Abstract
Objectives
To compare and describe type-specific characteristics of HPV16, HPV18 and HPV45 in cervical cancer with respect to 3′LCR methylation and disruption of E1/E2.
Methods
The methylation level of 137 cervical cancer samples (70 with HPV16, 37 with HPV18, and 30 with HPV45) of Brazilian patients was analyzed by pyrosequencing. PCR amplifications were performed to characterize E1 and E2 disruption as an episomal surrogate.
Results
The 3′LCR of HPV16 showed a higher methylation at all CpG sites (7%, 9%, 11%, 10% and 10%) than homologous HPV18 regions (4%, 5%. 6%, 9% and 5%) and HPV45 regions (7%, 7% and 5%). Presence of intact E1/E2 was associated with higher HPV16 and HPV18 methylation levels at all CpG sites (p < 0.05). Disruption of E1/E2 was more frequently found in HPV45 (97%) and HPV18 (84%) than in HPV16 DNA (30%). HPV16 disruption was more frequently found in E1 (48%) unlike HPV18, where it was found in E2 (61%). Concomitant disruption of E1/E2 was most frequent in HPV45 (72%).
Conclusions
The findings showed a higher methylation associated with intact E1/E2 for HPV16 and HPV18. The closely phylogenetic related HPV18 and HPV45 share a similar methylation level and the frequency of viral genome disruption.
Keywords: Human papillomavirus, Invasive cervical cancer, Methylation, Pyrosequencing, HPV genome, Viral genome integration
1. Introduction
The biology of human papillomaviruses (HPV) has been extensively described in the literature [1] in view of its association with specific cancer types, mainly cervical cancer (CC). Twelve mucosal genotypes (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58 and 59) have been characterized as high-risk HPV (HR-HPV) for cancer development [2] consequently to their high prevalence in CC and to the molecular evidence associated to carcinogenesis.
The HPV genome is a circular double-stranded DNA of approximately 8 Kilo-base pairs (kb) contained in an icosahedral capsid comprising three regions: (i) the E region, with genes coding for proteins predominantly expressed in early stages of infection (E1, E2, E4, E5, E6 and E7) and associated with viral DNA replication, regulation of HPV gene expression, control of cell cycle and oncogenesis, accounting for 4 kb of the viral genome; (ii) the L region, with genes encoding structural capsid proteins (L1 and L2) expressed in later stages of infection, comprising ~3Kb of the viral genome, and (iii) the Long Control Region (LCR), a non-coding region of approximately 1 kb which controls viral replication and transcription through DNA motifs recognized by DNA-binding proteins [3].
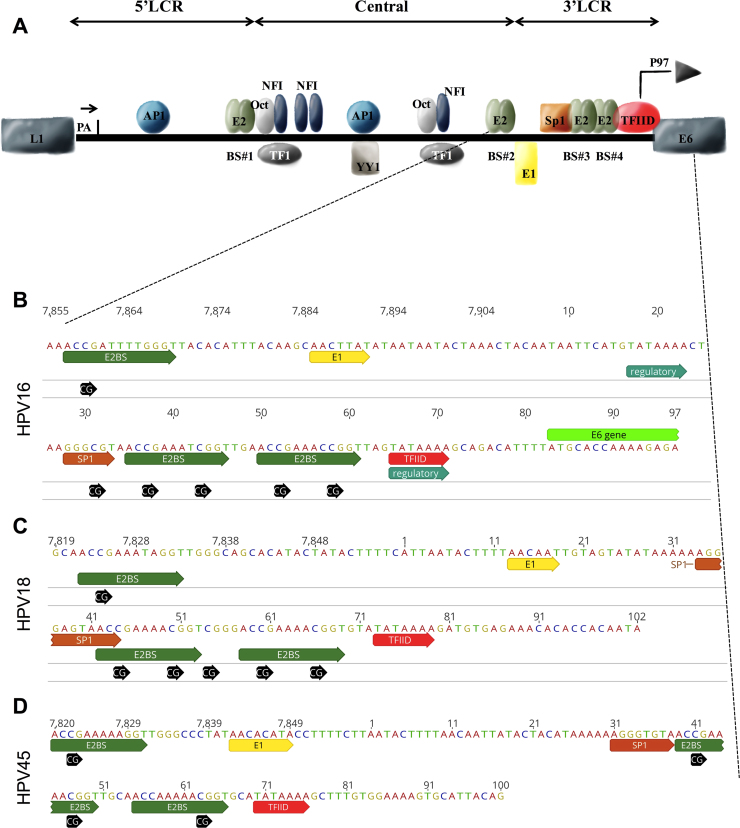
The LCR consists of three functionally separate segments, the 5′ segment (5′LCR), the central (or enhancer) and the 3′ segment (3′LCR or early promoter) (Fig. 1A) [3]. The 5′LCR is approximately 300 bp long and is located between the terminal L1 codon and the E2 protein binding site (named E2BS#1). This segment contains a nuclear matrix attachment region, a transcription termination region and polyadenylation sites [4]. The central, or enhancer segment, of approximately 400 bp, is flanked by two E2 binding sites (E2BS#1 and E2BS#2). Several cell transcription factors (i.e.: AP-1, NFI, YY1, Oct-1, TF-1, TEF-2, glucocorticoid and progesterone receptor) are capable of binding to this segment and transactivate HPV gene expression [5], [6]. The 3′LCR, of approximately 140 bp long and flanked by E2BS#2 and E6, is capable of controlling the expression of E6 and E7 viral oncogenes. This promoter has been well characterized in HPV16 and HPV18 (named P97 and P105, respectively), and is essential for immortalization of human keratinocytes by triggering massive production of E6 and E7 oncoproteins following HR-HPV infection. Moreover, the 3′LCR contains the origin of viral replication (overlapping with the E1 binding site), one Sp1 binding site, two E2 binding sites (E2BS#3 e E2BS#4), and one TATA box.
Fig. 1.
Schematic representation of HPV LCR. A, represents the three segments of HPV16 LCR (5′LCR, central or enhancer, and 3′LCR), considered a model of LCR for all HPVs. The 5′LCR contains the transcription termination signal, denoted ‘pA’. The central segment contains the majority of transcription factor binding sites. The 3′LCR contains the origin of replication and the E6/E7 promoter. B, C and D, represent the nucleotide sequence of 3′LCR of HPV16, HPV18 and HPV45, respectively, highlighting the binding motifs of E2, E1, Sp1, TFIID and all CpG sites within this segment. The Sp1 motif of HPV16 contains one CpG site in its core (B, GGGCGT). Differently no CpG is found in the Sp1 motif of HPV18 (C, GGGAGT) and HPV45 (D, GGGTGT). The E2BS#4 of HPV45 (D) has only one CpG site (nt 63), while HPV16 (B) and HPV18 (C) contain two CpG sites. (For information on motifs and reference genome data see references [3], [40], [47] and Papillomavirus Episteme database [26], respectively). All transcription factor binding sites are denoted by the abbreviation used in the text except for TEF-1 that is herein denoted as TF1.
The mechanisms of carcinogenesis induced by HR-HPV involve inhibition of p53 and pRB cell proteins by E6 and E7 oncoproteins, respectively [1], [7], [8], consequently to which, a severe chromosome instability is generated, favoring HPV DNA disruption and integration into the host genome. Generally, integration leads to increased expression and stability of E6 and E7 transcripts because disruption of the viral genome occurs either at or upstream of E2, inactivating this gene that encodes a dose-dependent transcriptional repressor of the early promoter in 3′LCR [9], [10], [11], [12], [13]. E2 activity is associated with displacement of the Sp1 and TFIID promoter activators from their respective binding sites. Thus, the absence or low level of E2 proteins results in overexpression of the E6 and E7 oncogenes and cancer progression.
Furthermore, it has been suggested that methylation at CpG sites in the 3′LCR of HPV16 in CC could trigger cancer development by modulating E2 protein activity when episomal HPV DNA is present [14]. Methylation at this segment has shown to be complex, with conflicting findings among CC samples; most available data being restricted to the HPV16 genotype [15], [16]. Moreover, clinical factors have also been found to affect methylation levels [17].
The specificity of HPV types in the etiology of cervical cancer shows a phylogenetic imprinting, with some members of the Alpha-papillomavirus genus associated with all cervical cancers [18]. Within this genus, however, specific species (α 1–11, 13 and 15) and even HPV genotypes show different pathogenicity, with HPV16 (belonging to the α 9 species) accounting for more than 60% of all CC worldwide [19]. Nevertheless, distinct biological characteristics between HPV genotypes have been identified among CC samples. Presence of viral DNA in the episomal state has been observed in approximately 40% of HPV16 + tumors, while higher frequency of episomal DNA has been detected in tumors associated with HPV52 and HPV58 infection (75% and 88% respectively) [20], [21]. Differently, most cervical cancers harboring HPV18 contained an integrated viral genome [21], [22]. Moreover, in addition to their frequency of integration, HPV16 differs from HPV18 with respect to CC progression. Reviews and meta-analyses have reported a lower frequency of HPV18 in pre-neoplastic lesions than in CC, suggesting a more rapid progression of HPV18 lesions [23], [24], [25]. Moreover, HPV18 + or HPV45 + CC (two different α7 species types) showed a higher proportion of adenocarcinomas than HPV16 + CC [23], [24], [25]. An additional difference between HPV genotypes accounts for the number of CpG sites along their genome, with 111 CpG sites in HPV16 vs 168 in HPV18 [26].
In this paper, we compare and describe type-specific characteristics of HPV16, HPV18 and HPV45 in CC samples with respect to 3′LCR methylation and disruption of E1/E2 open reading frames (ORFs). Moreover, we intent to verify whether methylation was associated with E1/E2 disruption in HPV18 and HPV45, as previously reported in HPV16 [17].
2. Material and methods
2.1. Samples
The study material consisted of 137 CC samples selected from a previously reported pool of 590 biopsies of invasive cervical cancer [27], 70 associated with HPV16, 37 with HPV18 and 30 with HPV45. Samples were collected at diagnosis from patients treated at the Instituto Nacional de Câncer (INCA - Rio de Janeiro, Brazil) referred for treatment between June 2011 and March 2014. The set of HPV16 + and HPV18 + samples was selected as previously reported [17] in according to their histopathological presentation while all HPV45 + tumors were included in view of their lower number in the pool with respect to HPV16 + and HPV18 + samples (Table 1). DNA isolation and HPV identification was as previously described [27]. Genomic DNA from HeLa, CaSki and SiHa cell lines was used as control for bisulfite treatment and pyrosequencing.
Table 1.
Characteristics of study group by HPV type.
| Characteristics (N) | Total | HPV16 | HPV18 | HPV45 |
|---|---|---|---|---|
| Overall population (N = 137) | 137 | 70 | 37 | 30 |
| Patient Age (yo) | ||||
| Mean; SD | 46; 13 | 45; 13 | 47; 13 | 49; 12 |
| Median | 45 | 43 | 45 | 47 |
| Tumor types | ||||
| ADN | 39 | 23 | 10 | 6 |
| SCC | 96 | 47 | 25 | 24 |
| No information | 2 | – | 2 | – |
| FIGO Stage | ||||
| I | 31 | 23 | 4 | 4 |
| II | 54 | 23 | 15 | 16 |
| III | 47 | 21 | 17 | 9 |
| IV | 5 | 3 | 1 | 1 |
| No information | – | – | – | – |
| Tumor Grade | ||||
| G1 | 12 | 6 | 3 | 3 |
| G2 | 81 | 45 | 18 | 18 |
| G3 | 25 | 11 | 10 | 4 |
| No information | 19 | 8 | 6 | 5 |
Note. ADC: adenocarcinoma, SCC: squamous cell carcinoma. N = number of samples. Yo: years old. FIGO: International Federation of Gynecology and Obstetrics.
All procedures were approved by the Ethics Committee of Instituto Nacional de Câncer (protocol CAAE 53398416.0.0000.5274). All patients signed an informed consent and filled an epidemiological questionnaire.
2.2. Sodium bisulfite treatment and PCR amplification
Sodium bisulfite treatment was carried out with EpiTect Bisulfite Kit (cat. no. 59104, Qiagen, Germany), with an input of 300–1500 ng of DNA and a final elution volume of 40 µL. Following treatment, regions covering 178 bp, 245 bp and 149 bp of the 3′LCR of HPV16, HPV18 and HPV 45, respectively, were PCR-amplified (Table 2). PCR was carried out in 30 µL mixtures containing 0.2 mM of each dNTP, 6 pmol of each primer, 1 U of Platinum Taq DNA Polymerase (Life Technologies, California, USA) and 1 × PCR buffer (67 mM Tris pH 8.8, 6.7 mM MgSO4, 16.6 mM (NH4)2SO4, and 10 mM 2-mercaptoethanol) [28]. PCR conditions consisted of 95°C for 6 min followed by 45 cycles of 95°C for 1 min, annealing (temperatures listed in Table 2) for 1 min, 72°C for 1 min, and one final extension step at 72°C for 5 min. Presence of amplified products was verified in 2% ultrapure agarose gels (Life Technologies, California, USA).
Table 2.
Primers for methylation analysis.
| Assay | Primers (5'-3') | CpG (nt position) | Amplicon size | T.A. |
|---|---|---|---|---|
| HPV16_3LCR* | forward 3'LCR: TTGTAAAATTGTATATGGGTGTG | 31, 37, 43, 52, 58 | 178 bp | 61°C |
| reverse 3'LCR (biotin): AAATCCTAAAACATTACAATTCTC | ||||
| sequencing (forward) 3'LCR: AATTTATGTATAAAATTAAGGG | ||||
| HPV18_3LCR | forward: ATTTTTAATATGAATTATAATATGATTAAG | 44, 50, 54, 60, 66 | 245 bp | 57°C |
| reverse (biotin): CACAAATCAAATAACTTATAAAATC | ||||
| sequencing (forward): GTAGTATATAAAAAAGGGAGTAA | ||||
| HPV45_3LCR | forward (biotin): GTTTATGTAATAGAAAAAGGTTGGGTTTTA | 41, 47, 63 | 149 bp | 62°C |
| reverse: CCTATAATACACTTTTCCACAAAACTT | ||||
| sequencing (reverse): ACTTTTCCACAAAACTTT |
2.3. Quantitation of DNA methylation by pyrosequencing
PCR products were submitted to pyrosequencing in a PyroMark Q24 platform (Qiagen, Hilden, Germany) following a standard protocol [17]. Briefly, streptavidin beads, PyroMark binding buffer (Qiagen, Hilden, Germany) and PCR products were mixed and incubated on a shaking platform. A Biotage Q24 Vacuum Prep Workstation was used for separating, denaturing and washing PCR products, which were subsequently added to a microtiter plate containing annealing buffer with sequencing primers (see Table 2). Primer annealing was carried out by incubation at 80⁰C for 2 min and cooling to room temperature before pyrosequencing. PyroGold reagents, including enzyme, substrate and nucleotides were used for the pyrosequencing reaction. Pyrograms were generated and analyzed with PyroMark Q24, v.2.0.6 (Qiagen, Hilden, Germany). Controls for hyper-methylation (with CaSki cells) and hypo-methylation (with SiHa and HeLa cells) were used as previously reported [29], [30].
2.4. E1 and E2 disruption
HPV integration into the host DNA genome frequently occurs within E1 or E2 disruption [31] resulting in suppression of E2 transcription. The presence of intact E1/E2 was identified by PCR amplification of overlapping fragments encompassing the E1 and E2 coding regions of HPV16, HPV18 and HPV45 using primers pairs listed in Table 3, with some of them previously published [32], [33]. PCR was carried out as described [17] with different annealing temperatures shown in Table 3. Negative reactions were repeated to confirm lack of amplification of target regions.
Table 3.
Primers for analysis of E1 and E2 gene disruption.
| Assay | Primers (5'-3') | TA | Nucleotide position | Amplicon size (bp) | ||
|---|---|---|---|---|---|---|
| HPV16 | E1a | forward: | CCATGGCTGATCCTGCAG | 61 °C | 863–1219 | 356 bp |
| reverse: | TCTCCTTTTTGCAGCTCT | |||||
| E1b | forward: | GACAGCGGGTATGGCAAT | 65 °C | 1254–1663 | 409 bp | |
| reverse: | CATTCCCCATGAACATGC | |||||
| E1c | forward: | AATAAATCAACGTGTTGCGATTGG | 65 °C | 1548–2084 | 536 bp | |
| reverse: | GTTTATAATGTCTACACATTGTTG | |||||
| E1d | forward: | GGATTGTGCAACAATGTG | 65 °C | 2072–2527 | 455 bp | |
| reverse: | TGGAGGGCATTTTAGTTG | |||||
| E1e | forward: | CAACTAAAATGCCCTCCA | 61 °C | 2529–2845 | 316 bp | |
| reverse: | CGCATGTGTTTCCAATAG | |||||
| E2a | forward: | CGAGGACAAGGAAAACGA | 65 °C | 2738–3189 | 451 bp | |
| reverse: | CTTGACCCTCTACCACAG | |||||
| E2b | forward: | GGTTTATATTATGTTCATGAAGG | 56 °C | 3220–3599 | 379 bp | |
| reverse: | TATGGGTGTAGTGTTACTATTACA | |||||
| E2c | forward: | GTAATAGTAACACTACACCCATA | 56 °C | 3596–3853 | 257 bp | |
| reverse: | GGATGCAGTATCAAGATTTG | |||||
| HPV18 | E1P1 | forward: | GGTGTGCATCCCAGCAGTAA | 59 °C | 888–1403 | 515 bp |
| reverse: | GCCGCCACTACATACATTGC | |||||
| E1P2 | forward: | GCGGCAATGTATGTAGTGGC | 59 °C | 1400–1908 | 508 bp | |
| reverse: | GCTGCAACACTACTTCGCAA | |||||
| E1P3 | forward: | TCAACCACCAAAATTGCGAAGT | 59 °C | 1877–2211 | 334 bp | |
| reverse: | TCGTTTTTGGGCTCGCCTAT | |||||
| E1P4 | forward: | GCAAACATTATAGGCGAGCCC | 59 °C | 2181–2546 | 365 bp | |
| reverse: | TGTCCAACACGTGGTCGTT | |||||
| E1P5 | forward: | GGTGGCCATGTTAGATGATGC | 59 °C | 2506–2895 | 389 bp | |
| reverse: | GATTTTGTCCTGCAACGCACT | |||||
| E2P1 | forward: | TCCAGATTAGATTTGCACGA | 61 °C | 2786–3192 | 407 bp | |
| reverse: | CAATTGTCTTTGTTGCCATC | |||||
| E2P2 | forward: | ATACAAAACCGAGGATTGGA | 61 °C | 3086–3388 | 303 bp | |
| reverse: | ACTTCCCACGTACCTGTGTT | |||||
| E2P3 | forward: | AACACAGGTACGTGGGAAGT | 61 °C | 3369–3739 | 371 bp | |
| reverse: | TTTCGCAATCTGTACCGTAA | |||||
| E2P4 | forward: | GACCTGTCAACCCACTTCT | 61 °C | 3598–3994 | 397 bp | |
| reverse: | ACATGGCAGCACACATACAT | |||||
| HPV45 | E1a | forward: | GGTGTAATGGCTGGTTCTTTGT | 55 °C | 881–1139 | 259 bp |
| reverse: | AATGGACTGTTTTCCTTGCTGC | |||||
| E1b | forward: | CAGTCCATTAGGGGAGCAGC | 57.5 °C | 1131–1775 | 645 bp | |
| reverse: | GCTGCAACACTACTTCGCAA | |||||
| E1c | forward: | AGCACATTGTTGCACGTACC | 57.5 °C | 1705–2144 | 440 bp | |
| reverse: | GGTCTCCAATCCCCACCTTC | |||||
| E1d | forward: | AAGGTGGGGATTGGAGACCC | 62 °C | 2126–2727 | 602 bp | |
| reverse: | AGGGATTCCTTCGGTGTCTG | |||||
| E1e | forward: | TTTGCACGAGGACGATGAAGA | 55 °C | 2685–2890 | 206 bp | |
| reverse: | CACCTGGTGGTTTAGTTTGGTAA | |||||
| E2a | forward: | GGACATGGTCCAGATTAGATTTGC | 55 °C | 2666–3068 | 403 bp | |
| reverse: | GCACGGTTTTACCGCCTTTT | |||||
| E2b | forward: | TACAGAACCGTCGCAGTGTT | 62 °C | 3025–3431 | 407 bp | |
| reverse: | TCTGGATGTGGGGTTTTGGG | |||||
| E2c | forward: | AGACAGCTACAACACGCCTC | 62 °C | 3359–3893 | 535 bp | |
| reverse: | TGCAGCACACATAAAGGCAC | |||||
Note. Bp, base pairs; primers covering E1 and E2 of HPV16 were designed by Vermont et al. (2007), and primers of E2 of HPV18 by Collins Constandinou-Williams et al. (2009).
2.5. Statistical analysis
The level of methylation at each CpG site per sample was estimated as the proportion of methylated cytosines, ranging from 0 (without methylation) to 100% (totally methylated). Comparisons of methylation levels were carried out with the Man-Whitney test between (i) homologous CpG sites (sites binding similar proteins) and (ii) individual CpG sites with and without E1/E2 disruption. All statistical analyzes and graphs were conducted with GraphPad Prism 7.
3. Results
3.1. Characteristics of study group
The clinical and pathology profiles of the 137 patients herein studied are summarized in Table 1. Age at diagnosis ranged from 19 to 80 years, with a mean of 46 (SD ± 13) and a median of 45. A higher proportion of squamous cell carcinomas than adenocarcinomas (approximately 2:1) were selected among HPV16 + and HPV18 + tumor samples while, among HPV45 + tumors, the proportion of SCC was higher (4:1) due to the limited number of available samples. In 77% of all tumors, FIGO staging was equal or above IIA, with 90% tumor grade G2 or G3 (Table 1).
3.2. Methylation at the 3′LCR of HPV DNA
Following bisulfite treatment, the methylation level of each CpG site per sample was estimated by pyrosequencing in the 3′LCR of HPV16 (nt 31, 37, 43, 52 and 58; GenBank: K02718.1), HPV18 (nt 44, 50, 54, 60 and 66; GenBank: X05015.1) and HPV45 (nt 41, 47 and 63; GenBank: X774479.1) (Fig. 1B to 1 D, respectively). Two samples (one HPV16 + and one HPV45 +) were excluded from analysis due to low quality of pyrosequencing data. The methylation levels of CpG sites per sample are listed in Supplemental Table 1.
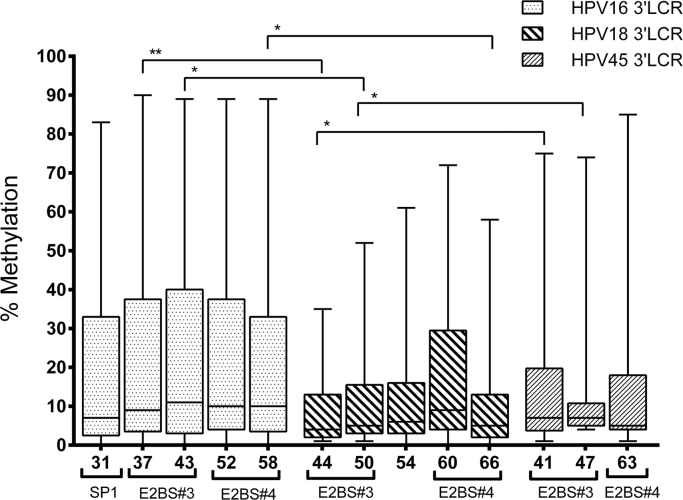
The CpG sites in the 3′LCR of HPV16 showed a higher median methylation (7%, 9%, 11%, 10% and 10%) than the homologous segment of HPV18 (4%, 5%, 6%, 9% and 5%) and HPV45 (7%, 7% and 5%). Moreover, HPV16 showed a wider range of methylation per sample at E2 binding sites (0–90%, in nt 37) than HPV18 (1–72%, in nt 60) and HPV45 (1–85%, in nt 63) (Fig. 2).
Fig. 2.
Methylation status of homologous CpG sites in the 3′LCR of HPV16, HPV18 and HPV45. The methylation level of each CpG site is represented by a box displaying upper and lower quartiles separated by the median line, and whisker plots. HPV16 displayed a higher level of methylation than HPV18 and HPV45, mainly at E2 binding sites (E2BS#3 and #4). This was statistically different when comparing the homologous CpG sites of HPV16 vs HPV18 (CpG 37 vs 44, p = 0.0018; CpG 43 vs 50, p = 0.0490; and 58 vs 66, p = 0.0109), and HPV18 vs HPV45 (CpG 44 vs CpG41). Analyses were performed with the Mann-Whitney U test.
Homologous CpG sites in E2 binding site motifs of each HPV genotype comprised: (i) nt 37, 44 and 41 of HPV16, HPV18 and HPV45, respectively, at the 5′ end of E2BS#3; (ii) nt 43, 50 and 47 of HPV16, HPV18 and HPV45, respectively, at the 3′ end of E2BS#3; (iii) nt 52 and 60 of HPV16 and HPV18, respectively, at the 5′ end of E2BS#4; and (iv) nt 58, 66 and 63 of HPV16, HPV18 and HPV45, respectively, at the 3′ end of E2BS#4 (Fig. 2). Significant differences were found in the methylation level of E2BS#3 CpG sites between HPV16 and HPV18 (p = 0.0018 and p = 0.0490), and in one E2BS#4 CpG site between HPV16 and HPV18 (p = 0.0109) (Fig. 2). Significant differences were also found between E2BS#3 CpG sites of HPV45 and HPV18 (p = 0.0029 and p = 0.0039) (Fig. 2).
A hypermethylation pattern was found at CpG sites in the 3′LCR of HPV16 in CaSki cells (90%, 86%, 97%, 92%, 98%) as well as a hypomethylation pattern in SiHa cells (2%, 3%, 2%, 4% and 5%). HPV18 + HeLa cells showed a pattern of hypomethylation (1%, 1%, 1%, 2% and 1%).
3.3. E1 and E2 gene integrity
PCR amplification covering E1 and E2 was used for detecting HPV disruption, a finding suggesting HPV integration into the host genome and lack of E2 expression. Disrupted E1 and E2 were found in 30% (21/69) of HPV16 + samples, 84% (31/37) of HPV18 + samples, and 97% (28/29) of HPV45 + samples.
In HPV16, the most frequent disruption occurred between nt 2529–3189 (67% - 14/21) encompassing the 3′ end of E1 and the 5′ end of E2 (HPV16REF; K02718.1). Forty-eight percent (10/21) of disruptions were exclusively found in E1 and 14% (3/21) in E2. Disruption in both E1 and E2 were found in 38% (8/21) of samples (Fig. 3A). In HPV18, the most frequent disruption occurred between nt 3369 and 3739 (90% − 28/31), a region inside E2 and overlapping the E4 ORF; nt 3418–3684 (HPV18REF; X05015.1) (Fig. 3B). E2 was more frequently disrupted (61% - 19/31), followed by disruption of both genes (35% − 11/31), and a single disruption in E1 (3% - 1/31) (Fig. 3B). In HPV45, the concomitant E1 and E2 disruption was the most frequent pattern (72% - 20/28) resulting in lack of amplification of a large extension of these genes; 46% of samples (13/28) showing loss of more than 1.7 Kb (between nt 2126 and 3893) (HPV45REF; GenBank accession number X74479.1) (Fig. 3C). Similarly, to HPV18, the most frequently missing amplicon (E2c) overlapped the E4 ORF (nt 3359–3893) (HPV45REF; X74479.1). Additionally, in 14% (4/28) of samples, losses were exclusively found in E1 and E2.
Fig. 3.
Map of disruptions sites inE1andE2of the 3′LCR of HPV per sample. A, E1 and E2 of HPV16 predominately showed intact genes (light orange color - I), with high disruption of E1. B, integrity of HPV18 E1 and E2, with most samples displaying disrupted DNA, predominately of E2. C displays E1 and E2 integrity of HPV45 showing significant loss of both genes and larger deletions. Black rectangles represent lack of PCR amplification; light orange indicate presence of amplification. D1/2 indicates E1 and E2 disruption; D1: exclusive E1 disruption; D2: exclusive E2 disruption; I: intact E1 and E2. E1a, E1b, E1c, E1d and E1e represent amplicons covering the E1 gene. E2a, E2b, E2c and E2d represent amplicons covering the E2 gene. The genome position of each amplification is described in Table 2.
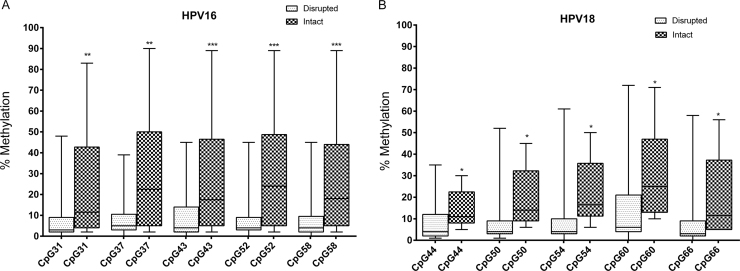
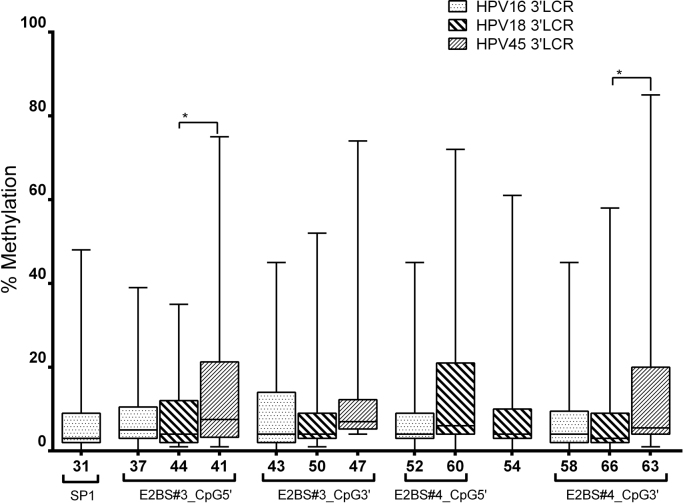
HPV16 + and HPV18 + samples with intact E1 and/or E2 showed higher median methylation levels in all 3′LCR sites (p < 0.05; Mann-Whitney U Test) than samples with disruptions (Fig. 4A and B). This analysis could not be performed with HPV45 + samples because only a single sample lacking E1/E2 disruption was detected. A comparison of the methylation level between homologous CpG sites in samples with E1/E2 disruption showed a higher methylation at E2BS#3 (nt 41) and E2BS#4 (nt 63) in HPV45 than in HPV18 (nt 44 and 66, respectively; p = 0.0163 and p = 0.0246, respectively; Supplemental Fig. 1).
Fig. 4.
3′LCR of HPV16 and HPV18 respective to E1 and E2 integrity. The methylation level of each CpG site is represented by a box displaying upper and lower quartiles separated by the median line, and whisker plots. A higher methylation at all CpG sites was found in samples with intact E1 and E2 than with disrupted genes (*p < 0.05; **p < 0.005 and ***p < 0.0005, Mann-Whitney Test).
4. Discussion
The present study compared HPV16, HPV18, and HPV45 (three HPV associated with high risk for cervical cancer development) with respect to 3′LCR methylation and E1/E2 integrity, a comparison not carried out to present. Methylation at CpG sites in the LCR of HPV16 has been extensively studied [34] and, in high-grade lesions and cervical tumors, a lower methylation level has been found in the LCR than in L1 or L2 [35], [36]. Other studies have pointed a wide range of methylation in the 3′LCR of HPV16 in clinical samples of CC [37], [38], suggesting a different regulation of HPV expression and a likely application for prognosis [38], [39]. Moreover, our previous study [17] suggested that patient aging may also contribute, independently, to increase methylation in the 3′LCR of HPV16, pointing to the complexity of HPV DNA methylation.
High methylation in the 3′LCR of HPV16 in cervical tumors has been associated with presence of episomal DNA [14] resulting in loss of negative control of transcription of E6 and E7 oncogenes by the E2 protein [40], [41]. To present, there is no evidence that this association (methylation vs integration) might occur in other HPV genotypes. The increased methylation level at the 3′LCR of HPV16 + and HPV18 + samples were herein associated with presence of intact E1/E2 genes (Fig. 4A and B), a finding suggestive for the presence of episomal viral DNA in these tumors. This association could not be assessed in HPV45 + samples due to their small number with intact E1/E2.
Furthermore, we observed a higher methylation level of HPV16 than in HPV18 and HPV45, particularly at CpG sites in E2BS#3 (Fig. 2), a finding that might be associated to different patterns of DNA disruption. A higher frequency (70%) of intact E1/E2 was observed in HPV16 + samples than in HPV18 + and HPV45 + samples (16% and 3%, respectively), where only few tumors showed intact E1/E2. The similar methylation pattern and E1/E2 disruption frequency of HPV18 and HPV45 might be associated with their close phylogenetic relationship; both belonging to species α 7, differently from HPV16 that belongs to species α 9 [42].
Differences in E1/E2 disruption patterns were also observed because E1 disruption was very frequent in HPV16 while HPV18 disruption occurred most frequently in E2. In HPV45, a different pattern was observed, with concomitant disruption frequently affecting both genes and with deletions of large viral genomic segments suggesting total loss of E1 and E2. A higher frequency of E2 disruption has been observed in HPV18 than in HPV16, although E1 disruptions were not assessed [32]. Differently, a high frequency of E1/E2 deletions have been reported in HPV16 + CC samples, with low-grade cervical intraepithelial neoplasia associated to exclusive E1 deletions [43]. In this study, we did not observe any association between disruption patterns and cancer staging (Supplemental Table 2).
The E4 ORF overlapping with the E2 ORF was the most frequently disrupted region in HPV18 and HPV45 DNA which also contains the highest numbers of CpG sites (N = 19 and N = 18, respectively). These findings might indicate that the methylation patterns of these CpG sites during early events of infection might affect the instability of HPV DNA.
Our findings highlight the importance of considering the frequency of specific disruptions when evaluating integration status. It is important to highlight that assessment of E1/E2 integrity by PCR has some limitations. Lack of E1/E2 amplicons confirms samples with disrupted E1/E2 viral genes, suggesting integration into host DNA in single copy HPV integration events. On the other hand, presence of intact E1/E2 did not exclude integration of multiple viral genomes (in tandem or multiple independent integrations) or the presence of concomitant forms (episomal and integrated). This approach, however, includes tumor samples in two groups: (i) unable to translate E2 (with only disrupted E1/E2 copies), and (ii) capable of translating E2, negatively regulating LCR promoter activity (intact E1/E2).
Different methods for detecting HPV DNA integration have been used; among which the most frequently ones were based on the integrity of a small E2 region by Real-Time PCR, usually estimated by the ratio between E2: E6 or E2: E7 copy number [44], [45]. More recently, an alternative approach carried out by HPV DNA capture with specific probes and NGS (next-generation sequencing) allowed a more precise identification of integration sites through detection of chimeric DNA (HPV DNA + host DNA). The few studies that used this approach reported a large diversity of host and HPV DNA breakpoints [22], [46] but, at present, it is still unclear whether breakpoints identified by NGS were relevant for cervical carcinogenesis.
The role of CpG methylation at upstream LCR E2 binding sites (encompassing the CpG sites (nt) 7453, 7459 and 7860 in HPV16; accession number AF125673) is still unclear, but there is evidence that CpG methylation might also affect progression from pre-cancer lesions to invasive cancer. In the proposed model for HPV16 [12], E2 binds with higher affinity to the upstream E2 binding sites (E2BS1 and E2BS2), activating transcription of p97 [12]. Additionally, cell lines transfected with plasmid constructs containing full LCR and methylated CpGs at E2BS1 (nt 7453 and 7459) presented a higher p97 promoter activity than with unmethylated constructs [48]. On the other side, significant differences in methylation level in invasive cancer were restricted to E2BS3 and E2BS4 (the targets of this work) between samples with episomal vs. integrated HPV16 viral genome, but not for the upstream E2BS1 and E2BS2 [14].
5. Conclusion
Our study showed a similar association between the methylation pattern at 3′LCR of HPV16 and HPV18 with the disruption of viral genome at E1/E2, reinforcing the importance of DNA methylation for E2 function. Additionally, we showed differences in the frequency of E1/E2 disruptions among the three HPV types herein studied, with the closely related HPV18 and HPV45 sharing a higher frequency of E1/E2 disruptions and the methylation level of the 3′LCR.
Acknowledgements
We greatly appreciate the excellent English revision assistance of Lisa Marie Zavesky and Dr. Hector N. Seuanez, and the computer graphic assistance in Fig. 1A of Caio Sant’anna Marinho.
Acknowledgments
Research funding
This study was financially supported by National Council for Scientific and Technological Development (CNPq) [grant numbers: 305873/2014-8; 573806/2008-0]; Carlos Chagas Filho Research Support Foundation (FAPERJ) [grant number: E26/170.026/2008]; The Ministry of Health and Brazilian National Cancer Institute (INCA).
Conflict of interest
None.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.pvr.2018.04.002.
Contributor Information
Sérgio Menezes Amaro-Filho, Email: sergioafilho@gmail.com.
Cláudia Bessa Pereira Chaves, Email: claudia.bessa67@gmail.com.
Shayany Pinto Felix, Email: shayany_shay@yahoo.com.br.
Diogo Lisbôa Basto, Email: diogolb89@gmail.com.
Liz Maria de Almeida, Email: lalmeida@inca.gov.br.
Miguel Angelo Martins Moreira, Email: miguelm@inca.gov.br, miguelm@inca.gov.
Appendix A. Supplementary material
Fig. S1.
Comparison of the methylation between homologous CpG sites at the 3′LCR of HPV in disrupted samples. A higher methylation in E2BS#3 and E2BS#4 of HPV45 (nt 41 and 63) was observed than in HPV18 (nt 44 and 66, respectively); p = 0.0163 and p = 0.0246, respectively. Homologous CpG sites in E2 binding site motifs of each HPV genotype comprised: (i) nt 37, 44 and 41 of HPV16, HPV18 and HPV45, respectively, at the 5′ end of E2BS#3; (ii) nt 43, 50 and 47 of HPV16, HPV18 and HPV45, respectively, at the 3′ end of E2BS#3; (iii) nt 52 and 60 of HPV16 and HPV18, respectively, at the 5′ end of E2BS#4; and (iv) nt 58, 66 and 63 of HPV16, HPV18 and HPV45, respectively, at the 3′ end of E2BS#4. Associations approaching borderline statistical significance: nt 47 vs 43 (p = 0.0668), nt 60 vs 52 (p = 0.0995) and 63 vs 58 (p = 0.0716).
Supplementary material
References
- 1.Doorbar J. Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci. 2006;110:525–541. doi: 10.1042/CS20050369. [DOI] [PubMed] [Google Scholar]
- 2.IARC Working group on the evaluation of carcinogenic risks to human. biological agents. A review of human carcinogens. IARC Monogr. Eval. Carcinog. Risks Hum. 2012;100B:1–441. [PMC free article] [PubMed] [Google Scholar]
- 3.Mark O′Connor S.-Y.C., Bernard Hans-Ulrich. Transcription Factor Binding Sites in the Long Control Region of Genital HPVs. In: Myers G., Bernard H.U., Delius H., Baker K., Icenogle J., Halpern A., Wheeler C., editors. Human Papillomaviruses, 1995 Compendium, Part III. Los Alamos National Laboratory; Los Alamos, NMex: 1995. pp. 21–40. [Google Scholar]
- 4.Tan S.H., Bartsch D., Schwarz E., Bernard H.U. Nuclear matrix attachment regions of human papillomavirus type 16 point toward conservation of these genomic elements in all genital papillomaviruses. J. Virol. 1998;72:3610–3622. doi: 10.1128/jvi.72.5.3610-3622.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernard H.U. Gene expression of genital human papillomaviruses and considerations on potential antiviral approaches. Antivir. Ther. 2002;7:219–237. [PubMed] [Google Scholar]
- 6.Desaintes C., Demeret C. Control of papillomavirus DNA replication and transcription. Semin. Cancer Biol. 1996;7:339–347. doi: 10.1006/scbi.1996.0043. [DOI] [PubMed] [Google Scholar]
- 7.Gage J.R., Meyers C., Wettstein F.O. The E7 proteins of the nononcogenic human papillomavirus type 6b (HPV-6b) and of the oncogenic HPV-16 differ in retinoblastoma protein binding and other properties. J. Virol. 1990;64:723–730. doi: 10.1128/jvi.64.2.723-730.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halbert C.L., Demers G.W., Galloway D.A. The E6 and E7 genes of human papillomavirus type 6 have weak immortalizing activity in human epithelial cells. J. Virol. 1992;66:2125–2134. doi: 10.1128/jvi.66.4.2125-2134.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dowhanick J.J., McBride A.A., Howley P.M. Suppression of cellular proliferation by the papillomavirus E2 protein. J. Virol. 1995;69:7791–7799. doi: 10.1128/jvi.69.12.7791-7799.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeon S., Allen-Hoffmann B.L., Lambert P.F. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 1995;69:2989–2997. doi: 10.1128/jvi.69.5.2989-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romanczuk H., Thierry F., Howley P.M. Mutational analysis of cis elements involved in E2 modulation of human papillomavirus type 16 P97 and type 18 P105 promoters. J. Virol. 1990;64:2849–2859. doi: 10.1128/jvi.64.6.2849-2859.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McBride A.A. The papillomavirus E2 proteins. Virology. 2013;445:57–79. doi: 10.1016/j.virol.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steger G., Corbach S. Dose-dependent regulation of the early promoter of human papillomavirus type 18 by the viral E2 protein. J. Virol. 1997;71:50–58. doi: 10.1128/jvi.71.1.50-58.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaiwongkot A., Vinokurova S., Pientong C., Ekalaksananan T., Kongyingyoes B., Kleebkaow P. Differential methylation of E2 binding sites in episomal and integrated HPV 16 genomes in preinvasive and invasive cervical lesions. Int. J. Cancer. 2013;132:2087–2094. doi: 10.1002/ijc.27906. [DOI] [PubMed] [Google Scholar]
- 15.Ding D.C., Chiang M.H., Lai H.C., Hsiung C.A., Hsieh C.Y., Chu T.Y. Methylation of the long control region of HPV16 is related to the severity of cervical neoplasia. Eur. J. Obstet. Gynecol. Reprod. Biol. 2009;147:215–220. doi: 10.1016/j.ejogrb.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 16.Mirabello L., Sun C., Ghosh A., Rodriguez A.C., Schiffman M., Wentzensen N. Methylation of human papillomavirus type 16 genome and risk of cervical precancer in a Costa Rican population. J. Natl. Cancer Inst. 2012;104:556–565. doi: 10.1093/jnci/djs135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filho S.M., Bertoni N., Brant A.C., Vidal J.P., Felix S.P., Cavalcanti S.M. Methylation at 3′LCR of HPV16 can be affected by patient age and disruption of E1 or E2 genes. Virus Res. 2017;232:48–53. doi: 10.1016/j.virusres.2017.01.022. [DOI] [PubMed] [Google Scholar]
- 18.Van Doorslaer K., Burk R.D. Evolution of human papillomavirus carcinogenicity. Adv. Virus Res. 2010;77:41–62. doi: 10.1016/B978-0-12-385034-8.00002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan P., Howell-Jones R., Li N., Bruni L., de Sanjose S., Franceschi S. Human papillomavirus types in 115,789 HPV-positive women: a meta-analysis from cervical infection to cancer. Int. J. Cancer. 2012;131:2349–2359. doi: 10.1002/ijc.27485. [DOI] [PubMed] [Google Scholar]
- 20.Cullen A.P., Reid R., Campion M., Lorincz A.T. Analysis of the physical state of different human papillomavirus DNAs in intraepithelial and invasive cervical neoplasm. J. Virol. 1991;65:606–612. doi: 10.1128/jvi.65.2.606-612.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho C.M., Chien T.Y., Huang S.H., Lee B.H., Chang S.F. Integrated human papillomavirus types 52 and 58 are infrequently found in cervical cancer, and high viral loads predict risk of cervical cancer. Gynecol. Oncol. 2006;102:54–60. doi: 10.1016/j.ygyno.2005.11.035. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y., Lu Z., Xu R., Ke Y. Comprehensive mapping of the human papillomavirus (HPV) DNA integration sites in cervical carcinomas by HPV capture technology. Oncotarget. 2016;7:5852–5864. doi: 10.18632/oncotarget.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clifford G., Franceschi S., Diaz M., Munoz N., Villa L.L. Chapter 3: HPV type-distribution in women with and without cervical neoplastic diseases. Vaccine. 2006;24(Suppl 3):26–34. doi: 10.1016/j.vaccine.2006.05.026. (S3) [DOI] [PubMed] [Google Scholar]
- 24.Clifford G.M., Rana R.K., Franceschi S., Smith J.S., Gough G., Pimenta J.M. Human papillomavirus genotype distribution in low-grade cervical lesions: comparison by geographic region and with cervical cancer. Cancer Epidemiol. Biomark. Prev. 2005;14:1157–1164. doi: 10.1158/1055-9965.EPI-04-0812. [DOI] [PubMed] [Google Scholar]
- 25.Clifford G.M., Smith J.S., Aguado T., Franceschi S. Comparison of HPV type distribution in high-grade cervical lesions and cervical cancer: a meta-analysis. Br. J. Cancer. 2003;89:101–105. doi: 10.1038/sj.bjc.6601024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.PaVE, The Papillomavirus Knowledge Source. 〈https://pave.niaid.nih.gov/〉; (Accessed 10 October 2017).
- 27.de Almeida L.M., Martins L.F.L., Pontes V.B., Correa F.M., Montenegro R.C., Pinto L.C. Human papillomavirus genotype distribution among cervical cancer patients prior to brazilian national HPV immunization program. J. Environ. Public Health. 2017;2017:1645074. doi: 10.1155/2017/1645074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kocher T.D., Thomas W.K., Meyer A., Edwards S.V., Paabo S., Villablanca F.X. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc. Natl. Acad. Sci. USA. 1989;86:6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Badal V., Chuang L.S., Tan E.H., Badal S., Villa L.L., Wheeler C.M. CpG methylation of human papillomavirus type 16 DNA in cervical cancer cell lines and in clinical specimens: genomic hypomethylation correlates with carcinogenic progression. J. Virol. 2003;77:6227–6234. doi: 10.1128/JVI.77.11.6227-6234.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandez A.F., Rosales C., Lopez-Nieva P., Grana O., Ballestar E., Ropero S. The dynamic DNA methylomes of double-stranded DNA viruses associated with human cancer. Genome Res. 2009;19:438–451. doi: 10.1101/gr.083550.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu B., Chotewutmontri S., Wolf S., Klos U., Schmitz M., Durst M. Multiplex identification of human papillomavirus 16 DNA integration sites in cervical carcinomas. PLOS One. 2013;8:e66693. doi: 10.1371/journal.pone.0066693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collins S.I., Constandinou-Williams C., Wen K., Young L.S., Roberts S., Murray P.G. Disruption of the E2 gene is a common and early event in the natural history of cervical human papillomavirus infection: a longitudinal cohort study. Cancer Res. 2009;69:3828–3832. doi: 10.1158/0008-5472.CAN-08-3099. [DOI] [PubMed] [Google Scholar]
- 33.Vernon S.D., Unger E.R., Miller D.L., Lee D.R., Reeves W.C. Association of human papillomavirus type 16 integration in the E2 gene with poor disease-free survival from cervical cancer. Int. J. Cancer. 1997;74:50–56. doi: 10.1002/(sici)1097-0215(19970220)74:1<50::aid-ijc9>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 34.Johannsen E., Lambert P.F. Epigenetics of human papillomaviruses. Virology. 2013;445:205–212. doi: 10.1016/j.virol.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirabello L., Schiffman M., Ghosh A., Rodriguez A.C., Vasiljevic N., Wentzensen N. Elevated methylation of HPV16 DNA is associated with the development of high grade cervical intraepithelial neoplasia. Int. J. Cancer. 2013;132:1412–1422. doi: 10.1002/ijc.27750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wentzensen N., Sun C., Ghosh A., Kinney W., Mirabello L., Wacholder S. Methylation of HPV18, HPV31, and HPV45 genomes and cervical intraepithelial neoplasia grade 3. J. Natl. Cancer Inst. 2012;104:1738–1749. doi: 10.1093/jnci/djs425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong D., Ye F., Lu W., Hu Y., Wan X., Chen Y. Methylation status of the long control region of HPV 16 in clinical cervical specimens. Mol. Med. Rep. 2008;1:555–560. [PubMed] [Google Scholar]
- 38.Leung T.W., Liu S.S., Leung R.C., Chu M.M., Cheung A.N., Ngan H.Y. HPV 16 E2 binding sites 1 and 2 become more methylated than E2 binding site 4 during cervical carcinogenesis. J. Med. Virol. 2015;87:1022–1033. doi: 10.1002/jmv.24129. [DOI] [PubMed] [Google Scholar]
- 39.Mazumder Indra D., Singh R.K., Mitra S., Dutta S., Chakraborty C., Basu P.S. Genetic and epigenetic changes of HPV16 in cervical cancer differentially regulate E6/E7 expression and associate with disease progression. Gynecol. Oncol. 2011;123:597–604. doi: 10.1016/j.ygyno.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 40.Tan S.H., Leong L.E., Walker P.A., Bernard H.U. The human papillomavirus type 16 E2 transcription factor binds with low cooperativity to two flanking sites and represses the E6 promoter through displacement of Sp1 and TFIID. J. Virol. 1994;68:6411–6420. doi: 10.1128/jvi.68.10.6411-6420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thain A., Jenkins O., Clarke A.R., Gaston K. CpG methylation directly inhibits binding of the human papillomavirus type 16 E2 protein to specific DNA sequences. J. Virol. 1996;70:7233–7235. doi: 10.1128/jvi.70.10.7233-7235.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schiffman M., Herrero R., Desalle R., Hildesheim A., Wacholder S., Rodriguez A.C. The carcinogenicity of human papillomavirus types reflects viral evolution. Virology. 2005;337:76–84. doi: 10.1016/j.virol.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 43.Tsakogiannis D., Gortsilas P., Kyriakopoulou Z., Ruether I.G., Dimitriou T.G., Orfanoudakis G. Sites of disruption within E1 and E2 genes of HPV16 and association with cervical dysplasia. J. Med. Virol. 2015;87:1973–1980. doi: 10.1002/jmv.24256. [DOI] [PubMed] [Google Scholar]
- 44.Cheung J.L., Cheung T.H., Yu M.Y., Chan P.K. Virological characteristics of cervical cancers carrying pure episomal form of HPV16 genome. Gynecol. Oncol. 2013;131:374–379. doi: 10.1016/j.ygyno.2013.08.026. [DOI] [PubMed] [Google Scholar]
- 45.Marongiu L., Godi A., Parry J.V., Beddows S. Human Papillomavirus 16, 18, 31 and 45 viral load, integration and methylation status stratified by cervical disease stage. BMC Cancer. 2014;14:384. doi: 10.1186/1471-2407-14-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holmes A., Lameiras S., Jeannot E., Marie Y., Castera L., Sastre-Garau X. Mechanistic signatures of HPV insertions in cervical carcinomas. NJP Genom. Med. 2016:16004. doi: 10.1038/npjgenmed.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoppe-Seyler F., Butz K. Activation of human papillomavirus type 18 E6-E7 oncogene expression by transcription factor Sp1. Nucleic Acids Res. 1992;20:6701–6706. doi: 10.1093/nar/20.24.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vinokurova S., von Knebel M. Doeberitz, Differential methylation of the HPV 16 upstream regulatory region during epithelial differentiation and neoplastic transformation. PLOS One. 2011;6:e24451. doi: 10.1371/journal.pone.0024451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material