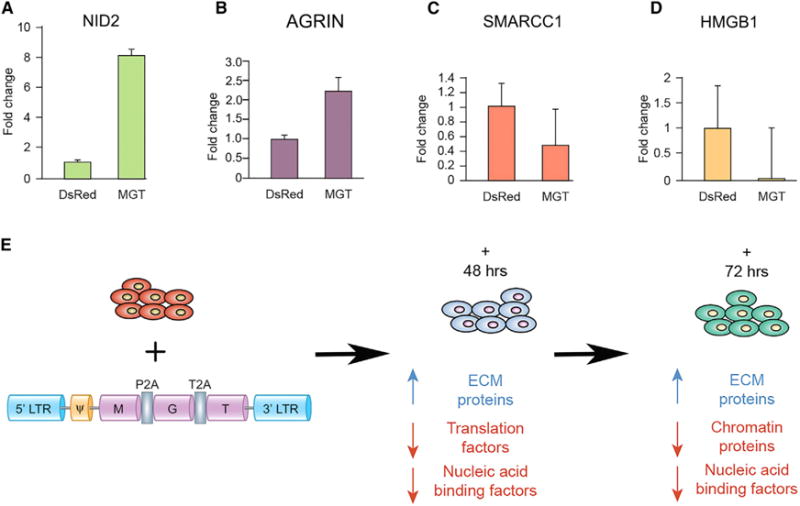
(A–D) Western blot analysis was performed on nuclear fractions of MEFs transduced with either MGT or DsRed and were probed with antibodies against representative proteins in each class. (A) Nid2 an extracellular matrix protein and (B) Agrin at 72 hr post-infection (C) SMARCC1 and (D) HMGB1, chromatin-associated proteins, at 48 post-transduction. Densitometry quantification reveals changes in levels of proteins examined are consistent with the TMT proteomic results. These changes were significant for both Nid2, Agrin, and SMARCC1 (p < 0.05).
(E) Schematic overview of infection and summary of the changes in relative protein abundances at 48 and 72 hr post-transduction. Data are represented as mean ± SEM.
Conclusions
The findings presented in this study demonstrate the presence of an orchestrated series of temporal steps during the initiating events of direct reprogramming. Our results further demonstrate that these temporal events are associated with the differential regulation of protein abundance (Figure 5E). Currently, the application of reprogramming of fibroblast to iCMs as an alternative strategy for cardiac regeneration is compromised in part by a mechanistic understanding of the relatively low efficiency of reprogramming associated with the initiating events. The protein pathways defined by these studies when manipulated genetically or biochemically should lead to alteration in the efficiency of reprogramming. Furthermore, because reprogramming of fibroblasts to iCM by MGT is direct and does not go through an embryonic state, comparing changes in protein levels reported in this study to that of normal cardiac program in mammals will reveal those pathways that are similar and dissimilar and thus, allow a basis for understanding how MGT bypasses these early essential events in development.
EXPERIMENTAL PROCEDURES
Mouse Lines
Transgenic mice harboring GFP under the control of the α-MHC promoter were used for mouse embryonic fibroblast (MEF) isolation (Ieda et al., 2010; Qian et al., 2013; Wang et al., 2015a). Animal care was provided in accordance with guidelines established by University of North Carolina, Chapel Hill.
Mouse Embryonic Fibroblast Preparation
Embryos from αMHC-GFP mice were isolated at E13.5. The head and red organs from these embryos were removed and the remainder of the embryo was minced with a sterile razor blade and digested in 1 mL of 0.05% trypsin/EDTA with 100 kunitz units of DNase I at 37°C for 15 min. Cells were then re-suspended in mouse embryonic fibroblast medium (DMEM with 10% FBS, and 50 U/50 μg/mL penicillin/streptomycin) and then plated onto a 0.1% gelatin-coated dish. After 2 hr, plates were washed with 1× PBS to remove unattached cells, and attached fibroblasts were used for subsequent experiments. The resulting mouse embryonic fibroblasts (MEFs) were maintained in growth medium containing DMEM with 10% FBS (Sigma) and 50 U/50 μg/mL penicillin/streptomycin.
Plasmids
To generate DsRed, DsRed was excised from pMXs-DsRed (Addgene 22,724) and cloned into pMXs-puro vector. pMX-MGT was generated as previously described (Wang et al., 2015a). Viral Packaging and Direct Reprogramming PlatE packaging cells were maintained in growth medium containing DMEM with 10% FBS, 50 U/50 μg/mL penicillin/streptomycin, and 5% Non-essential amino acids (Life Technologies). PlatE cells were seeded at 4–5 × 106 cells per 100 mm dish and transfected 24 hr later with the pMXs-MGT polycistronic construct or a pMXs-DsRed (Addgene, Cat. #22724) chosen for size similarity to MGT, using Nanofect transfection reagent according to the manufacturer’s recommendations (Alstem, Cat. #NF100). Supernatant was collected on days 4 and 5 post-transfection and filtered through 0.45 μm pore size cellulose acetate filter (Thermo Scientific). Retrovirus Precipitation Solution (Alstem, PEG4000) was added to virus-containing supernatant to concentrate the viral particles overnight at 4°C. Viruses were then resuspended by fibroblast medium with 4 μg/mL of polybrene (Life Technologies) and then added to target cells immediately. 24 hr after transduction, the virus containing medium was changed to iCM medium made up of a 4:1 ratio of DMEM to M199 with 10% FBS (Sigma). Cells were lysed and nuclear fractions were isolated (Nuclei EZ Prep, Sigma) at 48 and 72 hr post-transduction for proteomic analysis.
Western Blotting and Densitometry Analysis
Nuclear lysates were obtained from MEFs 48 and 72 hr post-transduction with MGT or DsRed Constructs using Nuclei EZ Prep Kit (Sigma, Cat. #nuc101). Whole cell lysate was obtained from cells using RIPA buffer. Samples were placed in LDS sample buffer with reducing agent (NuPAGE, Cat. #NP0007) and boiled at 75°C for 10 min before being run on a 4%–12% gradient polyacrylamide gel by electrophoresis (Invitrogen, Cat. #NP0322). Gels were transferred to a nitrocellulose membrane and blocked for 1 hr in 5% block (Bio-Rad, Cat. #170- 6404) dissolved in 1× Tris-buffered saline with Tween20 (BDH Chemicals, Poole Dorset, UK). Primary antibodies were incubated at (1:1,000) dilution overnight at 4°C. Primary antibodies used were LSD1 (Cell Signaling, Cat. #2184), eIF1α (Abcam, Cat. #ab177939), SMARCC1 (Novus, Cat. #NBP2-20415), MEF2C (Abcam, Cat. #ab64644), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Abcam, Cat. #ab181372). Appropriate HRP-conjugated secondary antibodies were used at (1:10,000) in 5% block and incubated for 1 hr at room temperature. Protein was detected using a chemiluminescent substrate (GE Healthcare, Cat. #RPN2109). Exact numbers of replicates are denoted in the figure legends.
Densitometry analysis was performed using Adobe Photoshop and values were normalized to a loading control.
Quantitative TMT-Based Proteomic Analysis of Reprogrammed MEFs
Enriched mouse embryonic fibroblast nuclei from 48 and 72 hr post-MGT transduction (N = 3 each) plus DsRed controls (N = 3) were solubilized in lysis buffer (50 mM ammonium bicarbonate [ABC] containing 2.5% sodium deoxycholate [NaDOC], pre-heated to 70°C) with alternate heating at 95°C for 5 min and bath sonication. Protein yields were determined by the BCA protein assay and samples were simultaneously reduced and alkylated with 20 mM TCEP and 20 mM chloroacetamide, respectively, for 20 min at 70°C. The reaction was quenched with 20 mM cysteine for 20 min at room temperature (RT). Aliquots of lysates (40 μg for experimental and 80 μg for controls) were diluted to 1% NaDOC, then digested in-solution with trypsin (Promega) in a 1:50 enzyme:protein ratio. Samples were incubated overnight at 37°C, then acidified to 1% trifluoroacetic acid. The deoxycholate (DOC) was removed by phase transfer using an equal volume of ethyl acetate (Greco et al., 2016). The aqueous phase was desalted using SDB-RPS StageTips (Kulak et al., 2014) concentrated to near dryness, and suspended in 50 mM HEPES pH 8.0 to ~1 μg/μL concentration. For each sample, peptides were labeled with either 0.1 mg (MGT) or 0.2 mg (DsRed) of TMT reagents (Pierce TMT6plex) for 1 hr at RT. The labeling was quenched with hydroxylamine (0.33%) for 10 min at RT. The TMT-labeled peptides from 72 hr MGT and 48 hr MGT samples (N = 3 each) were combined with equal peptide amounts of DsRed controls (N = 3) (see Figure 1A), generating two pooled TMT6plex experiments. The pooled samples were subjected to 2D StageTip fractionation. Briefly, the peptides were first separated into four fractions over C18-SCX StageTips with sequential elution (90 μL) in 0.05 M ammonium formate/20% acetonitrile (ACN), 0.05 M ammonium acetate/20% ACN, 0.05 M ammonium bicarbonate/20% ACN, and 0.1% ammonium hydroxide/20% ACN. Then, each fraction was diluted to 10% ACN, adjusted to 0.5% TFA, and further separated into four fractions over SDB-RPS StageTips with sequential elution (40 μL) in 0.15 M ammonium formate/0.5% formic acid (FA)/40% ACN, 0.2 M ammonium acetate/0.5% FA/60% ACN, and 5% ammonium hydroxide/80% ACN. The resulting samples were pooled into 13 fractions, were evaporated to near dryness by vacuum centrifugation, and suspended in ~10 μL of 1% FA/5% ACN.
Peptides (4 μL) were analyzed by nanoliquid chromatography-tandem mass spectrometry on a Dionex Ultimate 3000 UPLC coupled to an EASYSpray ion source and Orbitrap Velos ETD mass spectrometer (Thermo Fisher Scientific). Briefly, TMT-labeled peptides were separated over a 2 hr linear gradient (5% to 40% solvent B; solvent A: 0.1% FA, solvent B: 0.1% FA/97% ACN) by reverse phase chromatography (EASYSpray C18 column, 75 μm × 50 cm). As peptides eluted from the column, a data-dependent MS acquisition method was employed, which consisted of an analytical MS scan, measuring intact peptide ions, followed by up to 10 tandem MS fragmentation events (resolution: 15,000 at m/z = 400) on the most intense ions with dynamic exclusion enabled.
Informatics Analysis of TMT Proteomic Data
Tandem MS spectra collected across the 13 liquid chromatography-tandem mass spectrometry (LC-MS/MS) runs for 48 and 72 hr TMT 6plex experiments were analyzed by Proteome Discoverer (Thermo Fisher Scientific, v2.1.0.81). In summary, the two sets of 13 fractions were separately analyzed by a Processing workflow containing (1) the MSAmanda algorithm to generate peptide spectrum matches by database search of tandem MS spectra with static TMT modifications against the UniProt-SwissProt mouse reference proteome database, appended with common contaminants (2016–07, 16,810 sequences), (2) Percolator to calculate spectrum q-values based on reverse sequence database searches, and (3) a reporter ion quantifier module, which extracted TMT signal-to-noise values from MS/MS spectra. The resulting processed data were combined in the Consensus workflow, which defined peptide and protein filters to limit the false discovery rate to ≤1%, then assembled peptides into protein groups with strict parsimony and retained quantitative values from spectra that (1) were unique to the protein group, (2) had a co-isolation threshold of <50%, and (3) had an average S/N of >10. TMT S/N values were normalized between TMT reporter channels based on the total sum of the S/N values within each channel over all identified peptides. Protein abundances were calculated as the sum of normalized spectrum S/N values for each channel within each protein group. Protein groups and associated TMT abundances for each sample were exported to Excel. Only protein groups containing a minimum of 2 quantified unique spectra were considered for further analysis. The TMT abundance ratio for 48 or 72 hr MGT versus DsRed was calculated as the average abundance of MGT versus DsRed control (N = 3 each).
Network and GO Analysis of Quantitative MS Data
Primary gene symbols and respective abundance ratios were analyzed by Gene Set Enrichment Analysis (www.pantherdb.org) using the PANTHER protein class ontology (Mi et al., 2016). The proteins assigned to significantly regulated protein classes, evaluated by Bonferroni corrected p values, were analyzed and assembled into functional interaction networks using STRING (Szklarczyk et al., 2015). Networks were imported into Cytoscape (v3.4.0) (Shannon et al., 2003) for further visualization.
Supplementary Material
Highlights.
Global changes in protein levels exist at initiating stages of cardiac reprogramming
Functional signatures in protein networks are associated with steps of reprogramming
Extracellular matrix proteins are upregulated at the initiation stage
Chromatin-associated proteins are downregulated at the initiation stage
Acknowledgments
K.S., M.V., and F.L.C. are funded by NIH/NHLBI grants R01 HL112618 and R01 HL127640. I.M.C. is funded by NIH/NIGMS R01 GM114141.
Footnotes
DATA AND SOFTWARE AVAILABILITY
The accession number for the mass spectrometry proteomics data reported in this paper is PXD006152, deposited at the ProteomeXchange Consortium (Vizcaíno et al., 2014) via the PRIDE partner repository.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.01.047.
AUTHOR CONTRIBUTIONS
K.S., M.V., and L.W. performed all fibroblasts reprogramming experiments. K.S. performed all western blot analysis. T.M.G. conducted all TMT studies and performed large-scale bioinformatics analysis of all related data. I.M.C. provided additional proteomics expertise and L.Q. additional reprogramming expertise. F.L.C., I.M.C., and L.Q. designed the experiments. All authors discussed the results and wrote the manuscript.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Addis RC, Ifkovits JL, Pinto F, Kellam LD, Esteso P, Rentschler S, Christoforou N, Epstein JA, Gearhart JD. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J Mol Cell Cardiol. 2013;60:97–106. doi: 10.1016/j.yjmcc.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassat E, Mutlak YE, Genzelinakh A, Shadrin IY, Baruch Umansky K, Yifa O, Kain D, Rajchman D, Leach J, Riabov Bassat D, et al. The extracellular matrix protein agrin promotes heart regeneration in mice. Nature. 2017;547:179–184. doi: 10.1038/nature22978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaldi T, Straub T, Cox J, Kumar C, Becker PB, Mann M. Combined use of RNAi and quantitative proteomics to study gene function in Drosophila. Mol Cell. 2008;31:762–772. doi: 10.1016/j.molcel.2008.07.018. [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Njah K, Pobbati AV, Lim YB, Raju A, Lakshmanan M, Tergaonkar V, Lim CT, Hong W. Agrin as a mechanotransduction signal regulating YAP through the hippo pathway. Cell Rep. 2017;18:2464–2479. doi: 10.1016/j.celrep.2017.02.041. [DOI] [PubMed] [Google Scholar]
- Chen JX, Krane M, Deutsch MA, Wang L, Rav-Acha M, Gregoire S, Engels MC, Rajarajan K, Karra R, Abel ED, et al. Inefficient reprogramming of fibroblasts into cardiomyocytes using Gata4, Mef2c, and Tbx5. Circ Res. 2012;111:50–55. doi: 10.1161/CIRCRESAHA.112.270264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chick JM, Munger SC, Simecek P, Huttlin EL, Choi K, Gatti DM, Raghupathy N, Svenson KL, Churchill GA, Gygi SP. Defining the consequences of genetic variation on a proteome-wide scale. Nature. 2016;534:500–505. doi: 10.1038/nature18270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforou N, Chellappan M, Adler AF, Kirkton RD, Wu T, Addis RC, Bursac N, Leong KW. Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming. PLoS ONE. 2013;8:e63577. doi: 10.1371/journal.pone.0063577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Gherghe C, Liu D, Hamlett E, Srikantha L, Rodgers L, Regan JN, Rojas M, Willis M, Leask A, et al. Wnt1/bcatenin injury response activates the epicardium and cardiac fibroblasts to promote cardiac repair. EMBO J. 2012;31:429–442. doi: 10.1038/emboj.2011.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favier FB, Benoit H, Freyssenet D. Cellular and molecular events controlling skeletal muscle mass in response to altered use. Pflugers Arch. 2008;456:587–600. doi: 10.1007/s00424-007-0423-z. [DOI] [PubMed] [Google Scholar]
- Gjorevski N, Nelson CM. Bidirectional extracellular matrix signaling during tissue morphogenesis. Cytokine Growth Factor Rev. 2009;20:459–465. doi: 10.1016/j.cytogfr.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco TM, Guise AJ, Cristea IM. Determining the composition and stability of protein complexes using an integrated label-free and stable isotope labeling strategy. Methods Mol Biol. 2016;1410:39–63. doi: 10.1007/978-1-4939-3524-6_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happe CL, Engler AJ. Mechanical forces reshape differentiation cues that guide cardiomyogenesis. Circ Res. 2016;118:296–310. doi: 10.1161/CIRCRESAHA.115.305139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornberger LK, Singhroy S, Cavalle-Garrido T, Tsang W, Keeley F, Rabinovitch M. Synthesis of extracellular matrix and adhesion through beta(1) integrins are critical for fetal ventricular myocyte proliferation. Circ Res. 2000;87:508–515. doi: 10.1161/01.res.87.6.508. [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagawa K, Miyamoto K, Yamakawa H, Muraoka N, Sadahiro T, Umei T, Wada R, Katsumata Y, Kaneda R, Nakade K, et al. Induction of cardiomyocyte-like cells in infarct hearts by gene transfer of Gata4, Mef2c, and Tbx5. Circ Res. 2012;111:1147–1156. doi: 10.1161/CIRCRESAHA.112.271148. [DOI] [PubMed] [Google Scholar]
- Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, Zhang Z, Rosenberg P, Mirotsou M, Dzau VJ. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RK, Balasubramanian S, Kasiganesan H, Baicu CF, Zile MR, Kuppuswamy D. Beta3 integrin-mediated ubiquitination activates survival signaling during myocardial hypertrophy. FASEB J. 2009;23:2759–2771. doi: 10.1096/fj.08-127480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus A, Saga Y, Taketo MM, Tzahor E, Birchmeier W. Distinct roles of Wnt/beta-catenin and Bmp signaling during early cardiogenesis. Proc Natl Acad Sci USA. 2007;104:18531–18536. doi: 10.1073/pnas.0703113104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- Liu Z, Chen O, Zheng M, Wang L, Zhou Y, Yin C, Liu J, Qian L. Re-patterning of H3K27me3, H3K4me3 and DNA methylation during fibroblast conversion into induced cardiomyocytes. Stem Cell Res (Amst) 2016;16:507–518. doi: 10.1016/j.scr.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Wang L, Yin C, Liu J, Qian L. In vivo cardiac reprogramming using an optimal single polycistronic construct. Cardiovasc Res. 2015;108:217–219. doi: 10.1093/cvr/cvv223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathison M, Singh VP, Gersch RP, Ramirez MO, Cooney A, Kaminsky SM, Chiuchiolo MJ, Nasser A, Yang J, Crystal RG, et al. ‘‘Triplet’’ polycistronic vectors encoding Gata4, Mef2c, and Tbx5 enhances postinfarct ventricular functional improvement compared with singlet vectors. J Thorac Cardiovasc Surg. 2014;148:1656–1664. doi: 10.1016/j.jtcvs.2014.03.033. [DOI] [PubMed] [Google Scholar]
- McLeod LE, Proud CG. ATP depletion increases phosphorylation of elongation factor eEF2 in adult cardiomyocytes independently of inhibition of mTOR signalling. FEBS Lett. 2002;531:448–452. doi: 10.1016/s0014-5793(02)03582-2. [DOI] [PubMed] [Google Scholar]
- Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44(D1):D336–D342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka N, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Isomi M, Nakashima H, Akiyama M, Wada R, Inagawa K, et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014;33:1565–1581. doi: 10.15252/embj.201387605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam YJ, Song K, Luo X, Daniel E, Lambeth K, West K, Hill JA, DiMaio JM, Baker LA, Bassel-Duby R, Olson EN. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci USA. 2013;110:5588–5593. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie L, Wu G, Culley DE, Scholten JC, Zhang W. Integrative analysis of transcriptomic and proteomic data: challenges, solutions and applications. Crit Rev Biotechnol. 2007;27:63–75. doi: 10.1080/07388550701334212. [DOI] [PubMed] [Google Scholar]
- Oerlemans MI, Goumans MJ, van Middelaar B, Clevers H, Doevendans PA, Sluijter JP. Active Wnt signaling in response to cardiac injury. Basic Res Cardiol. 2010;105:631–641. doi: 10.1007/s00395-010-0100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa E, Saito Y, Harada M, Kamitani S, Kuwahara K, Miyamoto Y, Ishikawa M, Hamanaka I, Kajiyama N, Takahashi N, et al. Outside-in signalling of fibronectin stimulates cardiomyocyte hypertrophy in cultured neonatal rat ventricular myocytes. J Mol Cell Cardiol. 2000;32:765–776. doi: 10.1006/jmcc.2000.1119. [DOI] [PubMed] [Google Scholar]
- Protze S, Khattak S, Poulet C, Lindemann D, Tanaka EM, Ravens U. A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J Mol Cell Cardiol. 2012;53:323–332. doi: 10.1016/j.yjmcc.2012.04.010. [DOI] [PubMed] [Google Scholar]
- Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Berry EC, Fu JD, Ieda M, Srivastava D. Reprogramming of mouse fibroblasts into cardiomyocyte-like cells in vitro. Nat Protoc. 2013;8:1204–1215. doi: 10.1038/nprot.2013.067. [DOI] [PubMed] [Google Scholar]
- Ross RS, Borg TK. Integrins and the myocardium. Circ Res. 2001;88:1112–1119. doi: 10.1161/hh1101.091862. [DOI] [PubMed] [Google Scholar]
- Ross RS, Pham C, Shai SY, Goldhaber JI, Fenczik C, Glembotski CC, Ginsberg MH, Loftus JC. Beta1 integrins participate in the hypertrophic response of rat ventricular myocytes. Circ Res. 1998;82:1160–1172. doi: 10.1161/01.res.82.11.1160. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy SP, Grosberg A, Parker KK. The contribution of cellular mechanotransduction to cardiomyocyte form and function. Biomech Model Mechanobiol. 2012;11:1227–1239. doi: 10.1007/s10237-012-0419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo JL, Kahn M. The Wnt signaling pathway in cellular proliferation and differentiation: A tale of two coactivators. Adv Drug Deliv Rev. 2010;62:1149–1155. doi: 10.1016/j.addr.2010.09.012. [DOI] [PubMed] [Google Scholar]
- Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- Vizcaíno JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Ríos D, Dianes JA, Sun Z, Farrah T, Bandeira N, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada R, Muraoka N, Inagawa K, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Kaneda R, Suzuki T, Kamiya K, et al. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc Natl Acad Sci USA. 2013;110:12667–12672. doi: 10.1073/pnas.1304053110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu Z, Yin C, Asfour H, Chen O, Li Y, Bursac N, Liu J, Qian L. Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circ Res. 2015a;116:237–244. doi: 10.1161/CIRCRESAHA.116.305547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu Z, Yin C, Zhou Y, Liu J, Qian L. Improved generation of induced cardiomyocytes using a polycistronic construct expressing optimal ratio of Gata4, Mef2c and Tbx5. J Vis Exp. 2015b doi: 10.3791/53426. [DOI] [PMC free article] [PubMed]
- Yamakawa H, Muraoka N, Miyamoto K, Sadahiro T, Isomi M, Haginiwa S, Kojima H, Umei T, Akiyama M, Kuishi Y, et al. Fibroblast growth factors and vascular endothelial growth factor promote cardiac reprogramming under defined conditions. Stem Cell Reports. 2015;5:1128–1142. doi: 10.1016/j.stemcr.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Londono P, Cao Y, Sharpe EJ, Proenza C, O’Rourke R, Jones KL, Jeong MY, Walker LA, Buttrick PM, et al. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat Commun. 2015;6:8243. doi: 10.1038/ncomms9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Dickson ME, Kim MS, Bassel-Duby R, Olson EN. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc Natl Acad Sci USA. 2015;112:11864–11869. doi: 10.1073/pnas.1516237112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


