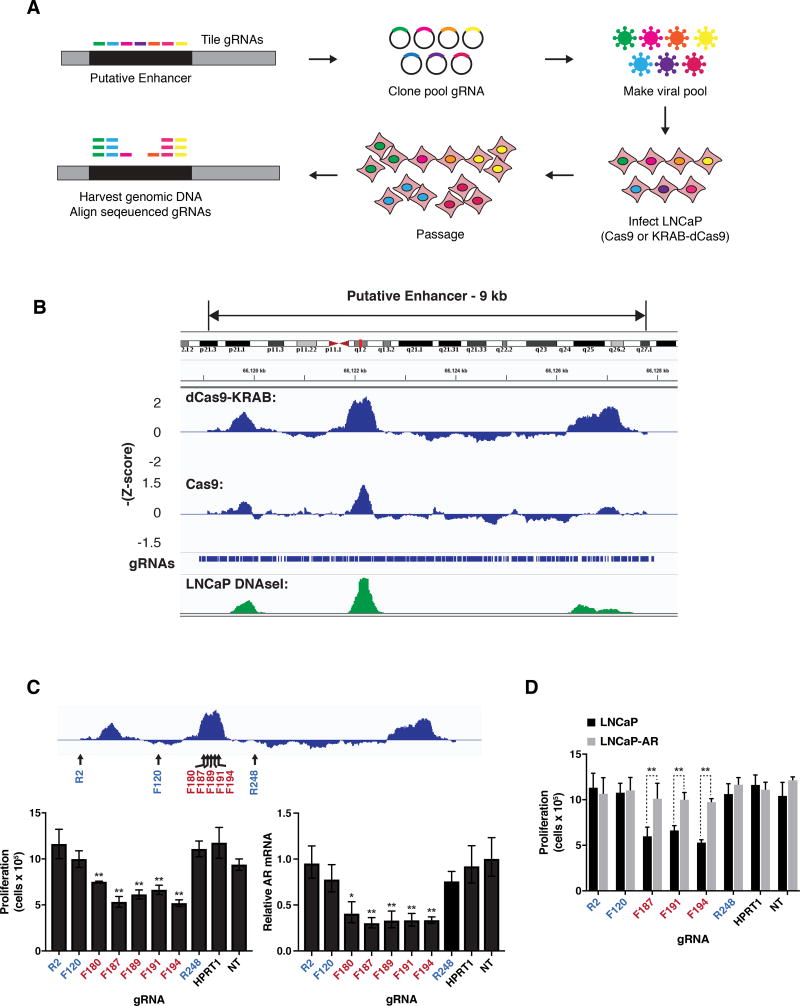
Figure 3. The AR enhancer is essential for cell viability in the LNCaP metastatic prostate cancer cell line.
(A) Design of pooled CRISPR screens to identify noncoding regions required for proliferation of LNCaP cells.
(B) Results of pooled CRISPR screens depicted as sliding window of average –Z-scores of adjacent gRNAs. The positions of the individual gRNAs in the library and LNCaP DNAseI hypersensitivity data from ENCODE are shown.
(C) Proliferation of LNCaP cells as measured by cell counting (left) and expression of AR as measured by qRT-PCR (right) after transduction of KRAB-dCas9 and indicated gRNAs. Positions of gRNAs targeting the 9 kb enhancer region are depicted relative to screen results (top). F indicates gRNA designed against the forward strand and R indicates reverse strand.
(D) Proliferation of LNCaP cells and LNCaP cells overexpressing exogenous AR (LNCaP-AR) as measured by cell counting after transduction with KRAB-dCas9 and indicated gRNAs.
For (C) and (D) transduction of gRNA targeting the unrelated gene HPRT1, and nontargeting gRNA (NT) were used as controls. Expression is normalized to nontargeting gRNA. Data represent the average and standard deviation of 3 biological replicates, and significance determined by Student’s t test. * p = 0.011, ** p< 0.01.
See also Figure S2.