Abstract
Background
Lead (Pb) is a widely used metal in modern industry and is regarded as a health hazard. Although lead-induced genotoxicity has been confirmed, the direct evidence that lead induces genotoxicity in human cells and its related mechanisms has not been fully elucidated. In this study, for the first time, we evaluated the genotoxicity induced by lead in human lymphoblastoid TK6 cells.
Material/Methods
The TK6 cells were incubated with various concentrations of Pb(Ac)2 for 6 h, 12 h, or 24 h. Cell viability was detected by CCK8 assay. Various biochemical markers were assessed by specific kits. Immunofluorescence assay was used to detect γ-H2AX foci formation. The promoter methylation was assessed by methylation-specific PCR. The protein levels were determined by Western blot assay.
Results
The results showed that after exposure to lead, cell viability was obviously decreased and γ-H2AX foci formation was significantly enhanced in TK6 cells. Moreover, the levels of 8-OHdG, ROS, MDA, and GSSG were increased, while the GSH level and SOD activity were decreased in lead-treated TK6 cells. The activation of the Nrf2-ARE signaling pathway was involved in lead-induced oxidative stress in TK6 cells. Finally, the expressions of DNA repair genes XRCC1, hOGG-1, BRCA1, and XPD were inhibited via enhancing their promoter methylation in TK6 cells after exposure to lead.
Conclusions
Taken together, our study provides the first published evidence that lead exposure results in DNA damage via promoting oxidative stress and the promoter methylation of DNA repair genes in human lymphoblastoid TK6 cells.
MeSH Keywords: DNA Methylation, Lead, Mutagenicity Tests, Oxidative Stress
Background
Lead is one of the most widely used metals in the world. It has distinctive physical and chemical properties, such as malleability, low melting point, and corrosion resistance. However, lead persists undegraded in the environment and cannot be metabolized in the body. As an occupational risk factors, accumulation of lead in the body is a serious threat to human health [1–3]. The most common routes of lead exposure are respiratory and digestive tracts, through which lead develops toxic effects on every organ and system of the body [4]. In recent years, it has been well demonstrated that lead or its compounds shows potential genotoxicity to various research subjects under different conditions [5–10], but few studies have explored lead-induced genotoxicity in human cells. TK6 human lymphoblasts with good differentiation ability, stable genome, and normal expression of P53, are widely used in genotoxicity studies [11–14] and can be useful for the in vitro study of human genotoxicity.
Epidemiological investigations indicated that the frequency of micronucleus and serum MDA level were significantly increased in workers exposed to lead, and the blood lead level was positively correlated with oxidative stress [15]. Sharma et al. showed that lead-induced overproduction of ROS can result in oxidative and anti-oxidative unbalance [16]. Moreover, the excessive amounts of ROS may cause DNA oxidative damage [17,18].
A feature common to the genotoxicity of various poisons is DNA damage, which can be repaired by multiple DNA repair genes, such as XRCC1, hOGG1, BRCA1, and XPD. The main types of DNA damage repair are base excision repair, nucleotide excision repair, and double-strand break repair [19]. Generally, the expression level of DNA repair gene is negatively correlated with its promoter methylation. It was reported that the promoter methylation of DNA repair genes can decrease the DNA damage repair capability [20]. The above research background suggests that oxidative damage and the promoter methylation of DNA repair genes may be involved in lead-induced genotoxicity in human TK6 cells.
In the present study, for the first time, we evaluated lead-induced genotoxicity and its potential molecular mechanisms in human TK6 cells.
Material and Methods
Drug
Lead acetate was obtained from Sigma Chemical Company and dissolved in deionized water at a stock concentration of 20 mM and stored at 4°C. The drug was diluted by deionized water into various concentrations and filtrated through a 0.22-μm membrane filter before use.
Cell culture
Human lymphoblastoid TK6 cells were provided by Professor Kuicheng Zheng (Fujian Center for Disease Control and Prevention, China). The TK6 cells were maintained in RPMI 1640 medium (Gibco, USA) supplemented by 10% heat-inactivated horse serum (Gibco, USA) at 37°C under a humidified atmosphere and 5% CO2.
CCK8 assay
TK6 cells (6×103 cells per well) were seeded in 96-well plates in 100 μl of culture medium. After cell attachment, various concentrations of lead acetate (0, 30, 60, 120, 240, 480, 960, 1920, and 3840 μM) in fresh medium were added to TK6 cells. The supernatant was discarded after incubation for 6, 12, and 24 h, then we added 100 μl RPMI 1640 medium and 10 μl CCK8 (Wanleibio, Shenyang, China) to the cells and incubated them for 4 h at 37°C. The optical density (OD) value was measured on a microplate reader (Bio-Tek, USA) at 450 nm. The formula for cell viability (%) was: cell viability (%)=OD in treatment group/OD in control group ×100%.
Immunofluorescence staining
To assess γ-H2AX foci formation in TK6 cells, immunofluorescence staining assay was performed. Briefly, cells were treated with lead acetate (0, 120, 240, and 480 μM) for 6, 12, and 24 h. The cells treated with 100 μM H2O2 for 10 min were used as a positive control. Then, the cells were collected and fixed onto slides. After washing with PBS, the slides were incubated with 0.1% tritonX-100 for 30 min at room temperature. The slides were washed with PBS and incubated with goat serum (Solarbio, China) for 15 min at room temperature. Thereafter, a primary polyclonal anti-H2AX antibody (1: 200, Proteintech, China) was added at 4°C overnight. Then, the slides were washed 3 times with PBS and incubated with Cy3-conjugated goat anti-rabbit secondary antibody (1: 200, Beyotime, China) for 1 h at room temperature. We used 4′,6-diamidino-2-phenylindole (DAPI) for nuclear counterstaining. The images were obtained under a fluorescence microscope (Zeiss, Germany) at a magnification of 400×. The integral optical density of γ-H2AX was analyzed by Image Pro-Plus software (Media Cybernetics, USA).
ELISA
The level of 8-OHdG in TK6 cells was assessed by ELISA assay. After being treated with lead acetate (0, 120, 240, and 480 μM) for 6, 12, and 24 h, the TK6 cells were lysed and subjected to ELISA assay using a commercially available kit (USCN, China) according to the manufacturer’s instructions. The 8-OHdG content was calculated through a standard curve.
Measurement of intracellular ROS
The intracellular ROS production was evaluated by DCFH-DA fluorescent probe. Briefly, TK6 cells were seeded into 6-well plates at a density of 1×106 cells per well. After being treated with lead acetate (0, 120, 240, 480 μM) for 6, 12, and 24 h, the cells were washed with PBS, incubated with DMEM containing DCFH-DA (1: 1000, Nanjing Jiancheng Bioengineering Institute, China) at 37°C for 20 min away from light. Then, the cells were washed with PBS and detected by flow cytometry.
MDA, GSH, GSSG, and SOD detections
TK6 cells were treated as detailed above and collected for further tests. The levels of MDA, GSH, and GSSG and activity of SOD were determined by commercially-available detection kits (Nanjing Jiancheng Bioengineering Institute, China) following the manufacturer’s instructions.
Quantitative real-time polymerase chain reaction (qPCR)
The mRNA expressions of Nrf2, HO-1, and NQO1 in TK6 cells were assessed by qPCR. Total RNAs were extracted using the RNApure quick extraction kit (Bio Teke, China). Then, the RNAs were reversed transcribed into cDNA using Super M-MLV reverse transcriptase (Bio Teke, China). SYBR Green quantitative PCR was performed using 2×Power Taq PCR MasterMix (Bio Teke, China) on an ExicyclerTM 96 Real-Time Quantitative Thermal Block (Bioneer, South Korea). The primer sequences are listed in Table 1. β-actin was used as an internal control. The relative expressions of genes were analyzed by the 2−ΔΔCT method.
Table 1.
Oligonucleotide primer sets for qPCR.
| Name | Sequence (5′-3′) | Length |
|---|---|---|
| Nrf2 F | GTCAGCGACGGAAAGAGTA | 19 |
| Nrf2 R | ACCTGGGAGTAGTTGGCA | 18 |
| NQO1 F | GCCGAGTCTGTTCTGGCTTAT | 21 |
| NQO1 R | TGGCAGCGTAAGTGTAAGCA | 20 |
| HO-1 F | CTCCGATGGGTCCTTACACTC | 21 |
| HO-1 R | CATAGGCTCCTTCCTCCTTTC | 21 |
| β-actin F | CTTAGTTGCGTTACACCCTTTCTTG | 25 |
| β-actin R | CTGTCACCTTCACCGTTCCAGTTT | 24 |
DNA extraction
TK6 cells were treated with various concentrations of lead acetate (0, 120, 240, and 480 μM) for 6, 12, and 24 h, and then the DNA from TK6 cells was extracted using the TIANamp Genomic DNA Kit (TIANGEN, China) according to the manufacturer’s instructions. To evaluate the concentration and purity of the extracted DNA, the OD260/OD280 ratio was detected by use of a NanoDrop ND-2000 ultramicrospectrophotometer (Thermo, USA).
Methylation-specific PCR (MS-PCR)
The methylation status of XRCC1, hOGG-1, BRCA1, and XPD was detected by MS-PCR. Briefly, 500 ng genomic DNA was modified with sodium bisulfite using the EZ DNA methylation kit (Zymo, USA) following the manufacturer’s instructions. The methylation PCR primers were designed using the MethPrimer online tool and synthesized by Sangon Biotech Company, Limited (Shanghai, China). The primers sequences are listed in Table 2. The MS-PCR was carried out using 2×Taq PCR MasterMix (BioTek, China). The thermocycling conditions were: denaturation at 95°C for 10 s, annealing at 55°C for 20 s, and extension at 72°C for 30 s, for 40 cycles. The MS-PCR products were subjected to 1.5% agarose gel electrophoresis. The MS-PCR results were also quantitatively analyzed by 2−ΔΔCt method.
Table 2.
Oligonucleotide primer sets for MS-PCR.
| Name | Sequence (5′-3′) | Length |
|---|---|---|
| hOGG-1(M) F | CGTTTATAGGTTTTGGGGGC | 20 |
| hOGG-1(M) R | CATACCTCGCCCTTTACGAA | 20 |
| hOGG-1(U)F | GTGTTTATAGGTTTTGGGGGT | 21 |
| hOGG-1(U) R | ACATACCTCACCCTTTACAAA | 21 |
| XRCC1(M)F | GAGATTTGTTAATTTTTTTCGC | 22 |
| XRCC1(M)R | AAACGTAAACGACTACGCTAA | 21 |
| XRCC1(U)F | AAAGAGATTTGTTAATTTTTTTTGT | 25 |
| XRCC1(U)R | CAAACATAAACAACTACACTAAAC | 24 |
| XPD(M)F | GACGTTTTTCGTATCGTTTTATTC | 24 |
| XPD(M)R | AAACCATTAACTAACCTACCCGTC | 24 |
| XPD(U)F | ATGTTTTTTGTATTGTTTTATTTGA | 25 |
| XPD(U)R | AACCATTAACTAACCTACCCATC | 23 |
| BRCA1(M)F | TCGTGGTAACGGAAAAGCGC | 20 |
| BRCA1(M)R | AAATCTCAACGAACTCACGCCG | 22 |
| BRCA1(U)F | TTGTGGTAATGGAAAAGTGT | 20 |
| BRCA1(U)R | AAATCTCAACAAACTCACACCA | 22 |
M – methylation; U – unmethylation; F – forward; R – reverse.
Western blot analysis
The protein levels in TK6 cells were assessed by Western blot assay. Briefly, TK6 cells from different treatment groups were lysed in RIPA Lysis Buffer (Wanleibio, China). Then, the protein concentration was determined using the BCA Protein Quantitation Kit (Wanleibio, China). Thereafter, 40 μg protein samples were separated on polypropylene acyl amine gel electrophoresis (PAGE), transferred to polyvinylidene difluoride (PVDF) membranes, and blocked by 5% skim milk for 1 h at room temperature. Then, the membranes were incubated with primary antibodies Nrf2 (1: 500, Sangon, China), HO-1 (1: 500, Wanleibio, China), NQO1 (1: 500, Wanleibio, China), XRCC1 (1: 500, Sangon, China), hOGG1 (1: 500, Sangon, China), BRCA1 (1: 500, Sangon, China), and XPD (1: 500, Sangon, China) at 4°C overnight. After incubation with the corresponding HRP-conjugated secondary antibodies (Wanleibio, China) at 37°C for 45 min, the bands were visualized by enhanced chemiluminescence (ECL) reagent.
Statistical analysis
All experimental data are shown as mean ± standard deviation (SD). One-way ANOVA was performed to compare differences among multiple groups using GraphPad Prism 5 software followed by Bonferroni’s multiple comparison test. Statistical differences were considered significant when p-value were less than 0.05.
Results
Effect of lead exposure on DNA damage of human TK6 cells
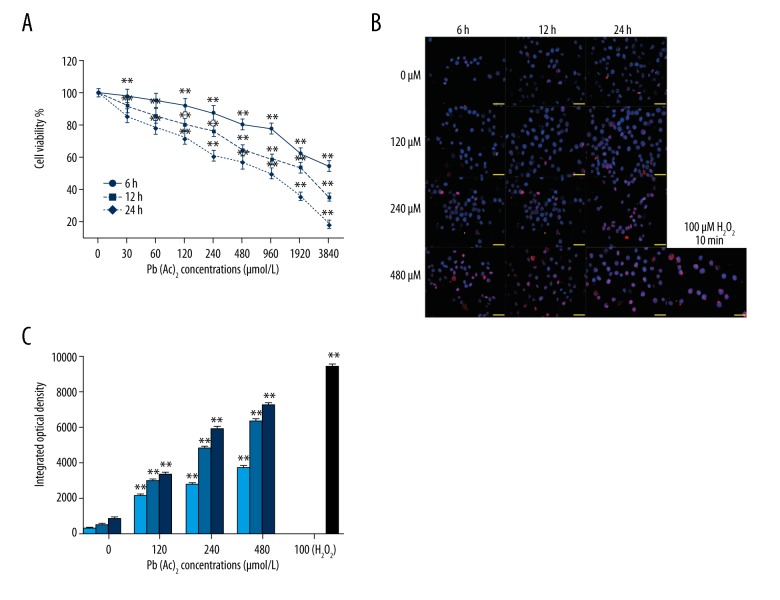
Firstly, the cytotoxicity of lead was evaluated in human TK6 cells by CCK8 assay. As shown in Figure 1A, cell viability was obviously inhibited by lead exposure in a concentration- and time-dependent manner. Moreover, DNA damage was assessed by immunofluorescence staining of γ-H2AX. The results showed that γ-H2AX foci formation was significantly promoted in TK6 cells with increased dosage or lead exposure time extension (Figure 1B). The integrated optical density of γ-H2AX-positive TK6 cells was concentration- or time-dependently increased by lead exposure (Figure 1C). The TK6 cells treated with H2O2 were used as positive control.
Figure 1.
Effect of lead exposure on DNA damage of TK6 cells. (A) Cell viability of TK6 cells at various time points and concentrations was determined by CCK8 assay. (B) The γ-H2AX foci formation in TK6 cells after exposure to lead was assessed by immunofluorescence staining assay at the indicated time points and concentrations. H2O2-treated cells were used as a positive control. (C) The quantitative analysis of integrated optical density values of γ-H2AX. The results are presented as mean ±SD (n=3). ** P<0.01, versus the corresponding cells treated with 0 μM Pb(Ac)2.
Effect of lead exposure on oxidative stress injury in human TK6 cells
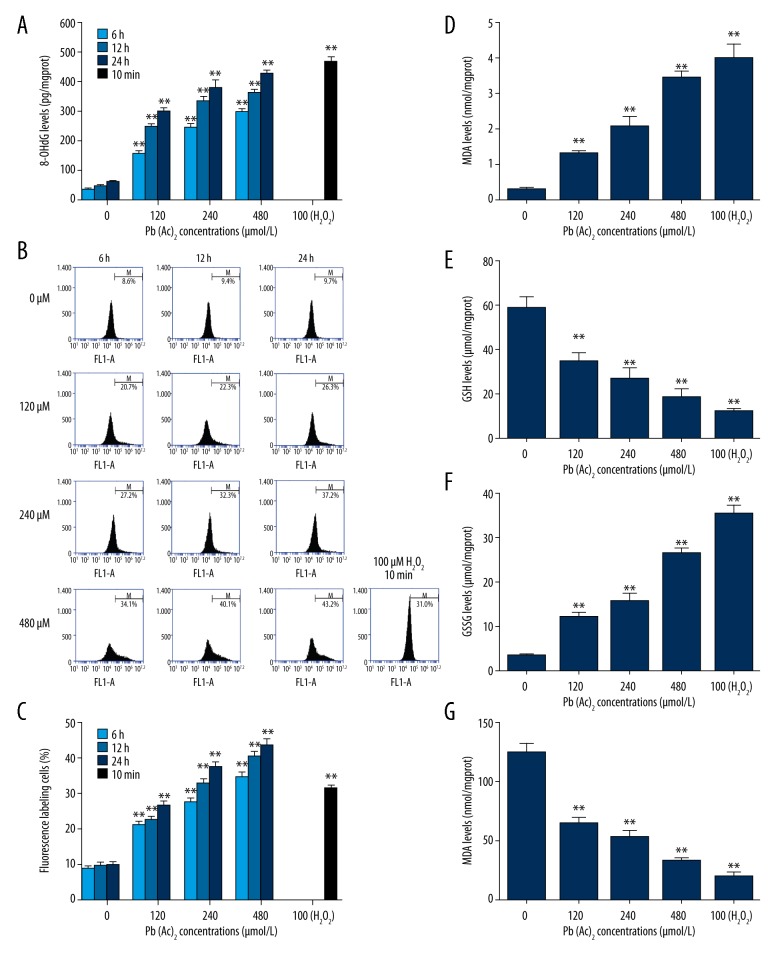
To evaluate the DNA oxidative damage, the 8-OHdG level in TK6 cells after exposure to lead was detected by ELISA assay. As presented in Figure 2A, there was a concentration- or time-dependent increase in 8-OHdG level in TK6 cells after exposure to lead. Subsequently, the ROS level in TK6 cells was assessed by flow cytometry. The results suggested that the ROS level was remarkably increased by lead exposure in TK6 cells (Figure 2B, 2C). Moreover, more oxidative stress markers or enzyme were investigated. As shown in Figure 2D–2G, the levels of MDA and GSSG were enhanced, while the level of GSH and activity of SOD were decreased in human TK6 cells after exposure to different concentrations of lead.
Figure 2.
Effect of lead exposure on oxidative stress injury in TK6 cells. (A) The 8-OHdG levels in TK6 cells after exposure to lead were detected by ELISA (n=3). (B) The ROS production in lead-treated TK6 cells was assessed by flow cytometry. (C) The percentage of ROS fluorescence labeling cells was quantitatively analyzed. The levels of MDA (D), GSH (E), GSSG (F), and activity of SOD (G) in TK6 cells after exposure to various concentrations of lead were determined by commercially available kits. The results are presented as mean ±SD (n=5, unless specified otherwise). ** P<0.01, versus the corresponding cells treated with 0 μM Pb(Ac)2.
Effect of lead exposure on Nrf2-ARE signaling pathway in human TK6 cells
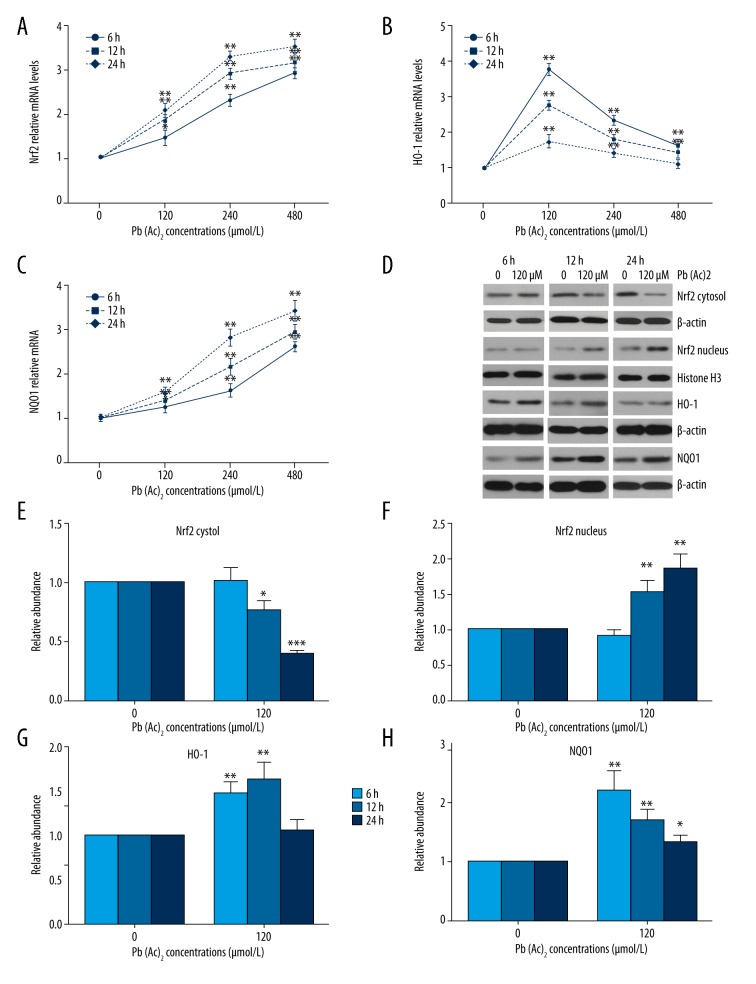
The Nrf2-ARE signaling pathway plays important roles in the regulation of cellular redox status [21], so we further evaluated the effect of lead exposure on the Nrf2-ARE signaling pathway. As shown in Figure 3A–3C, the mRNA expressions of Nrf2 and NQO1 were upregulated by lead exposure in a concentration- and time-dependant manner. The HO-1 mRNA expression first increased, then declined with the increase of lead concentration. Protein levels of Nrf2, HO-1, and NQO1 were also determined by Western blot assay and are shown in Figure 3D. The cytoplasmic Nrf2 level was downregulated, while the nuclear Nrf2 and NQO1 levels were upregulated by lead exposure with the extension of time. The protein level of HO-1 first increased, then decreased, which was consistent with the change in HO-1 mRNA expression.
Figure 3.
Effect of lead exposure on Nrf2-ARE signaling pathway in TK6 cells. (A–C) The mRNA expressions of Nrf2, HO-1, and NQO1 in TK6 cells were detected by qPCR assay. (D) The protein levels of cytoplasmic Nrf2, nuclear Nrf2, HO-1, and NQO1 in TK6 cells were assessed by Western blot assay. β-actin and histone H3 served as loading controls. (E–H) The quantitative analysis of the gray-scale values of the bands. The results are presented as mean ±SD (n=3). * P<0.05, ** P<0.01, versus the corresponding cells treated with 0 μM Pb(Ac)2.
Effect of lead exposure on promoter methylation of DNA repair genes
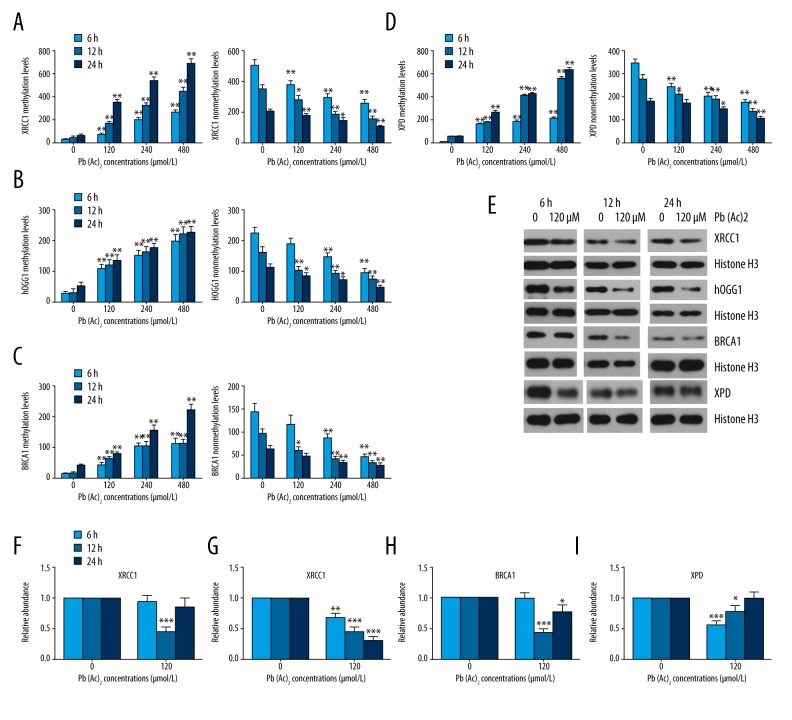
The methylation status of DNA repair genes was assessed by methylation-specific PCR. As shown in Figure 4A–4D, the methylation levels of XRCC1, hOGG-1, BRCA1, and XPD were significantly enhanced, whereas their unmethylation levels were obviously decreased with increasing time and concentration. Promoter methylation has been confirmed to repress gene transcription without changing the sequence, so we further assessed the protein levels of XRCC1, hOGG-1, BRCA1, and XPD. The results showed that lead exposure resulted in remarkable decreases in the protein levels of XRCC1 at 12 h; hOGG-1 at 6, 12, and 24 h; BRCA1 at 12 and 24 h; and XPD at 6 and 12 h in TK6 cells after exposure to lead.
Figure 4.
Effect of lead exposure on promoter methylation of DNA repair genes. (A–D) The methylation and nonmethylation levels of XRCC1, hOGG-1, BRCA1, and XPD in TK6 cells were assessed by methylation-specific PCR (MS-PCR) at the indicated time points and concentrations. (E) The protein levels of XRCC1, hOGG-1, BRCA1, and XPD in TK6 cells were detected by Western blot assay. Histone H3 was used as a loading control. (F–I) The quantitative analysis of the gray-scale values of the bands. The results are presented as mean ±SD (n=3). * P<0.05, ** P<0.01, *** P<0.001, versus the corresponding cells treated with 0 μM Pb(Ac)2.
Discussion
Genotoxicity is an important biological event in the processes of mutagenesis, teratogenesis [22], and carcinogenesis [23], which not only causes harm to parental health, but also to their offspring. This is the first study that observed the genotoxicity of lead in human TK6 cells and elucidated its potential molecular mechanisms. It has been reported that lead can induce genotoxicity in various species, including plants [24], humans [25], and polychaetes [26]. Unlike previous research, our study may provide a direct theoretical basis for possible prevention and control strategies for human exposure to lead.
The DNA double-strand breaks (DSBs) of eukaryotic cells can trigger the phosphorylation of histone H2AX, forming γ-H2AX [27]. There is a positive correlation between DSBs and γ-H2AX and γ-H2AX is an acknowledged marker for DSBs. The quantification of γ-H2AX has been widely used for evaluation of genotoxicity in many studies [28–30]. In addition, γ-H2AX is a DSB biomarker that reveals different types of DNA damage, notably DNA adducts and oxidative DNA lesions [31,32]. A recent study by Kopp et al. investigated the genotoxicity of 11 heavy metals, including lead, by γ-H2AX assay [31]; the γ-H2AX foci formation was detected by immunofluorescence staining. The results showed that lead exposure caused a time- and concentration-dependant increase in γ-H2AX foci formation in TK6 cells, which suggested that lead induced obvious DSBs and had potential genotoxicity to humans.
Oxidative stress damage can be triggered by lead exposure, and DNA damage is an important consequence of oxidative stress. Therefore, we further evaluated the role of lead in oxidative stress damage in TK6 cells. 8-OHdG is a molecular marker for DNA oxidative damage [33]. It has been reported that 8-OHdG concentration in the urine of people occupationally exposed to lead was significantly increased [34]. According to our results, the 8-OHdG level in TK6 cells was also enhanced by lead exposure, suggesting increased DNA oxidative damage. Furthermore, we assessed the levels of more oxidative stress-related markers or enzymes. ROS, largely produced by the stimulated cells, destroys the balance between oxidative and anti-oxidative systems and finally causes oxidative stress injury [35]. It is well documented that ROS can attack DNA and causes DNA damage [36]. MDA is a product of lipid peroxidation, which can reflect the degree of lipid peroxidation. GSH is an important antioxidant in the body, while GSSG is the oxidized state of GSH. The GSH level drop and the GSSG level rises when the body is challenged by oxidative stress. Therefore, the ratio of GSH/GSSG is a sensible index of oxidative stress state [37]. SOD is a key enzyme that can effectively scavenge superoxide free radicals and protect the body from oxidative stress damage [38]. In the present study, the ROS, MDA, and GSSG levels were increased and GSH level and SOD activity were decreased in TK6 cells after exposure to lead. These results suggest that oxidative stress is involved in lead-induced genotoxicity in TK6 cells.
To further evaluate the mechanisms of oxidative stress in lead-induced genotoxicity, we focused on the Nrf2-ARE signaling pathway. Nrf2 is an important transcription factor that regulates oxidative stress reaction and maintains the redox balance [39]. Under physiological conditions, Nrf2 is inactivated by binding with Kelch-like ECH-associated protein1 (Keap1) in the cytoplasm. Upon cellular stress, Nrf2 is separated from Keap1, translocates into the nucleus, and binds with antioxidant response element (ARE) to promote the expressions of its target antioxidant genes [40]. HO-1 and NQO1 are 2 representative target genes of the Nrf2-ARE pathway, which prevent oxidative stress injury [41]. In the present study, lead exposure promoted the transcriptional activation of Nrf2 and increased its target genes HO-1 and NQO1 expressions in TK6 cells.
DNA repair genes play pivotal roles in DNA damage repair processes, and their expressions are negatively associated with the promoter methylation level [42]. DNA methylation is an important part of epigenetics that has been discovered to produce lasting changes in gene expression without altering the DNA sequence. It has been found that the DNA methylation can affect DNA oxidative damage repair [43]. In the present study, we assessed the DNA methylation of 4 DNA repair genes: XRCC1, hOGG-1, BRCA1, and XPD. XRCC1 is an important component of the base excision repair and participates in the maintenance of genome integrity and stability [44]. hOGG-1 can specifically inhibit 8-OHdG level, which is a key enzyme that repairs DNA oxidative injury [45]. BRCA1, as a tumor suppressor gene, plays important roles in DNA damage repair [46]. XPD is a ATP-dependent DNA helicase, which takes part in nucleotide excision repair [47]. According to our results, the methylation levels of XRCC1, hOGG-1, BRCA1, and XPD were significantly increased in TK6 cells at 6, 12, and 24 h after exposure to lead. However, their protein levels were not decreased accordingly at any time points. This discrepancy may be due to several causes. First, protein expression is a complex process that is regulated by multiple factors, not limited to DNA methylation. In addition, there is a time difference in the processes of DNA methylation and corresponding protein expression.
The limitations of this study include the following. First, this study was carried out on only 1 human cell line, and the obtained results need to be verified in multiple human cell lines. Second, the role of these DNA repair proteins (XRCC1, hOGG-1, BRCA1, and XPD) in lead-induced genotoxicity is unclear and needs to be elucidated in the future.
Conclusions
Taken together, our results indicate that lead exposure decreased the cell viability, induced oxidative stress-mediated DNA damage via the Nrf2-ARE signaling pathway, and decreased the expressions of DNA repair genes via promoting their promoter methylation in human TK6 cells. Our study elucidates the genotoxic mechanisms of lead in human TK6 cells for the first time, which provides more direct and reliable theoretical evidence for the clinical prevention and treatment of human lead poisoning.
Footnotes
Conflict of interest
None.
Source of support: This study was supported by grants to Xiangquan Liu from the Young and Middle-aged Backbone Talent Training Project of Fujian Provincial Health Systems (No. 2014-ZQN-JC-35) and the Scientific Research Innovation Team Cultivation Project of Fuzhou Health and Family Planning Commission (No. 2016-S-wt12)
References
- 1.Chen L, Xu Z, Liu M, et al. Lead exposure assessment from study near a lead-acid battery factory in china. Sci Total Environ. 2012;429:191–98. doi: 10.1016/j.scitotenv.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 2.Zhang H, Wei K, Zhang M, et al. Assessing the mechanism of DNA damage induced by lead through direct and indirect interactions. J Photochem Photobiol B. 2014;136:46–53. doi: 10.1016/j.jphotobiol.2014.04.020. [DOI] [PubMed] [Google Scholar]
- 3.Brewster UC, Perazella MA. A review of chronic lead intoxication: An unrecognized cause of chronic kidney disease. Am J Med Sci. 2004;327:341–47. doi: 10.1097/00000441-200406000-00008. [DOI] [PubMed] [Google Scholar]
- 4.Pasha Shaik A, Sankar S, Reddy SC, et al. Lead-induced genotoxicity in lymphocytes from peripheral blood samples of humans: In vitro studies. Drug Chem Toxicol. 2006;29:111–24. doi: 10.1080/01480540500408739. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Leston J, Mendez J, Pasaro E, Laffon B. Genotoxic effects of lead: An updated review. Environ Int. 2010;36:623–36. doi: 10.1016/j.envint.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 6.Ustundag A, Behm C, Follmann W, et al. Protective effect of boric acid on lead- and cadmium-induced genotoxicity in v79 cells. Arch Toxicol. 2014;88:1281–89. doi: 10.1007/s00204-014-1235-5. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Leston J, Roma-Torres J, Vilares M, et al. Genotoxic effects of occupational exposure to lead and influence of polymorphisms in genes involved in lead toxicokinetics and in DNA repair. Environ Int. 2012;43:29–36. doi: 10.1016/j.envint.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Kasuba V, Rozgaj R, Milic M, et al. Evaluation of genotoxic effects of lead in pottery-glaze workers using micronucleus assay, alkaline comet assay and DNA diffusion assay. Int Arch Occup Environ Health. 2012;85:807–18. doi: 10.1007/s00420-011-0726-4. [DOI] [PubMed] [Google Scholar]
- 9.Nersesyan A, Kundi M, Waldherr M, et al. Results of micronucleus assays with individuals who are occupationally and environmentally exposed to mercury, lead and cadmium. Mutat Res. 2016;770:119–39. doi: 10.1016/j.mrrev.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Borghini A, Gianicolo EA, Andreassi MG. Usefulness of biomarkers as intermediate endpoints in health risks posed by occupational lead exposure. Int J Occup Med Environ Health. 2016;29:167–78. doi: 10.13075/ijomeh.1896.00417. [DOI] [PubMed] [Google Scholar]
- 11.Glover KP, Markell LK, Donner EM, Han X. Protein kinase c-activating tumor promoters modulate the DNA damage response in uvc-irradiated tk6 cells. Toxicol Lett. 2014;229:210–19. doi: 10.1016/j.toxlet.2014.06.030. [DOI] [PubMed] [Google Scholar]
- 12.Ji Z, Zhang L, Guo W, et al. The benzene metabolite, hydroquinone and etoposide both induce endoreduplication in human lymphoblastoid tk6 cells. Mutagenesis. 2009;24:367–72. doi: 10.1093/mutage/gep018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Recio L, Shepard KG, Hernandez LG, Kedderis GL. Dose-response assessment of naphthalene-induced genotoxicity and glutathione detoxication in human tk6 lymphoblasts. Toxicol Sci. 2012;126:405–12. doi: 10.1093/toxsci/kfs012. [DOI] [PubMed] [Google Scholar]
- 14.Olaharski AJ, Ji Z, Woo JY, et al. The histone deacetylase inhibitor trichostatin a has genotoxic effects in human lymphoblasts in vitro. Toxicol Sci. 2006;93:341–47. doi: 10.1093/toxsci/kfl068. [DOI] [PubMed] [Google Scholar]
- 15.Singh Z, Chadha P, Sharma S. Evaluation of oxidative stress and genotoxicity in battery manufacturing workers occupationally exposed to lead. Toxicol Int. 2013;20:95–100. doi: 10.4103/0971-6580.111550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma B, Singh S, Siddiqi NJ. Biomedical implications of heavy metals induced imbalances in redox systems. Biomed Res Int. 2014;2014:640754. doi: 10.1155/2014/640754. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Dutta S, Warshall C, Bandyopadhyay C, et al. Interactions between exosomes from breast cancer cells and primary mammary epithelial cells leads to generation of reactive oxygen species which induce DNA damage response, stabilization of p53 and autophagy in epithelial cells. PLoS One. 2014;9:e97580. doi: 10.1371/journal.pone.0097580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gastaldo J, Viau M, Bencokova Z, et al. Lead contamination results in late and slowly repairable DNA double-strand breaks and impacts upon the atm-dependent signaling pathways. Toxicol Lett. 2007;173:201–14. doi: 10.1016/j.toxlet.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 19.Spencer DM, Bilardi RA, Koch TH, et al. DNA repair in response to anthracycline-DNA adducts: A role for both homologous recombination and nucleotide excision repair. Mutat Res. 2008;638:110–21. doi: 10.1016/j.mrfmmm.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 20.Leng S, Stidley CA, Willink R, et al. Double-strand break damage and associated DNA repair genes predispose smokers to gene methylation. Cancer Res. 2008;68:3049–56. doi: 10.1158/0008-5472.CAN-07-6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng X, Li X, Xu C, et al. Schisandra sphenanthera extract (wuzhi tablet) protects against chronic-binge and acute alcohol-induced liver injury by regulating the nrf2-are pathway in mice. Acta Pharm Sin B. 2017;7:583–92. doi: 10.1016/j.apsb.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wells PG, Kim PM, Laposa RR, et al. Oxidative damage in chemical teratogenesis. Mutat Res. 1997;396:65–78. doi: 10.1016/s0027-5107(97)00175-9. [DOI] [PubMed] [Google Scholar]
- 23.Shelby MD. The genetic toxicity of human carcinogens and its implications. Mutat Res. 1988;204:3–15. doi: 10.1016/0165-1218(88)90113-9. [DOI] [PubMed] [Google Scholar]
- 24.Silva S, Silva P, Oliveira H, et al. Pb low doses induced genotoxicity in lactuca sativa plants. Plant Physiol Biochem. 2017;112:109–16. doi: 10.1016/j.plaphy.2016.12.026. [DOI] [PubMed] [Google Scholar]
- 25.Dobrakowski M, Pawlas N, Kasperczyk A, et al. Oxidative DNA damage and oxidative stress in lead-exposed workers. Hum Exp Toxicol. 2017;36:744–54. doi: 10.1177/0960327116665674. [DOI] [PubMed] [Google Scholar]
- 26.Singh N, Bhagat J, Ingole BS. Genotoxicity of two heavy metal compounds: Lead nitrate and cobalt chloride in polychaete perinereis cultrifera. Environ Monit Assess. 2017;189:308. doi: 10.1007/s10661-017-5993-4. [DOI] [PubMed] [Google Scholar]
- 27.Zhao X, Toyooka T, Ibuki Y. Silver ions enhance uvb-induced phosphorylation of histone h2ax. Environ Mol Mutagen. 2014;55:556–65. doi: 10.1002/em.21875. [DOI] [PubMed] [Google Scholar]
- 28.Hershman JM, France B, Hon K, Damoiseaux R. Direct quantification of gamma h2ax by cell-based high throughput screening for evaluation of genotoxicity of pesticides in a human thyroid cell lines. Environ Mol Mutagen. 2017;58:522–28. doi: 10.1002/em.22103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uboldi C, Orsiere T, Darolles C, et al. Poorly soluble cobalt oxide particles trigger genotoxicity via multiple pathways. Part Fibre Toxicol. 2016;13:5. doi: 10.1186/s12989-016-0118-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ando M, Yoshikawa K, Iwase Y, Ishiura S. Usefulness of monitoring gamma-h2ax and cell cycle arrest in hepg2 cells for estimating genotoxicity using a high-content analysis system. J Biomol Screen. 2014;19:1246–54. doi: 10.1177/1087057114541147. [DOI] [PubMed] [Google Scholar]
- 31.Kopp B, Zalko D, Audebert M. Genotoxicity of 11 heavy metals detected as food contaminants in two human cell lines. Environ Mol Mutagen. 2017 doi: 10.1002/em.22157. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 32.Ye B, Hou N, Xiao L, et al. Dynamic monitoring of oxidative DNA double-strand break and repair in cardiomyocytes. Cardiovasc Pathol. 2016;25:93–100. doi: 10.1016/j.carpath.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valavanidis A, Vlachogianni T, Fiotakis C. 8-hydroxy-2′-deoxyguanosine (8-ohdg): A critical biomarker of oxidative stress and carcinogenesis. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2009;27:120–39. doi: 10.1080/10590500902885684. [DOI] [PubMed] [Google Scholar]
- 34.Pawlas N, Olewinska E, Markiewicz-Gorka I, et al. Oxidative damage of DNA in subjects occupationally exposed to lead. Adv Clin Exp Med. 2017;26:939–45. doi: 10.17219/acem/64682. [DOI] [PubMed] [Google Scholar]
- 35.Pi J, Zhang Q, Fu J, et al. Ros signaling, oxidative stress and nrf2 in pancreatic beta-cell function. Toxicol Appl Pharmacol. 2010;244:77–83. doi: 10.1016/j.taap.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bisht S, Dada R. Oxidative stress: Major executioner in disease pathology, role in sperm DNA damage and preventive strategies. Front Biosci (Schol Ed) 2017;9:420–47. doi: 10.2741/s495. [DOI] [PubMed] [Google Scholar]
- 37.Collado R, Oliver I, Tormos C, et al. Early ros-mediated DNA damage and oxidative stress biomarkers in monoclonal b lymphocytosis. Cancer Lett. 2012;317:144–49. doi: 10.1016/j.canlet.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 38.Wu JQ, Kosten TR, Zhang XY. Free radicals, antioxidant defense systems, and schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2013;46:200–6. doi: 10.1016/j.pnpbp.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 39.Marhenke S, Lamle J, Buitrago-Molina LE, et al. Activation of nuclear factor e2-related factor 2 in hereditary tyrosinemia type 1 and its role in survival and tumor development. Hepatology. 2008;48:487–96. doi: 10.1002/hep.22391. [DOI] [PubMed] [Google Scholar]
- 40.Lu K, Alcivar AL, Ma J, et al. Nrf2 induction supporting breast cancer cell survival is enabled by oxidative stress-induced dpp3-keap1 interaction. Cancer Res. 2017;77:2881–92. doi: 10.1158/0008-5472.CAN-16-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satoh T, McKercher SR, Lipton SA. Nrf2/are-mediated antioxidant actions of pro-electrophilic drugs. Free Radic Biol Med. 2013;65:645–57. doi: 10.1016/j.freeradbiomed.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Thuijl HF, Mazor T, Johnson BE, et al. Evolution of DNA repair defects during malignant progression of low-grade gliomas after temozolomide treatment. Acta Neuropathol. 2015;129:597–607. doi: 10.1007/s00401-015-1403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng G, Fu Y, He C. Nucleic acid oxidation in DNA damage repair and epigenetics. Chem Rev. 2014;114:4602–20. doi: 10.1021/cr400432d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu J, Ma J, Zong HT, et al. Pharmacogenetic role of xrcc1 polymorphisms on the clinical outcome of gastric cancer patients with platinum-based chemotherapy: A systematic review and meta-analysis. Genet Mol Res. 2014;13:1438–46. doi: 10.4238/2014.March.6.2. [DOI] [PubMed] [Google Scholar]
- 45.Kuznetsov NA, Koval VV, Fedorova OS. Mechanism of recognition and repair of damaged DNA by human 8-oxoguanine DNA glycosylase hogg1. Biochemistry (Mosc) 2011;76:118–30. doi: 10.1134/s0006297911010123. [DOI] [PubMed] [Google Scholar]
- 46.Caestecker KW, Van de Walle GR. The role of brca1 in DNA double-strand repair: Past and present. Exp Cell Res. 2013;319:575–87. doi: 10.1016/j.yexcr.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 47.Wang F, Chang D, Hu FL, et al. DNA repair gene xpd polymorphisms and cancer risk: A meta-analysis based on 56 case-control studies. Cancer Epidemiol Biomarkers Prev. 2008;17:507–17. doi: 10.1158/1055-9965.EPI-07-2507. [DOI] [PubMed] [Google Scholar]