Summary
The innate immune response in the placenta depends on the ability of maternal immune cells and fetal trophoblast cells to detect and eliminate invading pathogens through germline‐encoded pattern recognition receptors (PRRs). In the present study, we analysed the transcripts and protein expression of interferon (IFN)‐inducible protein (IFI)16, melanoma differentiation‐associated protein 5 (MDA5), RIG‐I‐like receptor (RIG‐I) and Toll‐like receptor (TLR)‐3 in third‐trimester human placentas and investigated cytokine profiles generated during herpes simplex type 1 (HSV‐1) infection. Decidual and chorionic villous biopsies (38–42 weeks of gestation) were obtained from healthy women immediately after a caesarean section. The expression of the DDX58 (RIG‐I), IFIH1 (MDA5), IFI16 and TLR3 transcripts was measured using quantitative real‐time polymerase chain reaction (qRT–PCR). Extracellular cytokine and PRRs levels were then quantified by enzyme‐linked immunosorbent assays (ELISAs). All examined PRRs genes, including DDX58, IFIH1, IFI16 and TLR3, were expressed constitutively at the mRNA and protein levels in the placental biopsies. The concentration of the IFI16 protein was increased in HSV‐1‐infected decidual and chorionic villous explants compared to those of mock‐infected tissues (P = 0·029). Higher protein expression levels of RIG‐I in both the maternal and fetal parts of the placenta were found (P = 0·009 and P = 0·004, respectively). In addition, increased production of IFN‐β by HSV‐1‐infected tissues was noticed (P = 0·004 for decidua, P = 0·032 for chorionic villi). No significant differences in the IFN‐α, interleukin (IL)‐6 and IL‐8 levels were found. These results showed that HSV‐1 infection can enhance the expression of IFI16 and RIG‐I proteins in the human term placenta.
Keywords: cytokines, HSV‐1, IFI16, pattern‐recognition receptors, placenta
Introduction
Herpes simplex virus type 1 (HSV‐1) is a member of the Herpesviridae family and is one of the most common sexually transmitted viruses among adult women 1, 2. HSV‐1 has been associated traditionally with oral–facial infections, whereas HSV‐2 is associated with anogenital infection transmitted through sexual contact. In recent decades, the number of cases of genital herpes caused by HSV‐1 has increased 3, 4. A reduced seroprevalence of HSV‐1 and an increased frequency of oral sex have been suggested to be responsible for this trend. Peña et al. 4 found that genital HSV‐1 positivity rates were high in young women (47%) in the United States. The increasing acquisition rate of genital HSV‐1 among women of childbearing age suggests that more neonates may be exposed to the virus than in past decades. Although the mother‐to‐child transmission of HSV‐1 infection is rare 5, 6, it can result in severe disseminated neonatal disease and may lead to fetal abortion, congenital malformation and stillbirth 7, 8. The vast majority (85%) of mother‐to‐child HSV‐1 transmissions occur during the peripartum period. An additional 10% of infected newborns acquire the virus postnatally and the final 5% are infected with HSV‐1 in utero 9. The most common and serious HSV‐1 infection is associated with the first or second trimester of pregnancy 6, 8. Newborns with intrauterine HSV‐1 infection typically have a triad of clinical findings consisting of cutaneous manifestations (scarring, active lesions, aplasia cutis, hyper‐ or hypopigmentation and/or erythematous macular exanthem), neurological involvement (microcephaly, encephalomalacia, hydranencephaly and/or intracranial calcifications) and ophthalmological findings (chorioretinitis, microphthalmia and/or retinal dysplasia) 9, 10, 11, 12. An HSV‐1 infection acquired in either the peripartum or postpartum periods can be categorized as the most common disease of the skin, eyes and/or mouth (SEM) disease (45% of cases), central nervous system (CNS) disease, including meningitis or encephalitis (30% of cases) and disseminated disease that involves multiple organs (25% of cases) 11, 13. It is therefore critically important to identify the host anti‐viral factors that recognize and counteract viral infection in the placenta and to develop novel therapeutic strategies for improving the comfort and quality of life of affected children.
The interferon (IFN)‐mediated response is a first line of host defence against viral infections and is important for the activation of both innate and adaptive immunity 14, 15. The innate immune response is initiated by the activation of a broad range of sensors termed pattern recognition receptors (PRRs) by invading pathogens 16, 17. It was shown that HSV‐1 DNA and double‐stranded RNA (dsRNA), a by‐product of viral replication, are sensed by many types of PRRs, including RIG‐I‐like receptors (RLRs), IFN‐inducible protein 16 (IFI16), DNA‐dependent protein kinase (DNA‐PKR), DNA‐dependent activator of IFN‐regulatory factor (DAI), DDX41 (DEAD‐box helicase 41), cyclic guanosine monophosphate–adenosine monophosphate synthase (cGAS) and Toll‐like receptors 3 and 9 (TLR‐3 and TLR‐9) 18, 19, 20, 21, 22, 23, 24, 25. PRRs that specifically recognize DNA as a potential marker of viral infection trigger the IFN‐regulatory factor 3 (IRF‐3) pathway, leading to the induction of type I IFN 20, 26 and IFN‐stimulated genes (ISGs). Importantly, it was shown that adaptor protein stimulator of interferon genes (STING) is also essential for IFN‐β induction by DNA and by HSV‐1 27. However, little is known about the pattern of PRR expression during HSV‐1 infection in the human term placenta.
In this paper, we sought to identify and compare the expression patterns of the selected PRRs [TLR‐3, RIG‐I, melanoma differentiation‐associated protein 5 (MDA5) and IFI16] in term placentas in order to understand their capacity to recognize HSV‐1 infection as well as assess the cytokine levels after HSV‐1 stimulation ex vivo.
Materials and methods
Tissue collection
Placental biopsies were collected anonymously from healthy women (median age = 31·7; range = 27·3–36·9 years) undergoing elective caesarean section (38–42 weeks of gestation). Pregnancies complicated by maternal autoimmune or infectious diseases, diabetes and fetal abnormalities were excluded from the study. This study was conducted under a protocol approved by the institutional ethics committee of the Medical University of Lodz (RNN/120/09/KE) and was run in accordance with the Declaration of Helsinki. Written informed consent was obtained from all donors prior to their inclusion in the trial.
Decidual and chorionic villous explants cultures were performed and tested separately in each organ. Briefly, placental tissues were separated from surrounding membranes, washed several times in ice‐cold phosphate‐buffered saline (PBS; Sigma‐Aldrich Co. Ltd, Ayrshire, UK) until the supernatant was nearly free of blood, and minced into small pieces (approximately 2 mm3). The explants were either collected immediately at time zero or transferred into tissue culture plates and incubated in 2 ml of Dulbecco's modified Eagle's medium (DMEM; Sigma‐Aldrich) supplemented with 10% inactivated fetal bovine serum [fetal bovine serum (FBS); Sigma‐Aldrich], 2 mM L‐glutamine and 100 µg streptomycin‐100 U penicillin (Sigma‐Aldrich). Cultures were incubated for 48 h at 37°C in 5% CO2. After 24 and 48 h post‐infection (h.p.i.), the placental explants were harvested and tissue‐free supernatants were collected by centrifugation at 13 100 g for 10 min at 4°C and stored at −80°C until use.
Cell line and virus strain
The Vero cell line (ATCC CCL‐81; Manassas, VA, USA) was grown in Eagle's minimum essential medium (EMEM; Sigma‐Aldrich) containing 10% inactivated FBS and 2 mM L‐glutamine and 100 µg streptomycin‐100 U penicillin at 37°C in 5% CO2 until confluence. HSV‐1 strain MacIntyre (McIntyre, ATCC VR‐539) was cultured in the Vero cells with EMEM supplemented with 2% inactivated FBS and antibiotics. At 48 h.p.i., supernatants from infected Vero cells approaching > 80% cytopathic effect (CPE) were collected and used to infect the placental explants. The laboratory‐adapted strain of HSV‐1 was used for the inoculation of all placental explants and Vero cells. HSV‐1 stocks were prepared and the virus was quantitated using a standard plaque assay on Vero cells 20. Cellular debris was pelleted by centrifugation at 13 100 g for 10 min at 4°C, and the resulting supernatants were aliquot and stored at −80°C until use.
Viral infection
The explants were infected immediately with HSV‐1 [1 × 105 plaque‐forming units (PFU)/ml] in DMEM supplemented with 2% FBS and 2 mM L‐glutamine and 100 µg streptomycin‐100 U penicillin. Following 1 h incubation at room temperature, the inoculum was removed and the explants were washed three times with PBS. After washing, 2 ml of fresh culture medium were added and the explants were cultured for 48 h at 37°C in 5% CO2. Virus‐infected explants that were kept at 4°C for 24 h.p.i. served as a control sample of the starting level of the virus, while mock‐infected tissues served as a control. The tissue‐free supernatants were harvested at 24 and 48 h.p.i., centrifuged at 13 100 g for 10 min and stored at −80°C until analysis by enzyme‐linked immunosorbent assay (ELISA) and viral load measurement. As a positive control, permissive Vero cells inoculated with HSV‐1 at a multiplicity of infection (MOI) of 1 were provided.
DNA isolation
Genomic DNA was extracted from the specimens using a QIAamp DNA Mini Kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's recommendations. The extracted DNA was diluted in elution buffer to the optimal concentration for use as a template for PCR amplification and stored at −20°C until use. The DNA concentration and purity was determined using a NanoDrop 2000c UV‐Vis Spectrophotometer (Thermo Scientific, Waltham, MA, USA).
Assessment of HSV‐1 load
HSV‐1 DNA was detected and quantified in the placental tissues using the HSV‐1 polymerase chain reaction (PCR) kit (GeneProof a.s., Brno, Czech Republic), according to the manufacturer's instructions. The quantitative real‐time (qRT)–PCR was performed on the 7900HT Fast Real‐Time PCR system (Applied Biosystems, Foster City, CA, USA). A negative control without template DNA was included in every of amplification run.
Extraction of total RNA and cDNA synthesis
The decidual and chorionic villous tissues and the Vero cells were preserved in RNAlater (Ambion, Austin, TX, USA) at −80°C until further analysis. The total RNA was extracted using Tri Reagent (Ambion). First‐strand cDNA synthesis was performed using the high‐capacity cDNA reverse transcription kit (Applied Biosystems, according to the manufacturer's recommendations. The reverse transcription reaction was performed in a Veriti® 96‐well thermal cycler (Applied Biosystems). The concentration and purity of the RNA and cDNA were assessed using a NanoDrop 2000c UV‐Vis Spectrophotometer.
qRT–PCR analysis
Human DDX58, IFIH1, IFI16 and TLR3 mRNA expression was quantified by RT–PCR. The tyrosine 3‐monooxygenase/tryptophan 5‐monooxygenase activation protein, zeta polypeptide (YWHAZ) was selected for the normalization of target gene expression, as its expression levels were both highly stable and detectable in the human placenta 28. For each qRT–PCR, 50 ng cDNA was added to a mix containing gene‐specific primers 23, 29, 30, power SYBR Green PCR Master Mix (Applied Biosystems) and nuclease‐free water in a final volume of 25 µl. Amplification was performed in a 7900HT fast RT–PCR system under the following conditions: one cycle at 95°C for 10 min, followed by 50 cycles of 15 s at 95°C and 60 s at 60°C. All reactions were run in triplicate with a non‐template control. The qRT–PCR data were analysed using Data Assist version 3.0 software (Life Technologies, Grand Island, NY, USA), and the comparative Ct (ΔCt) method was used to calculate the relative gene expression.
ELISA
The tissue homogenates were prepared using the Mammalian Cell Lysis Kit (Sigma‐Aldrich), according to the manufacturer's instructions. The concentrations of IFI16, MDA5, RIG‐I and TLR‐3 proteins in tissue homogenates were determined using ELISA (EMELCA Biosciences, Breda, the Netherlands), according to the manufacturer's recommendations. The tissue culture supernatants were assayed for IFN‐α, IFN‐β (PBL Interferon Source, Piscataway, NJ, USA), IL‐6, C‐X‐C motif chemokine ligand 8 (CXCL8)/IL‐8 and tumour necrosis factor (TNF)‐α (R&D Systems, Minneapolis, MN, USA). The absorbance values of the samples and standards were read using a Benchmark Plus microplate reader (Bio‐Rad Laboratories, Hercules, CA, USA).
Statistical analysis
Statistical analyses were performed and graphs constructed using GraphPad Prism version 5.0 (GraphPad Software Inc., San Diego, CA, USA). All results are presented as the mean ± standard error of the mean (s.e.m.). The Mann–Whitney U‐test was used to assess any differences in the targeted genes and protein expression levels between the two groups (virus‐infected and mock‐infected tissues), while the Wilcoxon test was performed to compare two variables within the same group. A P‐value < 0·05 was considered significant.
Results
HSV‐1 replication in the placenta
To exclude the HSV‐1 infection of the placental samples, extracted DNA was subjected to qRT–PCR. None of the analysed samples expresses the HSV‐1 genome, indicating that the placentas were not infected with this virus in vivo. Hence, freshly obtained explants were infected with HSV‐1 strain McIntyre, and the viral load in decidual and chorionic villous explants after in‐vitro viral infection was studied. Isolated explants were relatively resistant to HSV‐1 infection. Viral replication was observed in four of eight placentas (50·0%) (Fig. 1). No significant difference between HSV‐1 replication in decidua and chorionic villi was found. All mock‐infected placental explants remained free of virus throughout the experiment.
Figure 1.
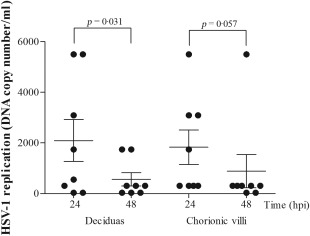
Comparison of the levels of herpes simplex type 1 (HSV‐1) replication in third‐trimester human placentas. Villous and decidual explants were infected with laboratory strain of HSV‐1 in vitro. HSV‐1 DNA was detected and quantified in the placental tissues using the HSV‐1 polymerase chain reaction (PCR) kit using quantitative real‐time (qRT)–PCR under the following conditions: denaturation at 95°C for 10 min, followed by 45 cycles of 95°C for 5 s and 60°C for 40 s.
IFI16 and TLR‐3 are expressed in term human chorionic villi and decidua
All examined PRRs genes, including DDX58 (RIG‐I), IFIH1 (MDA5), IFI16 (IFI16) and TLR3 (TLR‐3), were expressed constitutively at the mRNA and protein levels in the term decidual and chorionic villous tissues (Fig. 2a,b). A significantly higher level of IFI16 expression in HSV‐1‐infected decidual and chorionic villous explants compared to mock‐infected placentas was found at 48 h.p.i. (P = 0·029 and P < 0·001, respectively; Fig. 3a). Higher levels of TLR3 mRNA expression were also observed at 24 h.p.i. (P = 0·004 and P < 0·001, respectively; Fig. 3b). The DDX58 gene expression was lower in the virus‐infected explants at 24 h.p.i. (P = 0·028 for chorionic villi; Fig. 3c), whereas the levels of IFIH1 did not show any significant difference (P > 0·05; Fig. 3d).
Figure 2.
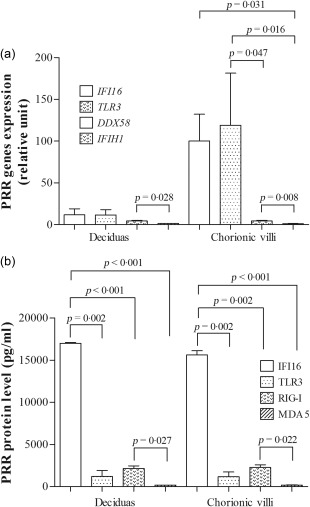
The constitutive pattern recognition receptors (PRRs) (a) mRNA and (b) protein expression (n = 8). The data are reported as the mean values ± standard error of the mean (s.e.m.). Statistically significant differences compared to mock‐infected placental explants are shown.
Figure 3.
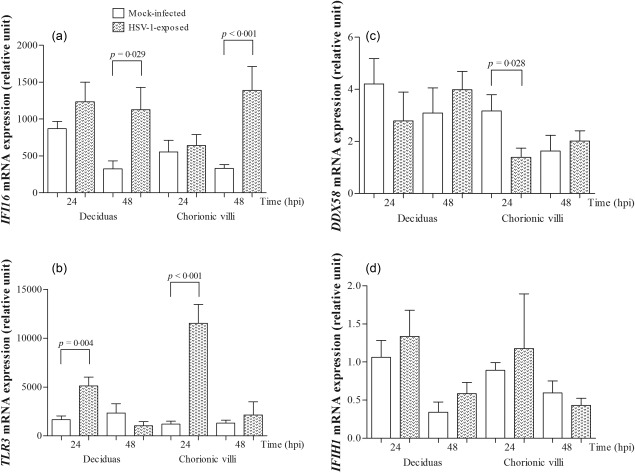
The mRNA expression levels of (a) IFI16, (b) TLR3, (c) DDX58 and (d) IFIH1 in third‐trimester mock‐infected and herpes simplex type 1 (HSV‐1)‐infected decidual and chorionic villi tissues (n = 8). Quantitative real‐time polymerase chain reaction (qRT–PCR) was performed using YWHAZ as a control gene. The data are reported as the mean values ± standard error of the mean (s.e.m.). Statistically significant differences compared to mock‐infected placental explants are shown. The Mann–Whitney two‐tailed test was used to assess statistically significant differences.
IFI16 and RIG‐I proteins are expressed during HSV‐1 infection
The level of IFI16 protein after HSV‐1 infection was higher than in mock‐infected tissues (P = 0·029; Fig. 4a). An increase in the TLR‐3 protein concentration was found in HSV‐1‐infected chorionic villi compared to mock‐infected explants at 48 h.p.i. (P = 0·028; Fig. 4b). Higher protein expression levels of RIG‐I upon virus infection were observed at 24 h.p.i. (P = 0·009 and P = 0·004 for decidua and chorionic villi, respectively; Fig. 4c). Our results revealed that all PRRs were expressed at both mRNA and protein levels in term human placenta samples.
Figure 4.
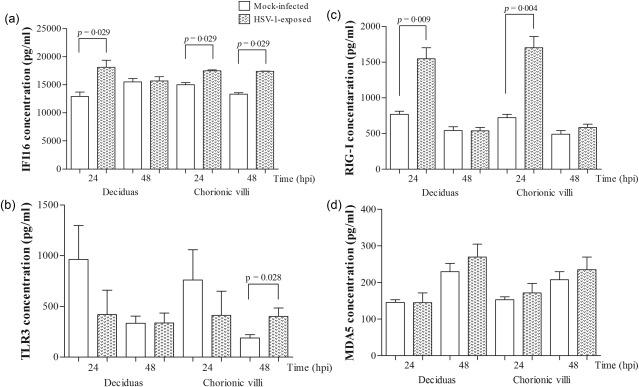
Differential expression of (a) IFI16, (b) Toll‐like receptor (TLR)‐3, (c) RIG‐I‐like receptor (RIG‐I) and (d) melanoma differentiation‐associated protein 5 (MDA5) in the placental tissue samples from mock‐infected and herpes simplex type 1 (HSV‐1)‐infected decidua and chorionic villi (n = 8). Data are reported as the mean values ± standard error of the mean (s.e.m.). The Mann–Whitney two‐tailed test was used to assess statistically significant differences.
IFN‐β is secreted in response to HSV‐1 infection
To examine whether PRRs contribute to the host response to virus infection, we measured the concentrations of IFN‐α, IFN‐β, IL‐6, IL‐8 and TNF‐α in supernatants from HSV‐1‐infected and mock‐infected explants. HSV‐1 induced IFN‐β production in both decidua and chorionic villi at 48 h.p.i. (P = 0·004 and P = 0·032, respectively; Fig. 5a). The significant decrease in the circulating TNF‐α levels was observed between mock‐infected and HSV‐1‐infected explants (P < 0·05) (Fig. 5b). The release of IFN‐α and ILs following HSV‐1 infection was unchanged (data not shown).
Figure 5.
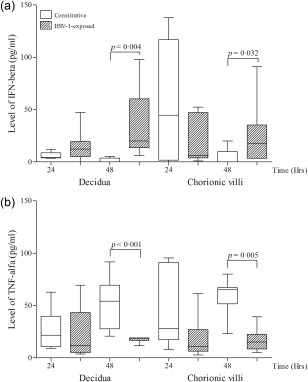
Analysis of the supernatants concentrations of (a) interferon (IFN)‐β and (b) tumour necrosis factor (TNF)‐α in decidual and chorionic villous tissue after herpes simplex type 1 (HSV‐1) infection. Decidual and chorionic villi explants were infected with 1 × 105 plaque‐forming units (PFU)/ml HSV‐1 (n = 8). Culture supernatants were harvested at 24 and 48 h post‐infection. The boxes indicate the 25th and 75th percentiles, while the bands near the middle indicate the median values. Data are expressed as the mean ± standard error of the mean (s.e.m.). The Mann–Whitney two‐tailed test was used to assess statistically significant differences.
Transcription of PRR genes in Vero cells after HSV‐1 infection
The next objective of this study was to determine which sensor is responsible for the HSV‐1 recognition in susceptible Vero cells. We found that the mRNA expression of TLR3, IFI16 and DDX58 increased after virus infection (P = 0·047 for TLR3; Fig. 6), while that of IFIH1 was blocked. We did not observe the induction of IFN in response to HSV‐1 infection in permissive Vero cells (P > 0·05; data not shown).
Figure 6.
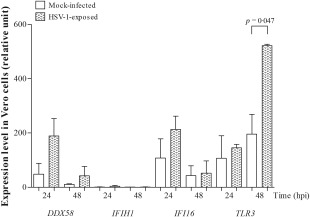
The relative expression of DDX58, IFIH1, IFI16 and TLR3 in mock‐infected and herpes simplex type 1 (HSV‐1)‐infected Vero cells (n = 4 independent experiments; passages 134–135). Quantitative real‐time polymerase chain reaction (qRT–PCR) was performed using YWHAZ as a housekeeping gene. The data are reported as the mean values ± standard error of the mean (s.e.m.). The Mann–Whitney two‐tailed test was used to assess statistically significant differences.
Discussion
PRRs are key components of the innate and adaptive immune responses and play a key role in the maternal–fetal interface 31, 32, 33, 34. We have shown that the IFI16 expression at both mRNA and protein levels was elevated significantly in third‐trimester human placentas infected with HSV‐1 ex vivo compared to mock‐infected explants. A higher level of RIG‐I protein expression in the tissues of fetal and maternal origin was observed. An enhanced IFN‐β production upon HSV‐1 infection was also noted, suggesting that these sensors activate downstream signalling in response to viral infection. To our knowledge, this is the first report that suggests that HSV‐1 triggered enhanced IFI16 and RIG‐I expression in the human term placenta.
IFI16 is a member of the human PYHIN family, and is an important nuclear dsDNA sensor that mediates the IFN‐β response 23, 35. It contains two C‐terminal DNA‐binding HIN domains and an N‐terminal pyrin domain that plays a role in regulating the inflammatory response and apoptosis 36, 37. DNA sensing by IFI16 is length‐dependent and GC content‐ and sequence‐independent 23. It was found that IFI16 binds fragments of dsDNA preferentially, which are approximately 150 base pairs (bp) long 38. Therefore, host DNA (10–20 bp long) is too short to activate this receptor. To date, the expression of IFI16 during HSV‐1 infection has been studied mainly in association with the innate immune response in other cells, including the corneal epithelium, human monocytic THP‐1 cells, primary human macrophages, neurones and primary human foreskin fibroblasts 23, 39, 40, 41, 42, 43, 44. We have shown that expression of IFI16 was induced during HSV‐1 infection in the tissues of fetal and maternal origin. An elevated IFN‐β production in supernatants of decidual and placental tissues following HSV‐1 treatment was also observed. It is well known that type I IFNs are important for mounting an anti‐viral response 43, 45. Hence, an increase in IFN‐β production in HSV‐1‐infected cultures suggests that the human term placenta initiate a classical anti‐viral response upon recognition of HSV‐1. As described previously, IFI16 can recognize HSV‐1 DNA both in the nucleus and in the cytoplasm to mediate type I IFNs via the STING/TBK1/IRF3 pathway and inflammatory cytokine responses via the nuclear factor kappa B (NF‐κB) pathway. We observed a significant decrease in TNF‐α production between supernatants of mock‐infected and HSV‐1 infected tissues that may be explained by the ability of HSV‐1 to suppress expression of proinflammatory cytokines by decreasing the stability of mRNA transcripts 46. It is known that proinflammatory immune responses are regulated tightly in the placenta to prevent immunological rejection of the fetal allograft and to decrease the incidence of stillbirth and preterm birth. We supposed that upon infection of placental explants with HSV‐1, IFI16 induces type I IFN through a pathway involving IFN‐stimulated genes activation. It also appears reasonable that endogenous cytokine restricts viral replication or there is an additional as‐yet undefined factor/s which blocks the replication machinery and proinflammatory responses.
Furthermore, we demonstrated that the viral infection of the placenta may also enhance the expression of RNA sensors, such as RIG‐I. It was described previously that RIG‐I acts in parallel with DAI in an RNA polymerase III‐dependent manner to initiate inflammatory and antiviral glial responses to HSV‐1 infection 47. An increase in the RIG‐I mRNA concentration and a trend towards an increased level of MDA5 mRNA in the blood of persistently infected with the bovine viral diarrhoea virus (BVDV) fetuses versus control fetuses were also described 48. These results showed that BVDV has the ability to cross the placenta and infect the fetus due to insufficient development of the fetal immune system and that RNA helicases RIG‐I and MDA5 can be induced in infected fetuses. RIG‐I, as well as MDA5, TLR‐3, TLR‐7 and TLR‐8, were also found to be expressed by the term placenta, choriodecidua and amnion in a second study 31. Our results may indicate that RIG‐I activate in chorionic villi and decidua the same intracellular signalling pathway as IFI16. We suggest that placental tissues possess a functional IFI16 as well as RIG‐I signalling systems, the activation of which can leads to IFN‐mediated anti‐HSV‐1 responses. In addition, the anti‐viral response was determined in Vero cells, known as deficient in production of type I IFNs. The results showed that HSV‐1 infection specifically enhanced expression of TLR‐3, IFI16 and DDX58 in Vero cells, whereas no IFN type I production was detected. This may indicate that recognition of HSV‐1 infection involves both TLR‐dependent and ‐independent mechanisms.
Expression levels between transcript and protein are not always correlated. Post‐transcriptional or post‐translational modifications, such as phosphorylation and ubiquitination, may be the reasons for differences in protein production with the corresponding mRNA level in tissue. Post‐translational management of proteins due to protein half‐life, experimental condition, protein degradation, etc. may also contribute more for the reverse result in response to the above. Moreover, the proteins may be long‐lived proteins which accumulate over time, while the mRNA turnover is speedy. In the present study, a low expression of IFI16 transcript and high expression of the protein in freshly isolated villous and decidual explants was found. We supposed that IFI16 mRNA becomes degraded, while its protein has a higher half‐life and remains in the cellular pool. Another possibility is that its mRNA is translated more preferentially to the protein. It is also possible that elevated expression of IFI16 in term placentas was associated with non‐necroptotic programmed cell death 49.
Some potential limitations of the present study must be acknowledged. First, the main limitation is that this study has small number of placental biopsies. Secondly, this study does not include the biopsies from intrauterine HSV‐1‐infected human placental tissues, as the availability of placental tissues with in‐utero HSV‐1 infection from pregnant women is limited. Thirdly, we did not isolate the specialized cells of the placenta (e.g. trophoblast), although we analysed the decidual and chorionic villi explants separately. Because the total concentration of proteins determined with the bicinchoninic acid (BCA) assay was very low, and we wanted to quantify the PRR proteins, we decided to use the ELISA assay. In addition, other putative DNA sensors, e.g. DAI, DDX41, cGAS and adaptor protein STING, were not included in this preliminary study. Hence, further studies will be needed to verify the importance of our observations.
In conclusion, the results of this study shed light upon the possible role of the PRR system in the detection of HSV‐1 infection and the activation of innate immunity in the human placenta. These findings suggest a possible role of IFI16 in the DNA‐induced immune response in the placenta, as well as RIG‐I as a cytoplasmic sensor for non‐self RNA. However, to confirm these hypotheses, future in‐depth and more advanced investigations will be needed to define the precise mechanisms by which PRRs modulate HSV‐1 pathogenesis in the placenta.
Disclosure
All authors state that they have nothing to disclose.
Acknowledgements
The authors are grateful all pregnant women who participating in this study. The authors thank to Edyta Jaworska for the technical support. This work was supported by the National Science Centre of Poland (Grant no. 2015/17/N/NZ6/02015) and the European Union Developmental Fund under Operational Programme Innovative Economy (Grant no. POIG.01.01.02–10‐107/09).
References
- 1. Bernstein DI, Bellamy AR, Hook EW III et al Epidemiology, clinical presentation, and antibody response to primary infection with herpes simplex virus type 1 and type 2 in young women. Clin Infect Dis 2013; 56:344–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Webber BJ, Pawlak MT, Jones NM, Tchandja JN, Foster GA. Sexually transmitted infections in U.S. Air Force recruits in basic military training. Med Surveill Monthly Rep 2016; 23:16–9. [PubMed] [Google Scholar]
- 3. Gupta R, Warren T, Wald A. Genital herpes. Lancet 2007; 370:2127–37. [DOI] [PubMed] [Google Scholar]
- 4. Peña KC, Adelson ME, Mordechai E, Blaho JA. Genital herpes simplex virus type 1 infection in women: detection in cervicovaginal specimens from gynecological practices in the United States. J Clin Microbiol 2010; 48:150–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anzivino E, Fioriti D, Mischitelli M et al Herpes simplex virus infection in pregnancy and in neonate: status of art of epidemiology, diagnosis, therapy and prevention. Virol J 2009; 6:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brown ZA, Wald A, Morrow RA, Selke S, Zeh J, Corey L. Effect of serologic status and cesarean delivery on transmission rates of herpes simplex virus from mother to infant. JAMA 2003; 289:203–9. [DOI] [PubMed] [Google Scholar]
- 7. Mercolini F, Verdi F, Eisendle K, Messner H, Staffler A. Congenital disseminated HSV‐1 infection in preterm twins after primary gingivostomatitis of the mother: case report and review of the literature. Z Geburtshilfe Neonatol 2014; 218:261–4. [DOI] [PubMed] [Google Scholar]
- 8. Syridou G, Spanakis N, Konstantinidou A et al Detection of cytomegalovirus, parvovirus B19 and herpes simplex viruses in cases of intrauterine fetal death: association with pathological findings. J Med Virol 2008; 80:1776–82. [DOI] [PubMed] [Google Scholar]
- 9. Kimberlin DW. Neonatal herpes simplex infection. Clin Microbiol Rev 2004; 17:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Allen UD, Robinson JL. Prevention and management of neonatal herpes simplex virus infections. Paediatr Child Health 2014; 19:201–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. James SH, Sheffield JS, Kimberlin DW. Mother‐to‐child transmission of herpes simplex virus. J Pediatric Infect Dis Soc 2014; 3:S19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Straface G, Selmin A, Zanardo V, De Santis M, Ercoli A, Scambia G. Herpes simplex virus infection in pregnancy. Infect Dis Obstet Gynecol 2012; 2012:385697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Corey L, Wald A. Maternal and neonatal HSV infections. N Engl J Med 2009; 361:1376–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gantt S, Muller WJ. The immunologic basis for severe neonatal herpes disease and potential strategies for therapeutic intervention. Clin Dev Immunol 2013; 2013:369172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Su C, Zhan G, Zheng C. Evasion of host antiviral innate immunity by HSV‐1, an update. Virol J 2016; 13:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paludan SR, Bowie AG, Horan KA, Fitzgerald KA. Recognition of herpesviruses by the innate immune system. Nat Rev Immunol 2011; 11:143–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Piret J, Boivin G. Innate immune response during herpes simplex virus encephalitis and development of immunomodulatory strategies. Rev Med Virol 2015; 25:300–19. [DOI] [PubMed] [Google Scholar]
- 18. Ferguson BJ, Mansur DS, Peters NE, Ren H, Smith GL. DNA‐PK is a DNA sensor for IRF‐3‐dependent innate immunity. Elife 2012; 1:e00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Melchjorsen J, Rintahaka J, Søby S et al Early innate recognition of herpes simplex virus in human primary macrophages is mediated via the MDA5/MAVS‐dependent and MDA5/MAVS/RNA polymerase III‐independent pathways. J Virol 2010; 84:11350–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rasmussen SB, Sørensen LN, Malmgaard L et al Type I interferon production during herpes simplex virus infection is controlled by cell‐type‐specific viral recognition through Toll‐like receptor 9, the mitochondrial antiviral signaling protein pathway, and novel recognition systems. J Virol 2007; 81:13315–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP‐AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013; 339:786–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takaoka A, Wang Z, Choi MK et al DAI (DLM‐1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 2007; 448:501–5. [DOI] [PubMed] [Google Scholar]
- 23. Unterholzner L, Keating SE, Baran M et al IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 2010; 11:997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double‐stranded RNA is produced by positive‐strand RNA viruses and DNA viruses but not in detectable amounts by negative‐strand RNA viruses. J Virol 2006; 80:5059–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol 2011; 12:959–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Almine JF, O'Hare CA, Dunphy G et al IFI16 and cGAS cooperate in the activation of STING during DNA sensing in human keratinocytes. Nat Commun 2017; 8:14392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA‐mediated, type I interferon‐dependent innate immunity. Nature 2009; 461:788–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meller M, Vadachkoria S, Luthy DA, Williams MA. Evaluation of housekeeping genes in placental comparative expression studies. Placenta 2005; 26:601–7. [DOI] [PubMed] [Google Scholar]
- 29. Komuro A, Horvath CM. RNA‐ and virus‐independent inhibition of antiviral signaling by RNA helicase LGP2. J Virol 2006; 80:12332–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Patni S, Wynen LP, Seager AL, Morgan G, White JO, Thornton CA. Expression and activity of Toll‐like receptors 1–9 in the human term placenta and changes associated with labor at term. Biol Reprod 2009; 80:243–8. [DOI] [PubMed] [Google Scholar]
- 31. Bryant AH, Menzies GE, Scott LM et al Human gestation‐associated tissues express functional cytosolic nucleic acid sensing pattern recognition receptors. Clin Exp Immunol 2017; 189:36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cardenas I, Mulla MJ, Myrtolli K et al Nod1 activation by bacterial iE‐DAP induces maternal‐fetal inflammation and preterm labor. J Immunol 2011; 187:980–6. [DOI] [PubMed] [Google Scholar]
- 33. Holmlund U, Cebers G, Dahlfors AR et al Expression and regulation of the pattern recognition receptors Toll‐like receptor‐2 and Toll‐like receptor‐4 in the human placenta. Immunology 2002; 107:145–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang YY, Chen H, Sun C et al Expression and functional characterization of NOD2 in decidual stromal cells isolated during the first trimester of pregnancy. PLOS ONE 2014; 9:e99612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Connolly DJ, Bowie AG. The emerging role of human PYHIN proteins in innate immunity: implications for health and disease. Biochem Pharmacol 2014; 92:405–14. [DOI] [PubMed] [Google Scholar]
- 36. Fairbrother WJ, Gordon NC, Humke EW et al The PYRIN domain: a member of the death domain‐fold superfamily. Protein Sci 2001; 10:1911–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jin T, Perry A, Jiang J et al Structures of the HIN domain: DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity 2012; 36:561–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gallucci S. An overview of the innate immune response to infectious and noninfectious stressors In: Amadori M, ed. The innate immune response to noninfectious stressors. Human and Animal Models. Brescia, Italy: Academic Press, 2016:1–24. [Google Scholar]
- 39. Conrady CD, Zheng M, Fitzgerald KA, Liu C, Carr DJ. Resistance to HSV‐1 infection in the epithelium resides with the novel innate sensor, IFI‐16. Mucosal Immunol 2012; 5:173–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Diner BA, Lum KK, Toettcher JE, Cristea IM. Viral DNA sensors IFI16 and cyclic GMP‐AMP synthase possess distinct functions in regulating viral gene expression, immune defenses, and apoptotic responses during herpesvirus infection. MBio 2016; 7:e01553–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Horan KA, Hansen K, Jakobsen MR et al Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. J Immunol 2013; 190:2311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Orzalli MH, DeLuca NA, Knipe DM. Nuclear IFI16 induction of IRF‐3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci USA 2012; 109:E3008–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rosato PC, Leib DA. Neuronal interferon signaling is required for protection against herpes simplex virus replication and pathogenesis. PLOS Pathog 2015; 11:e1005028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Søby S, Laursen RR, Ostergaard L, Melchjorsen J. HSV‐1‐induced chemokine expression via IFI16‐dependent and IFI16‐independent pathways in human monocyte‐derived macrophages. Herpesviridae 2012; 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Goodbourn S, Didcock L, Randall RE. Interferons: cell signaling, immune modulation, antiviral responses and virus countermeasures. J Gen Virol 2000; 81:2341–64. [DOI] [PubMed] [Google Scholar]
- 46. Mogensen TH, Melchjorsen J, Malmgaard L, Casola A, Paludan SR. Suppression of proinflammatory cytokine expression by herpes simplex virus type 1. J Virol 2004; 78:5883–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Crill EK, Furr‐Rogers SR, Marriott I. RIG‐I is required for VSV‐induced cytokine production by murine glia and acts in combination with DAI to initiate responses to HSV‐1. Glia 2015; 63:2168–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smirnova NP, Webb BT, Bielefeldt‐Ohmann H et al Development of fetal and placental innate immune responses during establishment of persistent infection with bovine viral diarrhea virus. Virus Res 2012; 167:329–36. [DOI] [PubMed] [Google Scholar]
- 49. Chu X, Chen W, Li N et al Cytosolic double‐stranded DNA induces nonnecroptotic programmed cell death in trophoblasts via IFI16. J Infect Dis 2014; 210:1476–86. [DOI] [PubMed] [Google Scholar]


