Abstract
Purpose
Neuropilin-1 (NRP1) as an isoform-specific receptor for vascular endothelial growth factor and placenta growth factor in endothelial cells has been demonstrated to be expressed in breast cancer cells where it plays functional roles in cell survival, invasion, and migration. We hypothesized that an expression of NRP1 in breast cancer tissues is associated with clinicopathological data of patients and expression of the tumor suppressor miR-206.
Patients and methods
We evaluated the expression of NRP1 in 48 invasive ductal carcinomas of the breast and their corresponding adjacent noncancerous tissues (ANCTs) by means of real-time polymerase chain reaction. We also extracted data on miR-206 gene expression from the same cohort of patients to evaluate the correlation between expression levels of miR-206 and NRP1. In addition, we quantified NRP1 protein levels using the enzyme-linked immunosorbent assay technique.
Results
No significant difference was found in NRP1 expression between tumoral tissues and ANCTs. We also assessed the associations between expression levels of NRP1 and clinicopathological data of patients and found no significant associations between NRP1 transcript levels and any characteristic. However, NRP1 protein concentrations were significantly higher in patients with lymph node involvement compared with those without lymph node involvement. No correlation was found between NRP1 and miR-206 expression levels.
Conclusion
NRP1 protein levels might be an indicator of metastasis potential in breast cancer. Future studies are needed to confirm these results in larger cohorts of patients.
Keywords: NRP1, neuropilin-1, breast cancer
Introduction
Breast cancer, as the most prevalent cancer among women,1 has been associated with a significant mortality rate.2 Despite much research,3–5 only few biomarkers or therapeutic targets have been approved, altering the ordinary practice of oncology.6 Therefore, continuous efforts are being made in the field of identification of biomarkers for this type of human malignancy. Neuropilin-1 (NRP1) as an isoform-specific receptor for vascular endothelial growth factor (VEGF) and placenta growth factor (PlGF) in endothelial cells has been demonstrated to be expressed in breast cancer cells where it plays functional roles in cell survival, invasion, and migration.7 In the context of cancer, NRP1 also regulates angiogenesis and the epithelial-to-mesenchymal transition (EMT) mechanisms through interactions with VEGF, and its receptors, PlGF and transforming growth factor β 1 (TGF-β1).8 Moreover, NRP1 has been shown to be a target of regulation by the tumor suppressor miR-206 in estrogen receptor-positive cell lines, MCF-7 and T47D. The inhibitory effects of miR-206 overexpression on the migration and invasive potentials of these cells are at least partly due to its suppressive effect on NRP1 expression.9 Taken together, these data suggest an oncogenic role for NRP1 in the pathogenesis of breast cancer and pinpoint it as a target of a functional tumor-suppressive microRNA (miRNA). Therefore, we designed a study to assess NRP1 expression at both RNA and protein levels in correlation with expression level of miR-206. We also extracted microarray gene expression data to compare those results with our expression data results.
Materials and methods
Patients’ samples
The expression study was performed on 48 tumoral tissues and their corresponding adjacent noncancerous tissues (ANCTs) obtained during surgery from patients with a definite diagnosis of invasive ductal carcinoma of the breast. Normal-appearing adjacent breast tissues were excised at least 2 cm away from the invasive tumor margin and were assessed by the pathologist to not contain tumoral cells. Tissues were obtained from Sina Hospital and Farmanieh Hospital (Tehran, Iran) during 2017. All patients signed an informed consent form. It was confirmed that the patients did not receive any kind of anticancer treatment before the surgery. The study protocol was approved by the ethical committee of Shahid Beheshti University of Medical Sciences (IR.SBMU.MSP.REC.1396.153).
RNA extraction and real-time polymerase chain reaction (PCR)
Total RNA was extracted from all samples using the TRIzol™ Reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the protocol provided by the company. cDNA was synthesized from extracted RNA samples using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Relative transcript levels of NRP1 were quantified using SYBR® Premix Ex Taq™ (TaKaRa, Otsu, Japan) on the Rotor Gene 6000 Corbett Real-Time PCR system. B2M gene was used as the normalizer. All experiments were carried out in duplicate. The nucleotide sequences of primers used for amplification of NRP1 and B2M are as follows: NRP1 forward primer: 5′-CTCCAACGGGGAAGACTGGA-3′, reverse primer: 5′-GTTGCAGGCTTGATTCGGAC-3′; B2M forward primer: 5′-AGATGAGTATGCCTGCCGTG-3′, reverse primer: 5′-GCGGCATCTTCAAACCTCCA-3′.
NRP1 tissue concentration
NRP1 tissue concentration was evaluated using the human NRP1 enzyme-linked immunosorbent assay (ELISA) kit (MyBiosource, San Diego, CA, USA) which uses the biotin double-antibody sandwich technology. The values of NRP1 are shown in pg/mL and defined as mean ± standard error of mean.
Extraction of microarray data
We retrieved GSE5926 and GSE5927 microarray gene expression data through application of the Gene Expression Omnibus repository at the National Center for Biotechnology Information archives.10 Expression of NRP1 gene was compared between breast cancer patients and normal samples using the GEO2R web tool (https://www.ncbi.nlm.nih.gov/geo/info/geo2r.html). Finally, we evaluated the correlations between expression of NRP1 and expression of miR-206 as a validated regulator of NRP1 in the mentioned microarray datasets.
Statistical analysis
R software (version 3.3.1; Vienna, Austria) was used for statistical analysis. Data were presented as mean ± standard deviation. Differences in NRP1 mRNA expression were evaluated using Student’s paired t-tests and Wilcoxon signed-rank test based on the presence or absence of normal distribution in data, respectively. The association of clinicopathological and demographic data with expression levels of NRP1 was evaluated using a chi-squared test, Student’s t-test and one-way analysis of variance. The value of 2−ΔΔCt was estimated to calculate the expression fold change for each gene. The cutoffs of the fold change values were calculated based on the median values of fold changes. Spearman’s rank correlation coefficient was used for evaluation of the correlations between NRP1 protein concentration and relative expression of its mRNA and miR-206. For all statistical tests, the level of significance was set at P<0.05.
Results
General demographic and clinical data of patients
The current study included 48 patients with invasive ductal carcinomas of the breast whose detailed demographic and clinical data are shown in Table 1.
Table 1.
General demographic and clinical data of patients
| Variables | Groups | Number of cases (%) |
|---|---|---|
| Age (years) | <50 | 21 (44) |
| ≥50 | 27 (56) | |
| Body mass index (kg/m2) | <25 | 2 (4) |
| 25–29 | 24 (50) | |
| >29 | 22 (46) | |
| Menopause status | Post | 37 (77) |
| Pre | 11 (23) | |
| Stage | I | 11 (23) |
| II | 18 (37.5) | |
| III | 16 (33) | |
| IV | 3 (6.5) | |
| Histological grade | I | 7 (14.5) |
| II | 25 (52) | |
| III | 16 (33.5) | |
| Mitotic rate | 1 | 19 (39.5) |
| 2 | 22 (46) | |
| 3 | 7 (14.5) | |
| Tumor size (cm) | <2 | 18 (37.5) |
| 2–5 | 22 (46) | |
| >5 | 8 (16.5) | |
| Lymph node involvement | Positive | 19 (39.5) |
| Negative | 29 (60.5) | |
| HER2 status | Positive | 36 (75) |
| Negative | 12 (25) | |
| ER status | Positive | 37 (77) |
| Negative | 11 (23) | |
| PR status | Positive | 35 (73) |
| Negative | 13 (27) |
Abbreviations: HER2, human epidermal growth factor receptor 2; ER, estrogen receptor; PR, progesterone receptor.
Relative expression of NRP1 in tumoral tissues compared with ANCTs
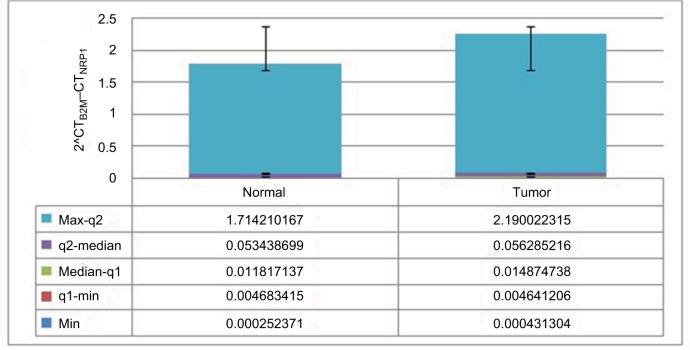
No significant difference was found in NRP1 expression between tumoral tissues and ANCTs (Figure 1).
Figure 1.
Relative expression of NRP1 in tumoral and ANCTs.
Abbreviation: NRP1, neuropilin-1; ANCTs, adjacent noncancerous tissues.
We also assessed the associations between expression levels of NRP1 and clinicopathological data of patients and found no significant associations between NRP1 transcript levels and any characteristic (Table 2).
Table 2.
Association of NRP1 transcript levels with patients’ clinicopathological data
| Variable | Number of cases | NRP1 high level | P-value |
|---|---|---|---|
| Age (years) | |||
| <50 | 21 (44%) | 11 | NS |
| ≥50 | 27 (56%) | 17 | |
| Body mass index (kg/m2) | |||
| 25–29 | 24 (50%) | 14 | NS |
| >29 | 22 (46%) | 13 | |
| Menopause status | |||
| Post | 37 (77%) | 21 | NS |
| Pre | 11 (23%) | 7 | |
| Stage | |||
| I | 11 (23%) | 4 | NS |
| II | 18 (37.5%) | 14 | |
| III | 16 (33%) | 6 | |
| IV | 3 (6.5%) | 2 | |
| Histological grade | |||
| I | 7 (14.5%) | 4 | NS |
| II | 25 (52%) | 15 | |
| III | 16 (33.5%) | 7 | |
| Mitotic rate | |||
| 1 | 19 (39.5%) | 10 | NS |
| 2 | 22 (46%) | 10 | |
| 3 | 7 (14.5%) | 6 | |
| Tumor size (cm) | |||
| ≤2 | 18 (37.5%) | 9 | NS |
| 2–5 | 22 (46%) | 12 | |
| ≥5 | 8 (16.5%) | 5 | |
| Lymph node involvement | |||
| Positive | 19 (39.5%) | 10 | NS |
| Negative | 29 (60.5%) | 16 | |
| HER2 | |||
| Positive | 25 (75%) | 16 | NS |
| Negative | 23 (25%) | 10 | |
| ER | |||
| Positive | 37 (77%) | 21 | NS |
| Negative | 11 (23%) | 5 | |
| PR | |||
| Positive | 35 (73%) | 19 | NS |
| Negative | 13 (27%) | 7 | |
| Breast cancer phenotype | |||
| Luminal A | 19 (39.5%) | 11 | 0.891 |
| Luminal B | 22 (46%) | 13 | |
| HER2 | 2 (4%) | 1 | |
| Triple negative | 5 (10.5%) | 3 |
Note: NRP1 high level was defined based on the cut-offs of the fold changes in tumoral tissue vs. the corresponding ANCT, and the median value of fold changes was set as cutoff.
Abbreviations: NRP1, neuropilin-1; ANCT, adjacent noncancerous tissue; HER2, human epidermal growth factor receptor 2; ER, estrogen receptor; PR, progesterone receptor; NS, not significant.
Assessment of NRP1 protein concentrations
NRP1 protein levels were quantified using a commercial ELISA kit in different subgroups of tumoral samples. Table 3 demonstrates the mean values of its expression in relation to patients’ clinicopathological data. NRP1 protein concentrations were significantly higher in patients with lymph node involvement compared with those without lymph node involvement (P=0.03). However, NRP1 protein concentrations were not associated with other clinicopathological or demographic data.
Table 3.
The association between NRP1 protein concentration and demographic and clinicopathological characteristics of patients
| Patient/tumor characteristics | NRP1 concentration (mean values ± standard error of mean, pg/mL) | P-value |
|---|---|---|
| Age (years) | ||
| <50 | 27.99±10.39 | 0.59 |
| ≥50 | 38.24±16.35 | |
| Body mass index (kg/m2) | ||
| 25–29 | 33.44±13.97 | 0.92 |
| >29 | 35.54±14.67 | |
| Menopause status | ||
| Post | 39.9±14.85 | 0.39 |
| Pre | 21.84±8.85 | |
| TNM stage | ||
| I | 15.71±7.77 | 0.39 |
| II | 20.50±11.52 | |
| III | 57.96±20.81 | |
| IV | 0 | |
| Histological grade | ||
| I | 0 | 0.78 |
| II | 31.62±13.13 | |
| III | 40.42±16.35 | |
| Mitotic rate | ||
| 1 | 28.90±14.54 | 0.49 |
| 2 | 27.89±14.43 | |
| 3 | 61.95±26.66 | |
| Tumor size (cm) | ||
| ≤2 | 21.61±14.8 | 0.607 |
| 2–5 | 46.02±17.02 | |
| ≥5 | 26.95±26.95 | |
| Lymph node involvement | ||
| Positive | 66.44±24.33 | 0.03 |
| Negative | 18.33±7.32 | |
| HER2 | ||
| Positive | 35.71±14.16 | 0.52 |
| Negative | 22.58±9.95 | |
| ER | ||
| Positive | 36.67±12.46 | 0.57 |
| Negative | 20.21±14.32 | |
| PR | ||
| Positive | 27.43±8.44 | 0.37 |
| Negative | 46.91±27.21 | |
| Breast cancer phenotype | ||
| Luminal A | 11.42±4.81 | 0.205 |
| Luminal B | 52.26±19.57 | |
| HER2 | 0 | |
| Triple negative | 0 |
Abbreviations: NRP1, neuropilin-1; HER2, human epidermal growth factor receptor 2; ER, estrogen receptor; PR, progesterone receptor.
Assessment of NRP1 expression in GSE5926 dataset
We also evaluated NRP1 expression levels in GSE5926 data-set and found significant downregulation of NRP1 in in situ tumors compared with normal sample (P=0.02). However, no significant difference was found in its expression between invasive and in situ tumors (P=0.35) or between invasive and normal tissues (P=0.11) (Table 4).
Table 4.
Relative expression of NRP1 in invasive, in situ, and normal samples included in the GSE5926 dataset
| Invasive vs. in situ | Invasive vs. normal | In situ vs. normal | |
|---|---|---|---|
| Fold change | 0.23 | −1.08 | −1.15 |
| P-value | 0.35 | 0.11 | 0.02 |
Abbreviation: NRP1, neuropilin-1.
We also compared NRP1 expression in different molecular subtypes of breast tumors and normal sample and found significant downregulation of NRP1 in human epidermal growth factor receptor 2-positive, basal-like, luminal A and luminal B subtypes compared with normal sample. However, NRP1 expression was not significantly different between normal-like breast tumor and normal samples (Table 5).
Table 5.
Relative expression of NRP1 in HER2+, basal-like, luminal A, luminal B, and normal-like subtypes compared with normal sample included in the GSE5926 dataset
| HER2+ vs. normal | Basal vs. normal | Luminal A vs. normal | Luminal B vs. normal | Normal-like vs. normal | |
|---|---|---|---|---|---|
| Fold change | −1.09 | −1.53 | −1.55 | −1.15 | −0.92 |
| P-value | 0.011 | 0.001 | 0.005 | 0.0002 | 0.14 |
Abbreviations: NRP1, neuropilin-1; HER2, human epidermal growth factor receptor 2.
Assessment of correlation between NRP1 and miR-206 expressions
We also retrieved data of miR-206 expression from the same cohort of patients (Seifi et al, unpublished data, 2017) and assessed the correlations between NRP1 expression levels and those of miR-206. No significant correlation was found between miR-206 and NRP1 transcript levels (Spearman’s correlation coefficient =−0.0004, P=0.92) or between miR-206 and NRP1 protein levels (Spearman’s correlation coefficient =−0.140, P=0.469). Also, the same analysis in the microarray datasets showed no correlation between transcript levels of miR-206 and NRP1 (Spearman’s correlation coefficient =−0.004, P=0.98). In addition, no correlation was found between mRNA and protein levels of NRP1 (Spearman’s correlation coefficient =−0.111, P=0.544).
Discussion
In this study, we compared the expression of NRP1 transcript between breast tumor tissues and ANCTs and found no significant difference in its expression levels between these sets of samples. We also retrieved gene expression data from GSE5926 dataset and found significant downregulation of NRP1 in in situ tumors compared with normal sample but no significant difference in its expression between invasive and in situ tumors or between invasive and normal tissues. Our results are in contrast with Ferrario et al’s study, which, through immunohistochemical staining of a breast tissue microarray, has shown significantly higher expression of NRP1 in malignant epithelium compared with benign and preinvasive breast lesions.7 Such inconsistency between our experimental results and the results of the mentioned study might be due to the difference in selection of control tissues (ANCTs vs. normal tissues). Aran et al recently assessed the transcriptomes of healthy tissues, ANCTs, and tumor tissues.11 They found that ANCTs show a distinctive intermediate expression profile between healthy and tumor tissues. More importantly, they observed activation of genes related to hypoxia, tumor necrosis factor α and TGF-β signaling, apoptosis, EMT, and angiogenesis in adjacent epithelium. Consequently, a relatively similar expres sion of NRP1 in tumoral tissues and ANCTs, as revealed in our study, as well as the similar expression of NRP1 in invasive and in situ tumors, as obtained from analysis of microarray dataset, might be explained by activation of the mentioned pathways in ANCTs. However, due to the low number of normal tissues in the GSE5926 dataset,12 the analysis between normal tissues and other types of tissues might be biased.
Noticeably, we found significantly higher expression of NRP1 in lymph node-positive patients compared with lymph node-negative patients. We also detected a trend toward higher NRP1 levels in higher stages and histological grades, but perhaps due to the small sample size, it did not reach the level of significance. Such observation is in line with previous assays which showed the role of another receptor for VEGF, NRP2, in cell migration and metastasis.13 Moreover, in vivo and in vitro studies have indicated the effectiveness of NRP1 inhibition in suppression of breast-cancer-related metastasis.14 More recently, higher plasma NRP1 levels were detected in breast cancer patients with advanced diseased nodes compared with patients with no nodal metastasis, those with less advanced nodal metastasis, and healthy subjects.8
Finally, we assessed the correlation between the mRNA and protein levels of NRP1 as well as those of NRP1 and miR-206 and found no correlation between any of them. The lack of correlation between mRNA and protein levels of a certain gene has been reported previously.15 A more recent study showed that mRNA and protein levels do not correlate well except when a gene-specific RNA-to-protein conversion factor is applied to improve the predictability of protein copy numbers from transcript levels.16 The dimensional and chronological alterations of mRNAs, along with the regional accessibility of resources for protein production, would affect the correlation between transcript and protein levels of a certain gene in many situations such as cancer.17
Despite the fact that NRP1 has been validated as a target of miR-206 in cell line studies,9 both in silico analysis of microarray data and assessment of our gene expression results showed no correlation between NRP1 and miR-206 levels. This observation can be explained by the inhibitory effect of several miRNAs on NRP1 expression. To provide evidence for such hypothesis, we used the miRWalk2.0 tool18 to obtain the list of miRNAs that target NRP1 based on evaluation of miRNA-binding sites within the complete sequence of NRP1, as well as search for experimentally verified miRNA–NRP1 interaction data. We found that numerous miRNAs including miR-181, miR-335, miR-124, miR-16, miR-186, miR-218, miR-338, and miR-320 target NRP1. Therefore, the expression level of NRP1 is influenced by numerous factors. Besides, in silico analysis of National Cancer Institute-60 expression data has revealed more negative correlations between miRNA and mRNA expression profiles in cases where the miRNA degrades the corresponding mRNA compared with situations when the miRNA inhibits the translation of its target gene.19 Therefore, the lack of correlation between miR-206 and NRP1 levels can be explained by the possible effect of miR-206 on translation of NRP1. Future studies are needed to elaborate the underlying mechanism.
Conclusion
This study provides evidence for superiority of protein levels over mRNA levels of NRP1 as a predictive marker in breast cancer. NRP1 protein levels might be an indicator of metastasis potential in breast cancer. Future studies are needed to confirm these results in larger cohorts of patients.
Acknowledgments
This work was financially supported by the Research Department of the School of Medicine Shahid Beheshti University of Medical Sciences (grant no. 9641). This article has been extracted from the thesis written by Mahnaz Seifi-Alan in School of Medicine, Shahid Beheshti University of Medical Sciences (Registration No. 105M).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Seifi-Alan M, Shamsi R, Ghafouri-Fard S, et al. Expression analysis of two cancer-testis genes, FBXO39 and TDRD4, in breast cancer tissues and cell lines. Asian Pac J Cancer Prev. 2014;14(11):6625–6629. doi: 10.7314/apjcp.2013.14.11.6625. [DOI] [PubMed] [Google Scholar]
- 2.Ghafouri-Fard S, Shamsi R, Seifi-Alan M, Javaheri M, Tabarestani S. Cancer-testis genes as candidates for immunotherapy in breast cancer. Immunotherapy. 2014;6(2):165–179. doi: 10.2217/imt.13.165. [DOI] [PubMed] [Google Scholar]
- 3.Dianatpour A, Faramarzi S, Geranpayeh L, Mirfakhraie R, Motevaseli E, Ghafouri-Fard S. Expression analysis of AFAP1-AS1 and AFAP1 in breast cancer. Cancer Biomark. 2018;22(1):49–54. doi: 10.3233/CBM-170831. [DOI] [PubMed] [Google Scholar]
- 4.Sarrafzadeh S, Geranpayeh L, Ghafouri-Fard S. Expression analysis of long non-coding PCAT-1 in breast cancer. Int J Hematol Oncol Stem Cell Res. 2017;11(3):185–191. [PMC free article] [PubMed] [Google Scholar]
- 5.Nikpayam E, Soudyab M, Tasharrofi B, et al. Expression analysis of long non-coding ATB and its putative target in breast cancer. Breast Dis. 2017;37(1):11–20. doi: 10.3233/BD-160264. [DOI] [PubMed] [Google Scholar]
- 6.Perez-Gracia JL, Sanmamed MF, Bosch A, et al. Strategies to design clinical studies to identify predictive biomarkers in cancer research. Cancer Treat Rev. 2017;53:79–97. doi: 10.1016/j.ctrv.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Ferrario C, Hostetter G, Bouchard A, Huneau M-C, Mamo A, Basik M. Expression of neuropilin-1 and related proteins in breast cancer. Proc Amer Assoc Cancer Res. 2006;47:5034. [Google Scholar]
- 8.Naik A, Al-Zeheimi N, Bakheit CS, et al. Neuropilin-1 associated molecules in the blood distinguish poor prognosis breast cancer: a cross-sectional study. Sci Rep. 2017;7(1):3301. doi: 10.1038/s41598-017-03280-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin K, Yin W, Wang Y, et al. MiR-206 suppresses epithelial mesenchymal transition by targeting TGF-beta signaling in estrogen receptor positive breast cancer cells. Oncotarget. 2016;7(17):24537–24548. doi: 10.18632/oncotarget.8233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrett T, Edgar R. Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Method Enzymol. 2006;411:352–369. doi: 10.1016/S0076-6879(06)11019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aran D, Camarda R, Odegaard J, et al. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat Commun. 2017;8:1077. doi: 10.1038/s41467-017-01027-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lesurf R, Aure MR, Mork HH, et al. Molecular features of subtype-specific progression from ductal carcinoma in situ to invasive breast cancer. Cell Rep. 2016;16(4):1166–1179. doi: 10.1016/j.celrep.2016.06.051. [DOI] [PubMed] [Google Scholar]
- 13.Yasuoka H, Kodama R, Tsujimoto M, et al. Neuropilin-2 expression in breast cancer: correlation with lymph node metastasis, poor prognosis, and regulation of CXCR4 expression. BMC Cancer. 2009;9:220. doi: 10.1186/1471-2407-9-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arpel A, Gamper C, Spenle C, et al. Inhibition of primary breast tumor growth and metastasis using a neuropilin-1 transmembrane domain interfering peptide. Oncotarget. 2016;7(34):54723–54732. doi: 10.18632/oncotarget.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kendrick N. A Gene’s mRNA Level Does not Usually Predict Its Protein Level. Madison, WI: Kendricklabs.com; 2014. [Google Scholar]
- 16.Edfors F, Danielsson F, Hallstrom BM, et al. Gene-specific correlation of RNA and protein levels in human cells and tissues. Mol Syst Biol. 2016;12(10):883. doi: 10.15252/msb.20167144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165(3):535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 18.Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(8):697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 19.Wang YP, Li KB. Correlation of expression profiles between microR-NAs and mRNA targets using NCI-60 data. BMC Genomics. 2009;10:218. doi: 10.1186/1471-2164-10-218. [DOI] [PMC free article] [PubMed] [Google Scholar]