Abstract
The human rhinovirus (hRV) is the causative agent of the common cold that often aggravates respiratory complications in patients with asthma or chronic obstructive pulmonary disease. The high rate of mutations and variety of serotypes are limiting the development of anti-hRV drugs, which emphasizes the need for the discovery of novel lead compounds. Previously, we identified antiviral compound 1 that we used here as the starting material for developing a novel compound series with high efficacy against hRV-A and -B. Improved metabolic stability was achieved by substituting an ester moiety with a 1,2,4-oxadiazole group. Specifically, compound 3k exhibited a high efficacy against hRV-B14, hRV-A21, and hRV-A71, with EC50 values of 66.0, 22.0, and 3.7 nM, respectively, and a relevant hepatic stability (59.6 and 40.7% compound remaining after 30 min in rat and human liver microsomes, respectively). An in vivo study demonstrated that 3k possessed a desirable pharmacokinetic profile with low systemic clearance (0.158 L·h–1·kg–1) and modest oral bioavailability (27.8%). Hence, 3k appears to be an interesting candidate for the development of antiviral lead compounds.
Keywords: Human rhinovirus, capsid-binding, antiviral compound, small molecule inhibitor, oxadiazole
The human rhinovirus (hRV), a member of the Enterovirus genus in the Picornaviradae family, is a persistent threat to public health. It is known to cause ∼60% of upper respiratory tract symptoms such as the common cold. Moreover, recent studies conducted with improved detection methods suggest that hRV infections can aggravate inflammatory illnesses such as asthma, chronic obstructive pulmonary disease, and otitis media.1−6 Analyses of viral specimens from pediatric patients with asthma exacerbations identified a high prevalence of hRV.7−9 A recent study also revealed that early life hRV wheezing illnesses increase the risk of asthma development at adolescence.10
More than 160 hRV serotypes have been identified and grouped into three species, hRV-A, -B, and -C, that are each divided into various subspecies.11 Like other picornaviruses, hRV has a positive-sense, single-stranded RNA genome packaged in an icosahedral capsid composed of four viral proteins (VP1 to VP4).12 The capsid features canyons around its 5-fold symmetry axes that contain binding sites for host receptors such as the intracellular adhesion molecule 1 (ICAM-1), which recognizes most of the hRV subspecies.13−15 The neutralizing epitopes, which are hypervariable among the hRV subspecies, are also located along the canyons.16−18 In addition, the canyons of several hRV subspecies harbor small molecules, the “pocket factors,” that are probably recovered from the host to facilitate receptor recognition by stabilizing the capsid structure.19 Receptor binding induces conformational changes in the capsid to promote its decomposition and enable the injection of the genome into the host cell.20 Viral RNA translation produces a single polyprotein, which is processed into its various parts such as 2A and 3C viral proteases that are crucial for maturation of viral proteins. Subsequent viral replication is accompanied by alterations in the host cell architecture, including rearrangements of the endoplasmic reticulum and Golgi secretory apparatus, although the specific steps vary among subspecies.21−23
Several drug candidates have been developed to combat hRV infections.24,25 Capsid-binding inhibitors include pleconaril and vapendavir that associate with the canyon to stabilize the capsid structure, thereby preventing viral genome intrusion.26,27 Inhibitors of 3C protease, such as rupintrivir and V-7404, block the maturation of viral proteins,28,29 whereas enviroxime, an inhibitor of viral protein 3A, prevents viral replication.30 However, there is a need for the discovery of novel anti-hRV agent candidates because current antiviral drugs are not approved for hRV treatment due to high treatment failure rates and significant side effects.
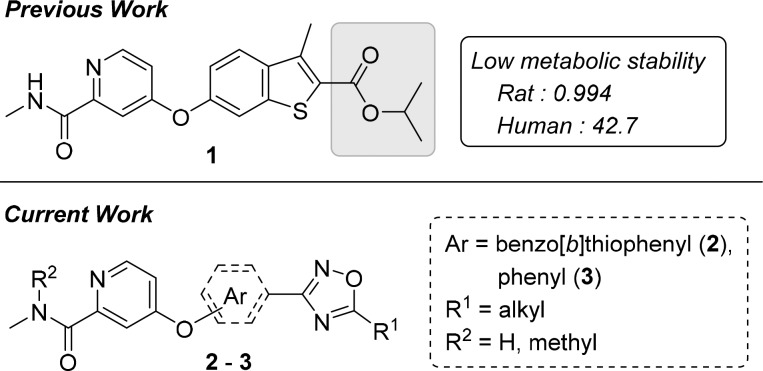
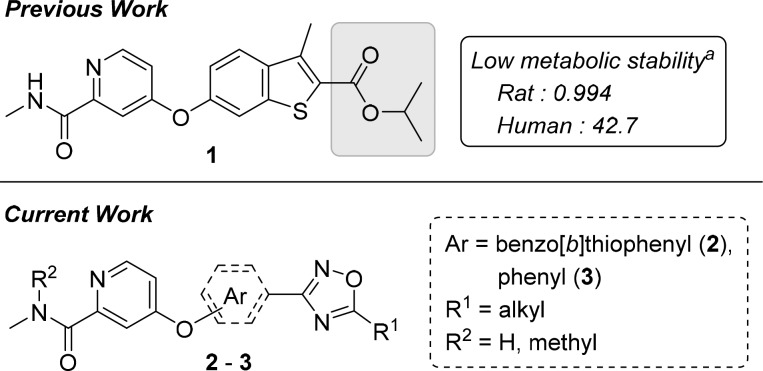
Recently, we revealed a novel series of small-molecule capsid-binding inhibitors with high effectiveness against replication of hRV-A and -B (Figure 1).31 An ester moiety and the high hydrophobicity of the inhibitors represented targets for optimization that may lead to improvements in metabolic stability and pharmacokinetics. Here, we hypothesized that these improvements can be achieved by substituting the ester with an oxadiazole moiety. This study describes new 3-aryl-1,2,4-oxadiazole derivatives that exhibit strong activity against hRV-B14, -A21, and -A71, along with significant metabolic stability and hydrophilicity.
Figure 1.
Previously discovered anti-hRV compound (1) and new derivatives (2 and 3). aLiver microsomal stability: amount remaining after 30 min (in percent).
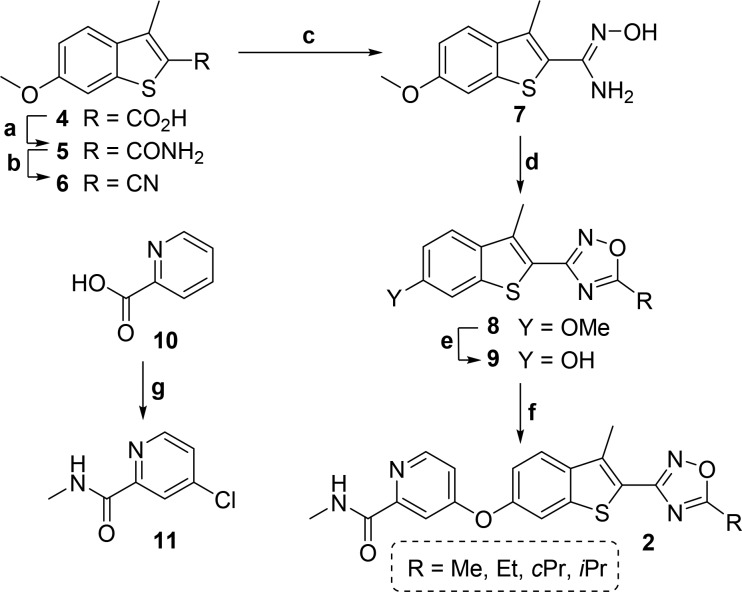
The oxadiazole moieties of the new derivatives were prepared from corresponding nitrile groups through cyclization of an N-hydroxyimidamide intermediate (Schemes 1 and 2). The benzo[b]thiophenyl oxadiazole compounds 2 were synthesized according to the synthetic route outlined in Scheme 1. A reference procedure was employed to synthesize benzo[b]thiophene core fragment 4, which was converted into amide 5 via the acid chloride intermediate using SOCl2. Amide 5 was further cyanated with trifluoroacetic anhydride (TFAA) and pyridine using chlorinating solvents. Addition of NH2OH in H2O–EtOH medium yielded the N′-hydroxyimidamide intermediate 7. Oxadiazole moieties with diverse alkyl chains were obtained by the cyclization using corresponding acid chlorides. Demethylation of the 6-methoxy mask with BBr3 exposed the phenolic hydroxyl group for subsequent coupling (9). The coupling partner, 4-chloro-N-methylpicolinamide (11), was generated from picolinic acid 10 by overchlorination in SOCl2 and DMF followed by amidation. Coupling of 9 and 11 under neat conditions at 150 °C yielded compounds 2 with one exception. The trifluoromethyl compound 2e was obtained by creating a precoupled amide from 11 and 6-hydroxy-3-methylbenzo[b]thiophene-2-carboxamide because of its instability under BBr3 demethylation condition (see Supporting Information for details).
Scheme 1. General Synthetic Route to Benzo[b]thiophene Compounds.
Reagents and conditions: (a) DMF, SOCl2, 80 °C, 4 h, then NH3, H2O, THF, 0 to 25 °C, 16 h; (b) TFAA, pyridine, ClCH2CH2Cl, 25 °C, 1.5 h; (c) NH2OH, H2O, EtOH, 90 °C, 48 h; (d) ROCl, pyridine, 0 to 120 °C, 16 h; (e) BBr3, CH2Cl2, −78 to 25 °C, 16 h; (f) 11, neat, 150 °C, 60 h; (g) DMF, SOCl2, 45 to 75 °C, 72 h, then CH3NH2, H2O, THF, 0 to 25 °C, 4 h.
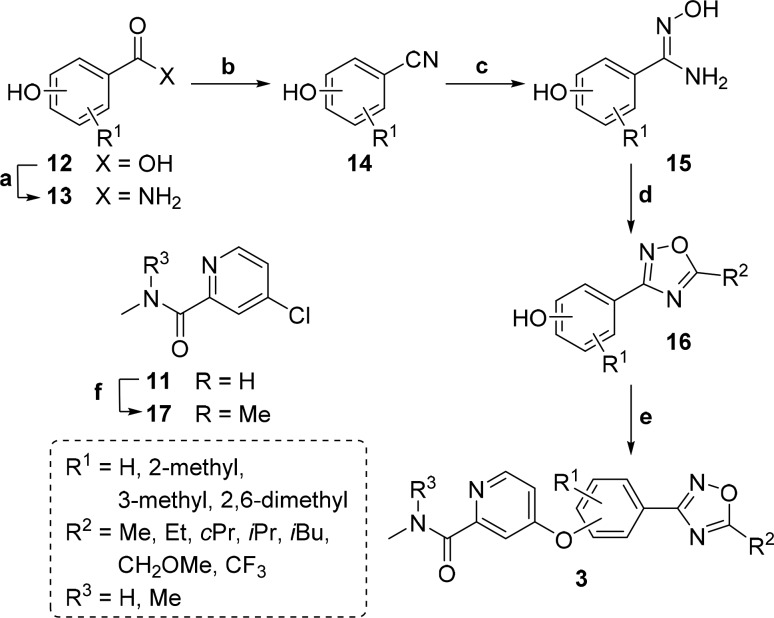
Scheme 2. General Synthetic Route to Phenyl Compounds.
Reagents and conditions: (a) DMF, SOCl2, 70 °C, 5 h, then NH3, H2O, THF, 0 to 25 °C, 16 h; (b) TFAA, pyridine, THF, 0 to 25 °C, 16 h; (c) NH2OH, H2O, EtOH, 90 °C, 16 h; (d) RCOCl or TFAA, pyridine, 0 to 120 °C, 16 h; (e) 11 or 17, neat, 150 °C, 90 h; (f) DMF, SOCl2, 45 to 75 °C, 72 h, then CH3NH2, H2O, THF, 0 to 25 °C, 4 h; (g) MeI, NaH, DMF, 0 to 25 °C, 3 h.
Although compound 2 derivatives possess a lower hydrophobicity than compound 1 because of the oxadiazole substitution, the new derivatives are extended in length, and additional alkyl length variations of two or more carbon atoms on the oxadiazole ring could significantly interfere in the interaction with one end of the viral capsid canyon. We assumed that changing the benzo[b]thiophene core in 2 to phenyl in 3 would not only further increase the hydrophilicity but also provide space for additional variations on the core fragment and the oxadiazole ring. Although the orientation of the oxadiazole ring became distorted, the anti-hRV activities were unlikely to be diminished because, in our previous study, the naphthyl analogs of 1 had also retained activity against hRV-A and -B species.
The synthetic route to phenyl oxadiazole compounds 3 is presented in Scheme 2. N′-Hydroxybenzimidamides 15 were obtained from corresponding 4- or 3-hydroxybenzonitriles 14 using NH2OH. Commercially unavailable benzonitriles were synthesized from hydroxybenzoic acids 12 via the amide intermediates 13. To obtain 14, cyanation of 13 was performed using TFAA and pyridine in THF because using the reagents in dichloroethane gave lower yields. Continuing the synthetic route by cyclization of 15 with TFAA or acyl chlorides yielded phenyl oxadiazole cores 16. 4-Chloro-N,N-dimethylpicolinamide 17 was prepared by methylating 11 using MeI and NaH. The phenolic group of 16 was coupled with 11 or 17 under neat conditions to obtain the desired compounds 3. Because of side reactions during oxadiazole ring formation, the pyridine analog 3s was synthesized using a synthetic route like in Scheme 1. The 6-(oxadiazolyl)naphthalen-2-ol counterpart 3t, however, was synthesized like the phenyl oxadiazole derivatives (see Supporting Information for the details).
The antiviral activities of oxadiazole compounds 2–3 against hRV-B14, -A21, and -A71 were assessed in H1HeLa cells using the MTT assay with pleconaril as a reference (Table 1). 5-Methyl-1,2,4-oxadiazole derivative 2a, a compound with a molecular volume similar to 1, exhibited activity against the three hRV strains at a nanomolar concentration range (entry 2). Longer and bulkier alkyl substitutions on position 5 of the oxadiazole ring resulted in significantly higher cytotoxicity and slightly improved anti-hRV activity (entries 3–5). Comparing the activities between ethyl-substituted 2b and cyclopropyl-substituted 2c, their activities were similar against hRV-B14, but 2c was much stronger against hRV-A21 than 2b, and approximately 50% less active against hRV-A71 (entries 3, 4). Assessing the differences between 2b and isopropyl-substituted 2d, the activity of 2d was lower against hRV-B14, higher against hRV-A21, and not different toward hRV-A71 (entry 5). 5-Trifluoromethyl derivative 2e had low cytotoxicity, but its activity toward the three hRV strains was the lowest among compound 2 variants, implying that electron-richness on the oxadiazole ring is critical for the anti-hRV activities (entry 6).
Table 1. Inhibitory Activities of Compounds 2 and 3 against hRVs.
CC50: cytotoxic concentration (μM) that reduced cell viability by 50%; measured in H1HeLa cells using MTT assay.
EC50: effective concentration (μM) inhibited hRV replication by 50%; measured in H1HeLa cells using MTT assay.
Anti-hRV activities and cytotoxicity of phenyl oxadiazole derivatives 3a–t were also assessed. The para-substituted phenyl variants 3a–d and 3g showed improved cytotoxicity but exhibited mostly lower activities than their respective benzo[b]thiophene counterparts (entries 7–10, 13). Specifically, 3a had much lower anti-hRV activities than 2a. Although the tendencies observed for the activities of compounds 3b–d were similar to their respective counterparts 2b–d, the activity of 3c was lower against hRV-21 than against hRV-71, which is the inverse activity pattern observed for 2c. Furthermore, the extension of the 5-alkyl substitution to isobutyl or methoxymethyl significantly reduced anti-hRV activities, suggesting that derivatives with 5-alkyl substitutions of more than two carbon atoms cannot properly fit into the viral capsid canyon (entries 11, 12). Like 2e, the CF3 derivative 3g exhibited a much lower antiviral activity than 3b–e (entry 13).
Based on our previous study, we hypothesized that an additional alkyl substitution on the phenyl core of 3 could enhance the interaction between the inhibitor and residue L25 of VP 3, one of the viral capsid constituents.31 Addition of a methyl group on the meta position toward oxadiazole preserved anti-hRV activities but increased cytotoxicity (entries 14–16). Specifically, an improved anti-hRV21 activity was observed for 3j, exhibiting EC50 values in the range between 2.5 and 73.0 nM against the three hRV species (entry 16). Further improvement was obtained by adding a methyl group in the ortho position toward oxadiazole, which significantly reduced the cytotoxicity (entry 17). Compound 3k exhibited EC50 values of 66, 22, and 3.7 nM against hRV-B14, -A21, and -A71, respectively. Although the dimethyl derivative 3l still exhibited high efficacy against hRV-A strains, the activity against hRV-B14 was strongly reduced along with a strong increase in cytotoxicity (entry 18). The N,N-dimethylpicolinamide analogs 3m–o also exhibited high cytotoxicity associated with CC50 values that were no longer above the EC50 values against hRV-A strains (entries 19–21). Variants with meta-substituted phenyl core 3p–r did not exhibit useful antiviral activity profiles, presumably, because these inhibitor molecules are bent, which may prevent the critical hydrophobic interaction between the oxadiazole moiety and the viral pocket (entries 22–24). In addition, the pyridine derivative 3s with low cytotoxicity showed only moderate antiviral activity (entry 25), whereas the naphthalene derivative 3t was effective against hRV-A71, but its cytotoxicity was too high for its other modest activities against hRV-B14 and -A21 (entry 26).
The Phase I metabolic stability of selected compounds (2d, 3j, 3k, and 3l) was investigated in rat and human liver microsomes (Table 2). The compounds showed higher stability in rat liver microsome than compound 1. Interestingly, 2d and 3l are more stable than 1 in human liver microsome. The most active and least cytotoxic compound 3k displayed improved Phase I metabolic stability to compound 1. To elucidate the pharmacokinetics of 3k, male Sprague–Dawley rats received doses of 5 and 10 mg·kg–1 via the intravenous and oral route, respectively. The 3k plasma concentration was determined using LC–MS/MS after sample deproteinization in acetonitrile (Figure 2). The plasma concentration–time data were analyzed by applying the noncompartmental method using Phoenix WinNonlin (v6.4; Pharsight Corp., Mountain View, CA, USA). After intravenous dosing, the AUCt value at 5 mg·kg–1 was 31.3 μg·h·mL–1 along with a low systemic clearance rate of 0.158 L·h–1·kg–1. After the oral administration, 3k was slowly absorbed and reached a Cmax of 3.9 μg·mL–1. Then, the 3k plasma concentrations declined with a terminal T1/2 of 3.2 h. The systemic exposure (AUCt) following an oral dose was 17.4 μg·h·mL–1 and the oral bioavailability was 27.8% (Table 3).
Table 2. In Vitro Liver Microsomal Phase I Stability (% Remaining after 30 min)a.
| compound | rat (%) | human (%) |
|---|---|---|
| 1 | 1.0 ± 0.1 | 42.7 ± 0.6 |
| 2d | 99.2 ± 0.3 | 69.8 ± 8.9 |
| 3j | 35.1 ± 6.3 | 9.9 ± 3.1 |
| 3k | 59.6 ± 5.7 | 40.7 ± 1.0 |
| 3l | 37.1 ± 2.6 | 61.1 ± 3.7 |
| buspirone | 0.1 ± 0.01 | 3.5 ± 0.5 |
Each value is presented as mean ± standard deviation of at least three independent experiments.
Figure 2.
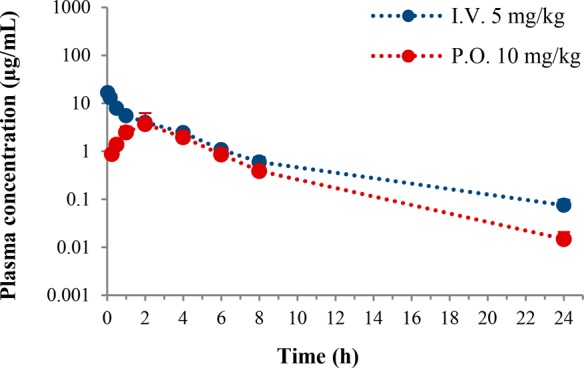
Plasma concentration–time profiles of 3k in male rats (n = 3).
Table 3. Pharmacokinetics of 3k in Male Ratsa.
| parameter | I.V., 5 mg·kg–1b | P.O., 10 mg·kg–1c |
|---|---|---|
| Tmax (h) | NA | 2.7 ± 1.2 |
| Cmax (μg·mL–1) | NA | 3.9 ± 2.4 |
| T1/2 (h) | 4.6 ± 1.1 | 3.2 ± 0.4 |
| AUCt (μg·h·mL–1) | 31.3 ± 2.5 | 17.4 ± 5.4 |
| AUC∞ (μg·h·mL–1) | 31.8 ± 2.8 | 17.4 ± 5.4 |
| CL (L·h–1·kg–1) | 0.158 ± 0.014 | NA |
| VSS (L·kg–1) | 0.611 ± 0.029 | NA |
| Ft (%) | NA | 27.8 |
Each value is presented as mean ± standard deviation of at least three independent experiments.
Parameters from intravenous administration.
Parameters from oral administration.
In this study, we used antiviral compound 1 as starting material for developing two novel compound series, derivatives 2 and 3, that we examined for their inhibitory activity against hRV-B14, -A21, and -A71. We showed that substituting the ester moiety of compound 1 by a 1,2,4-oxadiazole group in 2 and 3 created molecules with antiviral activity and significant metabolic stability. Several compounds were highly active with EC50 values in the nanomolar range against the three hRV species. Specifically, compound 3k displayed a high efficacy against hRV-B14, hRV-A21, and hRV-A71, with EC50 values of 66.0, 22.0, and 3.7 nM, respectively. In addition, the hepatic stability of 3k was better than compound 1 in rat microsomes and similar for both compounds in human microsomes. A pharmacokinetics analysis of 3k in rats demonstrated that the inhibitor had a low systemic clearance and a moderate oral bioavailability. Hence, 3k represents an interesting candidate for the development of novel antiviral lead compounds.
Acknowledgments
This study was supported by Korea Research Institute of Chemical Technology (Grant No. KK1703-C00, KK1703-E00, and KK1803-D00).
Glossary
ABBREVIATIONS
- AUCt
areas under the plasma concentration–time curve
- AUC∞
areas under the plasma concentration–time curve from time zero
- CL
total clearance from plasma
- Cmax
maximum plasma concentration
- DMF
dimethylformamide
- Ft
bioavailability
- hRV
human rhinovirus
- ICAM-1
intracellular adhesion molecule 1
- MTT
3-(4,5-dimethylthiazol-2yl)-2,5-diphenyltetrazolium bromide
- TFAA
trifluoroacetic anhydride
- THF
tetrahydrofuran
- Tmax
time of maximum drug concentration
- T1/2
terminal half-life
- VP
viral capsid protein
- Vss
steady-state volume of distribution
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.8b00134.
Detailed synthetic procedures and UPLC purity and characterization data for compounds 2–3, including the spectral copies of 1H and 13C NMR spectra (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- George S. N.; Garcha D. S.; Mackay A. J.; Patel A. R.; Singh R.; Sapsford R. J.; Donaldson G. C.; Wedzicha J. A. Human rhinovirus infection during naturally occurring COPD exacerbations. Eur. Respir. J. 2014, 44, 87–96. 10.1183/09031936.00223113. [DOI] [PubMed] [Google Scholar]
- Gern J. E. How rhinovirus infections cause exacerbations of asthma. Clin. Exp. Allergy 2015, 45, 32–42. 10.1111/cea.12428. [DOI] [PubMed] [Google Scholar]
- Johnston S. L.; Sanderson G.; Pattemore P. K.; Smith S.; Bardin P. G.; Bruce C. B.; Lambden P. R.; Tyrrell D. A.; Holgate S. T. Use of polymerase chain reaction for diagnosis of picornavirus infection in subjects with and without respiratory symptoms. J. Clin. Microbiol. 1993, 31, 111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallia P.; Message S. D.; Gielen V.; Contoli M.; Gray K.; Kebadze T.; Aniscenko J.; Laza-Stanca V.; Edwards M. R.; Slater L.; Papi A.; Stanciu L. A.; Kon O. M.; Johnson M.; Johnston S. L. Experimental rhinovirus infection as a human model of chronic obstructive pulmonary disease exacerbation. Am. J. Respir. Crit. Care Med. 2011, 183, 734–742. 10.1164/rccm.201006-0833OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Message S. D.; Laza-Stanca V.; Mallia P.; Parker H. L.; Zhu J.; Kebadze T.; Contoli M.; Sanderson G.; Kon O. M.; Papi A.; Jeffery P. K.; Stanciu L. A.; Johnston S. L. Rhinovirus-induced lower respiratory illness is increased in asthma and related to virus load and Th1/2 cytokine and IL-10 production. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 13562. 10.1073/pnas.0804181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppala E.; Sillanpaa S.; Nurminen N.; Huhtala H.; Toppari J.; Ilonen J.; Veijola R.; Knip M.; Sipila M.; Laranne J.; Oikarinen S.; Hyoty H. Human enterovirus and rhinovirus infections are associated with otitis media in a prospective birth cohort study. J. Clin. Virol. 2016, 85, 1–6. 10.1016/j.jcv.2016.10.010. [DOI] [PubMed] [Google Scholar]
- Heymann P. W.; Carper H. T.; Murphy D. D.; Platts-Mills T. A.; Patrie J.; McLaughlin A. P.; Erwin E. A.; Shaker M. S.; Hellems M.; Peerzada J.; Hayden F. G.; Hatley T. K.; Chamberlain R. Viral infections in relation to age, atopy, and season of admission among children hospitalized for wheezing. J. Allergy Clin. Immunol. 2004, 114, 239–247. 10.1016/j.jaci.2004.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetsuriani N.; Kazerouni N. N.; Erdman D. D.; Lu X.; Redd S. C.; Anderson L. J.; Teague W. G. Prevalence of viral respiratory tract infections in children with asthma. J. Allergy Clin. Immunol. 2007, 119, 314–321. 10.1016/j.jaci.2006.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlinson W. D.; Waliuzzaman Z.; Carter I. W.; Belessis Y. C.; Gilbert K. M.; Morton J. R. Asthma exacerbations in children associated with rhinovirus but not human metapneumovirus infection. J. Infect. Dis. 2003, 187, 1314–1318. 10.1086/368411. [DOI] [PubMed] [Google Scholar]
- Rubner F. J.; Jackson D. J.; Evans M. D.; Gangnon R. E.; Tisler C. J.; Pappas T. E.; Gern J. E.; Lemanske R. F. Jr Early life rhinovirus wheezing, allergic sensitization, and asthma risk at adolescence. J. Allergy Clin. Immunol. 2017, 139, 501–507. 10.1016/j.jaci.2016.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmenberg A. C.; Spiro D.; Kuzmickas R.; Wang S.; Djikeng A.; Rathe J. A.; Fraser-Liggett C. M.; Liggett S. B. Sequencing and Analyses of All Known Human Rhinovirus Genomes Reveal Structure and Evolution. Science 2009, 324, 55. 10.1126/science.1165557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M. G.; Arnold E.; Erickson J. W.; Frankenberger E. A.; Griffith J. P.; Hecht H. J.; Johnson J. E.; Kamer G.; Luo M.; Mosser A. G.; et al. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 1985, 317, 145–153. 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- Colonno R. J.; Condra J. H.; Mizutani S.; Callahan P. L.; Davies M. E.; Murcko M. A. Evidence for the direct involvement of the rhinovirus canyon in receptor binding. Proc. Natl. Acad. Sci. U. S. A. 1988, 85, 5449–5453. 10.1073/pnas.85.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer F.; Gruenberger M.; Kowalski H.; Machat H.; Huettinger M.; Kuechler E.; Blass D. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc. Natl. Acad. Sci. U. S. A. 1994, 91, 1839–1842. 10.1073/pnas.91.5.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staunton D. E.; Merluzzi V. J.; Rothlein R.; Barton R.; Marlin S. D.; Springer T. A. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell 1989, 56, 849–853. 10.1016/0092-8674(89)90689-2. [DOI] [PubMed] [Google Scholar]
- Glanville N.; Johnston S. L. Challenges in developing a cross-serotype rhinovirus vaccine. Curr. Opin. Virol. 2015, 11, 83–88. 10.1016/j.coviro.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Skern T.; Neubauer C.; Frasel L.; Grundler P.; Sommergruber W.; Zorn M.; Kuechler E.; Blaas D. A neutralizing epitope on human rhinovirus type 2 includes amino acid residues between 153 and 164 of virus capsid protein VP2. J. Gen. Virol. 1987, 68, 315–323. 10.1099/0022-1317-68-2-315. [DOI] [PubMed] [Google Scholar]
- Smith T. J.; Chase E. S.; Schmidt T. J.; Olson N. H.; Baker T. S. Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon. Nature 1996, 383, 350–354. 10.1038/383350a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. K.; Bothner B.; Smith T. J.; Siuzdak G. Antiviral agent blocks breathing of the common cold virus. Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 6774–6778. 10.1073/pnas.95.12.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisdorph N.; Thomas J. J.; Katpally U.; Chase E.; Harris K.; Siuzdak G.; Smith T. J. Human rhinovirus capsid dynamics is controlled by canyon flexibility. Virology 2003, 314, 34–44. 10.1016/S0042-6822(03)00452-5. [DOI] [PubMed] [Google Scholar]
- Belov G. A.; Ehrenfeld E. Involvement of cellular membrane traffic proteins in poliovirus replication. Cell Cycle 2007, 6, 36–38. 10.4161/cc.6.1.3683. [DOI] [PubMed] [Google Scholar]
- Mousnier A.; Swieboda D.; Pinto A.; Guedán A.; Rogers A. V.; Walton R.; Johnston S. L.; Solari R. Human Rhinovirus 16 Causes Golgi Apparatus Fragmentation without Blocking Protein Secretion. J. Virol. 2014, 88, 11671–11685. 10.1128/JVI.01170-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiner C. A.; Jackson W. T. Fragmentation of the Golgi apparatus provides replication membranes for human rhinovirus 1A. Virology 2010, 407, 185–195. 10.1016/j.virol.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma A. M.; Vliegen I.; De Clercq E.; Neyts J. Selective inhibitors of picornavirus replication. Med. Res. Rev. 2008, 28, 823–884. 10.1002/med.20125. [DOI] [PubMed] [Google Scholar]
- Patick A. K. Rhinovirus chemotherapy. Antiviral Res. 2006, 71, 391–396. 10.1016/j.antiviral.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diana G. D.; Rudewicz P.; Pevear D. C.; Nitz T. J.; Aldous S. C.; Aldous D. J.; Robinson D. T.; Draper T.; Dutko F. J.; Aldi C.; et al. Picornavirus inhibitors: trifluoromethyl substitution provides a global protective effect against hepatic metabolism. J. Med. Chem. 1995, 38, 1355–1371. 10.1021/jm00008a014. [DOI] [PubMed] [Google Scholar]
- Feil S. C.; Hamilton S.; Krippner G. Y.; Lin B.; Luttick A.; McConnell D. B.; Nearn R.; Parker M. W.; Ryan J.; Stanislawski P. C.; Tucker S. P.; Watson K. G.; Morton C. J. An Orally Available 3-Ethoxybenzisoxazole Capsid Binder with Clinical Activity against Human Rhinovirus. ACS Med. Chem. Lett. 2012, 3, 303–307. 10.1021/ml2002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragovich P. S.; Prins T. J.; Zhou R.; Webber S. E.; Marakovits J. T.; Fuhrman S. A.; Patick A. K.; Matthews D. A.; Lee C. A.; Ford C. E.; Burke B. J.; Rejto P. A.; Hendrickson T. F.; Tuntland T.; Brown E. L.; Meador J. W. 3rd; Ferre R. A.; Harr J. E.; Kosa M. B.; Worland S. T. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 4. Incorporation of P1 lactam moieties as L-glutamine replacements. J. Med. Chem. 1999, 42, 1213–1224. 10.1021/jm9805384. [DOI] [PubMed] [Google Scholar]
- Patick A. K.; Brothers M. A.; Maldonado F.; Binford S.; Maldonado O.; Fuhrman S.; Petersen A.; Smith G. J. 3rd; Zalman L. S.; Burns-Naas L. A.; Tran J. Q. In vitro antiviral activity and single-dose pharmacokinetics in humans of a novel, orally bioavailable inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 2005, 49, 2267–2275. 10.1128/AAC.49.6.2267-2275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikel J. H.; Paget C. J.; DeLong D. C.; Nelson J. D.; Wu C. Y.; Paschal J. W.; Dinner A.; Templeton R. J.; Chaney M. O.; Jones N. D.; Chamberlin J. W. Synthesis of syn and anti isomers of 6-[[(hydroxyimino)phenyl]methyl]-1-[(1-methylethyl)sulfonyl]-1H-benzimidazol-2-am ine. Inhibitors of rhinovirus multiplication. J. Med. Chem. 1980, 23, 368–372. 10.1021/jm00178a004. [DOI] [PubMed] [Google Scholar]
- Kim J.; Jung Y. K.; Kim C.; Shin J. S.; Scheers E.; Lee J. Y.; Han S. B.; Lee C. K.; Neyts J.; Ha J. D.; Jung Y. S. A Novel Series of Highly Potent Small Molecule Inhibitors of Rhinovirus Replication. J. Med. Chem. 2017, 60, 5472–5492. 10.1021/acs.jmedchem.7b00175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.