Abstract
Purpose
The present study aimed to analyze the endometrial and vaginal microbiome among a Japanese infertile population by sequencing and the impact of the endometrial and vaginal environment on implantation.
Methods
In total, 102 infertile (79 in vitro fertilization [IVF] and 23 non‐IVF) patients and seven healthy volunteers were recruited from August to December, 2017. Endometrial fluid and vaginal discharge samples for sequencing were collected by using an intrauterine insemination catheter. The bacterial status of the endometrium and vagina were analyzed.
Results
The Lactobacillus‐dominated microbiota (>90% Lactobacillus spp.) in the endometrium vs vagina was 38% (30/79) vs 44.3% (44/79) in the IVF patients, 73.9% (17/23) vs 73.9% (17/23) in the non‐IVF patients, and 85.7% (6/7) vs 85.7% (6/7) in the healthy volunteers. The percentage of endometrial Lactobacillus in the healthy volunteers was highly stable within the same menstrual cycle and even in the following cycle. The major taxonomies were Gardnerella, Streptococcus, Atopobium, Bifidobacterium, Sneathia, Prevotella, and Staphylococcus. Fifteen patients achieved pregnancy by a single vitrified‐warmed blastocyst transfer during this study; the median percentage of Lactobacillus in the pregnant women was 96.45 ± 33.61%.
Conclusion
A considerable percentage of non‐Lactobacillus‐dominated (NLD) microbiota was found in the endometrium of Japanese infertile women. Increasing the endometrial level of the Lactobacilli to >90% might favor the implantation outcome of NLD infertile patients.
Keywords: endometrial microbiota, infertile women, Japanese women, vaginal microbiota, volunteer
1. INTRODUCTION
Recurrent implantation failure (RIF) is a major issue of infertility that has not yet been fully investigated. It is determined when there are failed implantation cycles after several in vitro fertilization (IVF) attempts. Implantation failure is usually considered to have occurred after more than three cycles of IVF, with the transfer of morphologically good embryos,1 and there are several causes of RIF, such as pathologic alterations of the endometrial cavity, hydrosalpinx, embryonic aneuploidy, thrombophilias,2 and systemic factors like thyroid dysfunction. Although embryonic aneuploidy is likely to be the major contributor to human implantation failure, especially in cases with advanced maternal age,3 it has been reported that the proportion of euploid embryos failing to implant was ~40%,4 which may suggest the importance of the endometrium and its environment as another dominant factor for implantation failure.5
The uterine cavity has been considered to be sterile until recent studies using next‐generation sequencing of the 16S ribosomal RNA (rRNA) gene revealed the existence of an endometrial microbiota that is represented by Lactobacillus and other bacteria.6, 7, 8 Lactobacillus species generally dominate the vagina of healthy asymptomatic women and they presumably play key roles in preventing bacterial vaginosis and other urogenital diseases by lowering the environmental pH through lactic acid production.9 Recently, a study demonstrated that the endometrial microbiota that were analyzed by 16S rRNA gene sequencing have effects on implantation success.10 The bacterial status of the endometrium was defined as Lactobacillus‐dominated microbiota (>90% Lactobacillus spp.) or non‐Lactobacillus‐dominated microbiota (<90% Lactobacillus spp. with >10% of other bacteria), based on the composition of the microbiota in the endometrial fluid, comprised of 191 operational taxonomic units (OTUs). The endometrial microbiota was not hormonally regulated during the acquisition of endometrial receptivity and the presence of the non‐Lactobacillus‐dominated microbiota was associated with a significant decrease in the implantation, pregnancy, ongoing pregnancy, and live birth rates.10
The ethnic difference in the endometrial microbiota of patients who are undergoing assisted reproductive technology (ART) is still not fully investigated. This present study aimed to analyze the endometrial microbiome among a Japanese infertile population by sequencing and the impact of the endometrial environment on the outcomes of infertile patients.
2. MATERIALS AND METHODS
2.1. Patients and samples
In total, 79 IVF and 23 non‐IVF patients under 45 years old who agreed to undergo endometrial and vaginal microbiome analysis in the authors’ center from August, 2017 to December, 2017, were eligible for this study. Also, the endometrial and vaginal microbiomes of seven asymptomatic healthy volunteers were analyzed, and for five cases, the samples were collected in different cycles and different menstrual phases in order to analyze the changes in the endometrial and vaginal microbiota in the transition of menstrual cycles. All the patients were examined routinely by vaginal ultrasound in a sterile condition in order to confirm the menstrual phase of the patient. The participants had no complaint suggestive of vaginitis or endometritis.
This study was approved by the Institutional Review Board of Kyono ART Clinic Takanawa, Tokyo, Japan, on 29 July, 2017. All the patients who were involved in this study allowed the use of their medical record data for research in an unidentifiable manner. Written, informed consent was obtained from all the patients prior to sample collection.
2.2. Sample collection and DNA extraction
Paired samples from the vagina and endometrium were taken from the 79 IVF and 23 non‐IVF cases. Five out of the seven volunteers were examined more than once, resulting in 15 paired samples (Figure 1). Vaginal discharge specimens were collected with an OMNIgene VAGINAL accessory swab (DNA Genotek, Inc., Ottawa, ON, Canada). These were put into a 1 mL MMB collection tube containing stabilizing liquid for the microbiome (DNA Genotek, Inc.). After cleaning the mucous around the cervical os and the uterine cervix, endometrial fluid (EF) specimens were carefully aspirated with a intrauterine insemination (IUI) catheter (Kitazato Corporation, Shizuoka, Japan) with the utmost care not to touch the vaginal wall. These also were put into a 1 mL MMB collection tube (DNA Genotek, Inc.). The vaginal and endometrial samples were treated with proteinase K and lysozyme solution, according to the manufacturer's instructions. The genomic DNA was extracted by using an Agencourt Genfind v. 2 Blood & Serum DNA Isolation Kit (Beckman Coulter, Inc., Brea, CA, USA). The double‐stranded (ds)DNA concentration was quantified fluorometrically with a Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Figure 1.
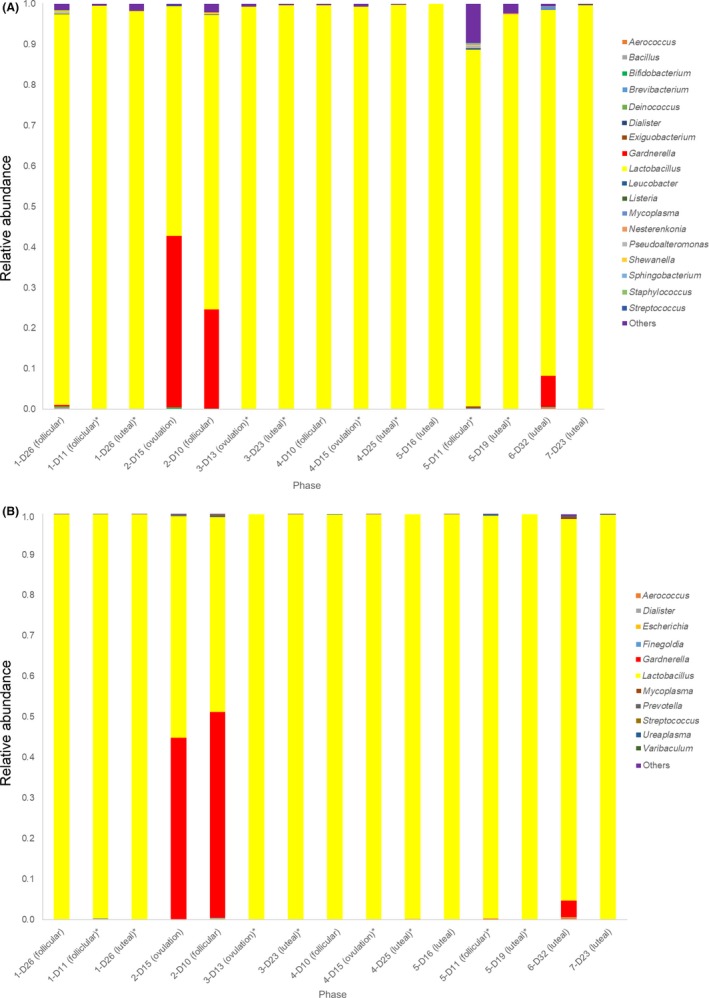
A, Endometrial microbiomes of the healthy volunteers. The same number represents the same participant. *, same menstrual cycle. B, The vaginal microbiomes of the healthy volunteers. The same number represents the same participant. *, same menstrual cycle
2.3. Polymerase chain reaction amplification and DNA sequencing
The variable region 4 (V4) hypervariable region of the bacterial 16S rRNA gene was amplified from the specimen's DNA by using a modified primer pair, 515f (5’‐TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGTGYCAGCMGCCGCGGTAA‐3’) and 806rB (5’‐GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGACTACNVGGGTWTCTAAT‐3’), with Illumina Nextera XT adapter overhang sequences (underlined).11 The universal bacterial primers to amplify the V1‐V2 region and V3‐V5 regions were 28f (5’‐TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGAGTTTGATCNTGGCTCAG‐3’) to 338r (5’‐GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTGCTGCCTCCCGTAGGAGT‐3’) and 357f (5’‐TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGAGGCAGCAG‐3’) to 926r (5’‐GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCCGTCAATTYMTTTRAGT‐3’), respectively. The polymerase chain reaction (PCR) amplification was performed, as previously described.10 The PCR was performed with 25 ng/μL DNA, 200 μmol/L each of the four deoxynucleotide triphosphates, 400 nmol/L of each primer, 2.5 U of FastStart HiFi polymerase, 4% of 20 mg/mL BSA (Sigma‐Aldrich, St. Louis, MO, USA), 0.5 mol/L betaine (Sigma‐Aldrich), and the appropriate buffer, with MgCl2 supplied by the manufacturer (Roche, Basel, Switzerland). Thermal cycling consisted of initial denaturation at 94°C for two minutes, followed by 30 cycles of denaturation at 94°C for 20 seconds, annealing at 50°C for 30 seconds, an extension at 72°C for one minute, and a final extension at 72°C for 5 minutes. Amplicon mixture was purified by using Agencourt AMPure XP (Beckman Coulter, Inc.). The purified PCR samples were multiplexed by using a dual‐index approach with the Nextera XT Index kit v. 2 (Illumina, Inc., San Diego, CA, USA), according to the Illumina 16S Metagenomic Sequencing Library Preparation protocol. Indexing the PCR was performed by using KAPA HiFi HotStart ReadyMix (Kapa Biosystems, Wilmington, MA, USA) in a 50 μL reaction volume, and subsequently, purification was performed with Agencourt AMPure XP beads. The final library was paired‐end sequenced at 2 bp × 200 bp or 2 bp × 300 bp using a MiSeq Reagent Kit v. 3 on the Illumina MiSeq platform, depending on the primer set. As the 16S rRNA gene target region had an immense impact on the analysis results, the V1‐V2, V3‐V5, and V4 variable regions that are commonly used in 16S rRNA sequencing from human samples were compared.12 In order to evaluate the representation of the microbial community, the ZymoBIOMICS Microbial Community Standard (Zymo Research, Irvine, CA, USA), containing a mixture of Pseudomonas, Escherichia, Salmonella, Lactobacillus, Enterococcus, Listeria, Bacillus, and two yeast species was used. All the species, except Salmonella, were observed by all the primer sets; however, the V1‐V2 and V3‐V5 amplicon sequencing failed to detect Salmonella (data not shown). To evaluate whether some target regions better represented the endometrial microbial community structure than other regions, three variable regions of 10 endometrial samples were sequenced. The detection of Gardnerella and Bifidobacterium was observed in the V3‐V5 and V4 amplicon sequencing, but it was absent in V1‐V2. From these results, a primer set that targeted the V4 region was used for the endometrial microbiome analysis.
2.4. Data analysis
The reads were merged by using EA‐Utils fastq‐join13 and a median merged sequence length of 291 bp was obtained. Quality control of the merged sequence was performed by using USEARCH v. 10.0.24014 to remove the PhiX (Illumina) reads, truncate the primer‐binding sequences, and discard the sequences with a <100 bp length and a sequence quality of <Q20. QIIME 1.9.115 was used with the default parameters for quality filtering, the chimera check, clustering sequences into OTUs, and the assignment of taxonomy. The sequences were clustered into OTUs by an open‐reference OTU‐picking strategy by using the UCLUST method, based on 97% sequence identity. The taxonomy was assigned to each OTU by using the Ribosomal Database Project classifier16 with a .50 confidence threshold against the Greengenes database v. 13_8.17 Low‐abundance taxa (.01%) were filtered from the OTU tables. All further analyses were performed at a rarefied depth of 5000 sequences per sample to correct for differences in the read depth across samples. As human specimens contains low bacterial DNA, background bacterial contamination critically affected the result.18 If the library concentration of an endometrial sample was as much as the blank control, UltraPure™ DNase/RNase‐Free Distilled Water (ThermoFisher Scientific, Inc.), the result of the microbial community was similar to the blank control. This similarity to the background microbiome makes it difficult to determine the presence of unique endometrial taxa; therefore, blank‐characteristic OTUs were subtracted to reduce the background noise, as in previous studies.19, 20 The following nine bacterial taxa that were found in a blank control and are known as reagent contaminations were excluded from the endometrial samples by using QIIME: Acinetobacter, Escherichia, Flavobacterium, Janthinobacterium, Methylobacterium, Pseudomonas, Rhodococcus, Sphingomonas, and Stenotrophomonas.
The profiles, bacterial status, and percentage of Lactobacilli in the endometrium and vagina of the patients were analyzed. Clinical pregnancy was defined as the confirmation of a gestational sac in the uterine cavity by ultrasound analysis.
2.5. Statistical analysis
The statistical analysis (using the software, StatMate V software [Tokyo, Japan]) was performed by using ANOVA and Kruskal‐Wallis tests for multiple comparison and the Mann‐Whitney U‐test, chi‐square analysis, or Fisher's exact test, where appropriate. A P‐value of <.05 was considered to be statistically significant.
3. RESULTS
3.1. Patient profiles
The average age of the 109 cases was 36.17 ± 4.51 years old (25‐44); 59 cases (54.1%) were multigravida and 17 cases (15.6%) were multipara; all cases were Asian. Of all the cases, 79 (72.5%) were enrolled in IVF treatment, 23 (21.1%) were non‐IVF cases, and seven cases were asymptomatic healthy volunteers. The past histories of failed vitrified‐warmed blastocyst transfers and total failed embryo transfers among the IVF group were 3.50 ± 3.15 cycles and 3.09 ± 3.09 cycles, respectively. There was no difference among the three groups in terms of their Body Mass Index, gravidity, and parity, although the non‐IVF patients were significantly younger than the IVF patients (33.22 ± 3.64 years vs 36.99 ± 4.22 years; P < .01). There was no difference in the duration of infertility between the non‐IVF patients and IVF patients (Table 1).
Table 1.
Characteristics of the three groups
| Characteristic | Healthy volunteers | Non‐IVF patients | IVF patients | P‐value |
|---|---|---|---|---|
| No. of patients | 7 | 23 | 79 | — |
| Age (years): mean ± SD | 36.57 ± 6.63a | 33.22 ± 3.64a | 36.99 ± 4.22a | <.01 |
| BMI: mean ± SD | 21.00 ± 2.57a | 20.92 ± 2.81a | 20.35 ± 3.00a | .66 |
| Duration of infertility (mo): median ± SD | — | 18.78 ± 10.77 | 20.77 ± 30.46 | .39 |
| Multigravida patients: N (%) | 3 (42.9)b | 8 (34.8)b | 48 (60.8)b | .08 |
| Multiparapatients: N (%) | 3 (42.9)c | 3 (13.0)c | 15 (19.0)c | .29 |
| Previous ET: mean ± SD | — | — | 3.50 ± 3.15 | — |
| Previous FBT: mean ± SD | — | — | 3.09 ± 3.09 | — |
| Sampling timing: follicular phase: N (%) | 2 (28.6)c | 18 (78.3)c | 38 (48.1)c | .08 |
| Ovulation phase: N (%) | 2 (28.6) | 2 (8.7) | 12 (15.2) | — |
| Luteal phase: N (%) | 3 (42.9) | 3 (13.0) | 29 (36.7) | — |
BMI, body mass index; ET, embryo transfer; FBT, frozen‐thawed blastocyst transfer; IVF, in vitro fertilization; SD, standard deviation.
ANOVA.
Mann‐Whitney's U‐test.
Chi‐square test.
The EF and the vaginal specimens were collected in the follicular phase, ovulation phase, or luteal phase of the menstrual cycles (Table 1). There was no statistical difference in the timing of sampling among the three groups; however, there was a tendency that more samples were collected in the follicular phase than in the ovulation and luteal phases of the non‐IVF patients (Table 1, P = .08).
3.2. Endometrial and vaginal microbial results of the asymptomatic healthy volunteers
The bacterial diversity in the endometrial communities was determined by pyrosequencing of V4 of the bacterial 16S rRNA genes. The median percentage of endometrial Lactobacilli and vaginal Lactobacilli of the seven healthy volunteers were 99.50 ± 15.85% and 99.80 ± 16.82%, respectively. Six out of the seven volunteers had a Lactobacillus‐dominated (LD) (>90%) endometrial microbial status (Table 2).
Table 2.
Percentages of Lactobacillus and Lactobacillus‐dominated (LD) (>90%) in the endometrium and vagina among the three groups
| Characteristic | Healthy volunteers | Non‐IVF patients | IVF patients |
|---|---|---|---|
| No. of patients | 7 | 23 | 79 |
| Endometrial lactobacilli (%): median ± SD | 99.50 ± 15.85a , b | 96.20 ± 34.61a , c | 63.90 ± 41.43a , b , c |
| Vaginal lactobacilli (%): median ± SD | 99.80 ± 16.82d | 99.40 ± 36.32d | 65.21 ± 43.70d |
| Patients with LD endometrium: N (%) | 6 (85.7)e | 17 (73.9)e | 30 (38.0)e |
| Patients with LD vagina: N (%) | 6 (85.7)f | 17 (73.9)f | 44 (44.3)f |
IVF, in vitro fertilization; SD, standard deviation.
P = .020 (Kruskal‐Wallis test).
P = .030 (Mann‐Whitney's U‐test).
P = .040 (Mann‐Whitney's U‐test).
P = .120 (Kruskal‐Wallis test).
P = .001 (chi‐square test).
P = .110 (chi‐square test).
Five out of the seven volunteers agreed to undergo re‐examination of the endometrial and vaginal microbiome in the following menstrual cycle and/or in a different phase of the same menstrual cycle in order to analyze the stability of the endometrial and vaginal microbiota. The percentage of the endometrial and vaginal Lactobacilli of the healthy volunteers was highly stable within the same menstrual cycle and was stable even in the following cycle (Figure 1A,B).
3.3. Endometrial and vaginal microbial results of the infertile patients
The median percentage of the endometrial Lactobacilli in the IVF patients was significantly lower than that of the non‐IVF patients and healthy volunteers (63.90 ± 41.43% vs 96.20 ± 34.61% vs 99.50 ± 15.85%; P = .02); also, the percentage of LD endometrial status was significantly lower in the IVF group, compared to the non‐IVF group and healthy volunteer group (38.0% vs 73.9% vs 85.7%; P = .001) (Table 2, Figure 2A). Also, the median percentage of vaginal Lactobacilli in the IVF patients was lower than that of the non‐IVF patients and healthy volunteers (65.21 ± 43.70% vs 99.40 ± 36.32% vs 99.80 ± 16.82%; P = .12), although the difference was not statistically significant (Table 2, Figure 2B). The great majority of patients with a non‐Lactobacillus‐dominated (NLD) (<90%) endometrial microbial status also had a NLD vaginal microbial status: 89.8% (44/49) in the IVF group, 100% (6/6) in the non‐IVF group, and 100% (1/1) in the healthy volunteers. There was no case with a NLD vaginal status that showed a LD endometrial status: those who had a NLD vaginal status also had a NLD endometrial status.
Figure 2.
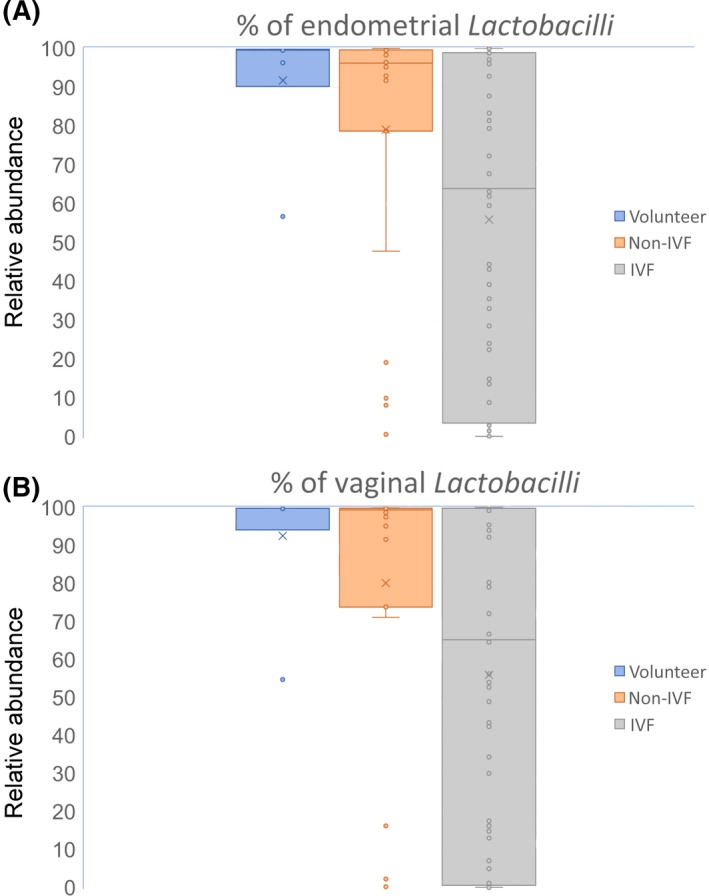
Percentages of (A) endometrial and (B) vaginal Lactobacilli in the healthy volunteers, non‐IVF patients, and IVF patients. IVF, in vitro fertilization
For the 79 IVF patients, in the subgroup analyses according to age and gravida or para status, there was no significant difference in the percentage of endometrial and vaginal Lactobacilli between those aged over 38 (n = 40) and those under 37 years old (n = 39) (Table 3). Also, there was no statistically significant difference in the percentage of endometrial and vaginal Lactobacilli among the nulligravida versus gravida‐nullipara versus para patients (70.00 ± 40.20% vs 59.10 ± 42.01% vs 44.40% ± 43.90%; P = .66) and (79.00% ± 41.26% vs 56.20% ± 44.28% vs 48.90% ± 47.91%; P = .68), respectively (Figure 3A,B).
Table 3.
Comparison between the ≥38 years old and the <38 years old in vitro fertilization patients
| Characteristic | ≥38 y | <38 y | P‐value |
|---|---|---|---|
| No. of patients | 40 | 39 | – |
| Age (years): mean ± SD | 40.60 ± 1.58a | 33.28 ± 2.48a | 1.65122E‐23 |
| Duration of infertility (mo): median ± SD | 24.56 ± 37.16b | 16.67 ± 20.70b | .32 |
| Multigravida patients: N (%) | 25 (62.5)c | 23 (59)c | .75 |
| Multiparapatients: N (%) | 7 (17.5)d | 8 (20.5)d | .96 |
| Previous ET: mean ± SD | 4.18 ± 3.49a | 2.59 ± 2.53a | .02 |
| Previous FBT: mean ± SD | 3.55 ± 3.46a | 2.43 ± 2.52a | .11 |
| Endometrial lactobacilli (%): median ± SD | 63.60 ± 41.21b | 63.90 ± 42.18b | .76 |
| Vaginal lactobacilli (%): median ± SD | 61.45 ± 43.29b | 65.21 ± 44.68b | .78 |
| Patients with LD endometrium: N (%) | 13 (32.5)c | 17 (45.6)c | .31 |
| Patients with LD vagina: N (%) | 17 (42.5)c | 18 (46.2)c | .74 |
ET, embryo transfer; FBT, frozen‐thawed blastocyst transfer; LD, Lactobacillus‐dominated (>90%); SD, standard deviation.
t test.
Mann‐Whitney's U‐test.
Chi‐square test.
Fisher's exact test.
Figure 3.
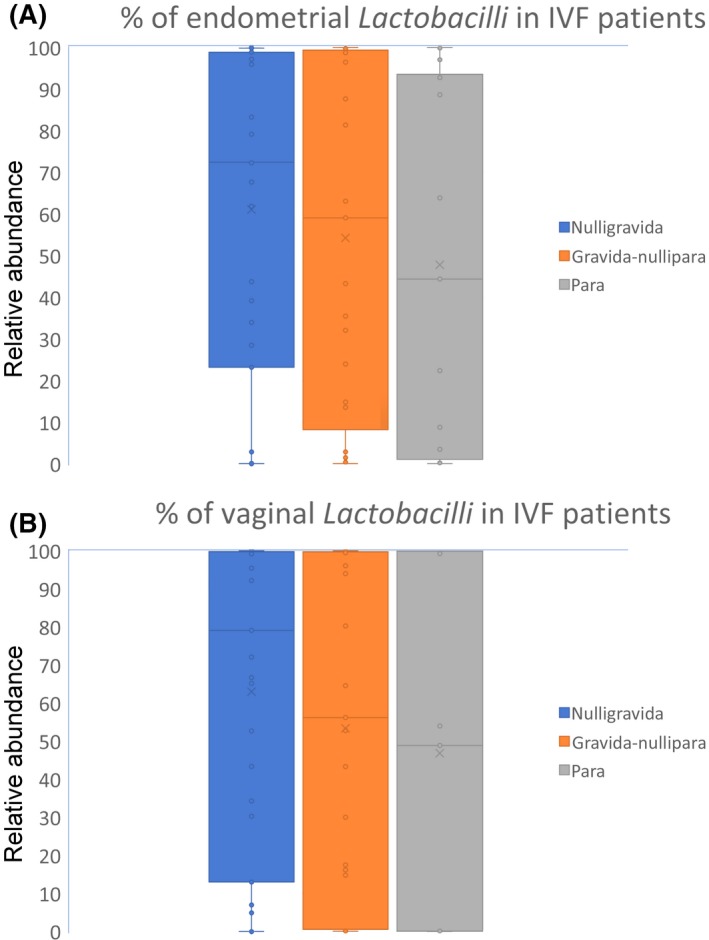
Percentages of (A) endometrial and (B) vaginal Lactobacilli in nulligravida, gravida‐nullipara, and para female IVF patients. IVF, in vitro fertilization
During this study period, 18 patients achieved pregnancy: three by natural conception and 15 by single vitrified‐warmed blastocyst transfer. The median percentages of the endometrial and vaginal Lactobacilli in those pregnant cases were 96.45% ± 33.61% and 97.80% ± 37.44%, respectively. There were seven NLD cases (six IVF and one non‐IVF) who achieved pregnancy (Figure 4); five cases are ongoing, one (No. 5) turned out to be an early miscarriage at 6 weeks of pregnancy, and one (No. 6) is lost to follow‐up. All seven cases conceived without any treatment intervention for the microbiome.
Figure 4.
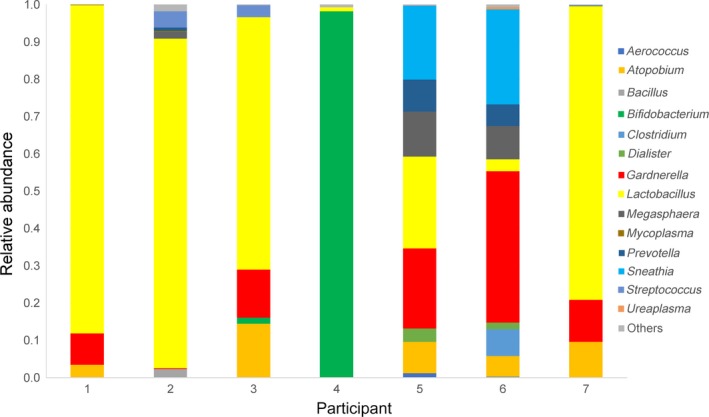
Seven pregnant cases with a non‐Lactobacillus‐dominated (NLD) endometrium. The relative proportion of the most abundant operational taxonomic units in the endometrium of the seven pregnant cases. The number below the graph shows each participant: No. 1‐6, IVF patient; No. 7, non‐IVF patient. IVF, in vitro fertilization
3.4. Bacterial communities in the endometrium of the non‐Lactobacillus‐dominated patients
The major taxonomies in the EF and vaginal specimens were Gardnerella, Streptococcus, Atopobium, Bifidobacterium, Sneathia, Prevotella, and Staphylococcus (Figure 5). In 28 (19 IVF, seven non‐IVF, two volunteers) of the women who were analyzed, there was a discordance in the pattern of the bacterial community that was detected in the endometrium, compared to that in the vagina (Figure 6).
Figure 5.
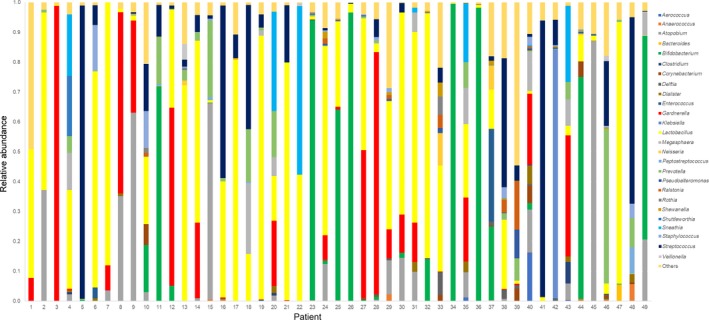
Relative proportion of the most abundant operational taxonomic units in the endometrium of the NLD IVF patients. The number below the graph shows each participant. IVF, in vitro fertilization; NLD, non‐Lactobacillus‐dominated
Figure 6.
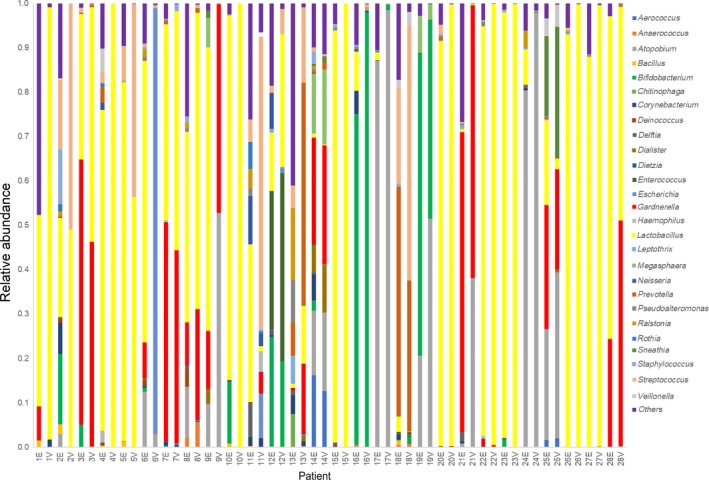
Patients with a discordance in the pattern of the endometrial or vaginal microbiome. E, endometrium; V, vagina
4. DISCUSSION
To the best of the authors’ knowledge, this study is the first of the endometrial and vaginal microbiota in infertile women and healthy female volunteers in Japan. This study found that the percentages of Lactobacilli in the endometrial microbiota of IVF patients, non‐IVF patients, and healthy volunteers were different. The Lactobacillus‐dominated microbiota (>90% Lactobacillus spp.) in the endometrium, compared to the vagina, was 38% (30/79) vs 44.3% (44/79) in the IVF patients, 73.9% (17/23) vs 73.9% (17/23) in the non‐IVF patients, and 85.7% (6/7) vs 85.7% (6/7) in the healthy volunteers. It was striking to find that 62% of the infertile patients who were undergoing IVF treatment had an abnormal endometrial bacterial status.10
Studies on the longitudinal dynamics of the vaginal microbiome in healthy reproductive‐aged women have shown that these communities remained relatively stable over 16 weeks.21 Although the number of cases was limited in this study, it was shown for the first time that the endometrial microbiome of the healthy women was highly stable, intercyclic and intracyclic.
The reason for the percentage of Lactobacilli being lower in the endometrium of the IVF patients, compared to that of the non‐IVF patients and healthy volunteers, could not be identified. However, it can be supposed that the reason might be from IVF procedures or the backgrounds of the IVF patients: a longer infertility period, seminal factor (male factor), frequent transvaginal examinations, past history of IUIs, frequent administration of antibiotics, hormonal fluctuations due to controlled ovarian stimulation, oocyte retrieval, embryo transfer, and the side‐effects of ovarian stimulation, such as ovarian hyperstimulation syndrome. The reason why the healthy volunteers showed a high percentage of LD endometrium might have been related to much less exposure to gynecological examinations or interventions. As the non‐IVF patients were significantly younger than the IVF patients (Table 1), the age factor might have influenced the outcome; however, in the subgroup analysis of the IVF patients (Table 3), there was no significant difference in the percentage of endometrial and vaginal Lactobacilli between those aged over 38 and those under 38 years old. Patient recruitment also might influence the outcome. In addition, a variation in the timing of sample collection (Table 1) might have influenced the microbiota, but considering the intercyclic and intracyclic stability of the endometrial and vaginal microbiome of the healthy volunteers (Figure 1), microbiomes might not differ largely by sampling timing. One study reported that the endometrial microbiota was highly stable during the acquisition of endometrial receptivity in the luteal phase.10
This study was different from the previous study,10 in terms of the ethnicity of the participants and the sampling timing. Also, an endometrial receptivity array was not performed simultaneously. In addition, the follow‐up period was too short to describe the reproductive outcomes of the LD and NLD patients, including the implantation, pregnancy, ongoing pregnancy, and live birth rates.
There are several limitations of the present study: a short follow‐up period and limited study number and not analyzing other aspects of gynecological histories, such as sexual contact, past oral contraceptive usage, miscarriage, and endometriosis. The pathological confirmation of chronic endometritis was not done routinely in this study. Also, the EF specimens might have contained some endocervix fluid, which was distinguished in one study,22 but the endometrial microbiota was not suspected to be carried over from the vaginal microbiota because there was a difference in the pattern of the bacterial community in the endometrium and vagina in 28 paired samples of the total number of women who were analyzed (Figure 6).
As reported previously,10 the major taxonomies in the EF and vaginal specimens in this study were Gardnerella, Streptococcus, Atopobium, Bifidobacterium, Sneathia, Prevotella, and Staphylococcus (Figure 5). One study reported that an adverse effect on pregnancy outcomes was more evident in patients who presented with high percentages of bacteria from the Gardnerella and Streptococcus genera and that the pregnancy outcome of IVF patients could be predicted by the relative abundance of Lactobacillus in the EF.10 In this preliminary study, the median percentage of endometrial Lactobacilli in 18 pregnant cases was 96.45% ± 33.61%, suggesting that a LD endometrium might favor implantation; meanwhile, there were seven NLD cases who conceived without intervention for endometrial microbiota. The follow‐up period in this study is too short to conclude which pathogen acts negatively on pregnancy outcomes. Meanwhile, there is a possibility that non‐Lactobacilli microbiotas are simply residents, not pathogens, of the endometrial cavity. Also, the mechanism of how the NLD microbial status affects the embryo's implantation is still not clear. It has been well known that a healthy vaginal status, which is associated with the presence of Lactobacillus spp.‐producing lactic acids and inducing the vaginal pH to ≤4.5, is suspected to prohibit the growth of pathogenic bacteria in healthy women,10, 23 but this might not be the case in the endometrium because there was no correlation between the pH levels in the EF and the endometrial microbiota10 and some kind of inflammatory response in the endometrium that was caused by the NLD microbiota could have affected the success of embryonic implantation.10 In addition, treatment for an endometrial NLD microbial status is still not established and remains an issue for the future. The administration of antibiotics only might not be helpful in creating a LD endometrium because the Lactobacilli also might be the target of some antibiotics; the concurrent administration of probiotic drugs containing Lactobacilli spp. would be expected to be one treatment option, as several studies using probiotics for treating bacterial vaginosis have been conducted.24, 25
Given the limitations of this pilot study, it is believed that for patients with unexplained RIF, there is a significance in searching for their endometrial microbial status, considering the percentage of abnormalities in the microbiome of the IVF patients, compared to the non‐IVF patients or healthy volunteers. Treatment for an endometrial NLD microbial status is still not established and remains an issue for the future, but by increasing the endometrial lactobacilli to >90% might favor the implantation outcome of the infertile patients with a NLD endometrial microbiota. By transferring euploid embryos in a personal window of implantation2 under the LD endometrium, much better pregnancy rates are expected.
DISCLOSURES
Conflict of interest: The authors declare no conflict of interest. Human rights statement and informed consent: This study was approved by the Institutional Review Board of Kyono ART Clinic and Kyono ART Clinic Takanawa, Japan. All the procedures were followed in accordance with the ethical standards of the responsible committees on human experimentation (institutional and national) and with the Helsinki Declaration of 1964 and its later amendments. Informed consent was obtained from all the patients to be included in the study. Animal studies: This article does not contain any study with animal participants that were performed by any of the authors.
ACKNOWLEDGEMENT
We thank the volunteers for participating in this study.
Kyono K, Hashimoto T, Nagai Y, Sakuraba Y. Analysis of endometrial microbiota by 16S ribosomal RNA gene sequencing among infertile patients: A single‐center pilot study. Reprod Med Biol. 2018;17:297–306. 10.1002/rmb2.12105
Kyono and Hashimoto contributed equally to this study.
REFERENCES
- 1. Simon A, Laufer N. Repeated implantation failure: clinical approach. Fertil Steril. 2012;97:1039‐1043. [DOI] [PubMed] [Google Scholar]
- 2. Ruiz‐Alonso M, Blesa D, Díaz‐Gimeno P, et al. The endometrial receptivity array for diagnosis and personalized embryo transfer as a treatment for patients with repeated implantation failure. Fertil Steril. 2013;100:818‐824. [DOI] [PubMed] [Google Scholar]
- 3. Kung A, Munne S, Bankowski B, Coates A, Wells D. Validation of next‐generation sequencing for comprehensive chromosome screening of embryos. Reprod Biomed Online. 2015;31:760‐769. [DOI] [PubMed] [Google Scholar]
- 4. Harton GL, Munne S, Surrey M, et al. Diminished effect of maternal age on implantation after preimplantation genetic diagnosis with array comparative genomic hybridization. Fertil Steril. 2013;100:1695‐1703. [DOI] [PubMed] [Google Scholar]
- 5. Fox C, Morin S, Jeong JW, Scott RT Jr, Lessey BA. Local and systemic factors and implantation: what is the evidence? Fertil Steril. 2016;105:873‐884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mitchell CM, Haick A, Nkwopara E, et al. Colonization of the upper genital tract by vaginal bacterial species in nonpregnant women. Am J Obstet Gynecol. 2015;212:611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Franasiak JM, Werner MD, Juneau CR, et al. Endometrial microbiome at the time of embryo transfer: next‐generation sequencing of the 16S ribosomal subunit. J Assist Reprod Genet. 2016;33:129‐136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen C, Song X, Wei W, et al. The microbiota continuum along the female reproductive tract and its relation to uterine‐related diseases. Nat Commun. 2017;8:875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive‐age women. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4680‐4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moreno I, Codoñer FM, Vilella F, et al. Evidence that the endometrial microbiota has an effect on implantation success or failure. Am J Obstet Gynecol. 2016;215:684‐703. [DOI] [PubMed] [Google Scholar]
- 11. Walters W, Hyde ER, Berg‐Lyons D, et al. Improved bacterial 16S rRNA gene (V4 and V4‐5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems. 2015;1:pii:e00009‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rintala A, Pietilä S, Munukka E, et al. Gut microbiota analysis results are highly dependent on the 16s RRNA gene target region, whereas the impact of DNA extraction is minor. J Biomol Tech. 2017;28:19‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aronesty E. Comparison of sequencing utility programs. Open Bioinforma J. 2013;7:1‐8. [Google Scholar]
- 14. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;272:2194‐2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high‐throughput community sequencing data. Nat Methods. 2010;7:335‐336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261‐5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McDonald D, Price MN, Goodrich J, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610‐618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence‐based microbiome analyses. BMC Biol. 2014;12:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Segal LN, Alekseyenko AV, Clemente JC, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 2013;1:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Urushiyama D, Suda W, Ohnishi E, et al. Microbiome profile of the amniotic fluid as a predictive biomarker of perinatal outcome. Sci Rep. 2017;7:12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gajer P, Brotman RM, Bai G, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med. 2012;4:132ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wee BA, Thomas M, Sweeney EL, et al. A retrospective pilot study to determine whether the reproductive tract microbiota differs between women with a history of infertility and fertile women. Aust N Z J Obstet Gynaecol. 2017; Dec 26. 10.1111/ajo.12754. [DOI] [PubMed] [Google Scholar]
- 23. Skarin A, Sylwan J. Vaginal lactobacilli inhibiting growth of Gardnerella vaginalis, Mobiluncus and other bacterial species cultured from vaginal content of women with bacterial vaginosis. Acta Pathol Microbiol Immunol Scand B. 1986;94:399‐403. [DOI] [PubMed] [Google Scholar]
- 24. LACTIN‐V study for recurrent bacterial vaginosis. ClinicalTrials.gov Web site. https://clinicaltrials.gov/ct2/show/NCT02766023; Published May 9, 2016. Updated October 13, 2017. Accessed March 22, 2018.
- 25. Fertility clinics treat abnormal vaginal bacteria aiming to improve reproductive outcome. Medical Xpress Web site. https://medicalxpress.com/news/2017-11-fertility-clinics-abnormal-vaginal-bacteria.html; Published November 27, 2017. Accessed March 22, 2018.


