Abstract
(1) Background. Non-small cell lung cancer (NSCLC) has a high mortality rate. MiRNAs have been found to be diagnostic biomarkers for NSCLC. However, controversial results exist. We conducted this meta-analysis to evaluate the diagnostic value of miRNAs for NSCLC. (2) Methods. Databases and reference lists were searched. Pooled sensitivity (SEN), specificity (SPE), and area under the curve (AUC) were applied to examine the general diagnostic efficacy, and subgroup analysis was also performed. (3) Results. Pooled SEN, SPE, and AUC were 85%, 88%, and 0.93, respectively, for 71 studies. Multiple miRNAs (AUC: 0.96) obtained higher diagnostic value than single miRNA (AUC: 0.86), and the same result was found for Caucasian population (AUC: 0.97) when compared with Asian (AUC: 0.91) and Caucasian/African population (AUC: 0.92). MiRNA had higher diagnostic efficacy when participants contained both smokers and nonsmokers (AUC is 0.95 for imbalanced group and 0.91 for balanced group) than when containing only smokers (AUC: 0.90). Meanwhile, AUC was 0.91 for both miR-21 and miR-210. (4) Conclusions. Multiple miRNAs such as miR-21 and miR-210 could be used as diagnostic tools for NSCLC, especially for the Caucasian and nonsmoking NSCLC.
1. Introduction
Lung cancer is the principal cause of cancer-associated deaths among males both in developed and in developing countries, and it has exceeded the breast cancer becoming the major cause of cancer-related deaths in females in the developed countries [1]. Non-small cell lung cancer (NSCLC) is a major type of lung cancer that is responsible for 85% lung cancer-associated deaths. Smoking has been recognized as a primary environmental risk factor of lung cancer. However, only a small number of smokers will develop into lung cancer patients.
MicroRNA is a group of 19–22 nucleotide, small, single-stranded, and conserved noncoding RNA that acts as a regulator of gene expression at both the posttranscriptional and the translational levels through acting on the 3′-untranslated region (UTR) of messenger RNA (mRNA) [2]. MiRNAs play important roles in various biological processes associated with the tumorigenesis such as the cellular proliferation, differentiation, metabolism, and apoptosis [3, 4]. It is available to isolate the miRNAs from the clinical specimens including the plasma, serum, sputum, and tissue. Meanwhile, it has a high stability. Due to these advantages, the miRNAs are increasingly becoming an ideal tool for the detection of NSCLC.
Recently, a series of articles have shown that different miRNAs might be applied to detect the NSCLC [5–7]. For example, miR-21, an oncogenic miRNA, has been shown to be overexpressed in lung cancer as well as other various human tumors [8]. Upregulation of miR-21 could promote the tumorigenesis of lung cancer through inhibiting the apoptosis process and negatively regulating the Ras/MEK/ERK signal pathway [9]. High miR-210 expression was correlated with the increased lymph node metastasis and a poor prognosis in patients with NSCLC [10]. Both these two, miR-21 and miR-210, have been explored to be used as diagnostic tools for NSCLC, no matter whether they are applied in combination with other miRNAs or alone [11–14]. However, as a result of the small sample sizes, the different miRNAs profiling, and the differences of the specimen and ethnicity, inconsistencies existed among studies that had examined the diagnostic value of miR-21, miR-210, and other miRNAs for NSCLC. Therefore, a meta-analysis was performed to assess the performance of miRNAs in the detection for NSCLC.
2. Materials and Methods
2.1. Search Strategy
Our meta-analysis was based on the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA). We searched PubMed, Google Scholar, Chinese National Knowledge Infrastructure (CNKI), Embase, and Medline to find all associated articles in order to investigate the potential utility of miRNAs as diagnostic tools for NSCLC. The combination of the Medical Subject Headings (MeSH) and the keywords (“lung neoplasm” OR “lung malignancy” OR “lung cancer”) AND (“miRNA” OR “microRNAs”) AND (“ROC curve” OR “sensitivity” OR “specificity” OR “diagnosis”) was used (updated to April 5, 2017). The reference lists of the reviews were also searched to obtain all the acceptable articles.
2.2. Study Selection
A series of criteria were applied for study inclusion and exclusion. For inclusion, the criteria were as follows: (1) patients with NSCLC; (2) the type of the controls being healthy controls (HC) or patients with benign pulmonary diseases (BPD); (3) assessing the diagnostic value of the miRNAs; (4) the possibility of extracting or calculating TP, FP, FN, and TN from the articles. For exclusion, the criteria were as follows: (1) studies that were duplicate publications, reviews, or unrelated; (2) studies without complete data.
2.3. Data Collection and Quality Assessment
Two authors collected the data independently as follows: the first author, publication year, and participant demographic characteristics (ethnicity, sample size, mean or median age, smoking status, the types of the controls, and the testing method of controls and cancer); types of the specimen; miRNA profiling and the data used for this meta-analysis (SEN, SPE, TP, FP, FN, and TN). The quality of these articles were assessed with the QUADAS-2 guidelines [15].
2.4. Statistical Analysis
All the statistical analyses were conducted by RevMan 5.3 (version 1.4) software and STATA 11.0 (STATA-Corp, College Station, TX, version 11.0) software. The heterogeneity among the selected studies was assessed through the Q test and the I2 value [16]. The P value for the Q test being less than 0.05 or the I2 ≥ 50 % demonstrated that there was heterogeneity among the included studies. The pooled SPE [TN/(FP+TN)], SEN [TP/(FN+TP)], diagnostic odds ratio (DOR) [PLR/NLR], the negative likelihood ratio (NLR) [(1-SPE)/SPE)], the positive likelihood ratio (PLR) [(SEN/(1-SEN)], and their 95% confidence intervals (95% CIs) were evaluated by a bivariate random-effect-regression model. The SROC curve was constructed and the AUC value was calculated too. A Fagan nomogram was also constructed to evaluate the clinical utility of miRNAs in the diagnosis of NSCLC. Subgroup analyses (grouped by miRNA profiling: single and multiple; smoking status: only smokers, smokers, and nonsmokers (imbalanced between groups), smokers and nonsmokers (balanced between groups), and unknown smoking status; specimen: serum, plasma, whole blood/blood cell, and not blood; ethnicity: Asian, Caucasian, and Caucasian/African; control-type: BPD, HC, and BPD/HC; stage: early stage and no early stage; and case number: large (≥ 50) and small (< 50)) and meta-regression analysis were used to identify the potential sources of the heterogeneity. The Deeks' funnel plot asymmetry test was also applied to explore the publication bias, with the P value less than 0.01 considered significant [17].
3. Results
3.1. Literature Search and the Studies' Characteristics
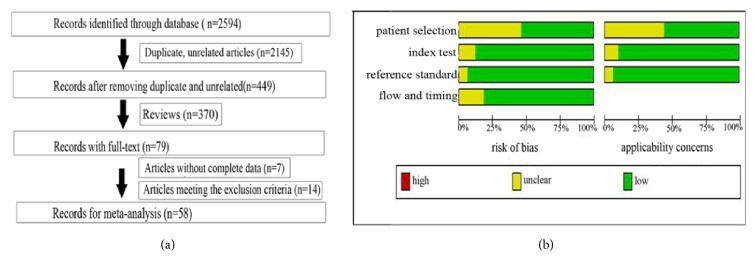
As shown in Figure 1(a), 2594 eligible articles were included, of which 2145 articles were removed as unrelated and duplicate articles. And then 370 reviews were also excluded, leaving 79 articles with full texts, and another 21 articles were then removed through carefully reading: 14 articles met the exclusion criteria and 7 articles did not have the complete data. Ultimately, 58 articles [5–7, 11–14, 18–68] with 71 studies published from 2009 to 2017 including 9,099 participants (5111 cases with NSCLC and 3988 controls from the healthy individuals and the patients with the benign pulmonary disease (BPD)) were included. The main characteristics of these 71 studies were shown in Table 1. Wang Y's article [7], Fan LH's article [52], Nadal E's article [45], Tang DF's article [32], Razzak R's article [14], Wang W's article [68], Yu L's article [19], and Xing LX's article [18] included 2 studies. Bediaga's article [28] included 3 studies, Wang C's article [46] included 4 studies, and the remaining articles [5, 6, 11–13, 20–27, 29–31, 33–44, 47–51, 53–67] included 1 study, respectively. Meanwhile, there were 18 studies [13, 14, 28, 31, 33, 45, 46, 48, 54, 56, 63, 65, 66] performed in Caucasian, 11 studies [18, 19, 21, 23, 27, 29, 30, 38, 44] performed in Caucasian/African, and 1 study [26] performed in African populations; the remaining studies were performed in Asian populations. A total of 50 studies detected the miRNAs in blood such as the whole blood, plasma, serum, and peripheral blood mononuclear cells (PBMC) [6, 7, 11, 20–24, 26, 29–35, 37, 39–42, 44–47, 49–54, 56–64, 66–68], while the remaining studies were detected in nonblood samples (7 tissue [5, 25, 28, 55, 68], 1 pleural effusion [43], 12 sputum [12, 14, 18, 19, 27, 36, 38, 48, 65], and 1 BAL [13]). We evaluated 45 studies for assessing the diagnostic value of multiple miRNAs and 26 studies [5, 6, 11, 20, 22, 24–26, 33–37, 39–41, 43, 47, 50, 51, 55, 57, 58, 60, 67, 68] of single miRNA.
Figure 1.
Flow chart of this meta-analysis of miRNAs in NSCLC detection (a) and the quality of these included articles according to the QUADAS-2 guidelines: proportion of articles with risk of bias (left) and proportion of articles with concerns regarding applicability (right) (b).
Table 1.
The main features of 71 included studies in this meta-analysis.
| Study ID | Ethnicity | Specimen | Case | Age | Control | Age | Type of control | Stage | MiRNA profiling | SEN (%) | SPE (%) | Reference miRNA | microRNA assay | Smoking status ∗ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | N | |||||||||||||
| Zhang H 2017 | Asian | plasma | 129 | 59.6 | 83 | 60.0 | HC | I-II | miR-145, miR-20a, miR-21, miR-223 | 81.8 | 90.1 | miR-16 | qRT-PCR | 3 |
| Halvorsen A 2016 | Caucasian | serum | 100 | 62.6 | 58 | 57.6 | HC | I-IV | miR-429, miR-205, miR-200b, miR-203, miR-12, miR-34b | 88.0 | 71.0 | miR-220, miR-19b, U6 | qRT-PCR | 3 |
| TaiMei C2016 | Asian | blood | 110 | 65.0 | 52 | 65.7 | HC | I-III | 20 miRNAs a | 89.1 | 100 | miR-159a, U6 | qRT-PCR | 4 |
| Su KL 2016 | Asian | plasma | 100 | NA | 100 | NA | HC | I-III | miR-195 | 78.0 | 86.0 | miR-39 | qRT-PCR | 3 |
| Zhu WY 2016 | Asian | plasma | 112 | 58.5 | 40 | 57.9 | HC | I-III | miR-182, miR-183, miR-210, miR-126 | 81.3 | 100 | U6 | qRT-PCR | 2 |
| Jiang LP 2016 | Asian | tissue | 154 | 54.9 | 63 | 57.8 | BPD | I-IV | miR-26b | 79.9 | 79.4 | U6 | qRT-PCR | 3 |
| Wang Y 2016 | Asian | plasma | 82 | NA | 91 | NA | HC | I-II | miR-532, miR-628, miR-425 | 91.5 | 97.8 | miR-39 | qRT-PCR | 4 |
| Wang Y 2016 | Asian | plasma | 36 | NA | 43 | NA | HC | I-II | miR-532, miR-628, miR-425 | 97.2 | 95.3 | miR-39 | qRT-PCR | 4 |
| Fan LH 2016 | Asian | serum | 94 | 60.5 | 58 | 58.1 | HC | I-III | miR-15b, miR-16, miR-20a | 86.2 | 91.4 | NA | qRT-PCR | 4 |
| Fan LH 2016 | Asian | serum | 70 | 59.7 | 54 | 58.0 | HC | I-III | miR-15b, miR-16, miR-20a | 94.3 | 94.2 | NA | FQDs | 4 |
| Sun L 2016 | Asian | plasma | 87 | 60.7 | 96 | 53.8 | HC,BPD | I-IV | miR-30a | 61.0 | 84.3 | U6 | qRT-PCR | 4 |
| Su Y 2016 | Asian | sputum | 144 | 66.3 | 171 | 65.2 | BPD | I | miR-21, miR-31, miR-210 | 81.5 | 85.9 | U6 | qRT-PCR | 1 |
| Gao X 2016 | Asian | plasma | 30 | 61.1 | 30 | 60.2 | HC | I | miR-324, miR-1285 | 93.3 | 90.0 | miR-39 | qRT-PCR | 4 |
| Wang X 2016 | Asian | plasma | 59 | 55.9 | 59 | 57.6 | BPD | I-III | miR-486 | 83.1 | 78.0 | miR-16 | qRT-PCR | 4 |
| Wei J 2016 | Asian | plasma | 63 | 61.0 | 30 | 57.0 | HC | I-IV | miR-21 | 76.2 | 70.0 | miR-16 | qRT-PCR | 3 |
| Razzak 2016 | Caucasian | sputum | 22 | 68 | 10 | 58 | HC,BPD | III-IV | miR-21, miR-210, miR-372 | 64 | 100 | U6 | qRT-PCR | 4 |
| Razzak 2016 | Caucasian | sputum | 21 | 70 | 10 | 58 | HC,BPD | I-II | miR-21, miR-210, miR-372 | 67 | 90 | U6 | qRT-PCR | 4 |
| Leidinger P 2016 | Caucasian | blood | 74 | NA | 20 | NA | HC | I-III | miR-720, miR-29c, miR-199a, miR-378a,let-7f | 91.0 | 98.0 | U24,U48 | qRT-PCR | 4 |
| Wang WZ 2016 | Asian | tissue | 15 | 57 | 16 | 58 | HC | I-IV | miR-182, miR-10a, miR-301b, miR-1244, miR-301a, miR-135b, miR-224, miR-21 | 93.3 | 93.8 | miR-16 | qRT-PCR | 4 |
| Wang WZ 2016 | Asian | serum | 54 | NA | 15 | NA | HC | I-IV | miR-1244 | 81.5 | 80 | miR-39 | qRT-PCR | 4 |
| Kim JL O 2015 | Caucasian | BAL | 21 | 70 | 10 | 59 | HC,BPD | I-II | miR-21, miR-143, miR-155, miR-210, miR-373 | 85.7 | 100 | U6 | qRT-PCR | 4 |
| Wang C 2015 | Asian | serum | 19 | 61.8 | 19 | 62.1 | HC | I-IV | miR-483, miR-193a, miR-25, miR-214, miR-7 | 100 | 84 | let-7d/g/i | qRT-PCR | 4 |
| Li WS 2015 | Asian | plasma | 11 | 59 | 11 | 55 | HC | I-III | mir-486 | 90.9 | 81.8 | miR-39, U44 | qRT-PCR | 4 |
| Wang C 2015 | Asian | serum | 63 | 61.9 | 63 | 59.7 | HC | I-IV | miR-483, miR-193a, miR-25, miR-214, miR-7 | 89.0 | 68.0 | let-7d/g/i | qRT-PCR | 4 |
| Wang C 2015 | Caucasian | serum | 108 | 67.2 | 56 | 63.7 | BPD | I-IV | miR-483, miR-193a, miR-25, miR-214, miR-7 | 95.0 | 95.0 | let-7d/g/i | qRT-PCR | 4 |
| Wang C 2015 | Caucasian | serum | 108 | 67.2 | 48 | 58.5 | HC | I-IV | miR-483, miR-193a, miR-25, miR-214, miR-7 | 95.0 | 84.0 | let-7d/g/i | qRT-PCR | 4 |
| Nadal E 2015 | Caucasian | serum | 70 | 67.5 | 22 | 67.0 | HC,BPD | I-III | miR-141, miR-200b, miR-193b | 96.0 | 95.0 | U6 | qRT-PCR | 2 |
| Nadal E 2015 | Caucasian | serum | 84 | 65.5 | 23 | 60.0 | HC,BPD | I-III | miR-141, miR-200b, miR-193b | 97.0 | 96.0 | U6 | qRT-PCR | 2 |
| Guo WG 2015 | Asian | plasma | 126 | NA | 50 | NA | HC | I-IV | mir-204 | 76.0 | 82.0 | U6 | qRT-PCR | 4 |
| Ma J 2015 | Caucasian, African | PBMC | 84 | 64.1 | 69 | 62.4 | BPD | I-IV | miR-19b, miR-29b | 72.6 | 82.6 | miR-423-3p | qRT-PCR | 2 |
| Li L 2015 | Asian | serum | 36 | 56.0 | 30 | 58.0 | HC,BPD | I-IV | miR-148a, miR-148b, miR-152 | 72.2 | 90.0 | U6 | qRT-PCR | 4 |
| Zhang XL 2015 | Asian | tissue | 125 | 61.0 | 125 | 61.0 | HC | I-IV | miR-141 | 64.8 | 64.8 | miR-191, miR-103 | qRT-PCR | 3 |
| Zhao W 2015 | Asian | serum | 80 | 57.6 | 60 | 55.4 | HC | NA | miR-21 | 73.8 | 71.7 | U6 | qRT-PCR | 4 |
| Wang RJ 2015 | Asian | serum | 70 | 64.4 | 70 | 63.7 | HC | NA | miR-145 | 92.8 | 61.4 | miR-39 | qRT-PCR | 3 |
| Yang JS 2015 | Asian | serum | 152 | NA | 300 | NA | HC | I-IV | miR-152, miR-148a, miR-148b, miR-21 | 96.0 | 91.0 | U6 | qRT-PCR | 3 |
| Xing LX 2015 | Caucasian | sputum | 67 | 66.4 | 69 | 64.9 | BPD | I-II | miR-21, miR-31, miR-210 | 82.1 | 88.4 | U6, miR-16 | qRT-PCR | 4 |
| Liu CM 2015 | Asian | Pleural effusion | 61 | 53.8 | 70 | 54.4 | BPD | NA | miR-192 | 61.3 | 79.5 | U6 | qRT-PCR | 2 |
| Dou HL 2015 | Asian | plasma | 120 | 63.2 | 360 | NA | HC | I-IV | miR-152 | 86.0 | 81.3 | U6 | digital PCR | 4 |
| Yang YL 2015 | Asian | PBMC | 74 | 62.5 | 52 | 61.8 | HC | I-IV | miR-10b | 86.5 | 76.9 | miR-16 | qRT-PCR | 3 |
| Li N 2014 | Caucasian, African | sputum | 35 | 68.9 | 40 | 65.7 | HC | I | miR-31, miR-210 | 65.7 | 85.0 | NA | qRT-PCR | 4 |
| Zhu W 2014 | Asian | serum | 70 | 59.0 | 48 | NA | HC | I-IV | miR-429 | 54.3 | 81.2 | U6,U48 | qRT-PCR | 4 |
| LI M 2014 | Asian | serum | 514 | NA | 54 | NA | HC | I-IV | miR-499 | 73.7 | 92.7 | miR-39 | qRT-PCR | 3 |
| Ulivi P 2013 | Caucasian | blood | 86 | 68.0 | 24 | 65.0 | HC | I-II | miR-328 | 70.0 | 83.0 | U38B,U58A | qRT-PCR | 4 |
| Bediaga 2013 | Caucasian | tissue | 45 | 66.4 | 45 | 66.4 | HC | I-IV | 8 miRNAs b | 100 | 97.8 | 4miRNAs c | qRT-PCR | 3 |
| Bediaga 2013 | Caucasian | tissue | 47 | 67.8 | 47 | 67.8 | HC | I-IV | 8 miRNAs b | 97.5 | 96.3 | 4miRNAs c | qRT-PCR | 3 |
| Bediaga 2013 | Caucasian | tissue | 22 | 68.4 | 22 | 68.4 | HC | I-IV | 8 miRNAs b | 100 | 95.0 | 4miRNAs c | qRT-PCR | 3 |
| Anjuman 2013 | Caucasian, African | sputum | 39 | 65.6 | 42 | 62.3 | BPD | I | miR-210, miR-31 | 61.5 | 90.5 | U6 | qRT-PCR | 4 |
| Tang DF 2013 | Asian | plasma | 62 | 64.8 | 60 | 66.0 | HC | I-III | miR-21, miR-145, miR-155 | 69.4 | 78.3 | U6 | qRT-PCR | 1 |
| Tang DF 2013 | Asian | plasma | 34 | 65.2 | 32 | 66.4 | HC | I-III | miR-21, miR-145, miR-155 | 76.5 | 81.3 | U6 | qRT-PCR | 1 |
| Mozzoni 2013 | Caucasian | plasma | 54 | 69.1 | 46 | 64.1 | BPD | I-III | miR-21, miR-486 | 87.0 | 86.5 | miR-16 | qRT-PCR | 4 |
| ZENG XL 2013 | Asian | PBMC | 64 | 58.9 | 26 | 54.4 | HC | I-IV | miR-143 | 75.0 | 92.3 | U6 | qRT-PCR | 4 |
| Yang XQ 2013 | Asian | sputum | 24 | 60.5 | 24 | 57.8 | BPD | I-IV | let-7a | 87.5 | 83.3 | U6 | digital PCR | 4 |
| Ma J 2013 | Caucasian, African | plasma | 36 | 66.7 | 38 | 64.6 | HC | I | miR-21, miR-335 | 71.8 | 80.6 | NA | qRT-PCR | 4 |
| Cazzoli R 2013 | Caucasian, African | plasma | 50 | 66.1 | 30 | 64.8 | BPD | I | miR-151a, miR-30a, miR-200b, miR-629, miR-100, miR-154 | 96.0 | 60.0 | let7a | qRT-PCR | 2 |
| Abd-E 2013 | African | Serum | 65 | 54.1 | 37 | 50.1 | HC | I-II | miR-182 | 100 | 86.5 | SNORD68 | qRT-PCR | 4 |
| Sanfiorenzo C 2013 | Caucasian | plasma | 52 | 65.1 | 10 | 68.9 | BPD | I-III | miR-152, miR-145, miR-199a, miR-24, miR-20a, miR-25 | 90.9 | 83.3 | miR-192, miR-16 | qRT-PCR | 4 |
| Roa Wilson H 2012 | Caucasian | sputum | 24 | 68.8 | 6 | 44.7 | HC | I-II | miR-21, miR-143, miR-155, miR-210, miR-372 | 83.3 | 100 | U6 | qRT-PCR | 4 |
| Li GJ 2012 | Asian | plasma | 16 | NA | 14 | NA | BPD | I | miR-494, miR-22, miR-200b | 85.3 | 94.5 | 18S | qRT-PCR | 4 |
| Ma YX 2012 | Asian | serum | 193 | NA | 110 | NA | HC | I-IV | miR-125b | 78.2 | 66.4 | NA | qRT-PCR | 4 |
| Hennessey P 2012 | Caucasian, African | serum | 55 | 68.2 | 75 | 65.7 | HC | I-IV | miR-15b, miR-27b | 100 | 84.0 | miR-16 | qRT-PCR | 4 |
| ZengXL 2012 | Asian | PBMC | 34 | NA | 26 | 54.4 | HC | I-IV | miR-150 | 87.5 | 69.2 | U6 | qRT-PCR | 4 |
| Zhao M 2012 | Asian | tissue | 55 | NA | 55 | NA | HC | I-IV | miR-29a | 49.1 | 85.5 | U6 | qRT-PCR | 3 |
| Shen J 2011 | Caucasian, African | plasma | 34 | 68.0 | 29 | 66.0 | HC | I-IV | miR-21, miR-126, miR-210, miR-486 | 91.7 | 96.6 | miR-16 | qRT-PCR | 1 |
| Jeong H 2011 | Asian | blood | 35 | 67.0 | 30 | 60.0 | HC | I-IV | let-7a | 90.3 | 90.3 | U6 | qRT-PCR | 4 |
| Wei J 2011 | Asian | plasma | 77 | 59.6 | 36 | 56.4 | HC | I-IV | miR-21 | 61.0 | 83.3 | miR-16 | qRT-PCR | 3 |
| Liu S 2011 | Asian | plasma | 130 | 53.1 | 170 | 57.5 | HC | I-III | miR-126 | 46.4 | 90 | NA | qRT-PCR | 3 |
| Yu L 2010 | Caucasian, African | sputum | 36 | 68.2 | 36 | 66.7 | HC | I | miR-486, miR-21, miR-200b, miR-375 | 80.6 | 91.7 | U6 | qRT-PCR | 4 |
| Yu L 2010 | Caucasian, African | sputum | 64 | 67.0 | 58 | 65.0 | HC | I-IV | miR-486, miR-21, miR-200b, miR-375 | 70.3 | 80.0 | U6 | qRT-PCR | 3 |
| Xing LX 2010 | Caucasian, African | sputum | 48 | 67.5 | 48 | 65.9 | HC | I | miR-205, miR-210, miR-708 | 73.0 | 96.0 | U6 | qRT-PCR | 4 |
| Xing LX 2010 | Caucasian, African | sputum | 67 | 68.0 | 55 | 65.0 | HC | I-IV | miR-205, miR-210, miR-708 | 72.0 | 95.0 | U6 | qRT-PCR | 3 |
| Keller Andreas 2009 | Caucasian | blood | 17 | 64.2 | 19 | 37.9 | HC | I-III | 24miRNAs d | 92.5 | 98.1 | NA | qRT-PCR | 4 |
amiR-451, miR-1290, miR-636, miR-30c, miR-22-3p, miR-19b, miR-486-5p, miR-20b, miR-93, miR-34b, miR-185, miR-126-5p, miR-93-3p, miR-1274a, miR-142-5p, miR-628-5p, miR-486-3p, miR-425, miR-645, miR-24; bmiR-96, miR-450a, miR-183, miR-9, miR-577, Let-7i, miR-27b and miR-34a; cmiR-26a, miR-140-5p, miR-195, miR-30b; dmiR-126, miR-423, miR-15a, let-7d, let-7i, miR-22, miR-98, miR-19a, miR-20b, miR-324, miR-574, miR-195, miR-25, let-7e, let-7c, let-7f, let-7a, let-7g, miR-140, miR-339, miR-361, miR-1283, miR-18a, miR-26b; ∗1: only smokers; 2: smokers and nonsmokers (smoking status was imbalanced between groups); 3: smokers and nonsmokers (smoking status was balanced between groups); 4: unknown smoking status.
N: number; HC: healthy control; BPD: benign pulmonary disease; miR: microRNA; SEN: sensitivity; SPE: specificity; FQDs: fluorescence quantum dots; BAL: bronchoalveolar lavage.
The quantitative real-time polymerase chain reaction (qRT-PCR) and digital polymerase chain reaction (digital PCR) were used in these studies to test the expression levels of different miRNAs, and the most common reference miRNAs were RNU6B, miR-39, and miR-16. Quality of the enrolled studies summarized in Figure 1(b) was generally good.
3.2. Pooled Diagnostic Performance
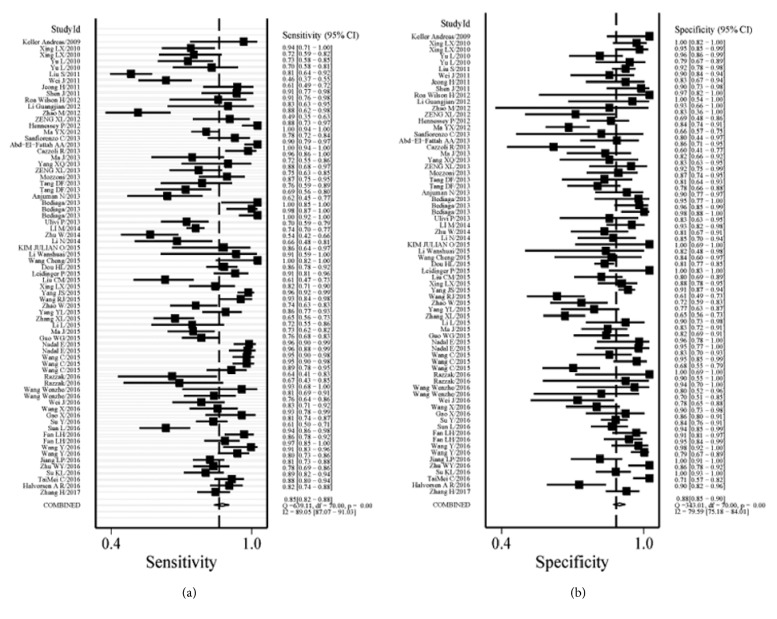
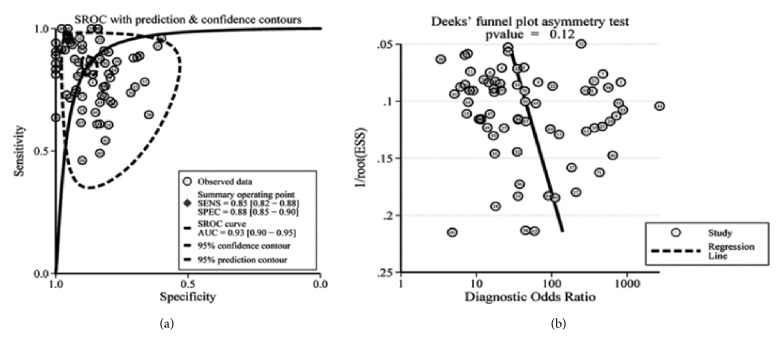
Significant heterogeneity was obtained since I2 values for SEN and SPE were 89.05% (95% CI: 87.07-91.03%) and 79.59% (95% CI: 75.18-84.01%), respectively. Therefore, a random-effect model was conducted for this study. Results indicated the pooled SEN and SPE for these 71 studies were 85% (95% CI: 82-88%) and 88% (95% CI: 85-90%), respectively (Figure 2). The PLR and NLR were 6.9 (95% CI: 5.6-8.4) and 0.17 (95% CI: 0.14-0.21), respectively (Figure 3), the DOR was 40 (95% CI: 28-58), and the AUC was 0.93 (95% CI: 0.90-0.95) (Figure 4(a)).
Figure 2.
Forest plots of SEN and SPE for the NSCLC diagnosis. Both the SEN and SPE of each study were shown by squares with the 95% confidence interval shown by the error bars.
Figure 3.
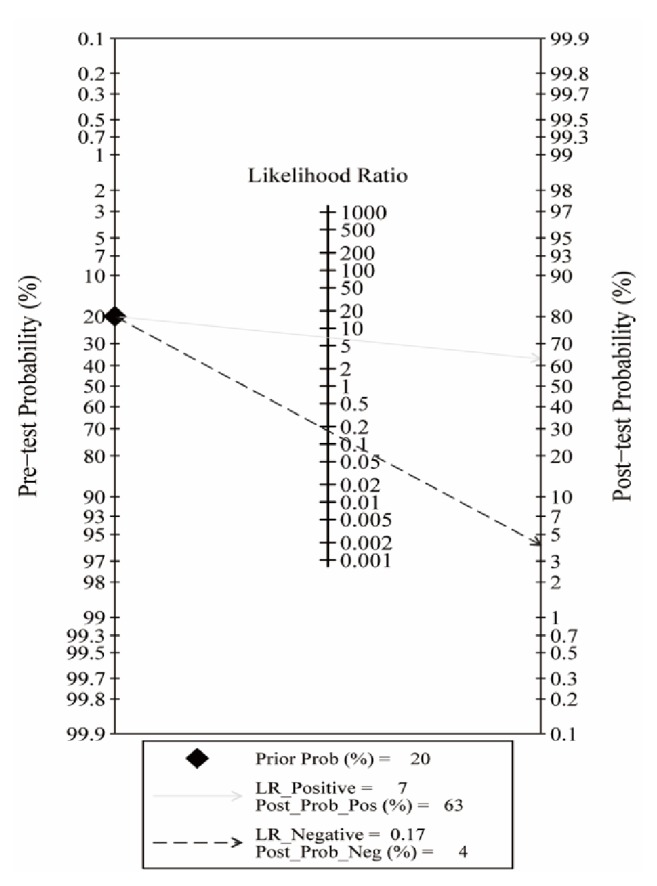
Fagan plot of PLR and NLR to evaluate the clinical utility of miRNAs for diagnosis of NSCLC.
Figure 4.
SROC curve of the miRNAs as diagnostic tools for NSCLC (a) and the Deeks' test for assessing the publication bias for miRNAs in the detection of NSCLC (b).
3.3. Publication Bias
Results of the Deeks' funnel plot asymmetry test showed that the publication bias did not exist in these studies as the funnel plot was symmetry (Figure 4(b)) and P value equaled 0.12.
3.4. Subgroup Analyses and Meta-Regression Analysis
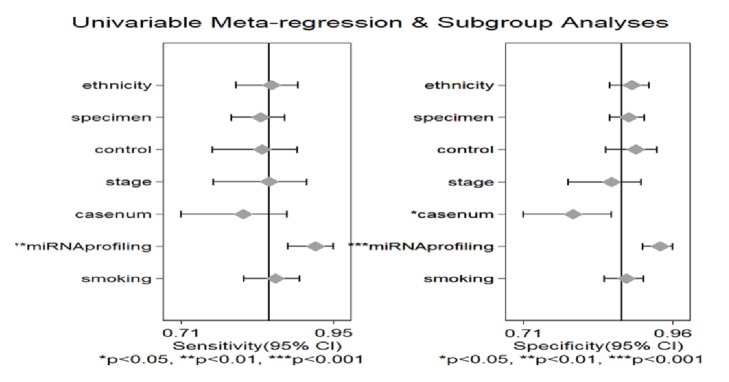
Results of the meta-regression analysis demonstrated that the heterogeneity might be explained by miRNA profiling (P < 0.001) and case number (P < 0.05) for SPE and by miRNA profiling (P < 0.01) for SEN as described in Figure 5. The subgroup analyses were also conducted and the results were presented in Table 2. For the subgroups of smoking status, compared with the subgroup of only smokers (SEN: 80% (95% CI: 70-87%), SPE: 86% (95% CI: 77-91%), and AUC: 0.90 (95% CI: 0.87-0.92)), miRNAs had a higher diagnostic efficacy in the subgroups of smokers and nonsmokers (SEN: 88% (95% CI: 74-95%), SPE: 90% (95% CI: 73-97%), and AUC: 0.95 (95% CI: 0.93-0.97) for imbalanced groups and SEN: 83% (95% CI: 74-90%), SPE: 86% (95% CI: 80-90%), and AUC: 0.91 (95% CI: 0.88-0.93) for balanced groups). Subgroup analysis by specimen showed that studies with serum samples exhibited higher diagnostic accuracy with SEN: 91% (95% CI: 86-95%), SPE: 85% (95% CI: 79-89%), and AUC: 0.94 (95% CI: 0.91-0.95) than studies with plasma samples with the SEN: 82% (95% CI: 76-87%), SPE: 87% (95% CI: 83-90%), and AUC: 0.92 (95% CI: 0.89-0.94) and not blooding samples with the SEN: 80% (95% CI: 72-86%), SPE: 89% (95% CI: 85-93%), and AUC: 0.92 (95% CI: 0.89-0.94), respectively. When compared with the large sample size, miRNA might be a better diagnostic tool for small sample size with SEN: 88% (95% CI: 82-92%), SPE: 91% (95% CI: 88-94%), and AUC: 0.95 (95% CI: 0.93-0.97). In the subgroups for the ethnicity, the miRNAs obtained a better diagnostic value in the Caucasian populations with the SEN: 91% (95% CI: 86-95%), SPE: 92% (95% CI: 87-96%), and AUC: 0.97 (95% CI: 0.95-0.98), respectively, when compared with the Asian populations with the SEN: 82% (95% CI: 77-85%), SPE: 86% (95% CI: 82-88%), and AUC: 0.91 (95% CI: 0.88-0.93), respectively, and the Caucasian/African populations with SEN: 85% (95% CI: 72-93%), SPE: 87% (95% CI: 81-91%), and AUC: 0.92 (95% CI: 0.89-0.94), respectively. In the subgroups of the miRNAs profiling, the multiple miRNAs had a higher accuracy for diagnosing the NSCLC with SEN: 88% (95% CI: 85-91%), SPE: 91% (95% CI: 88-93%), and AUC: 0.96 (95% CI: 0.93-0.97), respectively, when compared with the single miRNA with the SEN: 77% (95% CI: 71-82%), SPE: 80% (95% CI: 77-84%), and AUC: 0.86 (95% CI: 0.82-0.88), respectively. miRNAs had a higher value to distinguish the NSCLC patients from healthy individuals with the SEN: 86% (95% CI: 82-89%), SPE: 88% (95% CI: 85-91%), and AUC: 0.94 (95% CI: 0.91-0.95) than controls with benign pulmonary disease with SEN: 84% (95% CI: 77-89%), SPE: 84% (95% CI: 80-88%), and AUC: 0.90 (95% CI: 0.87-0.92). Compared with other miRNAs, miR-210 and miR-21 were more often used as diagnostic tools. However, they were usually associated with other miRNAs. The sensitivity, specificity, and AUC were, respectively, 77% (95% CI: 72-81%), 93% (95% CI: 88-96%), and 0.91(95% CI: 0.88-0.93) for miR-210 with other miRNAs. The sensitivity, specificity, and AUC of miR-21 with other miRNAs were, respectively, 82% (95% CI: 77-86%), 87% (95% CI: 84-89%), and 0.91 (95% CI: 0.88-0.93).
Figure 5.
Forest plots for the meta-regression analysis: SEN and SPE. The factors included miRNA profiling, smoking status, specimen, ethnicity, type of control, case number, and stage.
Table 2.
Subgroup analyses for the selected studies.
| Subgroups | No | SEN [95%CI] | SPE [95%CI] | PLR[95%CI] | NLR [95%CI] | DOR[95%CI] | AUC [95%CI] |
|---|---|---|---|---|---|---|---|
| MiR profiling | |||||||
| single | 26 | 0.77[0.71-0.82] | 0.80[0.77-0.84] | 3.9[3.3-4.7] | 0.28[0.22-0.36] | 14[10-20] | 0.86[0.82-0.88] |
| multiple | 45 | 0.88[0.85-0.91] | 0.91[0.88-0.93] | 10.0[7.5-13.3] | 0.13[0.10-0.17] | 79[50-126] | 0.96[0.93-0.97] |
| Smoking status | |||||||
| only smokers | 4 | 0.80[0.70-0.87] | 0.86[0.77-0.91] | 5.6[3.2-9.9] | 0.23[0.14-0.38] | 24[9-66] | 0.90[0.87-0.92] |
| S+NS (imbalanced) ∗ | 6 | 0.88[0.74-0.95] | 0.90[0.73-0.97] | 9.2[3.0-28.2] | 0.13[0.05-0.31] | 71[14-360] | 0.95[0.93-0.97] |
| S+NS (balanced) ∗ | 18 | 0.83[0.74-0.90] | 0.86[0.80-0.90] | 5.9[3.9-8.8] | 0.19[0.12-0.32] | 30[13-69] | 0.91[0.88-0.93] |
| unknown status | 43 | 0.86[0.82-0.89] | 0.88[0.85-0.91] | 7.3[5.7-9.4] | 0.16[0.12-0.21] | 46[30-70] | 0.93[0.91-0.95] |
| Specimen | |||||||
| plasma | 22 | 0.82[0.76-0.87] | 0.87[0.83-0.90] | 6.3[4.6-8.5] | 0.20[0.15-0.28] | 31[18-52] | 0.92[0.89-0.94] |
| serum | 19 | 0.91[0.86-0.95] | 0.85[0.79-0.89] | 6.1[4.3-8.5] | 0.10[0.06-0.17] | 60[28-128] | 0.94[0.91-0.95] |
| Whole blood/blood cell | 9 | 0.84[0.78-0.89] | 0.92[0.80-0.97] | 10.9[3.9-30.3] | 0.17[0.11-0.26] | 64[17-234] | 0.92[0.89-0.94] |
| not blood | 21 | 0.80[0.72-0.86] | 0.89[0.85-0.93] | 7.5[4.9-11.7] | 0.22[0.16-0.32] | 34[16-71] | 0.92[0.89-0.94] |
| Ethnicity | |||||||
| Asian | 41 | 0.82[0.77-0.85] | 0.86[0.82-0.88] | 5.7[4.5-7.2] | 0.21[0.17-0.27] | 27[18-40] | 0.91[0.88-0.93] |
| Caucasian | 18 | 0.91[0.86-0.95] | 0.92[0.87-0.96] | 12[7.0-20.4] | 0.09[0.06-0.15] | 127[54-302] | 0.97[0.95-0.98] |
| Caucasian/African | 12 | 0.85[0.72-0.93] | 0.87[0.81-0.91] | 6.6[4.6-9.4] | 0.17[0.09-0.33] | 39[17-88] | 0.92[0.89-0.94] |
| Control-type | |||||||
| BPD | 13 | 0.84[0.77-0.89] | 0.84[0.80-0.88] | 5.3[4.1-6.8] | 0.19[0.13-0.28] | 27[16-46] | 0.90[0.87-0.92] |
| HC | 50 | 0.86[0.82-0.89] | 0.88[0.85-0.91] | 7.4[5.7-9.5] | 0.16[0.12-0.21] | 47[30-74] | 0.94[0.91-0.95] |
| BPD, HC | 8 | 0.81[0.67-0.90] | 0.91[0.79-0.96] | 8.8[3.4-22.9] | 0.21[0.11-0.40] | 42[9-187] | 0.93[0.90-0.95] |
| Stage | |||||||
| I-II | 18 | 0.84[0.78-0.89] | 0.90[0.86-0.93] | 8.3[5.8-11.9] | 0.17[0.12-0.25] | 48[27-87] | 0.94[0.91-0.96] |
| I-IV | 50 | 0.86[0.82-0.89] | 0.88[0.84-0.90] | 6.5[5.4-8.7] | 0.16[0.13-0.22] | 42[27-66] | 0.93[0.90-0.95] |
| No. of cases | |||||||
| small | 25 | 0.88[0.82-0.92] | 0.91[0.88-0.94] | 10.0[7.1-14.2] | 0.14[0.09-0.21] | 74[38-143] | 0.95[0.93-0.97] |
| large | 46 | 0.84[0.79-0.87] | 0.86[0.82-0.88] | 5.8[4.6-7.2] | 0.19[0.15-0.24] | 31[20-46] | 0.91[0.89-0.94] |
| MiR-210 | 12 | 0.77[0.72-0.81] | 0.93[0.88-0.96] | 11.0[6.2-19.4] | 0.25[0.20-0.31] | 44[22-87] | 0.91[0.88-0.93] |
| MiR-21 | 16 | 0.82[0.77-0.86] | 0.87[0.84-0.89] | 6.3[5.0-8.1] | 0.21[0.15-0.28] | 31[19-50] | 0.91[0.88-0.93] |
No: the number of the studies; HC: healthy control; BPD: benign pulmonary disease; SEN: sensitivity; SPE: specificity; PLR: positive likelihood ratio; NLR:negative likelihood ratio; DOR: diagnostic odds ratio; AUC:area under the curve; no. of case: small (<50) and large (≥50).
∗ S: smokers; NS: nonsmokers; imbalanced: the smoking status was imbalanced between groups; balanced: the smoking status was balanced between groups.
4. Discussion
Due to the high mortality rate and low survival rate of NSCLC, there is an urgent need for the accurate detection method for the early detection of NSCLC especially for the nonsmoking NSCLC patients. Although miRNAs may have a high diagnostic accuracy according to the previous articles, the clinical utility of the miRNA for diagnosing NSCLC remains controversial. Compared with the previous meta-analyses [69–71], there were more studies and participants included in this meta-analysis. Our analysis showed the pooled SEN was 85% (95% CI: 82-88%), the pooled SPE was 88% (95% CI: 85-90%), and the AUC was 0.93 (95% CI: 0.90-0.95), suggesting that miRNAs had pretty high diagnostic value for NSCLC. Our results also showed that the pooled DOR was 40 (95% CI: 28-58), indicating that for an individual proved positive by miRNAs the chance of having NSCLC is 40 times higher than the negative ones. For the subgroup analyses, higher accuracy was observed in the multiple miRNA profiling when compared with the single miRNA, which was consistent with the previous conclusions [69–71]. MiRNAs might have a higher diagnostic efficacy for the nonsmoking NSCLC patients compared with the smoking ones. Meanwhile, differences were also observed among the Caucasian, Asian, and Caucasian/African populations. This result could be supported by the Wang H's article [71]. Furthermore, miRNAs from serum samples exhibited higher diagnostic value than miRNAs from other specimen. These results meant that combinations of various miRNAs may be better diagnostic tools than the single miRNA, and miRNA isolated from serum could have a higher diagnostic value for the Caucasian populations when compared with the Asian and Caucasian/African populations. Among the different multiple miRNAs, miR-210 and miR-21 associated with other miRNAs could be used for the detection of NSCLC. However, there were still some limitations that could not be neglected in this meta-analysis such as the heterogeneity among these 71 studies, the different methods in miRNA profiling, the possibility that some articles are missed or not published online.
5. Conclusions
Our meta-analysis showed the practicability of miRNAs for diagnosing NSCLC and demonstrated that the multiple miRNAs might have a relatively high diagnostic value for NSCLC compared with the single miRNA diagnosis. miR-210 and miR-21 could be used as effective tools through combining with other miRNAs. In addition, miRNAs, especially isolated from serum, had a better diagnostic accuracy in Caucasian populations than the Asian populations as well as the Caucasian/African populations. When compared with the smoking NSCLC patients, miRNAs might have a higher diagnostic efficacy for the nonsmoking ones. However, studies on the large samples are still demanded to verify our results.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (no. 81502878) and the Doctoral Research Project of Liaoning Province (no. 201601117).
Abbreviations
- miRNA:
MicroRNA
- NSCLC:
Non-small cell lung cancer
- SEN:
Sensitivity
- SPE:
Specificity
- SROC:
Summary receiver operating characteristic
- AUC:
The area under the SROC curve
- mRNA:
Messenger RNA
- 3′-UTR:
3′-untranslated region
- BPD:
Benign pulmonary disease
- HC:
Healthy Control
- CNKI:
Chinese national knowledge infrastructure
- PLR:
Positive likelihood ratio
- NLR:
Negative likelihood ratio
- DOR:
Diagnostic odds ratio
- TP:
True positive
- FP:
False positive
- FN:
False negative
- TN:
True negative
- qRT-PCR:
Quantitative real-time polymerase chain reaction
- PBMC:
Peripheral blood mononuclear cells
- PRISMA:
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- BAL:
Bronchoalveolar lavage.
Conflicts of Interest
The authors declare that there are no potential conflicts of interest.
Authors' Contributions
Baosen Zhou, Xuelian Li, and Min Jiang conceived and designed this study. Min Jiang and Xiaoying Li searched the literature and analyzed the data. Xiaowei Quan contributed to the analysis tools and the statistical analysis. Min Jiang and Baosen Zhou wrote and revised the paper.
References
- 1.Torre L. A., Bray F., Siegel R. L., Ferlay J., Lortet-Tieulent J. Global cancer statistics, 2012. CA: A Cancer Journal for Clinicians. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 3.Nana-Sinkam S. P., Croce C. M. Non-coding RNAs in cancer initiation and progression and as novel biomarkers. Molecular Oncology. 2011;5(6):483–491. doi: 10.1016/j.molonc.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calin G. A., Croce C. M. MicroRNA signatures in human cancers. Nature Reviews Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X., Li P., Rong M. MicroRNA-141 is a biomarker for progression of squamous cell carcinoma and adenocarcinoma of the lung: clinical analysis of 125 patients. The Tohoku Journal of Experimental Medicine. 2015;235(3):161–169. doi: 10.1620/tjem.235.161. [DOI] [PubMed] [Google Scholar]
- 6.Wang X., Zhi X., Zhang Y., An G., Feng G. Role of plasma MicroRNAs in the early diagnosis of non-small-cell lung cancers: a case-control study. Journal of Thoracic Disease. 2016;8(7):1645–1652. doi: 10.21037/jtd.2016.06.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y., Zhao H., Gao X., et al. Identification of a three-miRNA signature as a blood-borne diagnostic marker for early diagnosis of lung adenocarcinoma. Oncotarget . 2016;7(18):26070–26086. doi: 10.18632/oncotarget.8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang B., Zhang Q. The expression and clinical significance of circulating microRNA-21 in serum of five solid tumors. Journal of Cancer Research and Clinical Oncology. 2012;138(10):1659–1666. doi: 10.1007/s00432-012-1244-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatley M. E., Patrick D. M., Garcia M. R., et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18(3):282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osugi J., Kimura Y., Owada Y., et al. Prognostic Impact of Hypoxia-Inducible. Journal of Oncology. 2015;2015:1–8. doi: 10.1155/2015/316745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei J., Liu L.-K., Gao W., et al. Reduction of plasma MicroRNA-21 is associated with chemotherapeutic response in patients with non-small cell lung cancer. Chinese Journal of Cancer Research. 2011;23(2):123–128. doi: 10.1007/s11670-011-0123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su Y., Guarnera M. A., Fang H., Jiang F. Small non-coding RNA biomarkers in sputum for lung cancer diagnosis. Molecular Cancer. 2016;15(1, article no. 36) doi: 10.1186/s12943-016-0520-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J. O., Gazala S., Razzak R., et al. Non-small cell lung cancer detection using microRNA expression profiling of bronchoalveolar lavage fluid and sputum. Anticancer Research. 2015;35:1873–1880. [PubMed] [Google Scholar]
- 14.Razzak R., Bédard E., Kim J., et al. MicroRNA expression profiling of sputum for the detection of early and locally advanced non-small-cell lung cancer: a prospective case–control study. Current Oncology. 2016;23(2):p. 86. doi: 10.3747/co.23.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whiting P. F., Rutjes A. W. S., Westwood M. E., et al. Quadas-2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of Internal Medicine. 2011;155(8):529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 16.Higgins J. P. T., Thompson S. G., Deeks J. J., Altman D. G. Measuring inconsistency in meta-analyses. British Medical Journal. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deeks J. J., Macaskill P., Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. Journal of Clinical Epidemiology. 2005;58(9):882–893. doi: 10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 18.Xing L., Todd N. W., Yu L., Fang H., Jiang F. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Modern Pathology. 2010;23(8):1157–1164. doi: 10.1038/modpathol.2010.111. [DOI] [PubMed] [Google Scholar]
- 19.Yu L., Todd N. W., Xing L., et al. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. International Journal of Cancer. 2010;127(12):2870–2878. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeong H. C., Kim E. K., Lee J. H., Lee J. M., Nayoo H., Kim J. K. Aberrant expression of let-7a miRNA in the blood of non-small cell lung cancer patients. Molecular Medicine Reports. 2011;4(2):383–387. doi: 10.3892/mmr.2011.430. [DOI] [PubMed] [Google Scholar]
- 21.Shen J., Liu Z., Todd N. W., et al. Diagnosis of lung cancer in individuals with solitary pulmonary nodules by plasma microRNA biomarkers. BMC Cancer. 2011;11, article 374 doi: 10.1186/1471-2407-11-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S., Yang J. Application of plasma circulation microRNA - 126 to diagnosis of non - small cell lung cancer. Medical Journal of National Defending Forces in Southwest China. 2011;21:1280–1283. [Google Scholar]
- 23.Hennessey P. T., Sanford T., Choudhary A., et al. Serum microrna biomarkers for detection of non-small cell lung cancer. PLoS ONE. 2012;7(2) doi: 10.1371/journal.pone.0032307.e32307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuxia M., Zhennan T., Wei Z. Circulating miR-125b is a novel biomarker for screening non-small-cell lung cancer and predicts poor prognosis. Journal of Cancer Research and Clinical Oncology. 2012;138(12):2045–2050. doi: 10.1007/s00432-012-1285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao M., Shen X., Zhan P., Lv T., Liu H., Song Y. Expression of miRNA-29a in non-small cell lung cancer and clinical significance. Chinese Clinical Oncology. 2012;17:428–432. doi: 10.3969/j.issn.1009-0460.2012.05.010. [DOI] [Google Scholar]
- 26.Abd-El-Fattah A. A., Sadik N. A. H., Shaker O. G., Aboulftouh M. L. Differential microRNAs expression in serum of patients with lung cancer, pulmonary tuberculosis, and pneumonia. Cell Biochemistry and Biophysics. 2013;67(3):875–884. doi: 10.1007/s12013-013-9575-y. [DOI] [PubMed] [Google Scholar]
- 27.Anjuman N., Li N., Guarnera M., Stass S. A., Jiang F. Evaluation of lung flute in sputum samples for molecular analysis of lung cancer. linical and Translational Medicine. 2013;2, article 15 doi: 10.1186/2001-1326-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bediaga N. G., Davies M. P. A., Acha-Sagredo A., et al. A microRNA-based prediction algorithm for diagnosis of non-small lung cell carcinoma in minimal biopsy material. British Journal of Cancer. 2013;109(9):2404–2411. doi: 10.1038/bjc.2013.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cazzoli R., Buttitta F., di Nicola M., et al. MicroRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. Journal of Thoracic Oncology. 2013;8(9):1156–1162. doi: 10.1097/jto.0b013e318299ac32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma J., Li N., Guarnera M., Jiang F. Quantification of Plasma miRNAs by Digital PCR for Cancer Diagnosis. Biomarker Insights. 2013;8:p. BMI.S13154. doi: 10.4137/BMI.S13154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mozzoni P., Banda I., Goldoni M., et al. Plasma and EBC microRNAs as early biomarkers of non-small-cell lung cancer. Biomarkers. 2013;18(8):679–686. doi: 10.3109/1354750x.2013.845610. [DOI] [PubMed] [Google Scholar]
- 32.Tang D., Shen Y., Wang M., et al. Identification of plasma microRNAs as novel noninvasive biomarkers for early detection of lung cancer. European Journal of Cancer Prevention. 2013;22(6):540–548. doi: 10.1097/CEJ.0b013e32835f3be9. [DOI] [PubMed] [Google Scholar]
- 33.Ulivi P., Foschi G., Mengozzi M., et al. Peripheral blood miR-328 expression as a potential biomarker for the early diagnosis of NSCLC. International Journal of Molecular Sciences. 2013;14(5):10332–10342. doi: 10.3390/ijms140510332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng X. L., Zhang S. Y., Zheng J. F., Yuan H., Wang Y. Altered miR-143 and miR-150 expressions in peripheral blood mononuclear cells for diagnosis of non-small cell lung cancer. Chinese Medical Journal. 2013;126:4510–4516. [PubMed] [Google Scholar]
- 35.Wang Z., Zhang X., Bai H., et al. EML4-ALK Rearrangement and Its Clinical Significance in Chinese Patients with Advanced Non-Small Cell Lung Cancer. Oncology. 2012;83(5):248–256. doi: 10.1159/000341381. [DOI] [PubMed] [Google Scholar]
- 36.Yang X., Zhang Y., Sun B., et al. Diagnostic value of the detection of MicroRNAs in sputum of patients with non-small cell lung cancer. Journal of Clinical Pulmonary Medicine. 2013;18:226–229. doi: 10.3969/j.issn.1009-6663.2013.02.016. [DOI] [Google Scholar]
- 37.Li M., Zhang Q., Wu L., et al. Serum miR-499 as a novel diagnostic and prognostic biomarker in non-small cell lung cancer. Oncology Reports. 2014;31(4):1961–1967. doi: 10.3892/or.2014.3029. [DOI] [PubMed] [Google Scholar]
- 38.Li N., Ma J., Guarnera M. A., Fang H., Cai L., Jiang F. Digital PCR quantification of miRNAs in sputum for diagnosis of lung cancer. Journal of Cancer Research and Clinical Oncology. 2014;140(1):145–150. doi: 10.1007/s00432-013-1555-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu W., He J., Chen D., et al. Expression of miR-29c, miR-93, and miR-429 as potential biomarkers for detection of early stage non-small lung cancer. PLoS ONE. 2014;9(2) doi: 10.1371/journal.pone.0087780.e87780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dou H., Wang Y., Su G., Zhao S. Decreased plasma let-7c and miR-152 as noninvasive biomarker for non-small-cell lung cancer. International Journal of Clinical and Experimental Medicine. 2015;8:9291–9298. [PMC free article] [PubMed] [Google Scholar]
- 41.Guo W., Zhang Y., Zhang Y., et al. Decreased expression of MIR-204 in plasma is associated with a poor prognosis in patients with non-small cell lung cancer. International Journal of Molecular Medicine. 2015;36(6):1720–1726. doi: 10.3892/ijmm.2015.2388. [DOI] [PubMed] [Google Scholar]
- 42.Li L., Chen Y. Y., SQ Li., Huang C., Qin Y. Z. Expression of miR-148/152 family as potential biomarkers in non-small-cell lung cancer. Medical Science Monitor. 2015;21:1155–1161. doi: 10.12659/MSM.892940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu C., Wang C., Chen F., Yu H., Wang J., Yang J. Combined detection of pleural effusion cell DNA aneuploidy and microRNA-192 and its related factors in diagnosis of non-small cell lung cancer. China Journal of Modern Medicine. 2015;25:26–30. doi: 10.3969/j.issn.1005-8982.2015.32.005. [DOI] [Google Scholar]
- 44.Ma J., Lin Y., Zhan M., Mann D. L., Stass S. A., Jiang F. Differential miRNA expressions in peripheral blood mononuclear cells for diagnosis of lung cancer. Laboratory Investigation. 2015;95(10):1197–1206. doi: 10.1038/labinvest.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nadal E., Truini A., Nakata A., et al. A Novel Serum 4-microRNA Signature for Lung Cancer Detection. Scientific Reports. 2015;5 doi: 10.1038/srep12464.12464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang C., Ding M., Xia M., et al. A Five-miRNA Panel Identified From a Multicentric Case-control Study Serves as a Novel Diagnostic Tool for Ethnically Diverse Non-small-cell Lung Cancer Patients. EBioMedicine. 2015;2(10):1377–1385. doi: 10.1016/j.ebiom.2015.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang R. J., Zheng Y. H., Wang P., Zhang J. Z. Serum miR-125a-5p, miR-145 and miR-146a as diagnostic biomarkers in non-small cell lung cancer. International Journal of Clinical and Experimental Pathology. 2015;8:765–771. [PMC free article] [PubMed] [Google Scholar]
- 48.Xing L., Su J., Guarnera M. A., et al. Sputum microRNA biomarkers for identifying lung cancer in indeterminate solitary pulmonary nodules. Clinical Cancer Research. 2015;21(2):484–489. doi: 10.1158/1078-0432.CCR-14-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang J.-S., Li B.-J., Lu H.-W., et al. Serum miR-152, miR-148a, miR-148b, and miR-21 as novel biomarkers in non-small cell lung cancer screening. Tumor Biology. 2015;36(4):3035–3042. doi: 10.1007/s13277-014-2938-1. [DOI] [PubMed] [Google Scholar]
- 50.Yang Y.-L., Xu L.-P., Zhuo F.-L., Wang T.-Y. Prognostic value of microRNA-10b overexpression in peripheral blood mononuclear cells of nonsmall-cell lung cancer patients. Tumor Biology. 2015;36(9):7069–7075. doi: 10.1007/s13277-015-3366-6. [DOI] [PubMed] [Google Scholar]
- 51.Zhao W., Zhao J. J., Zhang L., et al. Serum miR-21 level: a potential diagnostic and prognostic biomarker for non-small cell lung cancer. International Journal of Clinical and Experimental Medicine. 2015;8:14759–14763. [PMC free article] [PubMed] [Google Scholar]
- 52.Fan L., Qi H., Teng J., et al. Identification of serum miRNAs by nano-quantum dots microarray as diagnostic biomarkers for early detection of non-small cell lung cancer. Tumor Biology. 2016;37(6):7777–7784. doi: 10.1007/s13277-015-4608-3. [DOI] [PubMed] [Google Scholar]
- 53.Gao X., Wang Y., Zhao H., et al. Plasma miR-324-3p and miR-1285 as diagnostic and prognostic biomarkers for early stage lung squamous cell carcinoma. Oncotarget . 2016;7(37) doi: 10.18632/oncotarget.11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Halvorsen A. R., Bjaanæs M., LeBlanc M., et al. A unique set of 6 circulating microRNAs for early detection of non-small cell lung cancer. Oncotarget . 2016;7(24):37250–37259. doi: 10.18632/oncotarget.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang L.-P., Zhu Z.-T., He C.-Y. Expression of miRNA-26b in the diagnosis and prognosis of patients with non-small-cell lung cancer. Future Oncology. 2016;12(9):1105–1115. doi: 10.2217/fon.16.21. [DOI] [PubMed] [Google Scholar]
- 56.Leidinger P., Brefort T., Backes C., et al. High-throughput qRT-PCR validation of blood microRNAs in non-small cell lung cancer. Oncotarget . 2016;7(4):4611–4623. doi: 10.18632/oncotarget.6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Su K., Zhang T., Wang Y., Hao G. Diagnostic and prognostic value of plasma microRNA-195 in patients with non-small cell lung cancer. World Journal of Surgical Oncology. 2016;14(1) doi: 10.1186/s12957-016-0980-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Sun L., Chen Y., Su Q., et al. Increased plasma miRNA-30a as a biomarker for non-small cell lung cancer. Medical Science Monitor. 2016;22:647–655. doi: 10.12659/MSM.897330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tai M. C., Yanagisawa K., Nakatochi M., et al. Blood-borne miRNA profile-based diagnostic classifier for lung adenocarcinoma. Scientific Reports. 2016;6(1) doi: 10.1038/srep31389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei J., Gao W., Zhu C.-J., et al. Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non-small cell lung cancer. Chinese Journal of Cancer. 2011;30(6):407–414. doi: 10.5732/cjc.010.10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu W., Zhou K., Zha Y., et al. Diagnostic value of serum miR-182, miR-183, miR-210, and miR-126 levels in patients with early-stage non-small cell lung cancer. PLoS ONE. 2016;11(4) doi: 10.1371/journal.pone.0153046.e0153046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang H., Mao F., Shen T., et al. Plasma miR-145, miR-20a, miR-21 and miR-223 as novel biomarkers for screening early-stage non-small cell lung cancer. Oncology Letters. 2017;13(2):669–676. doi: 10.3892/ol.2016.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Keller A., Leidinger P., Borries A., et al. MiRNAs in lung cancer - Studying complex fingerprints in patient's blood cells by microarray experiments. BMC Cancer. 2009;9, article no. 1471:p. 353. doi: 10.1186/1471-2407-9-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li G., Huang Y., He Y., et al. Study of miRNA Signatures as Biomarkers for Early Stage Nonsmoking Female Lung Cancer of Xuanwei Country of Yunnan Province. Cancer prevention and control research. 2012;39:802–806. doi: 10.3971/j.issn.1000-8578.2012.07.010. [DOI] [Google Scholar]
- 65.Roa W. H., Kim J. O., Razzak R., et al. Sputum microRNA profiling: a novel approach for the early detection of non-small cell lung cancer. Clinical & Investigative Medicine. 2012;35(5):E271–E281. doi: 10.25011/cim.v35i5.18700. [DOI] [PubMed] [Google Scholar]
- 66.Sanfiorenzo C., Ilie M. I., Belaid A., et al. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS ONE. 2013;8(1) doi: 10.1371/journal.pone.0054596.e54596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li W., Wang Y., Zhang Q., et al. MicroRNA-486 as a biomarker for early diagnosis and recurrence of non-small cell lung cancer. PLoS ONE. 2016;11(1) doi: 10.1371/journal.pone.0148589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang W., Li W., Ding M., et al. Identification of miRNAs as non-invasive biomarkers for early diagnosis of lung cancers. Tumor Biology. 2016;37(12):16287–16293. doi: 10.1007/s13277-016-5442-y. [DOI] [PubMed] [Google Scholar]
- 69.Chen L., Jin H. MicroRNAs as novel biomarkers in the diagnosis of non-small cell lung cancer: a meta-analysis based on 20 studies. Tumor Biology. 2014;35(9):9119–9129. doi: 10.1007/s13277-014-2188-2. [DOI] [PubMed] [Google Scholar]
- 70.Huang Y., Hu Q., Deng Z., Hang Y., Wang J., Wang K. Micrornas in body fluids as biomarkers for non-small cell lung cancer: a systematic review. Technology in Cancer Research & Treatment. 2014;13(3):277–287. doi: 10.7785/tcrt.2012.500377. [DOI] [PubMed] [Google Scholar]
- 71.Wang H., Wu S., Zhao L., Zhao J., Liu J., Wang Z. Clinical use of microRNAs as potential non-invasive biomarkers for detecting non-small cell lung cancer: A meta-analysis. Respirology. 2015;20(1):56–65. doi: 10.1111/resp.12444. [DOI] [PubMed] [Google Scholar]