SUMMARY
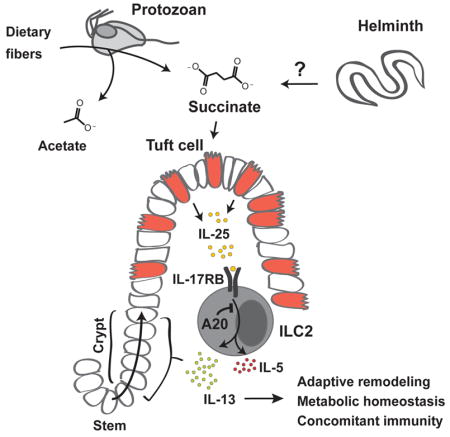
The small intestinal tuft cell-ILC2 circuit mediates epithelial responses to intestinal helminths and protists by tuft cell chemosensory-like sensing and IL-25-mediated activation of lamina propria ILC2s. Small intestine ILC2s constitutively express the IL-25 receptor, which is negatively regulated by A20 (Tnfaip3). A20-deficiency in ILC2s spontaneously triggers the circuit, and, unexpectedly, promotes adaptive small intestinal lengthening and remodeling. Circuit activation occurs upon weaning, and is enabled by dietary polysaccharides that render mice permissive for Tritrichomonas colonization, resulting in luminal accumulation of acetate and succinate, metabolites of the protist hydrogenosome. Tuft cells express GPR91, the succinate receptor, and dietary succinate, but not acetate, activates ILC2s via a tuft-, TRPM5-, and IL-25-dependent pathway. Also induced by parasitic helminths, circuit activation and small intestinal remodeling impairs infestation by new helminths, consistent with the phenomenon of concomitant immunity. We describe a metabolic sensing circuit that may have evolved to facilitate mutualistic responses to luminal pathosymbionts.
Keywords: Tuft cells, ILC2s, A20, epithelial remodeling, IL-25, TRPM5, eukaryotic pathosymbionts, succinate, succinate receptor, GPR91, hydrogenosome, Tritrichomonas, helminths, concomitant immunity
In Brief
Succinate produced by mutualistic colonizing protists in the small intestine activates a tuft cell - ILC2 circuit, leading to adaptive remodeling that increases surface area absorption and prevention of other infections.
INTRODUCTION
The small intestine is dedicated to nutrient acquisition aided by a complex microbiota and supported by large numbers of immune cells that regulate the balance between tolerance to food, maintenance of the microbiota, and defense against pathogens. Homeostasis at this interface is critical as revealed by increasingly prevalent autoinflammatory bowel diseases (Liu and Stappenbeck, 2016) and food allergies (Tordesillas et al., 2017). Better understanding of the interactions that mediate the dynamic interplay between diet, the microbiota, and the immune cell networks that maintain this barrier is needed to enable strategies to achieve therapeutic reconstruction of this essential mucosal organ.
Recently, several laboratories, including our own, identified a role for epithelial tuft cells in the intestinal response to helminths and protists (Gerbe et al., 2016; Howitt et al., 2016; von Moltke et al., 2016). Long recognized as a constituent of mucosal epithelia in vertebrates, tuft cells remained enigmatic, in part due to the rare frequency of these cells. The apical tuft, which is comprised of microvillus extensions into the lumen, suggested a sensory function, which was supported by the expression of components of the taste receptor signaling cascade in these cells (Sato, 2007). We showed that mucosal tuft cells constitutively express IL-25, and that these cells increase markedly in small intestinal epithelia after infection with helminths. Activation of tuft cell IL-25 after infection stimulated lamina propria ILC2s to secrete IL-13, which directly biased cell-fate decisions in epithelial stem cell progenitors, resulting in increased goblet and tuft cell frequencies that accompany the ‘weep-and sweep’ response to worms (von Moltke et al., 2016). An independent manuscript confirmed the role of tuft cells in this cytokine relay using Pou2f3-deficient mice, which lack tuft cells (Gerbe et al., 2016). A third group made the important discovery that an unappreciated protozoan pathosymbiont of the genus Tritrichomonas also elicited expansion of the tuft cell compartment via this ILC2-mediated epithelial circuit, which depended on TRPM5, a calcium-activated cation channel involved in G protein-coupled taste transduction and highly expressed in tuft cells (Howitt et al., 2016). Recognition of Tritrichomonas has uncovered roles for chronic colonization by these organisms in altering not only epithelial cell architecture of the small intestine, but also the ‘immunologic tone’ of the animals (Chudnovskiy et al., 2016; Escalante et al., 2016). Thus, these eukaryotic pathosymbionts, like certain keystone bacteria (e.g., segmented filamentous bacteria, Helicobacter spp.) can impact heterologous infectious or inflammatory processes due to effects on immune or epithelial tissues, or both, with the capacity to mediate tissue protection or pathology under certain conditions. The widespread prevalence of such adapted pathosymbiotic organisms in feral vertebrates, however, suggests a highly evolved mutualism. With this in mind, we undertook a more comprehensive study of factors that impact the small intestinal tuft cell – ILC2 circuit in mice.
RESULTS
Constitutive A20 expression in small intestine ILC2s restrains IL-25-mediated expansion of the tuft cell – ILC2 circuit
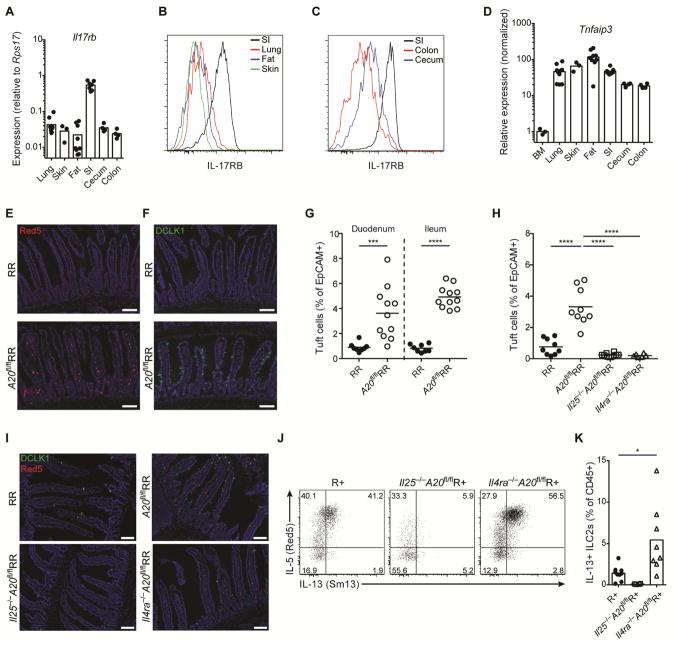
Characterization of peripheral ILC2 populations revealed heterogeneity in expression of IL-17RB, which with IL-17RA comprises the heterodimeric IL-25 receptor (Song et al., 2016). IL-17RB transcripts and surface expression were high in small intestinal ILC2s compared to ILC2s from other peripheral tissues, including fat, lung, skin or adjacent gastrointestinal tissues including cecum and colon (Fig. 1A–C). The signaling component of the IL-25 receptor, IL-17RA, and its adapter proteins are negatively regulated by A20 (Tnfaip3) (Garg et al., 2013). Indeed, A20 expression was constitutively high in ILC2s from peripheral tissues but not in bone marrow (Fig. 1D), consistent with a role in regulating homeostatic signaling. Further, constitutive high A20 expression was evident in small intestinal ILC2s regardless of age and in germ-free (GF) mice, and was down-regulated after infection with the potent circuit activator, N. brasiliensis, consistent with a dynamic role in circuit control (suppl. Fig. 1A).
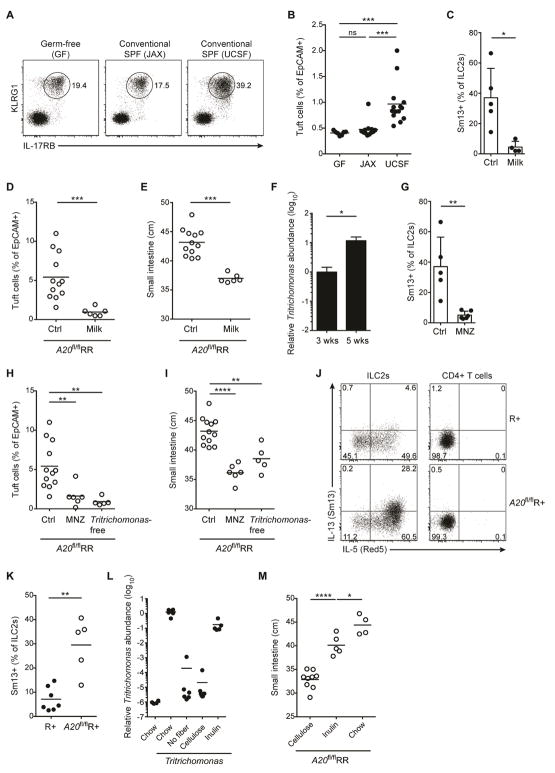
Figure 1. Constitutive high A20 expression in small intestine ILC2s restrains IL-25-mediated expansion of the tuft cell – ILC2 circuit.
(A–C) IL-17RB expression measured by qPCR (A) or flow cytometry (B and C) in ILC2s from indicated tissues. (D) A20 expression measured by qPCR in sorted ILC2s from indicated tissues, normalized to BM. (E and F) Presence of ILC2s (E) and tuft cells (F) in the small intestine (SI) of RR and A20flRR mice, visualized by expression of IL-5 (Red5 reporter) in red (E) and DCLK1 in green (F); DAPI in blue. (G and H) Quantification of tuft cells in SI by flow cytometry. (G) Tuft cell frequencies in proximal and distal SI of RR and A20flRR mice. (H) Percentages of tuft cells in SI of RR, A20flRR mice, Il25−/−A20flRR mice and Il4ra−/−A20flRR mice. (I) Presence of ILC2s and tuft cells in SI as in (E and F) from mice as in (H). (J) Flow cytometry analysis of IL-13 (Sm13 reporter) and IL-5 (Red5 reporter) expression by ILC2s gated on CD45+Lin-IL-17RB+KLRG1+ cells isolated from the SI of R+, Il25−/−A20flR+ and Il4ra−/−A20flR+ mice. (K) Percentages of IL-13+ ILC2s among CD45+ lamina propria cells as in (J). Data pooled from multiple independent experiments (A–D, G, H, K) or from one experiment representative of at least two independent experiments (E, F, I, J). Scale bars; 100 μm. *p < 0.05, ***p < 0.001, ****p < 0.0001; ns, not significant by Mann-Whitney U or one-way ANOVA.
To delete A20 in ILC2s, we crossed mice with loxP-flanked A20 alleles (Tavares et al., 2010) to IL-5 RFP-Cre (Red5) mice (Nussbaum et al., 2013), generating mice that are homozygous A20-floxed with one (A20flR+) or two alleles of Red5 (A20flRR) (suppl. Fig. 1B). A20-deficient ILC2s produced increased cytokines after stimulation in vitro, consistent with an inhibitory role for A20 in restraining ILC2 cytokine production (suppl. Fig. 1C). Unexpectedly, A20flRR mice had a marked increase in IL-5-expressing ILC2s in the lamina propria of small intestine as compared to control mice (Fig. 1E, suppl. Fig. 1D and E); A20flR+ mice were phenotypically comparable to A20flRR mice. Despite deletion of A20 from ILC2s in all tissues analyzed (suppl. Fig. 1B), effects on other tissue ILC2 numbers were much less dramatic; the numbers of ILC2s in the fat were unchanged and lung ILC2s were slightly reduced (suppl. Fig. 1F). The accumulation of ILC2s was accompanied by increased frequencies of DCLK1+ tuft cells along the entire small intestine (Fig. 1F and 1G), but not in other parts of the gastrointestinal tract, including the adjacent stomach, cecum and colon (suppl. Fig. 2A). Consistent with prior studies of small intestinal ILC2 activation (Gerbe et al., 2016; Moro et al., 2010; Neill et al., 2010; von Moltke et al., 2016), goblet cells were increased in A20flRR mice but enteroendocrine and Paneth cell numbers remained unchanged (suppl. Fig. 2B and C). Taken together, these data are consistent with spontaneous activation of the small intestinal tuft cell–ILC2 circuit in A20flRR and A20flR+ mice, and reminiscent of what we and others described in the context of intestinal nematode and parabasalid protozoan infection (Gerbe et al., 2016; Howitt et al., 2016; von Moltke et al., 2016).
The feed-forward nature of the tuft cell–ILC2 circuit is driven by IL-25 and IL-13, which are produced by the tuft cells and ILC2s, respectively. To confirm that these cytokines drive circuit activation in A20flRR mice, we crossed A20flRR mice to Il25−/− and Il4ra−/− mice. Indeed, tuft cell hyperplasia was abrogated in A20flRR mice deficient in IL-25 or IL-4Rα (Fig. 1H and 1I). Further, small intestine IL-5+ ILC2s were reduced in Il25−/−A20flRR and Il25−/−A20flR+ mice, indicating that IL-25 drives their accumulation in the absence of A20 (Fig. 1I–K). In contrast, elevated numbers of IL-5 and IL-13 reporter-positive ILC2s were present in Il4ra−/−A20flRR and Il4ra−/−A20flR+ mice (Fig. 1I–K), confirming the inability of IL-25-producing tuft cells to expand when the epithelial progenitor compartment is unresponsive to IL-13 (von Moltke et al., 2016). Thus, A20 functions as a cell-intrinsic brake on the response of small intestinal ILC2s to IL-25. Despite constitutively high expression of A20 in ILC2s from all peripheral tissues analyzed, the major effects of A20 deletion were restricted to small intestine.
Activation of the tuft cell – ILC2 circuit drives adaptive small intestine remodeling
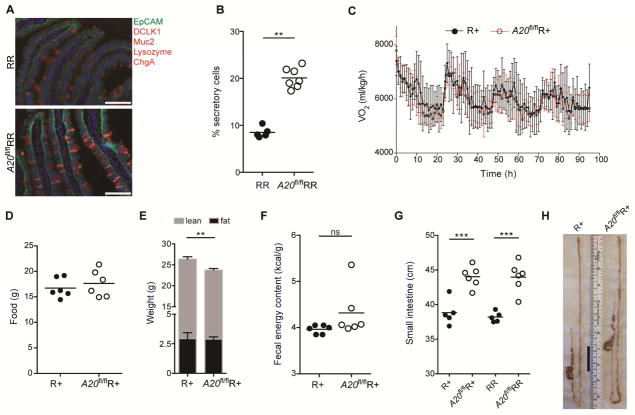
To assess the effects of small intestine ILC2 activation in A20flRR mice, we quantified the populations of secretory (tuft, goblet, enteroendocrine, Paneth cells) and absorptive enterocytes in the small intestine. As expected, the villus epithelium of A20flRR mice had a significantly increased frequency of secretory cells, with a corresponding decrease in absorptive enterocytes (Fig. 2A and 2B). Together with epithelial alterations, A20flRR mice developed morphological features associated with chronic helminth infection, including crypt hypertrophy and thickening of the surrounding muscularis layer (suppl. Fig. 2D and E) (Garside et al., 2000), consistent with generalized trophic effects restricted to small bowel tissues. To assess whether these changes caused metabolic alterations, we used metabolic cages to monitor parameters of intake, output and energy consumption. To avoid confounding effects introduced by the ablation of IL-5 in homozygous RR mice (Molofsky et al., 2013), we performed these experiments in A20flR+ mice. Unexpectedly, constitutive activation of the tuft cell – ILC2 circuit caused no significant alterations in total oxygen consumption (Fig. 2C) or food intake (Fig. 2D); body weight was slightly but significantly reduced as compared to control animals (Fig. 2E). Quantification of the caloric density of stool using bomb calorimetry as a measure of non-absorbed energy, however, showed no alterations in A20flR+ mice, consistent with comparable nutrient-extractive capacity of the intestine (Fig. 2F).
Figure 2. Energy balance in A20flRR mice is associated with an elongated small intestine.
(A, B) Quantification of secretory cells by immunofluorescence analysis of RR and A20flRR mice; secretory cell types in red identified by staining for Muc2 (goblet cells), DCLK1 (tuft cells), ChromograninA (ChgA, enteroendocrine cells) and lysozyme (Paneth cells), EpCAM in green, DAPI in blue. (C–F) Metabolic parameters of R+ and A20flR+ mice. (C) Oxygen consumption rate measured using CLAMS cages. (D) Food intake during a 4 day period. (E) Lean and adipose weight determined by EchoMRI. (F) Fecal energy content measured using bomb calorimetry. (G and H) Small intestine (SI) length (G) and representative image (H) of 8–12 week old R+, A20flR+, RR and A20flRR mice. The black bar indicates difference in SI length. Scale bars; 100 μm. Data from one experiment representative of at least three independent experiments (A–E, G, H) or from one experiment (F). (E, mean and s.e.m., n = 10–11). **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, not significant by Mann-Whitney U or one-way ANOVA.
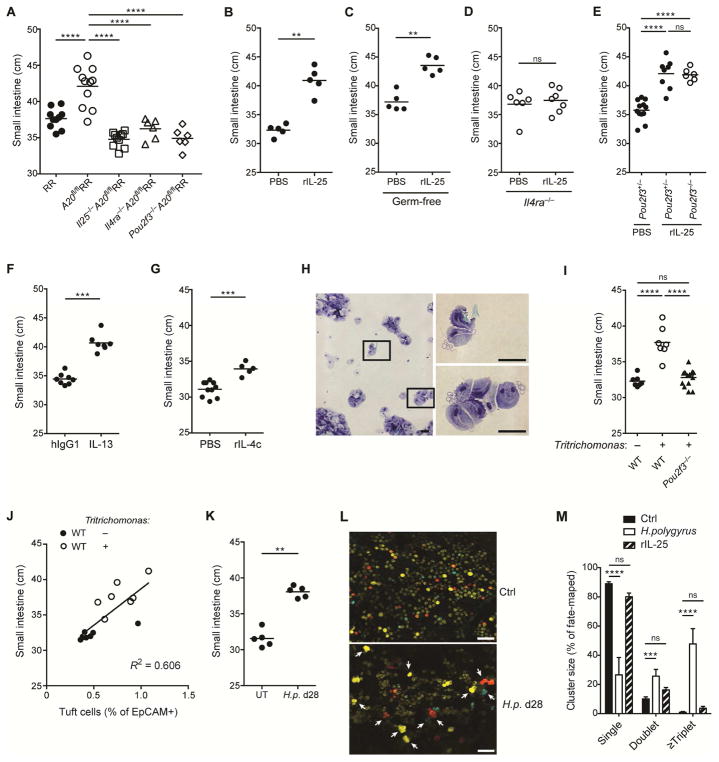
The maintenance of energy balance despite marked remodeling of the small intestine led us to consider mechanisms by which animals adapt to constitutive circuit activation. Strikingly, A20flR+ and A20flRR mice showed significant increases in small intestine length, suggesting adaptation driven by dispersal of relatively constant numbers of absorptive enterocytes over the increased surface area (Fig. 2G and 2H). The positive correlation between amplification of the circuit and small intestinal lengthening in A20flR+ and A20flRR mice was further supported by using an independent ILC2-specific A20-deletion strategy with YetCre-13 mice, which contain an IL-13 regulated Cre element (Price et al., 2010), which resulted in comparable phenotypes (suppl. Fig. 3A and 3B). Small intestine lengthening was absent in A20flRR mice crossed with animals deficient in IL-25, IL-4/IL-13 signaling or tuft cells (Pou2f3−/−), in which circuit amplification is abrogated (Fig. 3A). Small intestine lengthening was also significantly attenuated in A20flRR mice crossed to Trpm5-deficient mice, consistent with signaling through a tuft cell sensory pathway (suppl. Fig. 3C).
Figure 3. Activation of the tuft cell – ILC2 circuit drives small intestine lengthening.
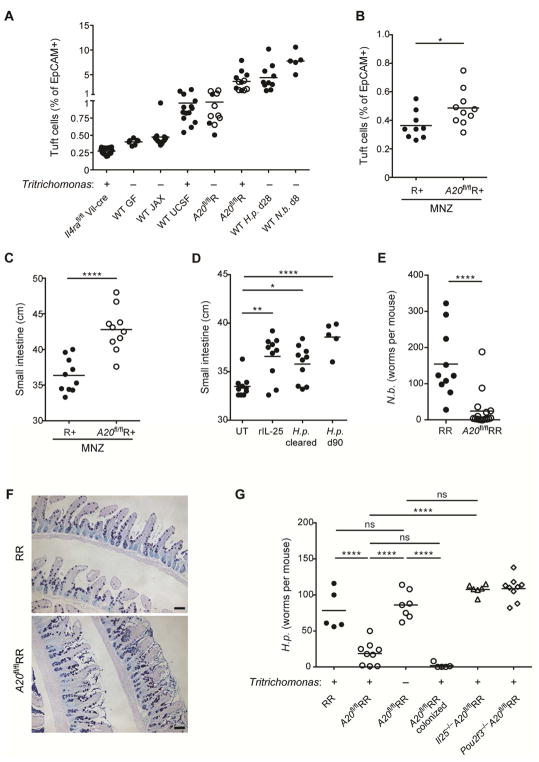
(A) Small intestine (SI) length in RR, A20flRR, Il25−/−A20flRR, Il4ra−/−A20flRR and Pou2f3−/−A20flRR mice. (B–E) SI length in (B) wild type, (C) C57BL/6 GF, (D) Il4ra−/− and (E) Pou2f3−/− mice after 4 weeks of serial treatment with rIL-25. (F) SI length of mice 5 weeks after hydrodynamic gene delivery with IL-13 or hIgG1 control plasmid. (G) SI length of mice treated with rIL-4 complexes for 4 weeks. (H) Cytospins stained with H&E. Micrographs show representative images of Tritrichomonas isolated from cecum of mice housed in UCSF vivarium. Scale bars; 20 μm. (I and J) Wild type (WT) mice were colonized with purified Tritrichomonas at 3 weeks of age. (I) SI length measured 8 weeks later and compared to age-matched Pou2f3−/− mice naturally colonized with Tritrichomonas. (J) SI length of wild type mice in (I) plotted against tuft cell frequency. (K) SI length in wild type mice 28 days after infection with H. polygyrus (H.p.). (L and M) Crypt fission in the proximal SI in Lgr5-CreERT2-EGFP x R26R-Confetti mice as described in methods. (L) Representive images from mice 28 days after infection with H. polygyrus. Lgr5 stem cells are pseudocolored in dim yellow (Lgr5-CreERT2-EGFP) and clonal crypts are randomly marked with bright yellow (YFP), red (RFP) or blue (CFP), driven from the R26R-Confetti locus. (M) Quantification of crypt fission of mice as in (L), or after 4 weeks of serial treatment with rIL-25. Scale bars; 200 μm. Data pooled from multiple independent experiments (A, C, D, F, M) or from one experiment representative of at least two independent experiments (B, E, G–I, L). (L, mean and s.e.m, n = 6–11). **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, not significant by Mann-Whitney U or one-way ANOVA.
To test whether amplification of the circuit was sufficient to induce adaptive small intestinal lengthening in wild-type mice, we used recombinant IL-25 to activate ILC2s. Activation of the circuit by 4 daily injections of IL-25, which directly activates small intestinal ILC2s to expand secretory epithelial lineages, increased the caloric density of stool, consistent with acutely decreased absorptive capacity (suppl. Fig. 3D). Chronic administration of IL-25 over four weeks, however, caused tuft cell expansion and significant lengthening of the small intestine as compared to vehicle control (Fig. 3B, suppl. Fig. 3E). Similar adaptation occurred following IL-25 administration to GF mice (Fig. 3C). Adaptive lengthening was absent when IL-25 was given to Il4ra−/− mice, but intact in Pou2f3−/− mice that lack tuft cells, and demonstrating a dominant effect mediated by stimulation with IL-13 (Fig. 3D and 3E, suppl. Fig. 3F). Hydrodynamic IL-13 overexpression and repeated treatment with rIL-4 complex similarly induced small intestinal lengthening (Fig. 3F and G). Taken together, these data suggest that amplification of the small intestinal tuft cell – ILC2 circuit is necessary and sufficient to induce adaptive remodeling of the small bowel.
Intestinal infections that activate the tuft cell – ILC2 circuit drive adaptive small intestine remodeling
We hypothesized that natural intestinal parasites of mice that activate the intestinal tuft cell – ILC2 circuit, such as Tritrichomonas spp. (Howitt et al., 2016) and H. polygyrus, would result in similar alterations. We isolated Tritrichomonas spp. (here designated Tritrichomonas) as confirmed by morphology and detection of 18S rRNA – ITS region on genomic DNA (Fig. 3H, Suppl. Fig. 3G), from cecal contents of colonized animals at UCSF to colonize 3-week old mice from The Jackson Laboratory (JAX) (suppl. Fig. 3H). Two months after colonization, the small bowel of Tritrichomonas-colonized mice was significantly longer as compared with non-colonized controls; lengthening did not occur in colonized Pou2f3−/− mice (Fig. 3I). Small intestine length correlated with the frequency of tuft cells and ILC2s (Fig. 3J, suppl. Fig. 3I). To extend these observations, we used another natural mouse parasite, H. polygyrus. This nematode induces marked activation of the tuft cell – ILC2 circuit (Howitt et al., 2016; von Moltke et al., 2016); tuft cell hyperplasia is maintained for as long as the luminal worms remain in the small intestine (suppl. Fig. 3J). Again, a significant increase in the length of the small intestine was present at 4 weeks post-infection (Fig. 3K). Thus, chronic amplification of the tuft cell – ILC2 circuit by eukaryotic pathosymbionts promotes small intestinal remodeling associated with adaption to the sustained increases in secretory-absorptive epithelial cell differentiation.
During development, the surface area of the intestine expands to increase absorptive capacity necessary to meet the metabolic requirements for growth, which can be mediated by a process of crypt fission resulting in duplication of stem cell niches required to nucleate additional villi. Crypt hypertrophy was apparent in A20flRR mice (Suppl Fig. 2D) and is a common feature of helminth infections (Garside et al., 2000). To assess whether activation of crypt fission contributes to expansion of the small intestine in mice with an amplified circuit, we performed lineage tracing in Lgr5+ intestinal stem cells using CreERT2-mediated induction of the multicolor reporter R26R-Confetti, in which crypt fission events are revealed by the appearance of two or more adjacent labeled clonal crypts bearing the same color. Consistent with previous reports (Snippert et al., 2010), crypt fission rate was low in the small intestine of adult mice (Fig. 3L and M). Mice infected with H. polygyrus, in contrast, demonstrated clonal expansion of crypts expressing the same mark, consistent with crypt fission, that was prominent in areas in proximity to the worms (Fig. 3L and M). Crypt fission was less obvious in mice treated with rIL-25, which may reflect additional pathways or the ease of identifying involved areas after worm infection (Fig. 3M). Amphiregulin, a member of the EGF family of growth factors, can promote epithelial proliferation after Trichuris muris infection (Zaiss et al., 2006). Although ILC2s can be an important source of amphiregulin (Gury-BenAri et al., 2016; Monticelli et al., 2011), Areg-deficent mice showed no diminution in small intestinal lengthening after H. polygyrus infection (suppl. Fig. 3K).
Weaning and post-natal differentiation of the tuft cell – ILC2 small intestinal circuit
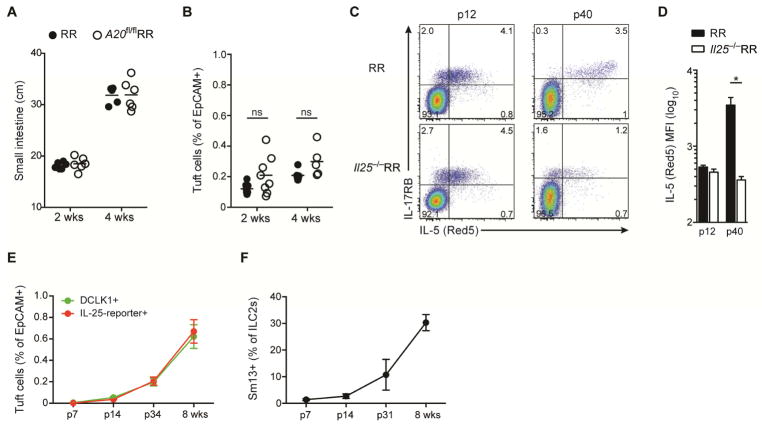
Because the small intestinal tissue undergoes marked alterations with weaning, we assessed when circuit activation and remodeling occur in A20flRR and control mice. We detected no differences in small intestinal length or tuft cell hyperplasia before (2 wks) and around weaning (4 wks), indicating that amplification of the tuft cell–ILC2 circuit occurs after weaning (Fig. 4A and 4B, Fig. 1). Notably, ILC2s were present, expressed IL-17RB, and became IL-5 reporter-positive as early as p12 (Fig. 4C), and similar to what we previously reported in lung ILC2s (Nussbaum et al., 2013). We noted, however, that levels of IL-5 expression by ILC2s increased significantly between 2 and 5 weeks of age. This increase was abrogated in Il25-deficient mice, consistent with ILC2 stimulation by IL-25 after weaning (Fig. 4C and 4D). IL-5 (Red5) expression was largely restricted to CD3−KLRG1+IL-17RB+ ILC2s, consistent with a dominant role for innate cells (Suppl. Fig. 4A). Using both IL-25 reporter expression and DCLK1 staining as markers, we found that tuft cells are extremely rare in the first 2 weeks of life, and begin to accumulate around and after weaning (Fig. 4E). IL-13 expression by ILC2s concomitantly increased after weaning (Fig. 4F), and was accompanied by IL-25-dependent changes in ILC2 marker expression, including decreases in Arg1 and Thy1, and increases in the inhibitory receptor, KLRG1 (suppl Fig. 4B and C). Thus, the basal tuft cell–ILC2 circuit is established post-weaning by a process dependent on IL-25 and negatively regulated by A20 in ILC2s.
Figure 4. Weaning leads to IL-25-dependent activation of the tuft cell – ILC2 small intestinal circuit.
(A and B) Small intestine (SI) length (A) and tuft cell frequencies (B) of RR and A20flRR mice at 2 and 4 weeks of age. (C) Expression of IL-17RB and IL-5-reporter (Red5) by cells gated on Lin- CD45+ cells isolated from SI of RR and Il25−/−RR mice at days 12 and 40 post-birth (p). (D) MFI of Red5 in ILC2s gated on Lin-CD45+IL-17RB+ cells of mice as in (C). (E and F) Frequencies of tuft cells (E) based on expression of DCLK1 or IL-25-reporter and of IL-13+ (Sm13 reporter) ILC2s (F) gated on CD45+Lin-IL-17RB+ cells quantified in SI by flow cytometry at the indicated time points post-birth (p). Data pooled from multiple independent experiments (A, B, E, F) or from one experiment representative of at least two independent experiments (C and D). Student’s t test was performed. (D–F, mean and s.e.m, n = 3–5). Statistical significance is indicated by *p < 0.05; ns, not significant.
Microbiota, diet, and pathosymbionts in priming and activation of the small intestine tuft cell – ILC2 circuit
Weaning is marked by changes from milk to solid food, and is accompanied by substantial alterations in energy expenditure and maturation of the microbiota (Sommer and Backhed, 2013). To assess the overall effects of the microbiota on tuft cell and ILC2 frequency in the small intestine, we analyzed adult GF mice. Using a number of surface markers, we could ascertain little differences in the status of ILC2s from GF mice as indicated by comparable expression of IL-17RB and KLRG1 (Fig. 5A and Suppl. Fig. 5A), and consistent with previous reports (Gury-BenAri et al., 2016); tuft cells were also comparable to those in naïve barrier-maintained mice but were increased in mice from UCSF vivarium that were colonized with Tritrichomonas (Fig. 5B). Due to the association with weaning, we explored the effects of milk on the intestinal tuft cell-ILC2 circuit. Feeding adult mice with pasteurized bovine milk as the lone dietary constituent led to reduced IL-13 expression by small intestinal ILC2s (Fig. 5C). Further, A20flRR mice that were kept only on milk diet after weaning had reduced tuft cell hyperplasia and small bowel lengthening (Fig. 5D and 5E). Notably, we detected an increased abundance of Tritrichomonas in the cecum following weaning (Fig. 5F), whereas Tritrichomonas were significantly depleted by placing colonized mice on a strict milk diet (Suppl. Fig. 5B), but not when milk was provided together with chow diet (Suppl. Fig. 5C), suggesting that dietary change facilitates colonization. Further, depletion of Tritrichomonas using metronidazole-treatment of colonized wild-type mice led to reduced IL-13 expression by small intestinal ILC2s (Fig. 5G). Continuous exposure to metronidazole starting from birth prevented tuft cell hyperplasia and small bowel lengthening in exposed and colonized A20flRR mice, whereas a cocktail of broad-spectrum antibiotics that substantially depletes the bacterial microbiota but not Tritrichomonas had no effect (Fig. 5H and 5I, Suppl. Fig. 5D). Likewise, tuft cell hyperplasia and small bowel lengthening were significantly reduced in A20flRR mice that were raised free of Tritrichomonas (Fig. 5H and 5I). IL-13 expression was induced in ILC2s of A20flR+ mice upon colonization with protists and preceded the accumulation of tuft cells and intestinal lengthening (Fig. 5J and 5K, suppl. Fig. 5E and 5F). Thus, the switch from milk to solid food corresponds with developmental priming of the tuft cell–ILC2 circuit mediated by widespread eukaryotic pathosymbionts, such as Tritrichomonas and helminths, but not bacteria, in concordance with prior reports (Gerbe et al., 2016; Howitt et al., 2016; von Moltke et al., 2016).
Figure 5. Dietary fibers facilitate Tritrichomonas colonization after weaning.
(A and B) The small intestine (SI) was isolated from GF mice, or wild-type mice that were offspring of JAX mice, which were bred without further manipulation (JAX) or after colonization with Tritrichomonas isolated from cecal content of mice in UCSF vivarium (UCSF). Expression of KLRG1 and IL-17RB by cells gated on Lin- CD45+ cells (A) and frequencies of tuft cells (B) analyzed by flow cytometry. (C) Adult IL-13-reporter (Sm13) mice fed bovine milk as the lone dietary constituent for 2 weeks and IL-13 expression by ILC2s in SI quantified by flow cytometry. (D and E) A20flRR mice fed bovine milk as the lone dietary constituent following weaning for 4 weeks and frequencies of tuft cells (D) and SI length (E) were measured. (F) Abundance of Tritrichomonas in cecal content of WT mice at 3 and 5 weeks post-birth quantified by qPCR. (G) Adult IL-13-reporter (Sm13) mice treated with metronidazole (MNZ) in drinking water for 2 weeks and IL-13 expression by ILC2s analyzed as in (C) and compared to the same control group. (H and I) A20flRR mice treated with metronidazole in drinking water starting at weaning and analyzed 6–8 weeks later. Tritrichomonas-free A20flRR mice established as described in methods. The frequencies of tuft cells (H) and SI length (I) measured as in (D, E) and compared to the same control groups. (J and K) Tritrichomonas-free R+ and A20flR+ mice colonized with Tritrichomonas after weaning and analyzed 7 days later. Expression of IL-5 (Red5) and IL-13 (Sm13) by ILC2s and CD3+CD4+ T cells in SI quantified by flow cytometry (J) and the frequencies of IL-13+ ILC2s shown (K). (L) WT mice fed chow or purified diets containing no fiber, 10% cellulose or 10% inulin. One day later mice were gavaged with 104 purified Tritrichomonas and the abundance of Tritrichomonas in the cecum analyzed after 14 days by qPCR. (M) A20flRR mice fed chow or purified diets containing 10% cellulose or 10% inulin after weaning for 6 weeks and the SI length measured. Data from one experiment representative of at least two independent experiments (A, C, G, J–M) or pooled from multiple independent experiments (B, D–F, H, I). F, mean and s.e.m.; n = 5–6. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, not significant by Mann-Whitney U or one-way ANOVA.
Parabasalid protozoa are common endobiotic symbionts of the intestinal tract in wood-digesting invertebrates such as termites, where the protists help to digest complex plant polysacharride fibers; persistent colonization across the social colony is actively maintained through anal-oral transmission by proctodeal trophallaxis (Brune, 2014). To address whether Tritrichomonas in coprophagic mice utilize complex dietary fibers, which are a major dietary constituent of feral and laboratory rodents, we fed mice isocaloric fiber-manipulated diets (Table S1). Notably, Tritrichomonas were heavily depleted when dietary fibers were absent or when cellulose was the sole fiber (Fig. 5L, suppl. Fig. 5G). In contrast, Tritrichomonas colonization became re-established when the oligofructan inulin was the sole dietary fiber (Fig. 5L, suppl. Fig. 5G). The fermentable fiber requirement for Tritrichomonas colonization and circuit activation was accompanied by abrogation of tuft cell hyperplasia and lengthening when A20flR+ mice were fed cellulose diet, but substantial reconstitution on inulin diet (Fig. 5M, suppl. Fig. 5H).
Products of Tritrichomonas metabolism activate the tuft cell – ILC2 circuit
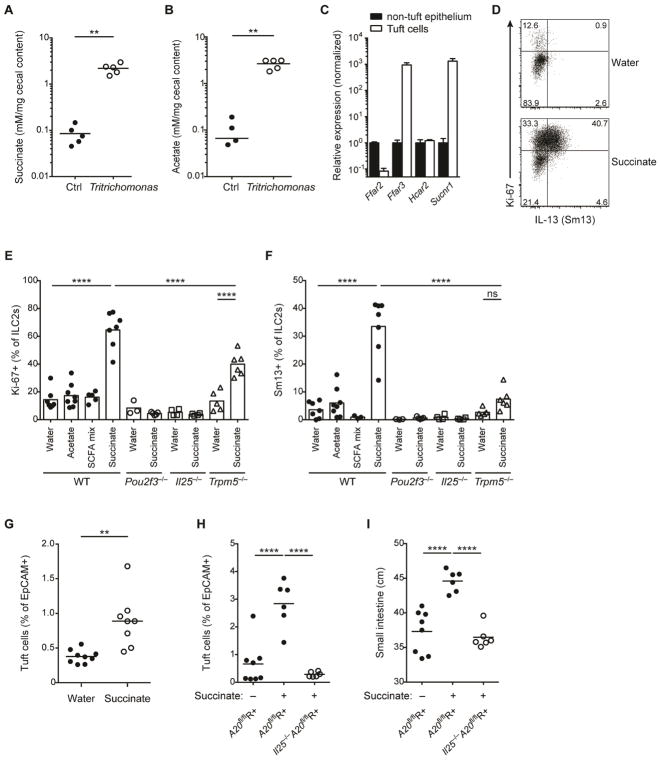
Fermentation of dietary fibers by the microbiota results in the generation of short-chain fatty acids (SCFAs) and other carboxylic acids, which impact many aspects of host physiology, including metabolism and immunity (Koh et al., 2016). Indeed, we detected increased cecal acetate in SPF mice colonized with Tritrichomonas (suppl. Fig. 6A). To assess whether these prevalent protists directly contribute to the production of these fermentation products, we monocolonized GF mice with Tritrichomonas. Notably, we found increased cecal acetate and succinate levels in GF mice colonized with Tritrichomonas (Fig. 6A and 6B), consistent with fermentative pyruvate oxidation in the protist hydrogenosomes (Muller et al., 2012). Because we noted high expression of G-protein coupled receptors for SCFAs (Ffar3) and succinate (Sucnr1) in tuft cells (Fig. 6C), we hypothesized that Tritrichomonas-derived metabolic end products might be sensed directly by tuft cells. Strikingly, succinate in drinking water induced potent proliferation and IL-13 expression in ILC2s (Fig. 6D–F), which was independent of bacterial microbiota as indicated using GF mice (suppl. Fig. 6B). In contrast, acetate or a mix of acetate, butyrate and propionate did not activate ILC2s (Fig. 6E and 6F). Notably, we detected no Sucnr1 expression in ILC2s (suppl. Fig. 6C). Further, succinate-mediated ILC2 activation required the presence of tuft cells and IL-25, and was significantly reduced in the absence of TRPM5 (Fig. 6E and 6F), demonstrating a crucial function in sensing luminal succinate and consistent with the role of the tuft cell – TRPM5 – IL-25 axis in detecting luminal pathosymbionts (Gerbe et al., 2016; Howitt et al., 2016; von Moltke et al., 2016) and with TRPM5-redundant pathways (Dutta Banik et al., 2018). The effects of luminal succinate were limited to ILC2s of the small intestine (suppl. Fig. 6D–F), consistent with highest expression of Sucnr1 by small intestinal tuft cells (suppl. Fig. 6G). Succinate-induced circuit activation resulted in increased tuft cell frequencies similar to colonization with Tritrichomonas (Fig. 6G), and was restricted by A20 in ILC2s, as indicated by pronounced tuft cell hyperplasia in succinate-treated (Tritrichomonas-free) A20flR+ mice that was IL-25-dependent (Fig. 6H). Importantly, succinate alone was sufficient to induce small intestinal lengthening in A20flR+ mice (Fig. 6I). Overall, our results identify the first luminal metabolite capable of activating the tuft cell – ILC2 circuit, and support the chemosensory role that was assigned to tuft cells decades ago.
Figure 6. Succinate is produced by Tritrichomonas and sufficient to activate the tuft cell – ILC2 circuit.
(A and B) GF mice monocolonized with Tritrichomonas and concentrations of acetate (A) and succinate (B) measured in the cecal content after 6 weeks. (C) Metabolite receptor mRNA expression quantified by qPCR in tuft cells versus other epithelial cells sorted from small intestine (SI) and normalized to levels in non-tuft epithelial cells. (D–F) Tritrichomonas-free IL-13-reporter (Sm13) mice were treated with succinate, acetate or a SCFA mix (acetate, butyrate, propionate) in drinking water for 4 days. Expression of Ki-67 and IL-13 by ILC2s in SI quantified by flow cytometry and representative dot plot from wild-type (WT) mice shown (D). Expression of Ki-67 (E) and IL-13 (F) by ILC2s from WT, Pou2f3−/−, Il25−/− and Trpm5−/− mice treated with indicated solutions. (G) Tritrichomonas-free WT mice treated with 100 mM succinate in drinking water for 10 days. Frequencies of tuft cells in SI by flow cytometry. (H and I) Tritrichomonas-free A20flR+ and Il25−/−A20flR+ mice treated with 100 mM succinate in drinking water for 25 days and frequencies of tuft cells (H) and SI length (I) analyzed. Data from one experiment (A, B) or from one experiment representative of at least two independent experiments (C, D, G–I) or pooled from multiple independent experiments (E, F). C, mean and s.e.m.; n = 3. **p < 0.01, ****p < 0.0001; ns, not significant by Mann-Whitney U or one-way ANOVA.
Activation of the tuft cell – ILC2 circuit contributes to concomitant immunity
The interdependence of development, diet and eukaryotic pathosymbionts in these lines of uncolonized, colonized (Tritrichomonas) and infected (H. polygyrus) mice allowed us to establish a hierarchical relationship between these luminal states and activation of the small intestinal ILC2-tuft cell circuit (Fig. 7A). To assess the stability of the small intestinal response, we cleared Tritrichomonas from the intestines of colonized adult A20flR+ mice. After two weeks, tuft cell frequencies had essentially normalized to levels that remained only slightly higher than those found in uncolonized control mice (Fig. 7B). Despite this, the numbers of small intestinal IL-5 reporter-positive ILC2s remained increased, even though their expression of IL-13 (as a dynamic marker of activation (Liang et al., 2011)) had become attenuated (suppl. Fig. 7A and 7B). Notably, the difference in small intestinal length was maintained, suggesting that adaptive remodeling was sustained and had become discordant from tuft cell hyperplasia (Fig. 7C). To confirm the generalizable nature of these findings, we determined that wild-type mice previously treated with recombinant IL-25 or infected with H. polygyrus, continued to have significant increases in small intestinal length up to 2 months later (Fig. 7D), at a time when the tuft cell composition of the small intestine had almost normalized (suppl. Fig. 7C).
Figure 7. The tuft cell – ILC2 circuit contributes to concomitant immunity.
(A) Small intestinal (SI) tuft cell frequencies in indicated mice with presence (+) or absence (−) of Tritrichomonas as indicated. Some data is repeated from previous figures for comparison. Wild-type (WT) mice as in Fig. 5: JAX, offsprings of mice from JAX; UCSF, offsprings of mice colonized with Tritrichomonas. A20flR pooled from A20flR+ (open circle) and A20flRR (closed circle) mice. WT mice analyzed at indicated time points after infection with H. polygyrus (H.p.) or N. brasiliensis (N.b.). (B and C) Adult R+ and A20flR+ mice treated with metronidazole (MNZ) in drinking water for 2 weeks and frequencies of tuft cells (B) and SI length (C) analyzed. (D) WT mice serially treated with rIL-25 or infected with H. polygyrus (H.p.) for 4 weeks followed by worm clearance (cleared) in indicated mice using pyrantel pamoate. SI length measured 2 months after the last rIL-25 administration or worm clearance. (E and F) Worm burden (E) and Alcian blue/PAS staining (F) in SI of RR and A20flRR mice 5 days after infection with N. brasiliensis. (G) SI worm counts of indicated mouse strains 13 days after infection with H. polygyrus. Presence (+) or absence (−) of Tritrichomonas indicated. One group of Tritrichomonas-free A20flRR mice was colonized with Tritrichomonas 2 weeks before the infection. Data pooled from multiple independent experiments (A–E) or from one experiment representative of at least two independent experiments (F, G).*p < 0.05, **p < 0.01, ****p < 0.0001; ns, not significant by Mann-Whitney U or one-way ANOVA.
We considered the hypothesis that epithelial re-wiring mediated by activation of the tuft cell – ILC2 circuit might contribute to concomitant immunity, the mechanism by which helminth-infected animals become resistant to new infection despite the inability to reject the primary infection (Smithers and Terry, 1969). Indeed, constitutive activation of the tuft cell – ILC2 circuit in Tritrichomonas-colonized A20flRR mice rendered animals resistant to infection by both N. brasiliensis and H. polygyrus (Fig. 7E–G), and was mediated by Tritrichomonas and dependent on IL-25 and tuft cells (Fig. 7G). Circuit activation in Rag1−/− mice by pre-treatment with IL-25 sigificantly reduced fecundity of N. brasiliensis suggesting that innate components are sufficient to mediate this response, and consistent with prior reports (Suppl. Fig. 7D) (Fallon et al., 2006). We also noted less robust infection with N. brasiliensis in mice previously infected and cleared of a heterologous parasites, H. polygyrus, in accordance with prior observations (Guo et al., 2015).
DISCUSSION
Our studies illuminate important findings relevant to small intestinal biology. First, although we and others called attention to expression of IL-25 by tuft cells in mucosal epithelia, we now link this with constitutive expression of the IL-25 receptor on small intestine ILC2s, and in contrast to ILC2s in other peripheral organs. This dedicated role for IL-25 in dynamically regulating small bowel homeostasis was further supported by the marked effect of ILC2-specific A20 deletion in sensitizing the small intestinal circuit with minimal effects on other tissue ILC2s. Second, our studies reveal a novel tripartite interplay between this circuit, diet and microbiota, which involved the protist-derived fermentative end-product succinate, which alone activates the circuit leading to adaptive intestinal remodeling, thus revealing a molecular signal promoting luminal detection. Third, an unsuspected component of the host response to chronic circuit activation consisted of small intestine lengthening and remodeling by a process associated with maintenance of systemic energy balance suggesting a conserved physiologic response that likely evolved to sustain host metabolic needs in the face of chronic parasite infestations, which are widespread among feral vertebrates. Lastly, the small intestinal physiologic changes, including tuft cell hyperplasia and small intestinal dynamic lengthening, although induced by viable luminal parasites themselves, impaired further intestinal infection by similar or even heterologous organisms, revealing an evolved host-parasite interaction that served to limit further colonization. Such détente is consistent with the phenomenon of concomitant immunity discovered almost fifty years ago (Smithers and Terry, 1969), and suggest that remodeling of the intestinal niche by IL-25 plays a key role in enabling a mutualistic state facilitating metabolic homeostasis in the host and a reproductive niche for the pathosymbiont.
Intriguingly, small intestine ILC2s constitutively express the IL-25 receptor, even in GF mice, revealing a dedicated role for these cells in setting the secretory-absorptive tone of the small intestine. In this way, intestinal ILC2s, which seed tissues during fetal development (Bando et al., 2015), resemble fetal-derived tissue macrophages (Mass et al., 2016) in their capacity to integrate into tissues and acquire organ-specific ability to sense anticipatory signals, like tuft cell IL-25, that appear postnatally. The activation of ILC2s in response to luminal eukaryotic pathosymbionts revealed heterogeneity in expression of cytokines like IL-5 and IL-13, and markers like Arginase 1 and Thy1 (Bando et al., 2013; Spits et al., 2013), consistent with observations using single-cell approaches (Gury-BenAri et al., 2016). IL-25 is a member of a family of IL-17 cytokines that occupy a key position at the interface of epithelia, innate immune cells and the microbiota (Song et al., 2016). These cytokines have deep evolutionary roots as assessed by sequence and functional analyses that indicate predominant expression by epithelia and immune cells at barriers that interface with microbes (Buckley et al., 2017; Han et al., 2015); dysregulation of these circuits can cause immunopathology (Amatya et al., 2017). Emerging roles for IL-17 family members in neural networks (Choi et al., 2016), including in nematodes (Chen et al., 2017), are consistent with broader roles in homeostasis. Amplification of the circuit in mice using exogenous IL-25 alone increased small intestinal length, as noted above, that was dependent on IL-4Rα signaling and bypassed the need for tuft cells, as shown in Pou2f3−/− mice. Intriguingly, crypt fission rates were increased in proximity to H. polygyrus, suggesting that small intestinal surface expansion might be mediated in part by multiplication of existing crypts. Notably, adaptive surface growth following small bowel resection is a well-described phenomenon in humans regulated by multiple hormones and growth factors (Warner, 2016), and roles for circuit amplification, perhaps involving additional tuft cell signals (Middelhoff et al., 2017) and nerve-epithelial interactions (Hayakawa et al., 2017), merit further study.
Intestinal immunity is influenced by complex interactions between dietary components and microbiota (Belkaid and Harrison, 2017). Although ILC2s occupy small intestine at birth, tuft cells appeared coincident with weaning. In our mouse facility, ILC2 activation and tuft cell expansion increased further in animals colonized by vertically-transmitted Tritrichomonas, corroborating observations from other SPF facilities (Chudnovskiy et al., 2016; Escalante et al., 2016; Howitt et al., 2016). These organisms are members of the ‘eukaryome’, including protists, fungi and helminths, that are widely prevalent in the vertebrate intestine of asymptomatic humans and animals (Lukes et al., 2015). The parabasalids represent an abundant group of flagellated endobiotic symbionts, commensals and parasites that commonly inhabit the digestive tract. Tritrichomonas has also been shown to promote IL-18-dependent Th1 and Th17 immunity in the colon under different conditions (Chudnovskiy et al., 2016; Escalante et al., 2016). Whether these observations reflect species-specific effects and/or tropism-related bias in immunomodulation remains unclear and requires further phylogenetic analysis of protists derived from various research facilities, which might reveal unappreciated diversity as indicated by early descriptions of enteric protozoa in wild rodents (Ring, 1959). We demonstrated a key role for dietary fiber in enabling Tritrichomonas colonization, which could be met by inulin but not cellulose, suggesting similar dietary functions as shown in termite parabasalid symbionts, which are critical for digestion of complex carbohydrates like lignocellulose (Brune, 2014). The capacity of Tritrichomonas to degrade recalcitrant complex polysaccharides is consistent with genomic replication of glycolytic enzymes (Barratt et al., 2016) and fermentative metabolism culminating in metabolic end-products of the mitochrondria-like hydrogenosomes, including acetate, succinate and hydrogen (Muller et al., 2012). Notably, other eukaryotic pathosymbionts, including helminths, share similar metabolic pathways (Muller et al., 2012). Important roles for dietary fiber and SCFA in vertebrate physiology are increasingly apparent, and unappreciated roles for prevalent parabasilid organisms in these metabolic interactions with host and bacteria will be of much interest (Koh et al., 2016).
We demonstrate that tuft cells express the succinate receptor, SUCNR1 (GPR91), at high levels as compared to other intestinal epithelia. Succinate, an end-product of Tritrichomonas metabolism, potently activated the circuit in a POU2F3-, IL-25- and TRPM5-dependent manner, consistent with a metabolite-triggered sensory role for tuft cells. Krebs cycle intermediates like succinate provide exquisite barometers of cellular energetic stability; small accumulations drive inflammatory programs in macrophages fueling the generation of mitochondrial reactive oxygen intermediates (Mills et al., 2016). Due to export and detection by GPR91, succinate has local and systemic signaling capacity with roles in multiple tissues, including kidney, heart, liver, retina and white adipose tissue, and has been implicated in tissue hypertrophy (Gilissen et al., 2016). Our results suggest that succinate sensing has evolved to detect certain pathosymbionts, such as Tritrichomonas, but also succinate-producing bacteria, which, depending on the context, might also be present. Tuft cell-mediated detection of succinate, a potential signal of hypoxic stress and injury (Gilissen et al., 2016), in order to activate ILC2s is consistent with the function of the latter in integrating tissue-derived homeostatic signals (von Moltke and Locksley, 2014), and might enable therapeutic strategies for restoring intestinal adaptation and regeneration in response to inflammation.
In addition to supporting metabolic homeostasis, circuit activation altered the intestinal niche such that further infections were attenuated. This state, termed concomitant immunity, was conceptualized in studies of schistosomiasis in monkeys 50 years ago (Smithers and Terry, 1969), and has been explained largely by adaptive immunity, antigenic variation amongst parasite stages, and Tregs. Our studies suggest that intestinal remodeling mediated by activation of the tuft cell – ILC2 circuit contributes to concomitant immunity by altering the intestinal niche, consistent with aspects of innate tissue memory, or ‘training’ (Netea and van der Meer, 2017). As such, effects on intestinal remodeling mediated by these eukaryotic pathosymbionts extend the concept of colonization resistance (Buffie and Pamer, 2013). Indeed, protective effects have been described across kingdoms as suggested by resistance of Tritrichomonas–colonized mice to Salmonella infection (Chudnovskiy et al., 2016). Our findings emphasize the deep evolutionary relationships of the ‘eukaryome’, including protozoa and helminths, in sustaining the resilience of the small intestine during development and dietary fluctuations, and suggest key roles for tuft cells, succinate and IL-25 in achieving this mutualistic state. We speculate that harnessing such organisms may enhance the capacity for extraction of nutrients from complex carbohydrates, and could contribute to the remarkable adaptability of the omnivorous gut.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Richard M. Locksley (richard.locksley@ucsf.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
A20-floxed mice (Tnfaip3tm2Ama) (Tavares et al., 2010) were provided by A. Ma and crossed with Il5R mice (Nussbaum et al., 2013) that express a dimer red fluorescent protein (tdTomato) linked by an internal ribosomal entry site (IRES) to a Cre element under the control of the Il5 locus resulting in conditional deletion of A20 in ILC2s (A20flR). B6.Trpm5−/− (B6.129P2-Trpm5tm1Dgen/J, 005848), Lgr5EGFP-CreERT2 (B6.129P2-Lgr5tm1(cre/ERT2)Cle/J; 008875) and wild-type (C57BL/6J; 000664) mice were purchased from Jackson Laboratories. R26R-Confetti (B6.129P2-Gt(ROSA)26Sortm1(CAG-Brainbow2.1)Cle/J) were provided by O. Klein. B6.Areg−/− (Aregtm1Dle) were provided by M. Conti. Arginase-1 reporter Yarg (B6.129S4-Arg1tm1Lky/J), B6.Il25−/−, IL-13 reporter Smart13 (B6.129S4(C)-Il13tm2.1Lky/J), Il13-cre (C.129S4(B6)-Il13tm1(YFP/cre)Lky/J), B6.Il4ra−/− and IL-25 reporter (Il25F25) mice were obtained or generated as described (Van Dyken et al., 2014) (Liang et al., 2011) (von Moltke et al., 2016). B6.Pou2f3−/− (C57BL/6N-Pou2f3tm1.1(KOMP)Vlcg) were provided by M. Anderson. Mice were maintained in the University of California San Francisco specific pathogen–free animal facility or UCSF Gnotobiotic Core Facility in accordance with the guidelines established by the Institutional Animal Care and Use Committee and Laboratory Animal Resource Center. SPF animals were housed in individually ventilated cage units that are changed every three weeks for maintenance cages and every other week for breeding cages; cage bottoms are covered with autoclaved bedding and nesting material for enrichment. Mice were fed irradiated food (PicoLab Mouse Diet 20, 5058M) and drink from autoclaved bottles or automatic watering system. All animals were manipulated using standard procedures including filtered air exchange stations, chlorine-based disinfection of gloves and work surfaces within manipulations with animals; personnel protection equipment (PPE) (disposable gowns, gloves, head caps, and shoe covers) is required to enter the facility. Mice aged 6–14 weeks were used for all experiments, except for specific long-term experiments. Experiments were carried out using age and gender matched groups. Unless otherwise noted, all strains of mice that were housed within the UCSF vivarium were positive for Tritrichomonas and was confirmed for experimental mice in cecal content by visual inspection or qPCR. Standard animal manipulation procedures prevented contamination of Tritrichomonas-free strains that were housed in the same room. To establish Tritrichomonas-free colonies mice were treated with drinking water supplemented with 2.5 g/l metronidazole and 1% sucrose for 2–3 weeks. WT C57BL/6J were originally obtained from Jackson Laboratories and maintained in the UCSF vivarium following colonization with Tritrichomonas or uncolonized.
METHOD DETAILS
Mouse infection and treatment
Mice were infected subcutaneously with 500 N. brasiliensis L3 or by oral gavage with 200 H. polygyrus L3, and were killed at the indicated time points to collect tissues for staining or to count intestinal worm burden, as described (von Moltke et al., 2016). To clear H. polygyrus mice were treated with a single dose of 2 mg pyrantel pamoate (Columbia Laboratories) by oral gavage. IL-25 (R&D Systems) was administered intraperitoneally in doses of 500 ng on four consecutive days during the first week followed by 3 doses per week for the subsequent 3 weeks. All injections were given in 200 μl. For the milk diet, whole cow’s milk (Costco Wholesale) was the only source of food and/or drink as indicated, and was replaced daily. Custom diets were formulated based on an open standard diet (D11112201; Research Diets, Inc.) and contained no fiber (D11112229), 10 % cellulose (D17030102) or 10% inulin (D11112226) (Table S1). For antibiotic-treatment experiments, mice received drinking water supplemented with ampicillin, metronidazole, neomycin (1 mg/ml each), and vancomycin (0.5 mg/ml) in 1% sucrose, or metronidazole (2.5 mg/ml) in 1% sucrose as indicated. For SCFA and succinate treatments, succinic acid (100 mM), sodium acetate (100 mM), or a SCFA mix (67.5 mM sodium acetate, 40 mM sodium butyrate, 25.9 mM sodium propionate) were provided in the drinking water and changed weekly. For hydrodynamic gene delivery, mouse IL-13 coding sequence was cloned into pLIVE in vivo expression vector (Mirus Bio). Plasmid DNA was purified using EndoFree Plasmid Mega Kit (Qiagen). Mice received hydrodynamic tail-vein injections of 1 μg plasmid DNA in a volume equal to 10% body weight (0.1 ml/g) within +/− 5 s. As a control, a hIgG1 expression vector was injected. rIL-4 complexes were generated by incubating 2 μg animal-free recombinant mouse IL-4 (PeproTech) with 10 μg InVivoPlus anti-mouse IL4 antibody (clone 11B11, Bio X Cell) for 30 min at room temperature, and administered i.p. as described for rIL-25. For GF experiments, fecal pellets were regularly analyzed for microbial contamination using PCR and standard culture techniques.
Intestine length
Immediately after euthanasia, the small intestine was cut distal to the stomach, carefully pulled out of the abdomen and detached from mesenteric tissue. After cutting proximal to the cecum, the small intestine length was measured by hanging vertically alongside a measuring tape.
Fixed tissue preparation and staining
For immunohistochemistry, tissues were flushed with PBS and fixed in 4% paraformaldehyde for 2–3 h at 4 °C followed by PBS wash and overnight incubation in 30% (w/v) sucrose. Intestines were coiled into a ‘Swiss roll’, tissues were embedded in Optimal Cutting Temperature Compound (Tissue-Tek) and stored at −80 °C before sectioning (8 μm) on a Cryostat (Leica). Staining was performed in Tris/NaCl blocking buffer (0.1 M Tris-HCl, 0.15 M NaCl, 5 μg/ml blocking reagent (Perkin Elmer), pH 7.5) as follows: 1 h 5% goat serum, 1 h primary antibody, 40 min secondary antibody, 5 min DAPI (Roche). For RFP and DCLK1 co-labelling experiments, slides were stained for RFP as described above, excluding the DAPI step. DCLK staining was as follows: 30 min rabbit IgG (Abcam, ab27472), 1 h anti-DCLK1-AF488, and 5 min DAPI. See Key Resources Table for a list of antibodies used in this study.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD3 PB (clone 17A2) | BioLegend | Cat# 100214 |
| Anti-mouse CD5 BV421 (clone 53-7.3) | BioLegend | Cat# 100629 |
| Anti-mouse/human CD11b PB (clone M1/70) | BioLegend | Cat# 101224 |
| Anti-mouse CD11c PB (clone N418) | BioLegend | Cat# 117322 |
| Anti-mouse CD19 PB (clone 6D5) | BioLegend | Cat# 115523 |
| Anti-mouse CD3 PE/Cy7 (clone 17A2) | BioLegend | Cat# 100220 |
| Anti-mouse FcεRIα PB (clone MAR-1) | BioLegend | Cat# 134314 |
| Anti-mouse TER-119 PB (clone TER-119) | BioLegend | Cat# 116232 |
| Anti-mouse Gr-1 PB(clone RB6-8C5) | BioLegend | Cat# 108430 |
| Anti-mouse NK-1.1 PB (clone PK136) | BioLegend | Cat# 108722 |
| Anti-mouse CD4 BV711 (clone RM4-5) | BioLegend | Cat# 100550 |
| Anti-mouse CD49b PB (clone DX5) | BioLegend | Cat# 108918 |
| Anti-mouse CD8α PB (clone 53-6.7) | BioLegend | Cat# 100725 |
| Anti-mouse TCR γ/δ BV421 (clone GL3) | BioLegend | Cat# 118120 |
| Anti-mouse CD326 (Ep-CAM) PerCP/Cy5.5 (clone G8.8) | BioLegend | Cat# 118220 |
| Anti-mouse F4/80 PB (clone BM8) | BioLegend | Cat# 123124 |
| Anti-mouse CD25 BV605 (clone PC61) | BioLegend | Cat# 102036 |
| Anti-mouse Thy1.2 BV785 (clone 30-H12) | BioLegend | Cat# 105331 |
| Anti-mouse CD45 BUV395 (clone 30-F11) | BD Biosciences | Cat# 565967 |
| Anti-mouse CD127 APC/eFluor780 (clone A7R34) | eBioscience | Cat# 47-1271-82 |
| Anti-mouse KLRG1 PerCP-eF710 (clone 2F1) | eBioscience | Cat# 17-5893 |
| Anti-mouse IL-17RB APC (clone 9B10) | BioLegend | Cat# 146308 |
| Anti-human CD4 PE (clone RPA-T4) | eBioscience | Cat# 12-0049 |
| Anti-human CD4 APC (clone RPA-T4) | eBioscience | Cat# 17-0049 |
| Anti-mouse Siglec F AF647 (clone E50-2440) | BD Biosciences | Cat# 562680 |
| Anti-mouse CD24 PE/Cy7 (clone M1/69) | BioLegend | Cat# 101822 |
| Anti-mouse Ki-67 FITC (SolA15) | eBioscience | Cat# 11-5698-82 |
| Anti-mouse Gata-3 PE (clone TWAJ) | eBioscience | Cat# 12-9966-42 |
| Anti-Mouse ROR gamma (t) APC (clone B2D) | eBioscience | Cat# 17-6981-82 |
| Anti-mouse CD3 AF488 (clone 17A2) | BioLegend | Cat# 100210 |
| Anti-mouse ST2 PE (clone DJ8) | MD Biosciences | Cat# 101001PE |
| Anti-mouse ST2 Biotin (clone DJ8) | MD Biosciences | Cat# 101001B |
| Anti-DsRed, rabbit polyclonal | Clontech | Cat# 632496 |
| Goat anti-Rabbit IgG (H+L)-AF555 | ThermoFisher | Cat# A-21428 |
| Anti-SP-1 Chromogranin A, rabbit polyclonal | Immunostar | Cat# 20085 |
| Anti-Muc2, rabbit polyclonal | Santa Cruz | Cat# sc-15334 |
| Anti-Lysozyme, rabbit polyclonal | DAKO | Cat# A009902 |
| Anti-DCLK1, rabbit polyclonal | Abcam | Cat# ab31704 |
| Anti-DCLK1 AF488, rabbit polyclonal | Abcam | Cat# ab203441 (custom) |
| Streptavidin APC | Biolegend | Cat# 405207 |
| Goat anti-Rabbit IgG (H+L)-AF488 | ThermoFisher | Cat# A-11070 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 4′,6-Diamidine-2′-phenylindole dihydrochloride (DAPI) | Roche | Cat# 10236276001 |
| Metronidazole | Sigma | Cat# M3761 |
| Succinic acid | Sigma | Cat# S3674 |
| Sodium acetate | Sigma | Cat# S2889 |
| Recombinant Mouse IL-17E Protein, CF | R&D Systems | Cat# 1399-IL/CF |
| Animal-free recombinant mouse IL-4 | PeproTech | Cat# AF-214-14 |
| InVivoPlus anti-mouse IL4 antibody | Bio X Cell | Cat# BP0045 |
| Pyrantel pamoate | Columbia Laboratories | N/A |
| Tamoxifen | Sigma | Cat# T5648 |
| Critical Commercial Assays | ||
| Succinate Assay Kit (Colorimetric) | Abcam | Cat# ab204718 |
| Experimental Models: Organisms/Strains | ||
| Mouse: wild-type: C57BL/6 | The Jackson Laboratory | Stock# 000664 |
| Mouse: A20fl: B6.Tnfaip3tm2Ama | Averil Ma, (Tavares et al., 2010) | N/A |
| Mouse: R: B6(C)-Il5tm1.1(icre)Lky/J | The Jackson Laboratory | Stock# 030926 |
| Mouse: Trpm5−/−: B6.129P2-Trpm5tm1Dgen/J | The Jackson Laboratory | Stock# 005848 |
| Mouse: Lgr5EGFP-CreERT2: B6.129P2-Lgr5tm1(cre/ERT2)Cle/J | The Jackson Laboratory | Stock# 008875 |
| Mouse: R26R-Confetti: B6.129P2-Gt(ROSA)26Sortm1(CAG-Brainbow2.1)Cle/J | The Jackson Laboratory | Stock# 017492 |
| Mouse: Areg−/−: B6.Aregtm1Dle | Marco Conti, (Luetteke et al., 1999) | N/A |
| Mouse: Yarg: B6.129S4-Arg1tm1Lky/J | The Jackson Laboratory | Stock# 015857 |
| Mouse: Il25−/−: B6.Il25tm1Anjm | Andrew N J McKenzie (Fallon et al., 2006) | N/A |
| Mouse: Smart13: B6.129S4(C)-Il13tm2.1Lky/J | The Jackson Laboratory | Stock# 031367 |
| Mouse: B6.YetCre-13: C.129S4(B6)-Il13tm1(YFP/cre)Lky/J | The Jackson Laboratory | Stock# 017353 |
| Mouse: B6.Il4ra−/−: BALB/c-Il4ratm1Sz/J | The Jackson Laboratory | Stock# 003514 |
| Mouse: IL-25 reporter | (von Moltke et al., 2016) | N/A |
| Mouse: B6.Pou2f3−/−: C57BL/6N-Pou2f3tm1.1(KOMP)Vlcg | Mark Anderson | N/A |
| Nippostrongylus brasiliensis | UCSF (R. Locksley) | N/A |
| Heligmosomoides polygyrus | UCSF (R. Locksley) | N/A |
| Oligonucleotides | ||
| Rps17 (fwd) 5′-CGCCATTATCCCCAGCAAG-3′ | Elim Biopharmaceuticals | N/A |
| Rps17 (rev) 5′-TGTCGGGATCCACCTCAATG-3′ | Elim Biopharmaceuticals | N/A |
| A20 (fwd) 5′-ACAGGACTTTGCTACGACAC-3′ | Elim Biopharmaceuticals | N/A |
| A20 (rev) 5′-CTGAGGATGTTGCTGAGGAC-3′ | Elim Biopharmaceuticals | N/A |
| Il17rb (fwd) 5′-GGCTGCCTAAACCACGTAATG-3′ | Elim Biopharmaceuticals | N/A |
| Il17rb (rev) 5′-CCCGTTGAATGAGAATCGTGT-3′ | Elim Biopharmaceuticals | N/A |
| Tritrichomonas spp. (fwd) 5′-AGAGGAAGGAGAAGTCGTAACAAGG-3′ | Elim Biopharmaceuticals | N/A |
| Tritrichomonas spp. (rev) 5′-CTCGTGTAAGAAGCCAAGACATCC-3′ | Elim Biopharmaceuticals | N/A |
| Eubacteria 16S (fwd) 5′-ACTCCTACGGGAGGCAGCAGT -3′ | Elim Biopharmaceuticals | N/A |
| Eubacteria 16S (rev) 5′-ATTACCGCGGCTGCTGGC-3′ | Elim Biopharmaceuticals | N/A |
| Recombinant DNA | ||
| pLIVE-hIgG1 | UCSF (R. Locksley) | N/A |
| pLIVE-IL13 | UCSF (R. Locksley) | N/A |
| Software and Algorithms | ||
| FlowJo v.9.9 | FlowJo | www.flowjo.com |
| Prism v.6.0 | GraphPad | www.graphpad.com |
| ImageJ v.1.49 | NIH | https://imagej.nih.gov/ij/ |
| Imaris | BitPlane | http://www.bitplane.com/imaris |
For goblet cell staining, intestinal tissues were fixed for 3 h in 10% buffered formalin (Fisher Scientific) at 4 °C before coiling into a ‘Swiss roll’ and returning to formalin. After 24 h, tissues were moved to 70% ethanol for storage. Tissue processing, paraffin embedding, and sectioning were performed by the UCSF Mouse Pathology Core. Periodic acid Schiff (PAS) and Alcian blue staining were performed as follows: cleared with xylenes (Fisher Scientific), rehydrated, 30 min in Alcian blue (Thermo Scientific), 5 min in periodic acid (Thermo Scientific), 15 min in Schiff reagent (Thermo Scientific), dehydrated, and mounted. Brightfield and fluorescent images were acquired with an AxioCam HR camera on an AxioImagerM2 upright microscope (Zeiss).
Single-cell tissue preparation
Lungs were perfused through the heart with cold PBS and harvested. Single-cell suspensions were prepared using a gentleMACS tissue dissociator (Miltenyi Biotec), running program lung_01 followed by incubation for 35 min at 37° C in RPMI-1640 containing 50 μg/ml Liberase TM (Roche) and 25 μg/ml DNase I (Roche), and then running gentleMACS program lung_02. The tissue was passed through 70-μm nylon filters, washed, and subjected to red blood cell lysis (PharmLyse; BD) before final suspension in FACS buffer (PBS, 3% FCS, 0.05% NaN3). Perigonadal visceral adipose tissue (VAT) was harvested and finely minced with scissors, followed by incubation for 40 min at 37° C in RPMI-1640 containing 100 μg/ml Liberase TM (Roche) and 50 μg/ml DNase I (Roche). The tissue was passed through 100-μm nylon filters, washed, and subjected to red blood cell lysis (PharmLyse; BD) before final suspension in FACS buffer. For lamina propria preparations, a 6 cm piece of small intestine was harvested, flushed with PBS, opened, and thoroughly cleaned with PBS.
Intestines were incubated for 20 min in 20 ml HBSS (Ca2+/Mg2+ free) supplemented with 2% fetal calf serum (FCS), 10 mM HEPES (UCSF Cell Culture Facility) and 5 mM DTT. Supernatants were discarded and intestines were incubated for 15 min in 10 ml HBSS (Ca2+/Mg2+ free) supplemented with 2% fetal calf serum (FCS), 10 mM HEPES (UCSF Cell Culture Facility) and 5 mM EDTA solution. This step was repeated twice using fresh solution. Next, intestines were incubated for 10 min in 20 ml HBSS (with Ca2+/Mg2+) supplemented with 3% FCS and 10 mM HEPES. After incubation, intestines were gently vortexed, cut into small pieces and incubated for 30 min in 5 ml HBSS (with Ca2+/Mg2+) supplemented with 3% FCS, 10 mM HEPES, 100 μg/ml Liberase TM (Roche) and 30 μg/ml DNase I (Roche). All the incubations were performed with gentle rocking at 37 °C. After digest, intestines were mechanically dissociated in GentleMACS C tubes (Miltenyi Biotec) using program m_intestine_01, passed through a 100 μm filter, and washed. The resulting cell pellet was resuspended in 5 ml 40% Percoll (Sigma-Aldrich), underlaid with 5 ml 90% Percoll and centrifuged at 2000 r.p.m. for 20 min at 20 °C. The 40/90 interphase of the Percoll gradient was harvested, washed, and stained for flow cytometry. EDTA washes were pooled and stained for epithelial cell analysis by flow cytometry.
Flow cytometry
Single-cell suspensions were incubated with Fc Block (Bio X Cell, 2.4G2) followed by staining with antibodies to surface markers. Lineage (Lin) cocktail included (CD3, CD4, CD5, CD8α, CD11b, CD11c, CD19, CD49b, NK-1.1, F4/80, TER-119, Gr-1, FcεRIα, TCR γ/δ). Exclusion of DAPI (4′,6-diamidine-2′-phenylindole dihydrochloride; Roche) identified live cells, which were enumerated with flow cytometric counting beads (CountBright Absolute; Life Technologies). Cells were kept on ice throughout staining procedure. See Supplementary Table 1 for a list of antibodies used in this study.
For intracellular DCLK1 staining, single-cell epithelial suspensions were incubated with Violet Live/Dead fixable stain (Life Technologies), fixed in 4% paraformaldehyde (Electron Microscopy Sciences) for 5–10 min, washed and stained in permeabilization buffer (ThermoFisher, 00-8333-56) with rabbit anti-DCLK1 (Abcam; ab31704; 1:4000) in perm/wash and rat anti-EpCAM-PerCP-Cy5.5 (Biolegend; G8.8; 1:300) for 20 min, followed by F(ab′)2 goat anti-rabbit IgG-Alexa Fluor 488 (Life Technologies; 1:4000) for 15 min. For intracellular staining of transcription factors, the FoxP3/Transcription Factor Staining Buffer Set (eBiosciences) was used according to manufacturer’s instructions.
Samples were analyzed on a LSRFortessa Dual (BD Biosciences) with five lasers (355 nm, 405 nm, 488 nm, 561 nm and 640 nm). Samples were FSC-A/SSC-A gated to exclude debris, FSC-H/FSC-A gated to select single cells, and gated to exclude DAPI+ dead cells. Data were analyzed with FlowJo 9 (Treestar).
Tuft and secretory cell quantification
Single-cell epithelial suspension were prepared as described, analyzed by flow cytometry and the frequency of tuft cells was calculated as number of DCLK1+ cells/total number of EPCAM+CD45− cells. Because the viability of epithelial cells was found to be very low when prepared from A20flR mice or worm-infected intestines, a shorter protocol was used when such samples were analyzed: 6 cm pieces of small intestine were harvested, flushed with PBS, opened, and thoroughly cleaned with PBS. Next tissues were cut into 0.5 cm pieces, gently vortexed in 20 ml PBS and incubated with gentle rocking for 15 min at 37 °C in 5 ml PBS (Ca2+/Mg2+ free) supplemented with 5% FCS, 10 mM HEPES, 1mM DTT and 4 mM EDTA. Samples were vortexed, centrifuged, resuspended in FACS buffer, passed through 100-μm nylon filters, washed, and stained. Alternatively, tuft cell frequency was quantified in frozen tissue sections by staining for DCLK1 as described earlier. A 4 × 4 grid of images was collected at ×100 magnification and the number of tuft cells was quantified on more than 25 villi using AxioVision software (Zeiss). Tuft cell numbers were calculated as DCLK1+ cells per mm villus length. For secretory cell quantification, tissue sections were stained for MUC2, LYZ1, CHRA, DCLK1 and EpCAM as described above. The number of cells positive for MUC2, LYZ1, CHRA or DCLK1 was assessed for 100 adjacent EpCAM+ cells per villus for 10 villi per sample.
Crypt fission
Crypt fission in the small intestine was assessed as described previously (Snippert et al., 2014). Briefly, Lgr5EGFP-CreERT2;R26R-Confetti mice were treated intraperitoneally with 4 mg of tamoxifen (Sigma-Aldrich) in 200 μl corn oil (Sigma-Aldrich) three times over the course of a week during which they were also kept on a tamoxifen diet (TD.130858; Envigo). 4 weeks after tamoxifen treatment the mice were infected with 200 H. polygyrus L3 and were killed on d28 post-infection to harvest small intestine. For whole-mount imaging, 1.5 cm pieces were cut open longitudinally, villi were scraped off using a coverslip and tissues were flushed with PBS before fixation in 4% paraformaldehyde at room temperature for 60 minutes. Next, parts were washed in PBS, transferred to a microscope slide with crypt bottoms oriented to the top and mounted in vectashield (Vector Laboratories). Confocal images were acquired using a Leica SP5 Inverted Confocal microscope. Images were processed using Imaris (Bitplane), ImageJ and Photoshop. At least 200 Lgr5EGFP+ crypts per sample were counted, of which the percentage of R26R-Confetti+ clones in singlet, doublet and triplet or more clusters was quantified.
Quantitative RT–PCR
Single-cell epithelial suspensions were isolated, stained as described earlier and sorted using a MoFlo XDP (Beckman Coulter). RNA was isolated using the Micro Plus RNeasy kit (Qiagen) and reverse transcribed using the SuperScript Vilo Master Mix (Life Technologies). The resulting cDNA was used as template for quantitative PCR (qPCR) with the Power SYBR Green reagent on a StepOnePlus cycler (Applied Biosystems). Transcripts were normalized to Rps17 (40S ribosomal protein S17) expression and relative expression shown as 2−ΔCt. Primer sequences (see also Key Resources Table): Rps17, 5′-CGCCATTATCCCCAGCAAG-3′, 5′-TGTCGGGATCCACCTCAATG-3′; A20, 5′-ACAGGACTTTGCTACGACAC-3′, 5′-CTGAGGATGTTGCTGAGGAC-3′; Il17rb, 5′-GGCTGCCTAAACCACGTAATG-3′, 5′-CCCGTTGAATGAGAATCGTGT-3′; Tritrichomonas, 5′-AGAGGAAGGAGAAGTCGTAACAAGG-3′, 5′-CTCGTGTAAGAAGCCAAGACATCC-3′; Eubacteria 16S, 5′-ACTCCTACGGGAGGCAGCAGT-3′, 5′-ATTACCGCGGCTGCTGGC-3′; Sucnr1 (PrimerBank 14161704a1), 5′-TCTTGTGAGAATTGGTTGGCAA-3′, 5′-CATCTCCATAGGTCCCCTTATCA-3′; Ffar2 (PrimerBank 22122727a1), 5′-CTTGATCCTCACGGCCTACAT-3′, 5′-CCAGGGTCAGATTAAGCAGGAG-3′; Ffar3 (PrimerBank 142345193c1), 5′-CTTCTTTCTTGGCAATTACTGGC-3′, 5′-CCGAAATGGTCAGGTTTAGCAA-3′; Hcar2 (PrimerBank 13507640a1), 5′-CTGGAGGTTCGGAGGCATC-3′, 5′-TCGCCATTTTTGGTCATCATGT-3′.
Tritrichomonas quantification
Genomic DNA was isolated from cecal content using QIamp Fast DNA Stool mini kit (Qiagen) according to the manufacturers instructions. Tritrichomonas gDNA was quantified by qPCR and normalized to Eubacteria 16S rRNA gene using the method 2−ΔCt (Tritrichomonas-16S).
Tritrichomonas purification
For Tritrichomonas isolation, the cecal content colonized mice was harvested, washed 3 times with 50 ml PBS, during which it was filtered twice through a 100 μm cell strainer and centrifuged at 1000 rpm for 10 min. Motile protists were enumerated using a hemocytometer. For GF colonization, PBS containing 100 mg/ml streptomycin, 100 U/ml penicillin and 50 μg/ml Vancomycin was used for all washing steps and protists were double sorted using a MoFlo XDP (Beckman Coulter) with a a 90 μm nozzle, spun down and resuspended in PBS at 100 Mio/ml. Culture of Tritrichomonas was performed as recently described (Chudnovskiy et al., 2016) with the following modifications. Briefly, cecal extract was generated by diluting cecal content isolated from Tritrichomonas-free mice with 25 volumes of PBS (1 g content per 25 ml PBS), stirred at 4 °C for 6 hr, spun down at 3500 rpm for 10 min and the supernatant was filtered through 100 μm strainer. The filtered cecal extract was used to resuspend the TrichoselTM broth (Becton Dickinson) according to the manufacturer’s protocol and titrated to pH 7 with NaOH. The medium was autoclaved prior to be supplemented with filter-sterilized 10% heat-inactivated horse serum (Gibco) and antibiotics including 100 μg/ml streptomycin, 100 U/ml penicillin (UCSF Cell Culture Facility), 50 μg/ml Vancomycin (Gold Biotechnology), 10 μg/ml Ciprofloxacin (Sigma), 20 μg/ml gentamicin and 0.5 μg/ml amphotericin B (ThermoFisher). Protists were inoculated at 2 Mio/ml in pre-equilibrated growth media and cultured in an anaerobic chamber (Coy) at 37 °C for 3 days on a MACSmix™ Tube Rotator (Miltenyi) at maximal speed. Then, the suspension was diluted in 50 ml PBS, centrifuged at 1000 rpm for 10 min and resuspended in PBS. Primary GF recipient mice received filter-sterilized drinking water supplemented with ampicillin, streptomycin, neomycin, bacitracin (1 mg/ml each), 125 μg/ml ciprofloxacin, 100 μg/ml ceftazimidine, 150 μg/ml gentamicin, 200 μg/ml amphotericin B (all from Sigma) and 0.5 mg/ml vancomycin (Gold Biotechnology) in 1% sucrose, which was started 3 days prior to inoculation with 2 Mio protists. 3 weeks later, absence of bacteria and fungi was confirmed using PCR and standard culture techniques, cecal content was harvested and diluted in PBS. 2 Mio protists were gavaged into 5–6 week old GF mice, which were again tested repeatedly for microbial contamination and analyzed 6 weeks later.
In vitro cell cultures
Sorted cells were cultured (37 °C, 5% CO2) at 3000–5000 cells/well in high-glucose DMEM supplemented as follows: 4.5 g/L glucose, 0.584 g/L l-glutamine, 3.7 g/L NaHCO3, 1× nonessential amino acids, 1× minimal essential vitamins, 0.116 g/L l-arginine HCl, 0.036 g/L l-asparagine, 0.006 g/L folic acid, 10% FBS, 100 mg/ml streptomycin, 100 U/ml penicillin, 10 mM Hepes, 1 mM Na pyruvate, 100 μM 2-ME, and 2 mM l-glutamine. Cells were stimulated for 3 days with 10 ng/ml IL-7 (R&D Systems) or 10 ng/ml IL-33 (R&D Systems), as indicated. At harvest, cell-free culture supernatants were collected for protein analysis, while cells were resuspended and stained for flow cytometric analysis.
Cytokine quantification
Cytokine levels in cell culture supernatant were measured using Cytokine Bead Array Flex Sets (BD) according to the manufacturer’s protocol and data was analyzed using Flow Cytometric Analysis Program (FCAP) Array software (BD).
Metabolic assays
Whole-animal metabolic analysis was performed using CLAMS cages (Comprehensive Laboratory Animals Monitoring System) per the manufacturer’s instructions (Columbus Instruments). In brief, animals were singly housed and measurements were taken every 12 min for 4 days, including oxygen consumption, carbon dioxide output, food consumption, water consumption, and three unique measures of movement. Respiratory exchange ratio and heat were calculated asVCO2/VO2 (RER) and VO2(3.815 + 1.232 × RER; heat), respectively. Heat, VO2, and VCO2 were all normalized to effective body mass Vxx = Vxx/[(weight(g)/mass unit)]0.75, per the manufacturer’s recommendations. To measure animal adiposity and lean mass, MRI was performed using an EchoMRI according to the manufacturer’s instructions (Echo Medical Systems LTD).
Fecal energy content
Fecal samples were collected and lyophilized to obtain dried mass. 100–200 mg of dried stool was pressed into a pellet using a pellet press. Gross energy content was measured using a isoperibol oxygen bomb calorimeter with a semimicro oxygen bomb (Models 6200 and 1109, respectively, from Parr Instrument Co.). The calorimeter energy equivalent factor was determined using benzoic acid standards.
Measurement of short-chain fatty acid and succinate concentrations
In order to measure cecal lumenal short chain fatty acids, a sample of cecal content was weighed and stored at −80 °C until analyzed. Samples and serially diluted standards were acidified using 6N HCl (1 volume of HCl to 5 volume of diluted sample) in the presence of 400 μM acetic acid-d4 and valeric acid (internal standards), then extracted with equal volume of ether. Solid samples (cecal content, fecal pellet) were lysed using a Qiagen TissueLyser II. The ether fraction was derivatized by MTBSTFA (1 to 10 volume of ether fraction) at room temperature for greater than 24 hrs, and used for GCMS for detection. Samples were run on Agilent Technologies 5977A GC/MSD system and short chain fatty acids were normalized to wet weight. For the succinate quantification in the cecal content, snap frozen samples were processed using a colorimetric succinate assay kit (Abcam, ab204718) according to the manufacturer’s instructions, including deproteinization and decolorization.
QUANTIFICATION AND STATISTICAL ANALYSIS
All experiments were performed using randomly assigned mice without investigator blinding. All data points and n values reflect biological replicates. No data were excluded. Where noted in the figures, statistical significance was calculated using two-tailed non-parametric Mann-Whitney U test was used for the comparison between two experimental groups or one-way ANOVA for multiple comparisons. Experimental groups included a minimum of three biological replicates. Intragroup variation was not assessed. All statistical analysis was performed using Prism 6 (GraphPad Software). Figures display means ± s.e.m. as indicated. No statistical methods were used to predetermine sample size.
Supplementary Material
Supplementary Table S1. The composition of the purified diets used in this study. Related to STAR Methods
(A) Expression of A20 was measured by qPCR in sorted ILC2s from the small intestine of naïve adult and 2 week old mice, GF mice, and from mice infected with H. polygyrus (d12) or N. brasiliensis (d4). (B) Quantification of A20 mRNA by qPCR in ILC2s from lung and visceral adipose tissue of RR and A20flRR mice. (C) Quantification of indicated cytokines in supernatant from lung ILC2s of RR and A20flRR mice cultured for 3d with IL-7 or IL-33; nd, not detected. (D and E) Small intestine of RR and A20flRR mice stained for Red5 RFP (IL-5 reporter, red) and CD3 (D) or Ki67 (E) (green). Arrowheads indicate examples of Red5 and Ki67 overlap. (F) Total numbers of ILC2s from lung and visceral adipose tissue of RR and A20flRR mice. Data are pooled from multiple independent experiments (A, F) or from one experiment representative of at least two independent experiments (B–E). C, mean and s.d., n = 4. **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, not significant by Mann-Whitney U test or one-way ANOVA.
(A) Indicated tissues of RR and A20flRR mice stained for DAPI (blue) and DCLK1 (green). (B) Small intestine of RR and A20flRR mice stained for goblet cells with periodic acid Schiff (PAS) Alcian blue. (C) Small intestine of RR and A20flRR mice stained for DAPI (blue) and MUC2, ChromograninA and or Lysozyme (green). (D and E) Proximal, intermediate and distal small intestine of RR and A20flRR mice stained with H&E (D) and quantification of smooth muscle area (E). Scale bars; (A–C; D, inset right) 100 μm, (D, left) 500 μm. Data are from one experiment representative of at least two independent experiments. E, mean and s.d., n = 5–6. *p < 0.05, **p < 0.01 by Mann-Whitney U test
(A and B) Tuft cells (A) and small intestine length (B) was quantified in A20flIl13Cre/+ mice and control A20+Il13Cre/Cre or A20flIl13+/+ mice. (C) Small intestine length was measured in A20flRR mice, Trpm5−/−A20flRR or Trpm5−/−A20flR+ mice, and Trpm5−/−A20fl or Trpm5+/−A20fl mice. (D) Wild-type mice were treated on 4 consecutive days with rIL-25. 2 days later, feces were collected from a 24 h period and fecal energy content was measured using bomb calorimetry. (E and F) Frequencies of tuft cells in the small intestine of wild type (E) and Pou2f3−/− (F) mice after 4 weeks of serial treatment with rIL-25. (G–I) Wild type mice were colonized with isolated Tritrichomonas at the age of 3 weeks and analyzed 8 weeks later. (G) Abundance of Tritrichomonas in cecal content was analyzed by qPCR. (H) Sections of cecum and ileum tissue and luminal content were stained with H&E. (I) Small intestine length plotted against frequencies of ILC2s. (J) Wild type mice were infected with H. polygyrus (H.p.) and small intestinal tuft cell frequencies were analyzed at indicated timepoints by flow cytometry. (K) Small intestine length was measured in Areg−/− or control (Areg+/+ and Areg+/−) mice that were untreated or 28 days after infection with H. polygyrus. Scale bars; 20 μm. Data are from one experiment (A), or from one experiment representative of at least two independent experiments (B, C, E, G–J) or pooled from multiple independent experiments (D, F, K). G, mean and s.e.m.; n = 7. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, not significant by Mann-Whitney U test or one-way ANOVA.
(A) Expression of CD3 and IL-5-reporter (Red5) by cells gated on CD45+ cells (left) and of IL-17RB and KLRG1 by gated subpopulation (colored outlined area) (right) isolated from the small intestine lamina propria of 6 weeks old RR mice. (B) Lamina propria cells were isolated from adult R+ mice. IL-17RB+KLRG1+ ILC2s were gated on Lin-CD45+ cells and the expression of IL-13-reporter (Sm13) and Arg1-reporter (Yarg) was measured by flow cytometry. (C) Expression of IL-17RB, Arg1-reporter (Yarg), Thy1 and KLRG1 by cells gated on Lin- CD45+ cells isolated from the small intestine lamina propria of RR and Il25−/−RR mice at days 12 and 40 post-birth (p). Data are from one experiment representative of more than three independent experiments.
(A) The small intestine was isolated from GF mice, or wild-type mice that were offsprings of mice originally obtained from JAX, which were bred without further manipulation (JAX) or after colonization with Tritrichomonas isolated from cecal content of mice within UCSF vivarium (UCSF). Expression of KLRG1, Thy1 and ST2 by cells gated on Lin- CD45+ lamina propria cells was analyzed by flow cytometry. (B and C) Adult mice were provided with bovine milk as the lone dietary constituent for 2 weeks (B) or together with chow diet (C), and abundance of Tritrichomonas in the cecal content was quantified by qPCR. (D) A20flRR mice were treated with a mix of antibiotics (1 mg/ml neomycin, metronidazole, ampicillin and 0.5 mg/ml vancomycin) in drinking water for 4 weeks and the frequencies of tuft cells in the small intestine was measured by flow cytometry. (E and F) Tritrichomonas-free R+ and A20flR+ mice were colonized with Tritrichomonas after weaning and the frequencies of tuft cells (E) and small intestine length (F) was measured 7 or 16 days later. (G) Wild-type mice were fed chow or purified diets containing no fiber, 10% cellulose or 10% inulin. One day later mice were gavaged with 104 purified Tritrichomonas and the number of protists in the cecal content were counted after 14 days using a hemocytometer. DL, detection limit. (H) A20flRR mice were fed chow or purified diets containing 10% cellulose or 10% inulin after weaning for 6 weeks and the frequencies of tuft cells were quantified by flow cytometry. Data are from one experiment representative of at least two independent experiments (A–G) or pooled from multiple independent experiments (H). B and C, mean and s.e.m.; B, n = 4; C, n = 4–5. *p < 0.05, **p < 0.01; ns, not significant by Mann-Whitney U test or one-way ANOVA.
(A) Concentrations of succinate in the cecal content of Tritrichomonas-colonized and uncolonized SPF mice. (B) GF C57BL/6J mice were treated with sterile-filtered 100 mM succinate in drinking water for 4 days and the expression of Ki-67 by ILC2s in the small intestine was quantified by flow cytometry. (C) Sucnr1 mRNA expression was quantified by qPCR in non-tuft epithelial cells, tuft cells and ILC2s sorted from the small intestine. n.d., not detected. (D–F) Tritrichomonas-free IL-13-reporter (Sm13) mice were treated with 100 mM succinate in drinking water for 4 days. (D) Expression of CD3 and IL-13 among CD45+ cells (left) and of IL-17RB and KLRG1 by gated subpopulations (indicated populations gated on colored outlined areas) (right) from the small intestine was quantified by flow cytometry and representative dot plots are shown. (E and F) Expression of Ki-67 (E) and IL-13 (F) by CD3+CD4+ T cells, Lin-RORg+ ILC3s and Lin-Gata3+ ILC2s isolated from indicated organs (SI, small intestine). (G) Sucnr1 mRNA expression was quantified by qPCR in tuft cells sorted from the indicated organs of IL-25-reporter mice. C and G, mean and s.e.m.; n = 6–7 (C), n = 3 (G). **p < 0.01, ****p < 0.0001; ns, not significant by Mann-Whitney U test or one-way ANOVA.
(A and B) Adult R+ and A20flR+ mice were treated with metronidazole (MNZ) in drinking water for 2 weeks and the frequencies of IL-5+ ILC2s (A) in the small intestine were analyzed by flow cytometry. (B) Expression of IL-5-reporter (Red5) and IL-13-reporter (Sm13) by KLRG1+IL-17RB+ ILC2s gated on Lin-CD45+ lamina propria cells. (C) Wild type mice serially treated with rIL-25 or infected with H. polygyrus (H.p.) for 4 weeks and worms were cleared using pyrantel pamoate. Small intestine length was measured 2 months after the last rIL-25 administration or worm clearance. (D) Rag1−/− were treated with rIL-25 for 2 weeks followed by infection with N. brasiliensis. Fecal egg numbers were quantified on d10. (E) Wild type mice were infected with H. polygyrus or sham-treated, and pyrantel pamoate was administered to all mice on day 28. One week later mice were infected with N. brasiliensis and intestinal worms were counted on day 6. Data are from pooled from multiple independent experiments (A, C) or from one experiment representative of at least two independent experiments (B, D, E). Statistical significance is indicated. *p < 0.05, **p < 0.01, ****p < 0.0001; ns, not significant by Mann-Whitney U test or one-way ANOVA.
Highlights.
Tuft cell IL-25 and IL-25R+ ILC2s drive A20-regulated small bowel adaptability
Circuit activation and small bowel hypertrophy maintain systemic energy balance
Succinate, a Tritrichomonas metabolite, triggers tuft cell chemosensation and IL-25
Circuit activation impedes new helminth infection and enables concomitant immunity
Acknowledgments
We thank Z. Wang, M. Ji and M. Consengco for technical expertise; members of the Locksley lab for discussions; O. Klein and M. Anderson for manuscript comments; C.J. Guo and M.A. Fischbach for assistance with SCFA measurements; J. Turnbaugh and UCSF Gnotobiotic Core Facility for GF mice; the UCSF BIDC for assistance with confocal microscopy; and the Mouse Metabolism Core at the UCSF DRC and NORC for assistance with metabolic experiments. This work was supported by grants AI26918, AI30663, HL128903, and HL122593 from the NIH, HHMI, and the SABRE Center at the UCSF. C.S. is supported by fellowships from the Swiss National Science Foundation (P2EZP3_162266; P300PA_171591). P.J.T. is a Nadia’s Gift Foundation Innovator supported by the Damon Runyon Cancer Research Foundation (DRR-42-16), the UCSF Program for Breakthrough Biomedical Research, and the Searle Scholars Program.
Footnotes
AUTHOR CONTRIBUTIONS
C.S. conceived the study, designed and performed experiments, analyzed data and wrote the manuscript with R.M.L.. C.E.O. designed and performed experiments, analyzed data and edited the manuscript. J.v.M. generated the A20flRR mice. H.-E.L. assisted with experiments. Q.Y.A. and P.J.T. assisted with energy balance analysis. S.R. and M.P. provided diets. A.M. provided A20fl mice. R.M.L. directed the study and wrote the paper with C.S.
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amatya N, Garg AV, Gaffen SL. IL-17 Signaling: The Yin and the Yang. Trends Immunol. 2017;38:310–322. doi: 10.1016/j.it.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando JK, Liang HE, Locksley RM. Identification and distribution of developing innate lymphoid cells in the fetal mouse intestine. Nat Immunol. 2015;16:153–160. doi: 10.1038/ni.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando JK, Nussbaum JC, Liang HE, Locksley RM. Type 2 innate lymphoid cells constitutively express arginase-I in the naive and inflamed lung. J Leukoc Biol. 2013;94:877–884. doi: 10.1189/jlb.0213084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt J, Gough R, Stark D, Ellis J. Bulky Trichomonad Genomes: Encoding a Swiss Army Knife. Trends Parasitol. 2016;32:783–797. doi: 10.1016/j.pt.2016.05.014. [DOI] [PubMed] [Google Scholar]
- Belkaid Y, Harrison OJ. Homeostatic Immunity and the Microbiota. Immunity. 2017;46:562–576. doi: 10.1016/j.immuni.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune A. Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol. 2014;12:168–180. doi: 10.1038/nrmicro3182. [DOI] [PubMed] [Google Scholar]
- Buckley KM, Ho ECH, Hibino T, Schrankel CS, Schuh NW, Wang G, Rast JP. IL17 factors are early regulators in the gut epithelium during inflammatory response to Vibrio in the sea urchin larva. Elife. 2017;6:860. doi: 10.7554/eLife.23481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Itakura E, Nelson GM, Sheng M, Laurent P, Fenk LA, Butcher RA, Hegde RS, de Bono M. IL-17 is a neuromodulator of Caenorhabditis elegans sensory responses. Nature. 2017;542:43–48. doi: 10.1038/nature20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi GB, Yim YS, Wong H, Kim S, Kim H, Kim SV, Hoeffer CA, Littman DR, Huh JR. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science. 2016;351:933–939. doi: 10.1126/science.aad0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudnovskiy A, Mortha A, Kana V, Kennard A, Ramirez JD, Rahman A, Remark R, Mogno I, Ng R, Gnjatic S, et al. Host-Protozoan Interactions Protect from Mucosal Infections through Activation of the Inflammasome. Cell. 2016;167:444–456. e414. doi: 10.1016/j.cell.2016.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta Banik D, Martin LE, Freichel M, Torregrossa AM, Medler KF. TRPM4 and TRPM5 are both required for normal signaling in taste receptor cells. Proc Natl Acad Sci U S A. 2018;115:E772–E781. doi: 10.1073/pnas.1718802115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante NK, Lemire P, Cruz Tleugabulova M, Prescott D, Mortha A, Streutker CJ, Girardin SE, Philpott DJ, Mallevaey T. The common mouse protozoa Tritrichomonas muris alters mucosal T cell homeostasis and colitis susceptibility. J Exp Med. 2016;213:2841–2850. doi: 10.1084/jem.20161776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon PG, Ballantyne SJ, Mangan NE, Barlow JL, Dasvarma A, Hewett DR, McIlgorm A, Jolin HE, McKenzie AN. Identification of an interleukin (IL)-25-dependent cell population that provides IL-4, IL-5, and IL-13 at the onset of helminth expulsion. J Exp Med. 2006;203:1105–1116. doi: 10.1084/jem.20051615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg AV, Ahmed M, Vallejo AN, Ma A, Gaffen SL. The deubiquitinase A20 mediates feedback inhibition of interleukin-17 receptor signaling. Sci Signal. 2013;6:ra44. doi: 10.1126/scisignal.2003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garside P, Kennedy MW, Wakelin D, Lawrence CE. Immunopathology of intestinal helminth infection. Parasite Immunol. 2000;22:605–612. doi: 10.1046/j.1365-3024.2000.00344.x. [DOI] [PubMed] [Google Scholar]
- Gerbe F, Sidot E, Smyth DJ, Ohmoto M, Matsumoto I, Dardalhon V, Cesses P, Garnier L, Pouzolles M, Brulin B, et al. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature. 2016;529:226–230. doi: 10.1038/nature16527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilissen J, Jouret F, Pirotte B, Hanson J. Insight into SUCNR1 (GPR91) structure and function. Pharmacol Ther. 2016;159:56–65. doi: 10.1016/j.pharmthera.2016.01.008. [DOI] [PubMed] [Google Scholar]
- Guo L, Huang Y, Chen X, Hu-Li J, Urban JF, Jr, Paul WE. Innate immunological function of TH2 cells in vivo. Nat Immunol. 2015;16:1051–1059. doi: 10.1038/ni.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gury-BenAri M, Thaiss CA, Serafini N, Winter DR, Giladi A, Lara-Astiaso D, Levy M, Salame TM, Weiner A, David E, et al. The Spectrum and Regulatory Landscape of Intestinal Innate Lymphoid Cells Are Shaped by the Microbiome. Cell. 2016;166:1231–1246. e1213. doi: 10.1016/j.cell.2016.07.043. [DOI] [PubMed] [Google Scholar]
- Han Q, Das S, Hirano M, Holland SJ, McCurley N, Guo P, Rosenberg CS, Boehm T, Cooper MD. Characterization of Lamprey IL-17 Family Members and Their Receptors. J Immunol. 2015;195:5440–5451. doi: 10.4049/jimmunol.1500892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa Y, Sakitani K, Konishi M, Asfaha S, Niikura R, Tomita H, Renz BW, et al. Nerve Growth Factor Promotes Gastric Tumorigenesis through Aberrant Cholinergic Signaling. Cancer Cell. 2017;31:21–34. doi: 10.1016/j.ccell.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt MR, Lavoie S, Michaud M, Blum AM, Tran SV, Weinstock JV, Gallini CA, Redding K, Margolskee RF, Osborne LC, et al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science. 2016;351:1329–1333. doi: 10.1126/science.aaf1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- Liang HE, Reinhardt RL, Bando JK, Sullivan BM, Ho IC, Locksley RM. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nat Immunol. 2011;13:58–66. doi: 10.1038/ni.2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TC, Stappenbeck TS. Genetics and Pathogenesis of Inflammatory Bowel Disease. Annu Rev Pathol. 2016;11:127–148. doi: 10.1146/annurev-pathol-012615-044152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukes J, Stensvold CR, Jirku-Pomajbikova K, Wegener Parfrey L. Are Human Intestinal Eukaryotes Beneficial or Commensals? PLoS Pathog. 2015;11:e1005039. doi: 10.1371/journal.ppat.1005039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mass E, Ballesteros I, Farlik M, Halbritter F, Gunther P, Crozet L, Jacome-Galarza CE, Handler K, Klughammer J, Kobayashi Y, et al. Specification of tissue-resident macrophages during organogenesis. Science. 2016;353:aaf4238–aaf4238. doi: 10.1126/science.aaf4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middelhoff M, Westphalen CB, Hayakawa Y, Yan KS, Gershon MD, Wang TC, Quante M. Dclk1-expressing tuft cells: critical modulators of the intestinal niche? Am J Physiol Gastrointest Liver Physiol. 2017;313:G285–G299. doi: 10.1152/ajpgi.00073.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EL, Kelly B, Logan A, Costa ASH, Varma M, Bryant CE, Tourlomousis P, Dabritz JHM, Gottlieb E, Latorre I, et al. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell. 2016;167:457–470. e413. doi: 10.1016/j.cell.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AB, Nussbaum JC, Liang HE, Van Dyken SJ, Cheng LE, Mohapatra A, Chawla A, Locksley RM. Innate lymphoid type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. J Exp Med. 2013;210:535–549. doi: 10.1084/jem.20121964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doering TA, Angelosanto JM, Laidlaw BJ, Yang CY, Sathaliyawala T, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol. 2011;12:1045–1054. doi: 10.1031/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, Furusawa J, Ohtani M, Fujii H, Koyasu S. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature. 2010;463:540–544. doi: 10.1038/nature08636. [DOI] [PubMed] [Google Scholar]
- Muller M, Mentel M, van Hellemond JJ, Henze K, Woehle C, Gould SB, Yu RY, van der Giezen M, Tielens AG, Martin WF. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev. 2012;76:444–495. doi: 10.1128/MMBR.05024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TK, Bucks C, Kane CM, Fallon PG, Pannell R, et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 2010;464:1367–1370. doi: 10.1038/nature08900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, van der Meer JW. Trained Immunity: An Ancient Way of Remembering. Cell Host Microbe. 2017;21:297–300. doi: 10.1016/j.chom.2017.02.003. [DOI] [PubMed] [Google Scholar]
- Nussbaum JC, Van Dyken SJ, von Moltke J, Cheng LE, Mohapatra A, Molofsky AB, Thornton EE, Krummel MF, Chawla A, Liang HE, et al. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature. 2013;502:245–248. doi: 10.1038/nature12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AE, Liang HE, Sullivan BM, Reinhardt RL, Eisley CJ, Erle DJ, Locksley RM. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc Natl Acad Sci U S A. 2010;107:11489–11494. doi: 10.1073/pnas.1003988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring M. Studies on the Parasitic Protozoa of Wild Mice from Berkshire with a Description of a New Species of Trichomonas. Proceedings of the Zoological Society of London. 1959;132:381–401. [Google Scholar]
- Sato A. Tuft cells. Anat Sci Int. 2007;82:187–199. doi: 10.1111/j.1447-073X.2007.00188.x. [DOI] [PubMed] [Google Scholar]
- Smithers SR, Terry RJ. Immunity in schistosomiasis. Ann N Y Acad Sci. 1969;160:826–840. doi: 10.1111/j.1749-6632.1969.tb15904.x. [DOI] [PubMed] [Google Scholar]
- Snippert HJ, Schepers AG, van Es JH, Simons BD, Clevers H. Biased competition between Lgr5 intestinal stem cells driven by oncogenic mutation induces clonal expansion. EMBO Rep. 2014;15:62–69. doi: 10.1002/embr.201337799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Sommer F, Backhed F. The gut microbiota--masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- Song X, He X, Li X, Qian Y. The roles and functional mechanisms of interleukin-17 family cytokines in mucosal immunity. Cell Mol Immunol. 2016;13:418–431. doi: 10.1038/cmi.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- Tavares RM, Turer EE, Liu CL, Advincula R, Scapini P, Rhee L, Barrera J, Lowell CA, Utz PJ, Malynn BA, et al. The ubiquitin modifying enzyme A20 restricts B cell survival and prevents autoimmunity. Immunity. 2010;33:181–191. doi: 10.1016/j.immuni.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordesillas L, Berin MC, Sampson HA. Immunology of Food Allergy. Immunity. 2017;47:32–50. doi: 10.1016/j.immuni.2017.07.004. [DOI] [PubMed] [Google Scholar]
- Van Dyken SJ, Mohapatra A, Nussbaum JC, Molofsky AB, Thornton EE, Ziegler SF, McKenzie AN, Krummel MF, Liang HE, Locksley RM. Chitin activates parallel immune modules that direct distinct inflammatory responses via innate lymphoid type 2 and gammadelta T cells. Immunity. 2014;40:414–424. doi: 10.1016/j.immuni.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J, Ji M, Liang HE, Locksley RM. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature. 2016;529:221–225. doi: 10.1038/nature16161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J, Locksley RM. I-L-C-2 it: type 2 immunity and group 2 innate lymphoid cells in homeostasis. Curr Opin Immunol. 2014;31:58–65. doi: 10.1016/j.coi.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner BW. The Pathogenesis of Resection-Associated Intestinal Adaptation. Cell Mol Gastroenterol Hepatol. 2016;2:429–438. doi: 10.1016/j.jcmgh.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss DM, Yang L, Shah PR, Kobie JJ, Urban JF, Mosmann TR. Amphiregulin, a TH2 cytokine enhancing resistance to nematodes. Science. 2006;314:1746. doi: 10.1126/science.1133715. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1. The composition of the purified diets used in this study. Related to STAR Methods
(A) Expression of A20 was measured by qPCR in sorted ILC2s from the small intestine of naïve adult and 2 week old mice, GF mice, and from mice infected with H. polygyrus (d12) or N. brasiliensis (d4). (B) Quantification of A20 mRNA by qPCR in ILC2s from lung and visceral adipose tissue of RR and A20flRR mice. (C) Quantification of indicated cytokines in supernatant from lung ILC2s of RR and A20flRR mice cultured for 3d with IL-7 or IL-33; nd, not detected. (D and E) Small intestine of RR and A20flRR mice stained for Red5 RFP (IL-5 reporter, red) and CD3 (D) or Ki67 (E) (green). Arrowheads indicate examples of Red5 and Ki67 overlap. (F) Total numbers of ILC2s from lung and visceral adipose tissue of RR and A20flRR mice. Data are pooled from multiple independent experiments (A, F) or from one experiment representative of at least two independent experiments (B–E). C, mean and s.d., n = 4. **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, not significant by Mann-Whitney U test or one-way ANOVA.
(A) Indicated tissues of RR and A20flRR mice stained for DAPI (blue) and DCLK1 (green). (B) Small intestine of RR and A20flRR mice stained for goblet cells with periodic acid Schiff (PAS) Alcian blue. (C) Small intestine of RR and A20flRR mice stained for DAPI (blue) and MUC2, ChromograninA and or Lysozyme (green). (D and E) Proximal, intermediate and distal small intestine of RR and A20flRR mice stained with H&E (D) and quantification of smooth muscle area (E). Scale bars; (A–C; D, inset right) 100 μm, (D, left) 500 μm. Data are from one experiment representative of at least two independent experiments. E, mean and s.d., n = 5–6. *p < 0.05, **p < 0.01 by Mann-Whitney U test
(A and B) Tuft cells (A) and small intestine length (B) was quantified in A20flIl13Cre/+ mice and control A20+Il13Cre/Cre or A20flIl13+/+ mice. (C) Small intestine length was measured in A20flRR mice, Trpm5−/−A20flRR or Trpm5−/−A20flR+ mice, and Trpm5−/−A20fl or Trpm5+/−A20fl mice. (D) Wild-type mice were treated on 4 consecutive days with rIL-25. 2 days later, feces were collected from a 24 h period and fecal energy content was measured using bomb calorimetry. (E and F) Frequencies of tuft cells in the small intestine of wild type (E) and Pou2f3−/− (F) mice after 4 weeks of serial treatment with rIL-25. (G–I) Wild type mice were colonized with isolated Tritrichomonas at the age of 3 weeks and analyzed 8 weeks later. (G) Abundance of Tritrichomonas in cecal content was analyzed by qPCR. (H) Sections of cecum and ileum tissue and luminal content were stained with H&E. (I) Small intestine length plotted against frequencies of ILC2s. (J) Wild type mice were infected with H. polygyrus (H.p.) and small intestinal tuft cell frequencies were analyzed at indicated timepoints by flow cytometry. (K) Small intestine length was measured in Areg−/− or control (Areg+/+ and Areg+/−) mice that were untreated or 28 days after infection with H. polygyrus. Scale bars; 20 μm. Data are from one experiment (A), or from one experiment representative of at least two independent experiments (B, C, E, G–J) or pooled from multiple independent experiments (D, F, K). G, mean and s.e.m.; n = 7. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, not significant by Mann-Whitney U test or one-way ANOVA.
(A) Expression of CD3 and IL-5-reporter (Red5) by cells gated on CD45+ cells (left) and of IL-17RB and KLRG1 by gated subpopulation (colored outlined area) (right) isolated from the small intestine lamina propria of 6 weeks old RR mice. (B) Lamina propria cells were isolated from adult R+ mice. IL-17RB+KLRG1+ ILC2s were gated on Lin-CD45+ cells and the expression of IL-13-reporter (Sm13) and Arg1-reporter (Yarg) was measured by flow cytometry. (C) Expression of IL-17RB, Arg1-reporter (Yarg), Thy1 and KLRG1 by cells gated on Lin- CD45+ cells isolated from the small intestine lamina propria of RR and Il25−/−RR mice at days 12 and 40 post-birth (p). Data are from one experiment representative of more than three independent experiments.
(A) The small intestine was isolated from GF mice, or wild-type mice that were offsprings of mice originally obtained from JAX, which were bred without further manipulation (JAX) or after colonization with Tritrichomonas isolated from cecal content of mice within UCSF vivarium (UCSF). Expression of KLRG1, Thy1 and ST2 by cells gated on Lin- CD45+ lamina propria cells was analyzed by flow cytometry. (B and C) Adult mice were provided with bovine milk as the lone dietary constituent for 2 weeks (B) or together with chow diet (C), and abundance of Tritrichomonas in the cecal content was quantified by qPCR. (D) A20flRR mice were treated with a mix of antibiotics (1 mg/ml neomycin, metronidazole, ampicillin and 0.5 mg/ml vancomycin) in drinking water for 4 weeks and the frequencies of tuft cells in the small intestine was measured by flow cytometry. (E and F) Tritrichomonas-free R+ and A20flR+ mice were colonized with Tritrichomonas after weaning and the frequencies of tuft cells (E) and small intestine length (F) was measured 7 or 16 days later. (G) Wild-type mice were fed chow or purified diets containing no fiber, 10% cellulose or 10% inulin. One day later mice were gavaged with 104 purified Tritrichomonas and the number of protists in the cecal content were counted after 14 days using a hemocytometer. DL, detection limit. (H) A20flRR mice were fed chow or purified diets containing 10% cellulose or 10% inulin after weaning for 6 weeks and the frequencies of tuft cells were quantified by flow cytometry. Data are from one experiment representative of at least two independent experiments (A–G) or pooled from multiple independent experiments (H). B and C, mean and s.e.m.; B, n = 4; C, n = 4–5. *p < 0.05, **p < 0.01; ns, not significant by Mann-Whitney U test or one-way ANOVA.
(A) Concentrations of succinate in the cecal content of Tritrichomonas-colonized and uncolonized SPF mice. (B) GF C57BL/6J mice were treated with sterile-filtered 100 mM succinate in drinking water for 4 days and the expression of Ki-67 by ILC2s in the small intestine was quantified by flow cytometry. (C) Sucnr1 mRNA expression was quantified by qPCR in non-tuft epithelial cells, tuft cells and ILC2s sorted from the small intestine. n.d., not detected. (D–F) Tritrichomonas-free IL-13-reporter (Sm13) mice were treated with 100 mM succinate in drinking water for 4 days. (D) Expression of CD3 and IL-13 among CD45+ cells (left) and of IL-17RB and KLRG1 by gated subpopulations (indicated populations gated on colored outlined areas) (right) from the small intestine was quantified by flow cytometry and representative dot plots are shown. (E and F) Expression of Ki-67 (E) and IL-13 (F) by CD3+CD4+ T cells, Lin-RORg+ ILC3s and Lin-Gata3+ ILC2s isolated from indicated organs (SI, small intestine). (G) Sucnr1 mRNA expression was quantified by qPCR in tuft cells sorted from the indicated organs of IL-25-reporter mice. C and G, mean and s.e.m.; n = 6–7 (C), n = 3 (G). **p < 0.01, ****p < 0.0001; ns, not significant by Mann-Whitney U test or one-way ANOVA.
(A and B) Adult R+ and A20flR+ mice were treated with metronidazole (MNZ) in drinking water for 2 weeks and the frequencies of IL-5+ ILC2s (A) in the small intestine were analyzed by flow cytometry. (B) Expression of IL-5-reporter (Red5) and IL-13-reporter (Sm13) by KLRG1+IL-17RB+ ILC2s gated on Lin-CD45+ lamina propria cells. (C) Wild type mice serially treated with rIL-25 or infected with H. polygyrus (H.p.) for 4 weeks and worms were cleared using pyrantel pamoate. Small intestine length was measured 2 months after the last rIL-25 administration or worm clearance. (D) Rag1−/− were treated with rIL-25 for 2 weeks followed by infection with N. brasiliensis. Fecal egg numbers were quantified on d10. (E) Wild type mice were infected with H. polygyrus or sham-treated, and pyrantel pamoate was administered to all mice on day 28. One week later mice were infected with N. brasiliensis and intestinal worms were counted on day 6. Data are from pooled from multiple independent experiments (A, C) or from one experiment representative of at least two independent experiments (B, D, E). Statistical significance is indicated. *p < 0.05, **p < 0.01, ****p < 0.0001; ns, not significant by Mann-Whitney U test or one-way ANOVA.