Abstract
In this protocol, we describe how to quantify starch in guard cells of Arabidopsis thaliana using the fluorophore propidium iodide and confocal laser scanning microscopy. This simple method enables monitoring, with unprecedented resolution, the dynamics of starch in guard cells.
Keywords: Starch, Guard cells, Arabidopsis, Stomatal opening, Propidium iodide
Background
Starch is a complex polymer of glucose and represents the most abundant form in which plants store carbohydrate. Starch serves different functions, according to the cell types from which it is derived, and the external environmental conditions. In guard cells, which border the stomatal pores that control water and carbon dioxide exchange with the environment, starch can be mobilized within minutes upon transition to light, helping to generate organic acids and sugars to increase guard cell turgor and promote stomatal opening. In mesophyll cells, starch typically accumulates gradually during the day and is degraded at night to support metabolism (Santelia and Lunn, 2017).
Because guard cells comprise only a minor fraction of the total leaf, it is difficult to measure starch quantitatively using conventional methods. Up until now, starch accumulation in guard cells has been mostly visualized by iodine staining. This technique can determine the presence/absence of starch but does not provide accurate, quantitative information.
Here, we describe a fluorescence-based imaging method to quantify starch in guard cells of Arabidopsis thaliana. This technique is based on the covalent labeling of cell wall material and other glucan substrates, including starch, with the fluorescent pseudo-Schiff reagent propidium iodide (PS-PI). Isolated epidermal peels are treated with periodic acid to oxidize the hydroxyl groups of the glucose units to aldehyde and ketone groups. The aldehyde groups (-CHO) can then react covalently with propidium iodide, resulting in samples with highly fluorescent glucans that are well suited for confocal laser scanning microscopy. The area of single starch granules within guard cell chloroplasts can be determined using digital imaging. We applied this method to assess the dynamics of starch content in guard cells of intact Arabidopsis leaves over the diurnal cycle, and to determine the impact of fusicoccin (a chemical activator of the proton pump) on starch amounts in guard cells of isolated epidermal peels fragments floating in stomatal opening buffer ( Horrer et al., 2016 ).
Our protocol is an adaptation of a previous mPS-PI staining technique ( Truernit et al., 2008 ). The original method was developed to image entire plant organs for three-dimensional reconstruction of their cellular organization. The main difference between the two methods is the incubation time with the propidium iodide solution, which is 1-2 h for entire plant organs, and only about 20-40 min for epidermal peels.
The technique described here is simple, accurate and highly reproducible. By facilitating the detailed quantification of starch amounts in guard cells, this method will increase the number of questions we will be able to answer about any aspect of guard cell starch metabolism.
Materials and reagents
Pipette tips (SARSTEDT)
12-well plate (Greiner Bio One International, catalog number: 665180)
Falcon tubes
Parafilm M (Bemis, catalog number: PM996)
Microscope slides (Thermo Fisher Scientific, Menzel-Gläser)
Coverslips (Thermo Fisher Scientific, Menzel-Gläser)
Kimtech Science precision wipes (KCWW, Kimberly-Clark, catalog number: 75512)
Square Petri dish (Greiner Bio One International, catalog number: 688102)
Arabidopsis thaliana ecotype Col-0
Methanol (Carl Roth, catalog number: 8388.4)
Ethanol (Reuss Chemie, catalog number: RC-A15-A)
Acetic acid
Periodic acid (Sigma-Aldrich, catalog number: P7875)
Sodium metabisulfite (Sigma-Aldrich, catalog number: S9000)
Hydrochloric acid (Carl Roth, catalog number: 4625.1)
Propidium iodide (Sigma-Aldrich, catalog number: 81845)
Chloral hydrate (Sigma-Aldrich, catalog number: 15307-R)
Glycerol (Carl Roth, catalog number: 3783.1)
Gum arabic (Carl Roth, catalog number: 4159.3)
Fixative solution (see Recipes)
Schiff Reagent (see Recipes)
Chloral hydrate solution (see Recipes)
Hoyer’s solution (see Recipes)
Equipment
Glass beaker
Precision tweezers (RubisTech, catalog number: 5-SA RT)
Pipettes (Gilson)
Fume hood (Renggli AG)
Refrigerator (Liebherr)
Oven (Ehret)
Green light LED lamp (In-house built)
-
Confocal laser scanning microscope (Leica Microsystems, model: Leica TCS SP5)
Note: This product has been discontinued. Any confocal laser scanning microscope can be used.
Computer
Software
ImageJ (NIH USA, version 1.8, https://imagej.nih.gov/ij/)
Procedure
-
Epidermal peel harvest from Arabidopsis thaliana
Label a 12-well plate according to the defined time points and genotypes, and add 1 ml of fixative solution (see Recipes) into each labeled well.
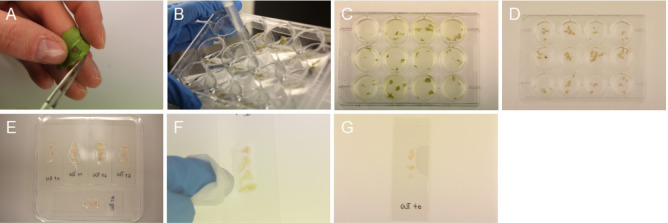
Collect epidermal peels from the abaxial side of the 5th or 6th leaf using precision tweezers (Figure 1A; Video 1). Usually, 4 replicates from 4 individual plants are harvested per time point and genotype. One well contains 4 epidermal peels. See Notes 1-3 for further details.
Incubate the 12-well plate containing the collected epidermal peels for at least 12 h at 4 °C in the dark. The samples can be stored in fixative solution up to 4 weeks at 4 °C. The epidermal peels are fragile and require careful handling from now onwards. See Note 4.
-
mPS-PI staining
Perform Steps B2-B8 in a fume hood.
Remove the fixative solution from the wells using a P1000 pipette (Figure 1B). Collect the waste in a glass beaker and dispose it into the halogenated liquid waste at the end of the protocol.
Wash the samples by adding 1 ml of dH2O and slowly shaking the plate on the bench with circular movements. Remove the dH2O.
Destain the epidermal peels by adding 1 ml of 80% ethanol and incubating at 65 °C for 5-15 min (Figure 1C). Remove the ethanol and repeat washing step (Step B3).
Incubate the samples at room temperature (20 °C, RT) for 1 h in 1 ml of fixative solution. Remove the fixative solution and repeat washing step (Step B3).
Add 1 ml of 1% periodic acid solution to the epidermal peels and incubate the plate for 40 min at RT. Make sure to cover the peels fully with the solution. See Note 5. Remove the periodic acid solution and repeat washing step (Step B3). Proceed carefully as peels are extremely fragile after this step.
Stain the epidermal peels by adding 500 µl Schiff reagent (see Recipes) and 50 µl propidium iodide solution (1 mg·ml-1) for 20-40 min at RT. Make sure to cover the plant tissues fully with the solution. The samples should appear pinkish after this step (Figure 1D). Remove the propidium iodide solution and collect it separately in a Falcon tube. Propidium iodide waste should be treated like ethidium bromide waste.
Destain the samples in 1 ml of dH2O for 20-30 min at RT.
-
Microscope slide preparation
Label microscope slides according to the defined time points and genotypes.
Perform Steps C3-C4 and C6-C7 in a fume hood.
Add 70 µl of chloral hydrate solution (see Recipes) onto every microscope slide. Be careful to distribute the solution at the center of the slide to avoid leaking from the borders of the slide.
Carefully transfer the stained epidermal peels onto the microscope slide containing the chloral hydrate solution using precision tweezers. Place all four replicates next to each other (as shown in Figure 1E). Peels should be as flat as possible on the slide. Be careful; the peels will rupture easily.
Without placing a coverslip, transfer the microscope slides into square Petri dishes and incubate them at RT in the dark for 24 h.
Remove as much chloral hydrate solution as possible from the borders of the samples using Kimtech wipes (Figure 1F). Do not touch the samples with the wipes.
Place 2-3 drops of Hoyer’s solution (see Recipes) onto the epidermal peels and add a coverslip. Apply gentle pressure on the coverslip to assure that the peels are flat and even on the microscope slide.
Transfer the slides back to the square Petri dishes and store them at RT in the dark for at least 3 days to allow the mountant to set (Figure 1G). See Note 6. The slides can be stored for several months in the dark.
-
Imaging
Image acquisition is achieved with a Confocal Laser Scanning Microscope.
Set microscope as shown in Table 1.
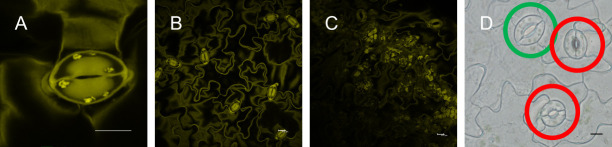
Acquire 20 images of individual stomata (Figure 2A; 5 images per replicate) per microscope slide. Acquire pictures of stomata only from mesophyll-free parts (Figures 2B and 2C) of the epidermal peels. Image stomata always with the same zoom factor (6x, Figure 2A) for data comparison. See Note 7.
Figure 1. Harvest and staining of epidermal peels.
A. Epidermal peel from the abaxial side of the leaf; B. Removal of solutions; C. Destaining of epidermal peels; D. Pinkish plant tissue; E. Microscope slide containing chloral hydrate solution and epidermal peels; F. Removal of chloral hydrate solution; G. Final slide.
Video 1. Collection of epidermal peels from the abaxial side of an Arabidopsis leaf.

This video shows the procedure for collecting epidermal peels from Arabidopsis leaves that are clean from mesophyll cells contamination.
Table 1. Standard microscope settings for guard cell starch imaging.
| Microscope settings | Standard values |
|---|---|
| Argon laser | 5% |
| Objective | 63x, glycerol |
| Excitation | 488 nm |
| Detector | HyD3 |
| Emission filter | 610-640 nm |
| Format | 1,024 x 1,024 pixel |
| Zoom | 6x (Figure 2A) |
Figure 2. Imaging of guard cell starch granules.
A. Single stomata (scale bar = 10 µm); B. Mesophyll cell-free epidermal peel (scale bar = 10 µm); C. Mesophyll cell contamination of epidermal peel (scale bar = 10 µm); D. Fully (green circle) and not fully (red circle) developed stomata (scale bar = 10 µm).
Data analysis
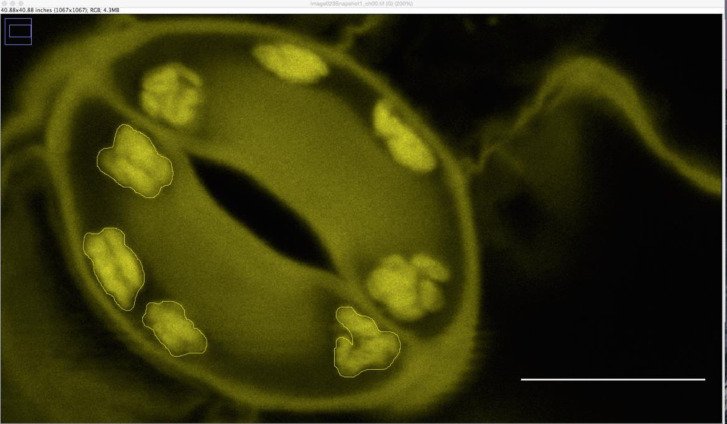
Starch granule area (in µm2) is determined using ImageJ version 1.8 (NIH USA, http://rsbweb.nih.gov/ij/) by encircling the granule area (Figure 3). A suitable scale is drawn onto the first picture using the software provided by the microscope manufacturer. Guard cell starch granule area is measured for each guard cell individually, totally collecting at least 40 values per time point and genotype. Starch content can then be expressed as an average of these values. Repeat the same experimental setup at least 3 times on independent plant material to obtain a final data set.
Figure 3. Outlined guard cell starch granule area in ImageJ.
Scale bar = 5 µm.
Notes
The harvest time of epidermal peels per time point and genotype should not be longer than 1-2 min. In case of harvest of multiple genotypes, harvest one replicate per genotype. Harvest the additional replicates following the same sequence to ensure similar treatment of all genotypes.
In case of harvest in the dark (e.g., during the night), any green light source can be used to collect the epidermal peels.
To obtain a suitable epidermal peel for guard cell starch staining, it is important that a large fraction of the epidermal peel is mesophyll-free.
Make sure that the peels do not dry out while they are stored after the harvest and during the staining procedure.
Be careful in handling periodic acid as this compound is toxic.
The microscope slides are ready for imaging as soon as the coverslip is not movable anymore.
Acquire images only for fully developed stomata (Figure 2D).
Recipes
-
Fixative solution
50% methanol
10% acetic acid
Make up to 500 ml with dH2O
Note: Fixative solution can be stored for long-term usage at 4 °C.
-
Schiff reagent
1.9 g sodium metabisulfite
3 ml 5 N HCl
97 ml dH2O
Note: Store the Schiff reagent at 4 °C for up to 2 months.
-
Chloral hydrate solution
40 g chloral hydrate
10 ml glycerol
20 ml dH2O
Note: Store the chloral hydrate solution at 4 °C for long-term usage.
-
Hoyer’s solution
30 g gum arabic
200 g chloral hydrate
20 g glycerol
50 ml dH2O
Note: Gum arabic is added to the water in a beaker with a magnet bar, which is placed on a shaker in a fume hood overnight to dissolve it. Chloral hydrate is then added in tiny amounts into the beaker using a funnel in a fume hood. Let it dissolve with shaking. Lastly, the glycerol is added. Allow the solution to set for a couple of days before use. Store the solution at RT in the dark. The solution stays turbid. Do not shake it.
Acknowledgments
This work was supported by the Swiss National Science Foundation (SNSF-Grant 31003A_166539) and the ETH Zürich. The authors would like to acknowledge Mario Coiro for initial help in setting up the pseudo-Schiff propidium iodide staining method; Daniel Horrer and Diana Pazmino for adapting the method to epidermal peels. This protocol is adapted from Truernit et al. (2008). The authors declare no conflicts of interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Horrer D., Flutsch S., Pazmino D., Matthews J. S., Thalmann M., Nigro A., Leonhardt N., Lawson T. and Santelia D.(2016). Blue light induces a distinct starch degradation pathway in guard cells for stomatal opening. Curr Biol 26(3): 362-370. [DOI] [PubMed] [Google Scholar]
- 2. Santelia D. and Lunn J. E.(2017). Transitory starch metabolism in guard cells: unique features for a unique function. Plant Physiol 174(2): 539-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Truernit E., Bauby H., Dubreucq B., Grandjean O., Runions J., Barthelemy J. and Palauqui J. C.(2008). High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis . Plant Cell 20(6): 1494-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]