Abstract
Wheat is one of the most important cereals used worldwide in the form of a range of products. Crop landraces have been an immense source of diversity for the breeders. In the present study, 517 Indian wheat landraces have been observed for the difference in bread making quality by assessing allelic behaviour of high molecular weight-glutenin subunits (HMW-GS). A total of 33 Glu-1 alleles (3 at Glu-A1, 15 at Glu-B1 and 15 at Glu-D1) were detected in wheat landraces. Allelic frequency of HMW-GS allelic band pattern null, 17 + 18, 2 + 12 (24.75%) was found to be the highest. Allelic frequency of HMW-GS allele null (68.27%) at Glu-A1, 17 + 18 (49.14%) at Glu-B1, and 2 + 12 (72.81%) at Glu-D1 was found to be the highest Five Novel alleles were identified at Glu-D1 locus, 12*, 12.1, 12.1*, 12.2 and 12.3. As Glu-D1 has highest quality contribution as compared to Glu-A1 and Glu-B1, reporting novel alleles at Glu-D1 represents that genetic variability available for selection is increased and it will provide tools for breeders to further improve dough properties and bread making quality.
Electronic supplementary material
The online version of this article (10.1007/s13197-018-3259-y) contains supplementary material, which is available to authorized users.
Keywords: Genetic diversity, Glu-A1, Glu-B1, Glu-D1, Novel alleles, SDS-PAGE
Introduction
Improved varieties of crop tolerant to biotic and abiotic stresses have been restricted due to narrow genetic background. Earlier farmers grew genetic blended cultivars (landraces) on a very large scale and thereby extended genotypes possess effective genetic make-up against various plant diseases. It is realized now that the use of genetically varied varieties is an effective strategy for minimizing genetic vulnerability. Depending on their geographical regions, landraces had specific genetic background that can be used in genetic research program. In addition; landraces are important genetic resources that improve gene pools of modern cultivars by introducing new alleles.
Wheat is one of the most important cereals used worldwide, feeding about half of the world population and providing 20% of total protein and calories in human nutrition (Gupta and MacRitchie 1991). Wheat landraces are a dynamic population of cultivated wheat that has historical origin, distinct identity and lacks formal crop improvement and are important because they are genetically diverse, locally adapted (genetically adapted to withstand local climate, disease and pests, even cultural practices) and associated with traditional farming systems (hence often called farmer’s variety). Genetic diversity determines their potential for improved efficiency and hence their use for breeding and allele scoring as they have chances for presence of novel allele, which eventually will provide tools for breeders to further improve dough properties and gluten quality (Katyal et al. 2017). Wheat’s dough and flour have unique physical properties for making bread, cakes, biscuits, pasta and noodles (Kaur et al. 2014). Wheat quality is largely determined by storage proteins present in endosperm of wheat grain. End product quality depends on quality and quantity of wheat proteins. Wheat proteins were first classified, by Osborne (1907), into four main fractions according to their characteristics: Albumins (dissolve in water and coagulate on heat), Globulins (dissolve in neutral salt solution), Glidains/Prolamins (dissolve in 75% alcohol), and Glutenins/Glutelins (do not dissolve in alcohol, but dissolve in diluted acid or diluted base).
Roughly 85% of total protein content within wheat endosperm is gluten, responsible for unique viscoelastic properties of wheat dough. The major components of gluten are monomeric gliadins and polymeric glutenins. The composition and amount of these components, particularly HMW-GS, are associated with dough making quality and baking quality of wheat flour (Li Vigni et al. 2013). Glutenins are responsible for elasticity of dough and gliadins for extensibility of dough essential for bread-making (Payne et al. 1984). Glutenins are classified as high molecular weight-glutenin subunits (HMW-GS) located at Glu-1 loci and low molecular weight-glutenin subunits (LMW-GS) located at Glu-3 loci. The effects of the Glu-1 and Glu-3 alleles in a wider range of genotypes are needed before their use in predicting dough properties can be fully justified. A better understanding of the effect of individual alleles on quality parameters will provide clearer information for the breadmaking quality breeders. The genes for HMW-GS are encoded on the long arms of chromosomes 1A, 1B, and 1D at the Glu-A1, Glu-B1, and Glu-D1 loci, respectively (Payne 1987). The genes for LMW-GS occur on the short arms of chromosomes 1A, 1B, and 1D at the Glu-A3, Glu-B3 and Glu-D3 loci which are tightly linked to the Gli-1 locus (Singh and Shepherd 1988). HMW-GS and LMW-GS differ on the basis of amino acid composition, molecular weight and also in their structures (Žilic et al. 2011). The molecular weight of LMW-GS ranges from 23 to 68 kDa and from 77 to 160 kDa for HMW-GS (Veraverbeke et al. 2000). The HMW-GS of wheat proteins are quantitatively minor, but are important functionally in the process of bread making. The relationships between bread making quality and HMW-GS were studied as the presence or absence of subunits or as the quantity of a subunit affecting quality and the role of HMW-GS and LMW-GS together in improving bread making quality. Correlations and genetic studies of HMW-GS established subunits considering both positive (5 + 10) and negative (2 + 12) effects on bread making quality. In general, null at Glu-A1 locus, 6 + 8 subunit encoded at Glu-B1 and 2 + 12 at Glu-D1 are negatively related with the quality parameters. HMW-GS scoring system has been developed as the sum of the contributions of each of the three HMW-GS loci (Payne et al. 1987).
The present study investigated various Indian wheat landraces to find the frequency of occurrence of different alleles of HMW-GS and to find a novel allele of HMW-GS in wheat.
Materials and methods
Wheat samples
517 Indian wheat landraces were taken from the National Bureau of Plant Genetic Resources (NBPGR).
Extraction of HMW-GS proteins
Total seed protein was extracted from 10 mg of wheat flour by using method of Singh et al. 1991 with 0.5 ml of 0.05 M Tris-HCI buffer (pH 8.0) having 0.2% SDS and 5 M Urea. To this extraction buffer, 5 μl of 2-mercaptoethanol was added at the time of use, thoroughly mixed and centrifuged for 5 min. After that 35 µl of dye (containing bromophenol blue and glycerol) was added to each sample and tubes were centrifuged at 8000 rpm for 10 min. Finally, 5 µl of samples were loaded into the wells of SDS-PAGE gel (Lawrence and Shepherd 1980).
SDS-PAGE
The proteins extracted in the supernatant were size fractionated using SDS-PAGE (9.5% Acrylamide) using Laemmli buffer system (Laemmli 1970) and stained with Coomassie Brilliant Blue (CBB) dye (10% (v/v) acetic acid, 0.006% (w/v) Coomassie dye and 90% ddH2O). Gels were run at 60 mA for an hour, then at 120 mA for 4 h (Lawrence and Shepherd 1980). Destaining was done with 4% NaCl. After destaining, gels were photographed by placing them on white light. The bands of HMW-GS were read using the standardized HMW-GS methodology and the nomenclature described by Payne and Lawrence (1983).
Results
High level of HMW-GS polymorphism was displayed in results. 61 electrophoretic profiles were observed in analysis of 517 genotypes of Indian wheat landraces (Supplementary Table 1). A total of 33 Glu-1 alleles (3 at Glu-A1, 15 at Glu-B1 and 15 at Glu-D1) were detected in Indian wheat landraces. The dominant band pattern of HMW-GS was null, 17 + 18, 2 + 12 (24.75%). Three HMW-GS alleles namely, null, 1 and 2* were detected on the loci Glu-A1. The frequency of two active HMW-GS allele 1 and 2* were 2.32 and 29.4%, respectively. Null allele encoding no subunit was observed as the most frequent HMW-GS allele (68.27%) encoded by locus Glu-A1 (Fig. 1a). There was more variation at Glu-B1 and Glu-D1 loci as compared to Glu-A1. Fifteen HMW-GS alleles, seven in subunit pairs i.e., 13 + 16, 14 + 15, 17 + 18, 20x + 20y, 7 + 11, 7 + 8, & 7 + 9 and eight single/rare subunits i.e., 6, 7, 8, 9, 16, 19, 20, 21, were found on the loci Glu-B1. Subunit 17 + 18 was observed as the most frequent HMW-GS allele (49.14%) encoded by locus Glu-B1 (Fig. 1b).
Fig. 1.
HMW-GS allelic frequency in Indian wheat landraces, a Glu-A1 locus, b Glu-B1, c Glu-D1 locus
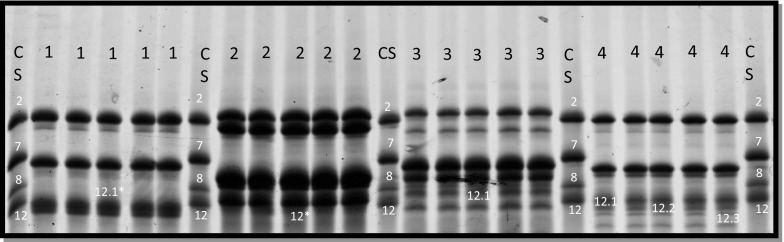
Fifteen alleles, four subunit pairs i.e., 2 + 10, 2 + 12, 5 + 10, and 5 + 12, and six single/rare alleles i.e., 2, 5, 10, 11, 12, and null, and five novel alleles were detected on Glu-D1 locus. HMW-GS allele 2 + 12 was the most abundant (72.81%) on Glu-D1 locus in Indian wheat landraces (Fig. 1c). Five novel alleles were detected on Glu-D1 loci, namely 12*, 12.1, 12.1*, 12.2 and 12.3, named on the basis of their mobility on SDS-PAGE (Fig. 2). Further elaboration of results to quantify allelic variation by wheat genomes showed minimum contribution by A genome (9.09%) and maximum as well as an equal contribution by B genome and D genome (45.45%). According to Payne et al. (1987), individual scores of HMW-GS alleles were summed to calculate quality (Glu) score of Indian wheat landraces (Supplementary Table 1). Based on HMW-GS composition among Indian wheat landraces, the quality score results showed higher variation among landraces with generally low score. The quality scores lies in the range of 1 to 10 with an average of 5. Only one landrace had a good quality score of 9 (0.19%). 79 landraces had quality score of 8 (15.28%). 17 landraces had a medium quality score of 7 (3.28%). Low quality score 6 was more frequent 52.8% in this investigation. Other lower quality scores i.e., 5,4,3,2, and 1 appeared with frequency of 0.38, 22.43, 2.51, 2.51, and 0.58%, respectively (Fig. 3).
Fig. 2.
SDS PAGE analysis of Indian wheat landraces (LR) showing novel alleles, CS Chinese spring. 1 = 1734-IC-104541B, 2 = 305-IC-75353C, 3 = 305-IC-75353D, 4 = 1014-IC-82326
Fig. 3.
Percentage of quality scores detected in Indian wheat landraces
Discussion
The most important source of food protein is cereals, including wheat. The major factor influencing the technological quality of wheat flour is the quantity and quality of seed storage proteins (Payne and Lawrence 1983). High molecular weight glutenin subunits (HMW-GS) influence the quality of seed in a positive and negative way. HMW-GS localized on Glu-D1 namely, 5 + 10 has a positive effect and 2 + 12 subunit has a negative effect on technological quality of wheat grain (Autran et al. 1990).
Earlier, some researchers monitored storage proteins composition of bread wheat ancestors and detected new and rare combination of HMW-GS alleles, which confirmed the hypothesis about close correlation of composition of HMW-GS alleles and origin of bread wheat genotypes (Sun et al. 2006). Juhász et al. (2003) investigated old genotypes of Bánkúti 1201 originated from Hungary to study influence of environment on HMW-GS composition and recognized multiline character and heterogeneity of this genotype which is typically shown by landraces. These results indicate needs for detail study of old genotypes and landraces as potential donors of specific features which can be utilized in bread wheat breeding program.
SDS-PAGE analysis of wheat glutenin proteins is used to identify the phenotypic effect of individual alleles of each gene. Here focus was done on detection of novel alleles by variance in electrophoretic spectra of individual HMW-GS in relationship to technological quality of 517 Indian wheat landraces. 61 different allelic combinations were observed in 517 Indian wheat landraces with a total of 33 Glu-1 alleles (Supplementary Table 1), depicting the presence of high allelic variation of HMW-GS responsible for differences in bread-making properties as well as providing tools for breeding future varieties. Storage protein being the direct expression of its genotype and is expected to be a cultivar constant element, thus it can provide a useful aid to cultivar identification.
In present study, Glu-A1 contributed three alleles Null, 1 and 2 with dominancy of null allele (68.28%) (Fig. 1a) which was in contrast with previous reports (Chaparzadeh et al. 2008; Goutam et al. 2015). Frequency of allele 1 was lowest (2.32%) and frequency of allele 2* was 29.4%. In contrast to our results, some researchers mentioned absence of allele 1 in their wheat genotypes (Chaparzadeh et al. 2008). The reason behind these differences may be due to the difference in germplasm used or lesser number of varieties used previously. Glu-Al allele 1 has positive contribution to wheat quality; hence quality of local varieties might be improved by introgressing allele 1 into their genetic background by continued efforts of hybridization and selection procedures. In this study high frequency of null allele was observed in Indian landraces which was in accordance to the findings of Mohibullah et al. 2015 but in contrary with researchers who estimated very few null alleles in Soviat wheat varieties (Morgunov et al. 1990).
Glu-B1 showed higher level of polymorphism by contributing 15 different types of allelic variants (Fig. 1b). The most frequent pattern was 17 + 18 (49.14%) in accordance with findings of Shitre et al. 2016. Similar to some previous reports, single/rare subunit 21 was detected in the present study (Dong et al. 2009). Subunit 20 was found in ninety-five varieties (17.95%) and was tenable with findings that reported subunit 20 as more frequent allele of Glu-B1 (Carrillo et al. 1995). However, present study results were contrary with findings of researchers who reported a lower frequency of subunit 20 in different wheat genotypes (Yan et al. 2007). In agreement with previous reports, here lower frequency of 13 + 16 alleles were observed (Mandonlakani et al. 2008). A low frequency of subunits 20x + 20y and 13 + 16 was found which was in agreement with previous studies who reported very low frequency, between 1% and 3% of these two subunit pairs (Payne and Lawrence 1983; Tohver 2007). In some reports, subunit pairs 20x +20y (x and y subunits) and 13 + 16 have not been found at all (Fang et al. 2009). However, some researchers reported higher frequency of subunit pairs 20x + 20y and 13 + 16 specifically in Protuguese wheat landraces (Rodriguez-Quijano et al. 1990). Other researchers also reported high frequency (88%) in Triticum aestivum ssp. spelta from North Spain (Caballero et al. 2004).
Glu-D1 locus also showed higher level of polymorphism by attributing 15 different types of allelic variants (Fig. 1c). Wheat grain flour quality is most influenced by alleles localized on Glu-D1 loci (Kolster 1991). We observed that HMW-GS allele 2 + 12 which have the negative effect on wheat flour quality was the most abundant (72.81%) in Indian wheat landraces and in agreement with other studies (Shitre et al. 2016). Again in agreement with results of some other reports, we reported a lower frequency of 5 + 10 (0.76%) (Chaparzadeh et al. 2008). Few rare subunit pair 5 + 12, and rare single subunits 10, 12, 5, 2 and 11 were also detected having an unknown quality contribution. A detailed study to access their effects on wheat quality must be conducted in the future. Some rare Null alleles were also detected at Glu-D1 locus which was in agreement with findings of Payne and Seekings (1994).
Five novel alleles were detected at Glu-D1 locus which were named according to their mobility as 12*, 12.1, 12.1*, 12.2 and 12.3 (Fig. 2). The electrophoretic mobility of subunit 12.1 was faster than that of subunit 12 while subunit 22.2 and 12.3 was found to be of higher mobility to 12.1 and 12.2 simultaneously.12* represents subunit of higher intensity but same mobility as of subunit 12. Reporting novel alleles at the Glu-D1 locus symbolizes that the genetic variation available for selection is enhanced and identification of novel alleles from the huge wheat gene pool will provide tools for breeders to further improve dough properties and gluten quality. The resulting knowledge will lead to better conservation of wheat genetic resources.
Seed storage proteins are product of many genes present in genome of species and their analysis can provide valuable information about evolutionary relationships and genomic complexity of species. The allelic variation dissection into genomic contribution revealed that Glu-B1 is a source of genetic variability, but highest quality contribution is through Glu-D1. Our study showed equal contribution of Glu-B1 and Glu-D1 towards genetic variance. Similarly, Chao et al. (2007) reported a ranking of B > D>A among US wheat genotypes, while Sheoran et al. (2015) identified highest mean number of alleles in the B genome followed by A and D genome,
Wheat flour’s bread-making quality is a complex trait influenced by environment. However, HMW-GS composition of wheat storage proteins can possibly predict technological quality of wheat flour by calculation of Glu score (Supplementary Table 1) (Lasztity 2003). Highest possible Glu score calculated value is 10 and lowest calculated value is 8 (for good technological quality). Some researchers investigated suitability of Glu-score as a universal tool for simple and rapid prediction of qualitative parameters of wheat genotypes (He et al. 2005). Present results showed a low frequency of good quality alleles (15.47%), hence generally low bread making quality of Indian wheat landraces which is in accordance with other reports which also suggested generally low bread making quality of the evaluated wheat germplasm (Schuster et al. 1997). The lower proportion of good quality score indicates landraces have been developed without complete knowledge of glutenin subunits and their effect on bread making quality.
In conclusion, present study revealed allelic variations among HMW-GS that strongly contributes towards high bread making quality. Five novel alleles were detected i.e., 12*, 12.1, 12.1*, 12.2, and 12.3 (Fig. 2) at Glu-D1 locus, having highest quality contributing alleles as compared to Glu-A1 and Glu-B1, which can provide tools for breeders to produce new wheat cultivar. New wheat cultivar production is costly, time consuming and demands a lot of effort. HMW-GS can serve as biochemical markers to breeders for screening wheat germplasm with high bread making quality, developing uniformity and improving heterogenous cultivars selecting best genotypes. The genotype having good quality alleles could be combined through various breeding procedures for producing high quality wheat.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to the ICAR Network Project on Transgenics in Crops (NPTC) for providing funding support. The authors would furthermore like to thank National Beaureu of Plant genetic resources, Delhi for providing wheat germplasm.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s13197-018-3259-y) contains supplementary material, which is available to authorized users.
References
- Autran JC, Laiegnelet B, Morel MH. Characterization and qualification of low molecular weight glutenins in durum wheats. Biochemie. 1990;69:669–711. doi: 10.1016/0300-9084(87)90191-x. [DOI] [PubMed] [Google Scholar]
- Caballero L, Martín LM, Alvarez JB. Genetic variability of the low-molecular-weight glutenin subunits in spelt wheat (Triticum aestivum ssp. spelta L. em Thell.) Theor Appl Genet. 2004;108:914–919. doi: 10.1007/s00122-003-1501-z. [DOI] [PubMed] [Google Scholar]
- Carrillo JM, Vázquez JF, Ruiz M. Variability for glutenin proteins in Spanish durum wheat landraces. Opt Mediter Ser A. 1995;22:143–147. [Google Scholar]
- Chao S, Zhang W, Dubcovskyand J, Sorrel M. Evaluation of genetic diversity and genomewide linkage disequilibrium among US wheat (Triticum aestivum L.) germplasm representing different market classes. Crop Sci. 2007;47:1018–1030. doi: 10.2135/cropsci2006.06.0434. [DOI] [Google Scholar]
- Chaparzadeh N, Sofalian O, Javanmard A, Hejazi MS, Zarandi L. Study of glutenin subunits in some wheat landraces from northwest of Iran by SDS-PAGE technique. Int J Agric Biol. 2008;10:101–104. [Google Scholar]
- Dong K, Hao CY, Wang AL, Cai MH, Yan YM. Characterization of HMW glutenin subunits in bread and tetraploid wheats by reversed-phase high-performance liquid chromatography. Cereal Res Commun. 2009;37:65–73. doi: 10.1556/CRC.37.2009.1.8. [DOI] [Google Scholar]
- Fang J, Liu Y, Luo J, Wang Y, Shewry PR, He G. Allelic variation and genetic diversity of high molecular weight glutenin subunit in Chinese endemic wheats (Triticum aestivum L.) Euphytica. 2009;166:177. doi: 10.1007/s10681-008-9812-4. [DOI] [Google Scholar]
- Goutam U, Tiwari R, Gupta RK, Kukreja S, Chaudhury A. Allelic variations of functional markers for high molecular weight glutenin genes in Indian wheat (Triticum aestivum L.) cultivars and their correlation with bread loaf volume. Indian J Plant Physiol. 2015;20:97–102. doi: 10.1007/s40502-015-0141-z. [DOI] [Google Scholar]
- Gupta RB, MacRitchie F. A rapid one-step one-dimensional SDS-PAGE procedure for analysis of subunit composition of glutenin in wheat. J Cereal Sci. 1991;14:105–109. doi: 10.1016/S0733-5210(09)80130-6. [DOI] [Google Scholar]
- He ZH, Liu L, Xia XC, Liu JJ, Peňa RJ. Composition of HMW and LMW glutenin subunits and their effects on dough properties, pan bread and noodle quality of chinese bread wheats. Cereal Chem. 2005;82:345–350. doi: 10.1094/CC-82-0345. [DOI] [Google Scholar]
- Juhász A, Larroque OR, Tamás L, Hsam SLK. Bánkúti 1201—an old Hungarian wheat variety with special storage protein composition. Theor Appl Genet. 2003;107:697–704. doi: 10.1007/s00122-003-1292-2. [DOI] [PubMed] [Google Scholar]
- Katyal M, Singh N, Virdi AS, Kaur A, Chopra N, Ahlawat AK, Singh AM. Extraordinarily soft, medium–hard and hard Indian wheat varieties: composition, protein profile, dough and baking properties. Food Res Int. 2017;100:306–317. doi: 10.1016/j.foodres.2017.08.050. [DOI] [PubMed] [Google Scholar]
- Kaur A, Singh N, Kaur S, Ahlawat AK, Singh AM. Relationships of flour solvent retention capacity, secondary structure and rheological properties with the cookie making characteristics of wheat cultivars. Food Chem. 2014;158:48–55. doi: 10.1016/j.foodchem.2014.02.096. [DOI] [PubMed] [Google Scholar]
- Kolster P (1991) High molecular weight glutenin subunits of wheat qualitative and quantitative variation in relation to BMQ. Ph.D. thesis, Agriculture University, Wageningen, Netherlands
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lasztity R. Prediction of wheat quality—success and doubts. Period Polytech Chem. 2003;46:39–49. [Google Scholar]
- Lawrence GJ, Shepherd KW. Variation in glutenin protein subunits of wheat. Aust J Biol Sci. 1980;2:221–234. doi: 10.1071/BI9800221. [DOI] [Google Scholar]
- Li Vigni M, Baschieri C, Marchetti Cocchi M. RP-HPLC and chemometrics for wheat flour protein characterisation in an industrial bread-making process monitoring context. Food Chem. 2013;139:553–562. doi: 10.1016/j.foodchem.2013.01.085. [DOI] [PubMed] [Google Scholar]
- Mandonlakani BA, Gomarian M, Shahnejat-Bushehri AA. Identification of parents for bread making quality improvement in bread wheat based on RAPD and seed storage protein (HMW) marker. Pak J Biol Sci. 2008;9:497. [Google Scholar]
- Mohibullah M, Rabbani MA, Munir M, Marwat SK, Gangohi S, Iqbal M, Amin A, Khakwani AA, Rashid M, Ullah I. Estimation of genetic divergence based on different quantitative traits and allelic variation in bread wheat (Triticum aestivum L.) germplasm through SDS-page techniques. Am Eurasian J Agric Environ Sci. 2015;15:1940. [Google Scholar]
- Morgunov AI, Rogers WJ, Sayers EJ, Metakovsky EV. The high-molecular-weight glutenin subunit composition of Soviet wheat varieties. Euphytica. 1990;51:41–52. doi: 10.1007/BF00022891. [DOI] [Google Scholar]
- Osborne TB. The Proteins of the Wheat Kernel. Washington: Carnegie Institution of Washington; 1907. [Google Scholar]
- Payne PI. Genetics of wheat storage proteins and the effect of allelic variation on bread-making quality. Ann Rev Plant Physiol. 1987;38:141–153. doi: 10.1146/annurev.pp.38.060187.001041. [DOI] [Google Scholar]
- Payne PI, Lawrence GJ. Catalogue of alleles for the complex gene loci, Glu-A1, Glu-B1, Glu D1, which code for high molecular weight subunits of glutenin in hexaploid wheat. Cereal Res Commun. 1983;11:29–35. [Google Scholar]
- Payne PI, Seekings JA (1994) Glu-D1 wheat flour blends. US patent no. 5308635 A
- Payne PI, Holt LM, Jackson EA, Law CN. Wheat storage proteins: their genetics and their potential for manipulation by plant breeding. Philos Trans R Soc Lond B. 1984;304:359–379. doi: 10.1098/rstb.1984.0031. [DOI] [Google Scholar]
- Payne PI, Nightingale MA, Krattiger AF, Holt LM. The relationship between HMW glutenin subunit composition and the bread-making quality of British-grown wheat varieties. J Sci Food Agric. 1987;40:51–65. doi: 10.1002/jsfa.2740400108. [DOI] [Google Scholar]
- Rodriguez-Quijano M, Vazquez JF, Carrillo JM. Variation of high-molecular-weight glutenin subunits in Spanish landraces of Triticum aestivum ssp. vulgare and ssp. spelta. J Genet Breed. 1990;44:121–126. [Google Scholar]
- Schuster I, Souza D, Cardoso MA, Sediyama AA, Moreira CS. Correlation between high molecular weight gluten subunits composition and bread making quality in Brazilian wheat. Brazil J Genet. 1997;20:1. doi: 10.1590/S0100-84551997000100001. [DOI] [Google Scholar]
- Sheoran S, Sharma P, Singh V, Pawar S, Sharma D, Jain N, Kumar R, Thakur V, Pandey GC, Malik R, Tiwari R. Assessment of genetic diversity in elite wheat genotypes using simple sequence repeat and quality protein markers. J Wheat Res. 2015;7:18–26. [Google Scholar]
- Shitre AS, Bakshi S, Gadekar DA, Padhye AP, Das BK. Characterization of high molecular weight glutenin subunits of wheat genotypes. Electron J Plant Breed. 2016;7:282–290. doi: 10.5958/0975-928X.2016.00036.3. [DOI] [Google Scholar]
- Singh NK, Shepherd KW. Linkage mapping of genes controlling endosperm storage proteins in wheat. 1 genes on the short arm of group 1 chromosomes. Theor Appl Genet. 1988;75:628–641. doi: 10.1007/BF00289132. [DOI] [Google Scholar]
- Singh NK, Shepherd KW, Cornish GB. A simplified SDS—PAGE procedure for separating LMW subunits of glutenin. J Cereal Sci. 1991;14(3):203–208. doi: 10.1016/S0733-5210(09)80039-8. [DOI] [Google Scholar]
- Sun X, Hu S, Liu X, Qian W. Characterization of the HMW glutenin subunits from Aegilopssearsii L. and identification of a novel variant HMW glutenin subunit. Theor Appl Genet. 2006;113:631–641. doi: 10.1007/s00122-006-0327-x. [DOI] [PubMed] [Google Scholar]
- Tohver M. High molecular weight (HMW) glutenin subunit composition of some Nordic and middle European wheats. Genet Res Crop Evol. 2007;54:67–81. doi: 10.1007/s10722-005-1885-5. [DOI] [Google Scholar]
- Veraverbeke WS, Larroque OR, Békés F, Delcour JA. In vitro polymerization of wheat glutenin subunits with inorganic oxidizing agents. I. Comparison of single-step and stepwise oxidations of high molecular weight glutenin subunits. Cereal Chem. 2000;77(5):582–588. doi: 10.1094/CCHEM.2000.77.5.582. [DOI] [Google Scholar]
- Yan ZH, Dai SF, Liu DC, Wei YM, Zheng YL. Allelic variation of high molecular weight glutenin sub units in hexaploid wheat landraces of Tibet, China. Int J Agric Res. 2007;2:838. doi: 10.3923/ijar.2007.838.843. [DOI] [Google Scholar]
- Žilić S, Barać M, Pešić M, Dodig D. Characterization of proteins from grain of different bread and durum wheat genotypes. Int J Mol Sci. 2011;12:5878–5894. doi: 10.3390/ijms12095878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.