Abstract
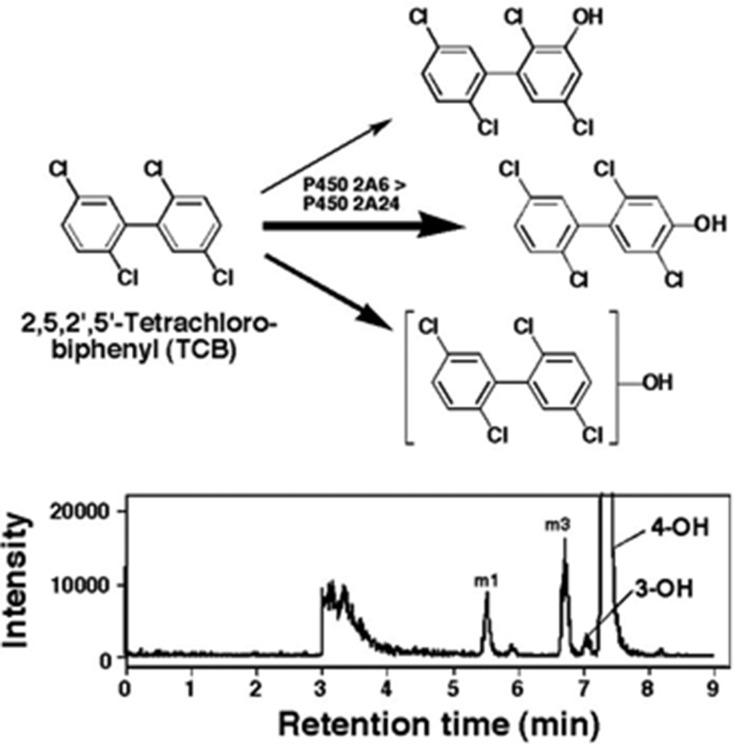
2,5,2′,5′-Tetrachlorobiphenyl (TCB) induced type I binding spectra with cytochrome P450 (P450) 2A6 and 2A13, with Ks values of 9.4 and 0.51 µM, respectively. However, CYP2A6 oxidized 2,5,2′,5′-TCB to form 4-hydroxylated products at a much higher rate (∼1.0 minute−1) than CYP2A13 (∼0.02 minute−1) based on analysis by liquid chromatography–tandem mass spectrometry. Formation of 4-hydroxy-2,5,2′,5′-TCB by CYP2A6 was greater than that of 3-hydroxy-2,5,2′,5′-TCB and three other hydroxylated products. Several human P450 enzymes, including CYP1A1, 1A2, 1B1, 2B6, 2D6, 2E1, 2C9, and 3A4, did not show any detectable activities in oxidizing 2,5,2′,5′-TCB. Cynomolgus monkey CYP2A24, which shows 95% amino acid identity to human CYP2A6, catalyzed 4-hydroxylation of 2,5,2′,5′-TCB at a higher rate (∼0.3 minute−1) than CYP2A26 (93% identity to CYP2A6, ∼0.13 minute−1) and CYP2A23 (94% identity to CYP2A13, ∼0.008 minute−1). None of these human and monkey CYP2A enzymes were catalytically active in oxidizing other TCB congeners, such as 2,4,3′,4′-, 3,4,3′,4′-, and 3,5,3′,5′-TCB. Molecular docking analysis suggested that there are different orientations of interaction of 2,5,2′,5′-TCB with the active sites (over the heme) of human and monkey CYP2A enzymes, and that ligand interaction energies (U values) of bound protein-ligand complexes show structural relationships of interaction of TCBs and other ligands with active sites of CYP2A enzymes. Catalytic differences in human and monkey CYP2A enzymes in the oxidation of 2,5,2′,5′-TCB are suggested to be due to amino acid changes at substrate recognition sites, i.e., V110L, I209S, I300F, V365M, S369G, and R372H, based on the comparison of primary sequences.
Introduction
Commercial polychlorinated biphenyl (PCB) mixtures have been widely used in electrical transformers, electrical switches, sealants, plasticizers, rubbers, adhesives, carbonless copy papers, paints, inks, and dust control agents (McFarland and Clarke, 1989; Mills et al., 2007; International Agency for Research on Cancer, 2015). Yusho disease occurred in western Japan in 1968 following the ingestion of rice oil contaminated with PCBs and polychlorinated dibenzofurans. Also, a similar incident in Taiwan was referred to as Yu-cheng disease in 1979 (Kuratsune et al., 1972; Kunita et al., 1984). After cessation of production and use of commercial PCBs after 1970, the levels of PCBs in human tissues and milk decreased yearly in Japan (Yakushiji et al., 1979, 1984). However, due to their strong persistent nature, PCBs have been shown to be present at significant levels in environmental and biologic samples (Kunita et al., 1984; Yakushiji et al., 1984). Konishi et al. (2006) determined the levels of PCB congeners in human milk in Osaka and found that 12 major PCBs, including three hepta-, two hexa-, four penta-, and four tetrachlorobiphenyl congeners, decreased between 1973 and 2000, but the rate of elimination depends on the individual PCB congeners determined, probably due to their susceptibilities to being chemically degraded and enzymatically metabolized (Akutsu et al., 2005; Todaka et al., 2008; Arnich et al., 2009).
2,5,2′,5′-Tetrachlorobiphenyl (#PCB52) (Mills et al., 2007; International Agency for Research on Cancer, 2015) (Fig. 1) is one of the PCB congeners identified in human milk and in foodstuffs at significant levels, and its biologic activities and metabolism by xenobiotic-metabolizing enzymes has been extensively studied in experimental animal models (Yoshimura et al., 1975; Preston et al., 1983; Borlakoglu et al., 1991; Ishida et al., 1991; Safe, 1993; Koga et al., 1995a). Cytochrome P450 (P450) has been shown to be the major enzyme responsible for the oxidation of this tetrachlorobiphenyl (TCB), and there are species-related differences in the metabolism by different forms of P450s in rats, mice, guinea pigs, hamsters, and rabbits (Preston et al., 1983; Borlakoglu et al., 1991; Koga et al., 1996). However, little is known about the roles of human P450 enzymes in the metabolism of 2,5,2′,5′-TCB. Our previous studies have shown that 2,5,2′,5′-TCB induces type I binding spectra with CYP2A13 and 2A6 and is able to inhibit coumarin 7-hydroxylation activities catalyzed by CYP2A6 and 2A13, indicating that these P450 enzymes may participate in the metabolism of this TCB congener (Shimada et al., 2013).
Fig. 1.
Structure of chemicals used in this study.
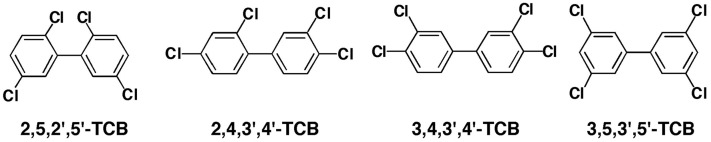
In this study, we determined whether human CYP2A6 and 2A13 and other human P450 enzymes are able to oxidize 2,5,2′,5′-TCB to oxygenated products, and how homologous monkey CYP2A enzymes oxidize this TCB. We used recombinant human and monkey P450 enzymes expressed in Escherichia coli and in microsomes of Trichoplusia ni cells and analyzed product formation with gas chromatography–mass spectrometry (GC-MS) and liquid chromatography–tandem mass spectrometry (LC-MS/MS) analyses. Molecular docking simulation was used to probe the interaction of 2,5,2′,5′-TCB with active sites (over the heme) of these CYP2A enzymes. Other TCB isomers, including 2,4,3′,4′-, 3,4,3′,4′-, and 3,5,3′,5′-TCBs, which have been shown to be contaminants in the environment (Akutsu et al., 2005; Konishi et al., 2006; Todaka et al., 2008), were also studied for their oxidation by these P450 enzymes.
Materials and Methods
Chemicals.
2,5,2′,5′-TCB (Fig. 1) and 4-hydroxy-2,5,2′,5′-TCB were synthesized as reported previously (Yoshimura et al., 1975; Ishida et al., 1991; Koga et al., 1995a). 3-Hydroxy-2,5,2′,5′-TCB was isolated from the feces of rats treated i.p. with 2,5,2′,5′-TCB at a single dose of 200 mg/kg of body weight according to a method reported previously (Hanioka et al., 1991). 2,4,3′,4′-, 3,4,3′,4′-, and 3,5,3′,5′-TCB (Fig. 1) and the metabolites 3-hydroxy-2,5,3′,4′-TCB, a mixture of 3-hydroxy- and 4-hydroxy-2,5,3′,4′-TCB, 5-hydroxy-2,4,3′,4′-TCB, 4-hydroxy-2,3,3′,4′-TCB (a metabolite of 2,4,3′,4′-TCB), and 4-hydroxy-3,5,3′,5′-TCB were obtained from the sources described previously (Koga et al., 1989, 1992, 1994).
Other chemicals and reagents used in this study were obtained from the sources described previously or were of the highest quality commercially available (Shimada et al., 2013, 2015, 2016b).
Enzymes.
Expression in E. coli and purification of human P450 enzymes were described previously (Parikh et al., 1997; Shimada et al., 2009, 2011, 2013). Bacterial bicistronic CYP1A2, 1B1, 2A6, 2A13, 2C9, and 3A4 membranes in which human NADPH-P450 reductase was coexpressed were prepared, and the E. coli membranes were suspended in 10 mM Tris-HCl buffer (pH 7.4) containing 1.0 mM EDTA and 20% glycerol (v/v) as previously described (Sandhu et al., 1993, 1994; Guengerich, 2015). CYP1A1, 2A6, 2A13, 2C9, 3A4, NADPH-P450 reductase, and cytochrome b5 (b5) were purified from membranes of recombinant E. coli as described elsewhere (Sandhu et al., 1993, 1994; Shimada et al., 2013; Guengerich, 2015).
Expression of cynomolgus monkey CYP2A23, 2A24, and 2A26 in E. coli together with human NADPH-P450 reductase was done as described previously (Uehara et al., 2014, 2015).
Recombinant CYP2B6, 2E1, and 2D6, expressed in microsomes of T. ni cells infected with a baculovirus containing human P450 and NADPH-P450 reductase cDNA inserts, were obtained from GENTEST (Woburn, MA). P450 contents in these microsomes were based on the values in the data sheets provided by the manufacturer.
Spectral Binding Titrations.
Purified CYP2A6 and 2A13 enzymes were diluted to 1.0 µM in 0.10 M potassium phosphate buffer (pH 7.4) containing 20% glycerol (v/v), and binding spectra were recorded with subsequent additions of 2,5,2′,5′-TCB and other TCB congeners in a JASCO V-550 or an OLIS-Aminco DW2a spectrophotometer (On-Line Instrument Systems, Bogart, GA) as described previously (Shimada et al., 2009, 2011, 2013). Spectral dissociation constants (Ks) were estimated using GraphPad Prism software (GraphPad Software, San Diego, CA), using either hyperbolic plots or quadratic fits for tight binding.
Oxidation of 2,5,2′,5′-TCB and Other TCB Isomers.
Oxidative metabolism of 2,5,2′,5′-TCB and other TCB isomers by P450 enzymes was determined in a standard incubation mixture (0.25 ml) containing bicistronic P450s (50 pmol P450 in E. coli membranes or 10 pmol P450s in microsomes of T. ni cells coexpressing human NADPH-P450 reductase), 50 µM each TCB isomer, and an NADPH-generating system consisting of 0.5 mM NADP+, 5 mM glucose 6-phosphate, and 0.5 units of yeast glucose 6-phosphate dehydrogenase/ml (Shimada et al., 2015, 2016b). [The PCBs and other chemicals were dissolved in (CH3)2SO as 10 mM stock solutions and diluted into aqueous solution, with the final solvent concentration ≤0.5% (v/v).]
In reconstitution experiments, P450 membranes were replaced by purified P450 (50 pmol), NADPH-P450 reductase (100 pmol), b5 (100 pmol) (when required), and l-α-dilauroyl-syn-glycero-3-phosphocholine (50 µg) as described previously (Shimada et al., 2015, 2016b). Our previous studies have suggested that more than 2-fold excess NADPH-P450 reductase over P450 is required to account for full catalytic activities for drug oxidations in reconstituted systems, although the ratio of expression of the reductase and P450 is not always as high in bicistronic systems (Shimada et al., 2000; Yamazaki et al., 2002). Incubations were carried out at 37°C, following a preincubation time of 1 minute. Reactions were terminated by adding 0.25 ml of cold CH3OH and then extracted with 0.5 ml of a CHCl3-ethyl acetate mixture [1:1 (v/v)] twice. The organic layers were subjected to filtration using Disposable Syringe Filters (Cellulose Acetate Membrane, 3 mm × 0.20 µm, code 2012-003; Iwaki, Osaka, Japan), and the filtrates (10 µl) were used for analysis with GC-MS and LC-MS/MS.
LC-MS/MS Analysis of 2,5,2′,5′-TCB Metabolites.
LC-MS/MS analyses were performed using a liquid chromatograph (ACQUITY UPLC I-Class system; Waters, Milford, MA) coupled to a tandem quadruple mass spectrometer (Xevo TQ-S; Waters) (Kakimoto et al., 2015). Chromatographic separation was performed on an octadecylsilane (C18) column (CORTECS C18, 100 × 2.1–mm i.d., 1.6 μm; Waters) or an RP-amide column (Ascentis Express, 100 × 2.1–mm i.d., 2 μm; Sigma-Aldrich, St. Louis, MO). The temperature of the C18 column was maintained at 45°C, and gradient elution [CH3CN; 35% in water to 75% (v/v) over 10 minutes] was performed at a flow rate of 0.25 ml/min. The temperature of the RP-amide column was maintained at 7°C, and gradient elution [acetonitrile; 75% in water to 95% (v/v) over 10 minutes; hold for 2 minutes] was used at a flow rate of 0.15 ml/min.
MS/MS analysis was performed on a negative electrospray ionization mode with a capillary voltage of 3000 V, cone voltage of −10 V, and collision energy of 20 eV. The selected reaction-monitoring mode was used to quantify the m/z 306.9 to 270.9 transitions for identification of monohydroxylated metabolites and for further verification (the m/z 306.9 to 234.8 transition was also determined). The product ion m/z 306.9 was scanned following elution from a C18 column; the same collision energy of the selected reaction-monitoring mode was used for obtaining the product ion mass spectrum.
GC/MS Analysis of TCB Isomers.
GC-MS was performed with a gas chromatograph (7890A series; Agilent Technologies, Santa Clara, CA) coupled with a mass spectrometer (Quattro micro GC; Waters) using a capillary column (VF-5ms, 30 m × 0.25–mm i.d., 0.25-μm film thickness; Agilent Technologies); the column temperature program was as follows: 60°C (held for 1 minute) to 140°C at 20°C/min, and to 260°C at 8°C/min, then to 300°C (held for 5 minutes) at 40°C/min; carrier gas, helium was set at 1.0 ml/min. The MS conditions used were as follows: electron ionization mode; electron energy, 70 eV; and ion source temperature, 250°C. The selected ion-monitoring mode of m/z 292 was used to quantify TCB concentration, and for further verification, the m/z 290 was also used. Mass Lynx (version 4.1) was used to control the LC-MS/MS and GC-MS/MS systems and to acquire and process the data.
Other Assays.
P450 and protein contents and coumarin 7-hydroxylation activities (CYP2A6 and 2A13) were determined by methods described previously (Omura and Sato, 1964; Brown et al., 1989).
Docking Simulations in Human P450 Enzymes.
Crystal structures of CYP2A6 bound to coumarin [Protein Data Bank (PDB) 1Z10], pilocarpine (PDB 3T3R), and nicotine (PDB 4EJJ), and CYP2A13 bound to nicotine (PDB 4EJG), 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) (PDB 4EJH), indole (PDB 2P85), and pilocarpine (PDB 3T3S) have been reported and were used in this study (Yano et al., 2005; Smith et al., 2007; DeVore et al., 2009, 2012; DeVore and Scott, 2012a; Shimada et al., 2016a). Simulations were carried out after removing each ligand from these P450 structures using the MMFF94x force field described in the MOE software (version 2015.10; Computing Group, Montreal, Canada) as previously described (Shimada et al., 2010, 2011, 2013). Crystal structures of monkey CYP2A23, 2A24, and 2A26 have not yet been reported, and these P450 primary sequences were aligned with human CYP2A6 (PDB 1Z10) for CYP2A24, and with CYP2A13 (PDB 4EJG) for CYP2A23 and 2A26 using MOE software for modeling of three-dimensional structures (Chemical Computing Group, Inc., Montreal, Canada). Ligand-interaction energies (U values) were obtained by use of the ASE dock program in the MOE software. A lower U value is an indication of a higher interaction between a chemical and the enzyme.
Kinetic Analysis.
Kinetic parameters were estimated by nonlinear regression analysis of hyperbolic plots using the program KaleidaGraph (Synergy Software, Reading, PA) or GraphPad Prism (GraphPad Software, La Jolla, CA).
Results
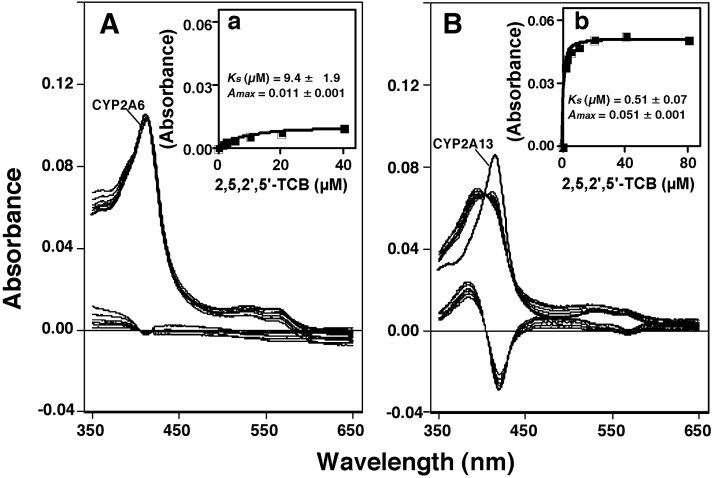
Spectral Interactions of 2,5,2′,5′-TCB with Purified CYP2A6 and 2A13.
We first examined the spectral interaction of 2,5,2′,5′-TCB with purified CYP2A6 and 2A13 and found that CYP2A13 interacted more strongly with TCB to produce type I binding spectra than CYP2A6 did (Fig. 2). The Ks value was 0.51 µM for CYP2A13, whereas that for CYP2A6 was 9.4 µM. The type I spectral changes (Ks values) of interaction of CYP2A13 with 2,4,3′,4′-, 3,4,3′,4′-, and 3,5,3′,5′-TCB were >80, 54, and 13.6 µM, respectively, and such spectral interactions of these TCBs with CYP2A6 were not found.
Fig. 2.
Spectral interaction of 2,5,2′,5′-TCB with CYP2A6 and 2A13. The absolute spectra were first obtained, and then this was repeated after adding 2,5,2′,5′-TCB to a 1 µM concentration of each P450 enzyme (A and B, top). Difference spectra were then obtained by subtracting the P450 spectra in the absence of 2,5,2′,5′-TCB from the P450 spectra in the presence of 2,5,2′,5′-TCB (bottom part of the figures). Spectral dissociation constants (Ks) were estimated from hyperbolic plots using GraphPad Prism software (GraphPad Software) (a and b).
GC-MS Analysis of Metabolism of 2,5,2′,5′-TCB by CYP2A6 and 2A13.
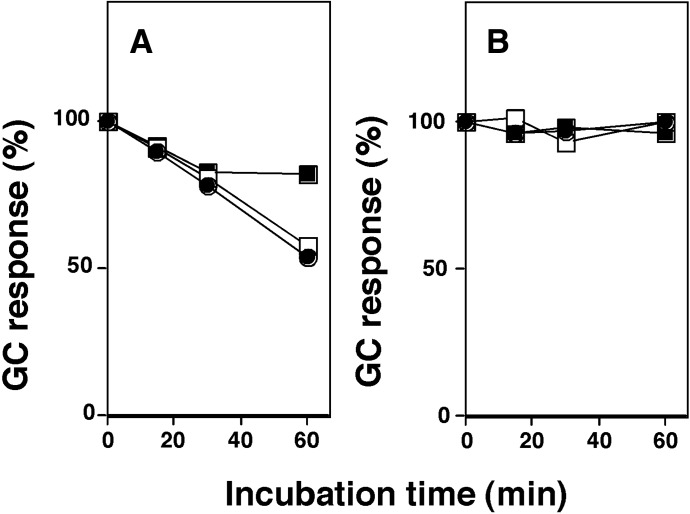
2,5,2′,5′-TCB was incubated with CYP2A6 and 2A13 in the presence of an NADPH-generating system, and the parent compound and its metabolites were extracted with CH3OH and then with CHCl3-ethyl acetate mixture [1:1 (v/v)]. The organic layer, after the centrifugation, was submitted directly to GC-MS to determine the changes in TCB concentration during the metabolism (note that TCB levels were decreased when the extracts were evaporated to dryness under a nitrogen atmosphere). Incubation with CYP2A6, but not CYP2A13, caused decreases in TCB concentration in a time-dependent manner, and the decreases were more marked at lower substrate concentrations, resulting in 50% decreases in TCB during 60-minute incubation (Fig. 3, A and B).
Fig. 3.
Effects of incubation time on the levels of 2,5,2′,5′-TCB after metabolism with CYP2A6 (A) and 2A13 (B). TCB at 12.5 µM (closed circles), 25 µM (open squares), and 50 µM (closed squares) were incubated with 0.05 µM CYP2A6 (A) or 2A13 (B) for different periods of time, and after incubation, the remaining TCB was extracted with CH3OH and then with a CHCl3–ethyl acetate mixture [1:1 (v/v)]. The organic layer, after the centrifugation, was submitted directly for GC-MS to determine the levels of TCB. Results are expressed as means of duplicate determinations.
Metabolism of 2,5,2′,5′-TCB by CYP2A6.
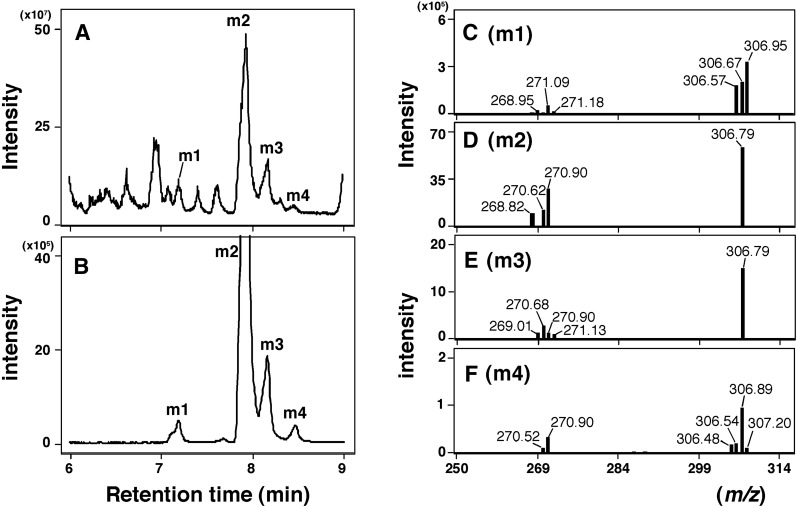
Since the aforementioned results suggested that CYP2A6 was more active than CYP2A13 in metabolizing 2,5,2′,5′-TCB, LC-MS/MS analysis (with a CORTECS C18 column) was carried out to identify the oxidative metabolites (Fig. 4). We first searched the products with a molecular mass between m/z 250 and m/z 350 and found several peaks in which four oxidative metabolites (namely, m1, m2, m3, and m4) were suggested to be monooxygenated products with (molecular ions) m/z 307 (Fig. 4, A and B). We also searched whether dioxygenated products were formed during metabolism of TCB with CYP2A6 and found that none with a molecular mass of m/z 323 were formed (results not shown).
Fig. 4.
LC-MS/MS analysis of metabolism of 2,5,2′,5′-TCB by CYP2A6. Product formation was determined by LC (CORTECS C18 column) on analysis of total ion scans between m/z 250 and 350 (A) and selected reaction-monitoring (m/z 306.9 > 270.9) (B). Major metabolites were tentatively termed m1, m2, m3, and m4 in (A) and (B). The product ion mass spectra of metabolites m1 (C), m2 (D), m3 (E), and m4 (F) were also determined.
Four peaks—m1, m2, m3, and m4—with m/z 307 were further analyzed with product ion scanning to determine the fragmentation ion spectrum (Fig. 4, C–F). Elimination of HCl from the products m1, m2, m3, and m4 gave new peaks of m/z 271 in these cases (Fig. 4, C–F).
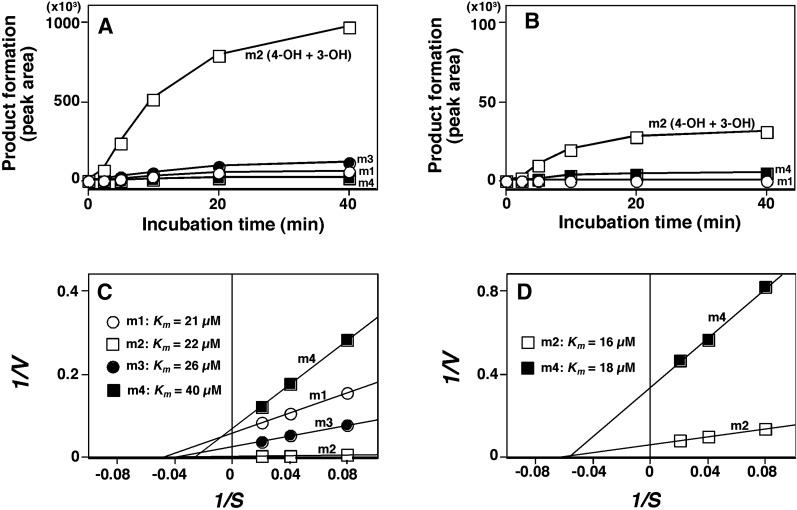
Time-dependent formation of four oxygenated products (m1, m2, m3, and m4) was analyzed on incubation of TCB with CYP2A6 and 2A13 (Fig. 5, A and B); in the case of CYP2A13, we detected only two products, m2 and m4. With both P450 enzymes examined, m2 was the most abundant, and we found that CYP2A6 was about 40-fold more active in forming m2 than CYP2A13. (Product m2 was later identified to be a mixture of 3- and 4-hydroxylated TCB products.) Kinetic analysis indicated that Km values for the formation of m1, m2, m3, and m4 by CYP2A6 were 21, 22, 26, and 40 µM, and the Km values for m2 and m4 by CYP2A13 were 16 and 18 µM, respectively (Fig. 5, C and D).
Fig. 5.
Effects of incubation time (A and B) and kinetic analysis (C and D) of oxidation of 2,5,2′,5′-TCB by CYP2A6 (A and C) and 2A13 (B and D). 2,5,2′,5′-TCB (at 25 µM) was incubated with CYP2A6 (A) and 2A13 (B) for indicated periods of time, and the metabolites formed were analyzed with LC-MS/MS. Note that the values of vertical lines are different in (A) and (B) (10-fold scale difference) since CYP2A6 gave higher rates of formation of TCB metabolites than CYP2A13. Lineweaver-Burk plots of oxidation of 2,5,2′,5′-TCB by CYP2A6 (C) and 2A13 (D) and Km values of formation of m1, m2 (mixture of 3- and 4-hydroxy products), m3, and m4 by CYP2A6 and those of m2 and m4 by CYP2A13 are indicated in the figures. Results are expressed as means of two to three experiments.
The effects of b5 on CYP2A6- and 2A13-dependent oxidation of 2,5,2′,5′-TCB in reconstituted monooxygenase systems containing human NADPH-P450 reductase and l-α-1,2-dilauroyl-sn-glycero-3-phosphocholine were examined, and none of the activities were enhanced with b5 were observed in our assay conditions (results not shown).
Oxidation of 2,5,2′,5′-TCB by Human P450 Enzymes and Monkey CYP2A Enzymes.
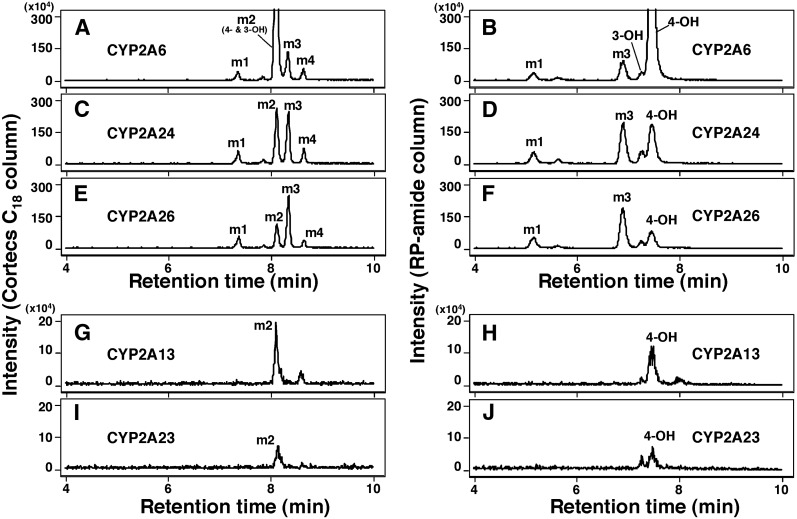
We compared product formation of 2,5,2′,5′-TCB by human CYP2A6 and 2A13 and monkey CYP2A23, 2A24, and 2A26 by using two LC columns, a CORTECS C18 column (Fig. 6, A, C, E, G, and I) and an RP-amide column (Fig. 6, B, D, F, H, and J). The former column did not separate 3- and 4-hydroxylated products (showing a single peak at the position of m2) (Fig. 6, A, C, E, G, and I). However, the RP-amide column was used to successfully identify the 4- and 3-hydroxy products; the formation of 4-hydroxy product was greater than the 3-OH product with all of the human and monkey CYP2A enzymes examined (Fig. 6, B, D, F, H, and J). CYP2A6 produced 4-hydroxy-TCB at the highest rate, and CYP2A24 and 2A26 produced metabolite m3 at higher rates than CYP2A6. Both human CYP2A13 and monkey CYP2A23 had low catalytic activities for oxidizing this TCB.
Fig. 6.
Oxidation of 2,5,2′,5′-TCB by CYP2A6 (A and B), CYP2A24 (C and D), CYP2A26 (E and F), CYP2A13 (G and H), and CYP2A23 (I and J) on analysis with LC-MS/MS using a CORTECS C18 column (A, C, E, G, and I) and RP-amide column (B, D, F, H, and J). P450 and 2,5,2′,5′-TCB concentrations used were 0.1 and 50 µM, respectively, and incubation time was 60 minutes.
Ten human P450 enzymes (CYP1A1, 1A2, 1B1, 2A6, 2A13, 2B6, 2D6, 2E1, 2C9, and 3A4) and three monkey CYP2A enzymes were compared with regard to their catalytic activities in oxidizing 2,5,2′,5′-TCB (Table 1). Separate experiments with GC-MS were also performed to investigate the disappearance of the substrate 2,5,2′,5′-TCB after metabolism by P450s (incubating for 60 minutes), and it was found that CYP2A6, 2A24, and 2A26 (but not other P450s) caused decreases in TCB concentration by about 42, 24, and 24%, respectively (Table 1). LC-MS/MS analysis showed that, of 10 human P450s examined, only CYP2A6 and 2A13 catalyzed the oxidation of 2,5,2′,5′-TCB at significant levels. Formation of 4-hydroxy-2,5,2′,5′-TCB by CYP2A6 and 2A13 was 34- and 15-fold higher, respectively, than that of 3-hydroxy product (Table 1), and the former enzyme was more active than CYP2A13. Other human P450 enzymes did not show any detectable activities in oxidizing 2,5,2′,5-TCB. Among the three monkey CYP2A enzymes, CYP2A24 was the most active, followed by CYP2A26, and CYP2A23 was least active in catalyzing the oxidation of 2,5,2′,5′-TCB (Table 1). The ratio of formation of 4- to 3-hydroxylated products with monkey P450 2A enzymes was lower than those with human CYP2A6 and 2A13 (Table 1).
TABLE 1.
Oxidation of 2,5,2′,5′-TCB by human and monkey P450s
| P450 | Substrate Disappearancea | Product Formation |
Ratio of 4-OH/3-OH | |
|---|---|---|---|---|
| 4-OH-TCB | 3-OH-TCB | |||
| Human | % in 60 min | nmol/min/nmol P450 | nmol/min/nmol P450 | |
| CYP1A1b | <1.0 | <0.001 | <0.0001 | — |
| CYP1A2c | <1.0 | <0.001 | <0.0001 | — |
| CYP1B1*3c | <1.0 | <0.001 | <0.0001 | — |
| CYP2A6c | 42 ± 3.1 | 0.98 ± 0.11 | 0.029 ± 0.0032 | 34 |
| CYP2A13c | <1.0 | 0.017 ± 0.0041 | 0.0011 ± 0.003 | 15 |
| CYP2B6d | <1.0 | <0.001 | <0.0001 | — |
| CYP2D6d | <1.0 | <0.001 | <0.0001 | — |
| CYP2E1d | <1.0 | <0.001 | <0.0001 | — |
| CYP2C9c | <1.0 | <0.001 | <0.0001 | — |
| CYP3A4c | <1.0 | <0.001 | <0.0001 | — |
| Monkey | ||||
| CYP2A23c | <1.0 | 0.0076 ± 0.0011 | 0.0020 ± 0.0005 | 3.8 |
| CYP2A24c | 24 ± 4.4 | 0.29 ± 0.032 | 0.042 ± 0.0048 | 6.9 |
| CYP2A26c | 24 ± 3.1 | 0.13 ± 0.021 | 0.021 ± 0.0031 | 6.2 |
GC-MS analysis of 2,5,2′,5′-TCB after metabolism for 60 minutes. Results are expressed as the means ± S.D. (n = 3–5) (these values were not analyzed for statistical significance because a large number of different comparisons were made).
A reconstituted system containing CYP1A1 and NADPH-P450 reductase.
E. coli membranes coexpressing P450 and NADPH-P450 reductase.
Baculo enzyme system expressing P450 and NADPH-P450 reductase.
Oxidation of 2,4,3′,4′-, 3,4,3′,4′-, and 3,5,3′,5′-TCB by Human and Monkey P450s.
Because these results suggest that human and monkey CYP2A enzymes are active in catalyzing 2,5,2′,5′-TCB, we re-examined metabolism of 2,4,3′,4′-, 3,4,3′,4′-, and 3,5,3′,5′-TCB by these enzymes. Neither human CYP2A6 and 2A13 nor monkey CYP2A23, 2A24, or 2A26 oxidized these three TCB isomers when the chromatographic profiles of incubates were compared in the presence and absence of an NADPH-generating system (results not shown). We also studied the metabolism of 2,4,3′,4′-TCB by human CYP1A1, 1A2, 1B1, and 3A4; 3,4,3′,4′-TCB by human CYP1A1, 1A2, and 1B1; and 3,5,3′,5′-TCB by human CYP1A1, 1A2, 1B1, 2B6, and 3A4, and found that these three TCB isomers were not oxidized by these P450 enzymes (results not shown).
Docking Simulations of Interactions of 2,5,2′,5′-TCB with Human and Monkey CYP2A Enzymes.
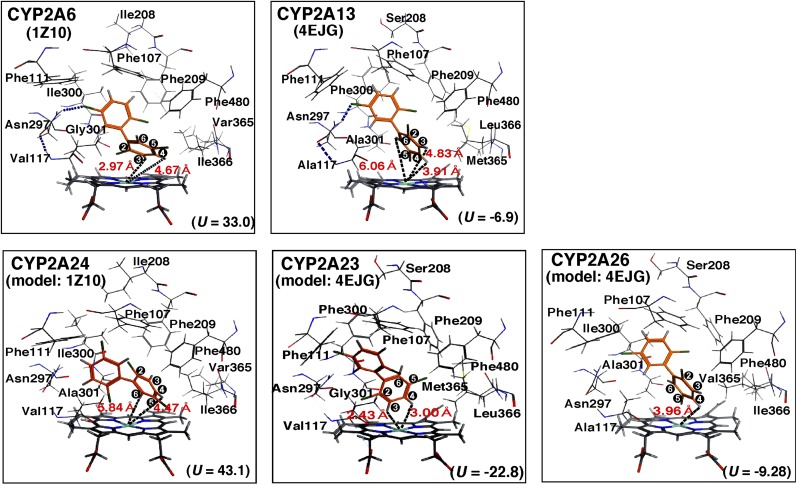
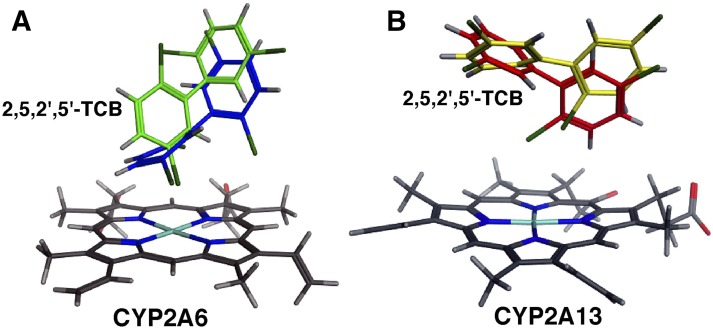
We compared the molecular interactions of 2,5,2′,5′-TCB with human and monkey CYP2A enzymes using docking simulation as described in Materials and Methods (Figs. 7). Several amino acid residues in substrate-recognition sites (SRSs) (Gotoh, 1992) were indicated in the figure. Crystal structures of CYP2A6 1Z10 and CYP2A13 4EJG were used to see the interaction of 2,5,2′,5′-TCB with human CYP2A6 and 2A13, respectively, and were used for alignment of monkey CYP2A23, 2A24, and 2A26. Ligand-interaction energies (U values) of bound protein-ligand complexes obtained for the interaction of 2,5,2′,5′-TCB with CYP2A6 and 2A13 were 33.0 and −6.9, respectively, and those with CYP2A24, 2A23, and 2A26 were 43.1, −22.8, and −9.29, respectively. The possible interaction of Asn297 with the C5′ atom of 2,5,2′,5′-TCB was found in CYP2A6 and 2A13, and the distance between C3 and C4 of the TCB and the Fe in the heme of CYP2A6 and 2A13 was calculated to be 2.97 and 4.67 Å and 6.06 and 3.91 Å, respectively (Fig. 7). The distance between the C6 atom of TCB and the Fe in the heme of CYP2A13 was 4.83 Å. The position of Gly301 in CYP2A6 was somewhat different from that of Ala301 in CYP2A13 around the TCB molecule.
Fig. 7.
Molecular docking simulation of interaction of 2,5,2′,5′-TCB with human CYP2A6 and 2A13 and monkey CYP2A24, 2A23, and 2A26. Distance between chemical sites of TCB and the centers (Fe) of the heme of CYP2A enzymes are indicated in red. Positions of carbon atoms in 2,5,2′,5′-TCB are shown in black.
Our results showed that C3 and C4 positions of 2,5,2′,5′-TCB interacted with (the heme of) CYP2A6, whereas C4 and C6 interacted with (the heme of) CYP2A24 (Fig. 7). The distribution of amino acids surrounding the TCB was not so different in CYP2A6 and 2A24, except that a possible hydrogen bond interaction between Asn297 and TCB was found in the case of CYP2A26. Molecular docking analyses of CYP2A23 and 2A26 were aligned using CYP2A13 4EJG as a model structure, and it was found that C3 and C4 of TCB interacted with (the heme of) CYP2A24, but interaction with the C4 position was found in CYP2A26.
Correlation of U Values in Interaction of Human and Monkey CYP2A Enzymes with CYP2A Ligands and Four TCBs.
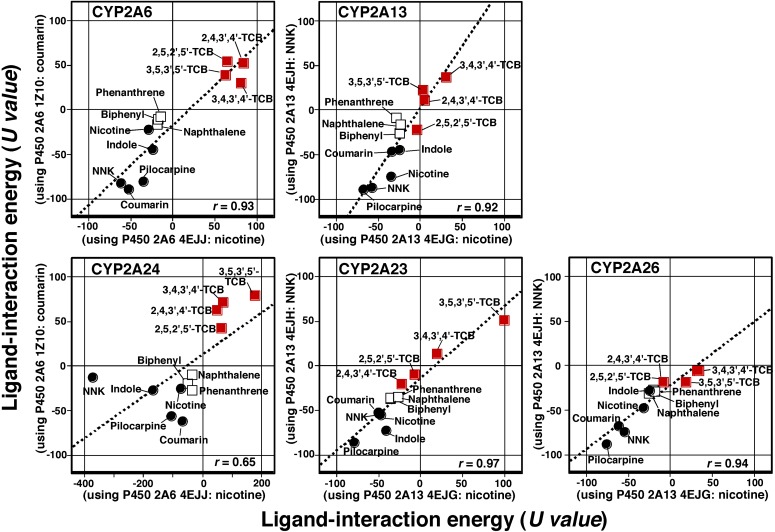
We found that there were different orientations in the molecular interaction of 2,5,2′,5′.-TCB over the heme of CYP2A6 and 2A13 when examined with CYP2A6 1Z10 and 4EJJ for CYP2A6 (Fig. 8A) and CYP2A13 4EJH and 4EJG for CYP2A13 (Fig. 8B). We compared ligand-interaction energies (U values) with the molecular structures of CYP2A6 4EJJ and 1Z10 in the cases of CYP2A6 and 2A24 and CYP2A13 4EJG and 4EJH in the cases of CYP2A13, 2A23, and 2A26 (Fig. 9). Ligands used for the comparison were four TCBs (2,4,3′,4′-, 2,5,2′,5′-, 3,4,3′,4′-, and 3,5,3′,5′-TCBs), five known CYP2A ligands (coumarin, nicotine, NNK, indole, and pilocarpine), and three chemicals (naphthalene, phenanthrene, and biphenyl) that we recently found to be good substrates of CYP2A13 and 2A6 (Shimada et al., 2016a). There were good correlations of interaction for a total of 12 chemicals with CYP2A6 and 2A13 using two reported molecular structures, except that the correlation coefficient was not as high (r = 0.66) in CYP2A24 when we used CYP2A6 1Z10 and 4EJJ for alignment of molecular structure of the enzyme.
Fig. 8.
Docking simulation of 2,5,2′,5′-TCB (shown in green and blue) to CYP2A6 1Z10 (coumarin-bound form) and CYP2A6 4EJJ (nicotine-bound form), respectively (A), and 2,5,2′,5′-TCB (shown in yellow and red) to CYP2A13 4EJH (NNK-bound form) and CYP2A13 4EJG (nicotine-bound form), respectively (B).
Fig. 9.
Correlation of ligand-interaction energies (U values) using molecular docking with CYP2A6 1Z10 (coumarin-bound form) and CYP2A6 4EJJ (nicotine-bound form) and CYP2A13 4EJG (nicotine-bound form) and CYP2A13 4EJH (NNK-bound form). Typical CYP2A substrates (coumarin, nicotine, pilocarpine, indole, and NNK) and naphthalene, phenanthrene, and biphenyl derivatives used in our recent paper (Shimada et al., 2016a) were included for correlative comparison. The correlation coefficient (r) was determined with all of these chemicals, including four TCB congeners. Similarly, correlation of U values in monkey CYP2A24, 2A23, and 2A26 enzymes and these chemicals are also shown.
By comparing the interaction of human CYP2A6 and 2A13 with four TCB congeners used in this study, the lowest U values obtained in these cases were determined when interactions of CYP2A13, 2A23, and 2A26 with 2,5,2′,5′-TCB were examined, and with the latter two P450s, the U values were low in interaction with 2,4,3′,4′-TCB (Fig. 9). Correlation coefficients of U values obtained with CYP2A6, 2A13, 2A23, 2A24, and 2A26 using crystal structures of 1Z10 and 4EJJ for CYP2A6 and 2A24 and of 4EJG and 4EJH for CYP2A13, 2A23, and 2A26 with 12 ligands were also determined (Supplemental Table 1).
Discussion
Our present results showed that CYP2A6 is an important enzyme in the oxidation of 2,5,2′,5′-TCB to form 4-hydroxy-2,5,2′,5′-TCB as a major metabolite and three to four metabolites as minor ones, including 3-hydroxy-2,5,2′,5′-TCB, and that CYP2A13 had very low activities in oxidizing 2,5,2′,5′-TCB as compared with CYP2A6. We also found decreases in the concentration of 2,5,2′,5′-TCB in the reaction mixture on GC-MS analysis when it was incubated with CYP2A6, but not CYP2A13. These two human CYP2A enzymes were not active in oxidizing 2,4,3′,4′-, 3,4,3′,4′-, and 3,5,3′,5′-TCB, as judged by the results obtained with our LC-MS/MS system. Other human P450 enzymes (including CYP1A1, 1A2, 1B1, 2B6, 2C9, and 3A4) did not catalyze the oxidation of 2,4,3′,4′-, 3,4,3′,4′-, and 3,5,3′,5′-TCB, as well as 2,5,2′,5′-TCB. In addition, we found that monkey CYP2A24, which shows 95% amino acid identity to CYP2A6, catalyzed the oxidation of 2,5,2′,5′-TCB to form 4-hydroxy- and 3-hydroxy-2,5,2′,5′-TCB at rates of ∼0.3 minute−1 and ∼0.04 minute−1, respectively. Another monkey P450, CYP2A26 (93% amino acid identity to CYP2A6) (Emoto et al., 2013), catalyzed the 4-hydroxylation of 2,5,2′,5′-TCB at a rate about one-half that catalyzed by CYP2A24. The previous two monkey P450 2A enzymes showed substrate disappearance on analysis with GC-MS (Table 1). Monkey CYP2A23, whose amino acid identity to CYP2A13 is 94% (Emoto et al., 2013), was not very active; the activities were comparable to those catalyzed by human CYP2A13. When we compared ligand-interaction energies (U values) with human and monkey CYP2A enzymes using the reported crystal structures of CYP2A6 and 2A13 (Yano et al., 2005; Smith et al., 2007; DeVore and Scott, 2012), we obtained good correlation coefficients with the 12 chemicals used in this study (Fig. 9; Supplemental Table 1), indicating that the structures of human and monkey CYP2A enzymes are similar, and that small amino acid changes in the molecular structures may define how these P450 2A enzymes catalyze the oxidation of 2,5,2′,5′-TCB. Phylogenic trees of human and monkey CYP2A enzymes revealed changes in the catalytic specificity of oxidation of ligands such as 2,5,2′,5′-TCB (Supplemental Fig. 1).
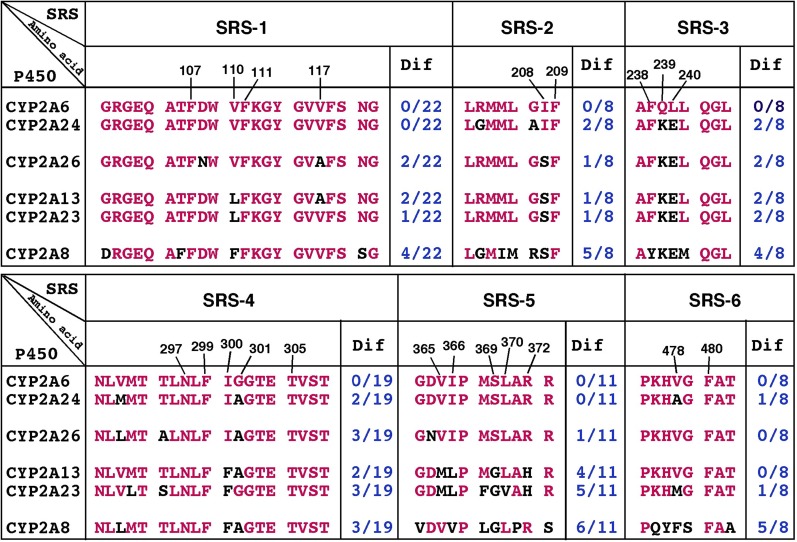
Because human CYP2A6 and monkey CYP2A24 (and CYP2A26) were more active in catalyzing the oxidation of 2,5,2′,5′-TCB than CYP2A13 and 2A23, we compared amino acid differences in SRSs (Gotoh, 1992) in these human and monkey CYP2A enzymes (Fig. 10). The differences in amino acids V110L in SRS1; I208S in SRS2; Q239K and L240E in SRS3; I300F in SRS4; V365M, I366L, and S369G in SRS5; and R372H in SRS6 were noted between CYP2A6/2A24 and CYP2A13/2A24, indicating that these residues are important in catalyzing 2,5,2′,5′-TCB, as has been reported previously using human CYP2A variants and their various ligands (He et al., 2004; Sansen et al., 2007; DeVore et al., 2009, 2012; DeVore and Scott, 2012). It is also interesting to note that there is a difference in G207A in CYP2A6 and 2A24; 4-hydroxylation of 2,5,2′,5′-TCB by the former P450 was about 3-fold higher than CYP2A24. The importance of Gly207 has been reported in mouse coumarin hydroxylation catalyzed by P450coh (Juvonen et al., 1993). Small differences in amino acid sequences in monkey CYP2A24 and 2A26 were also noted (Fig. 10) and are suggested to cause catalytic differences in these P450s.
Fig. 10.
Differences in amino acid sequences in SRS-1 through SRS-6 regions of CYP2A6, 2A24, 2A26, 2A13, 2A23, and 2A8. Amino acid residues of P450s that differed from those of CYP2A6 are indicated in black characters.
In rats, 2,5,2′,5′-TCB has been shown to be oxidized to 3-hydroxy-2,5,2′,5′-TCB as a major metabolite in vivo (Yoshimura et al., 1975; Ishida et al., 1991; Koga et al., 1995a,b), and this reaction is catalyzed by CYP2B1 and 2B2 in vitro (Preston et al., 1983; Ishida et al., 1991; Matsusue et al., 1996). Guinea pig CYP2B18, hamster P450 4-hydroxy-1-(3-pyridyl)-1-butanone-1, and rabbit CYP2B4 have also been suggested to participate in the 3-hydroxylation of 2,5,2′,5′-TCB in liver microsomes in vitro (Koga et al., 1995a,b, 1998; Ohta et al., 2009). Interestingly, hamster CYP2A8 catalyzes the oxidation of 2,5,2′,5′-TCB to form 4-hydroxy-2,5,2′,5′-TCB at a rate of 0.022 minute−1 (Koga et al., 1996); this study found that the turnover number is similar to that catalyzed by human CYP2A13 (0.017 minute−1) but not that catalyzed by CYP2A6 (∼1.0 minute−1). In fact, the amino acid identity of CYP2A8 to CYP2A13 and 2A6 has been found to be only 76 and 74%, respectively (Emoto et al., 2013), and the 2A subfamily comparisons have been shown to be different in human and hamster CYP2A enzymes (Supplemental Fig. 1). In addition, there are differences in amino acid sequences in the SRS regions between human and hamster CYP2A enzymes (Fig. 10).
2,5,2′,5′-TCB induced type I binding spectra with CYP2A6 and 2A13, with Ks values of 9.4 and 0.51 µM, respectively, although the former enzyme catalyzed oxidation of 2,5,2′,5′-TCB at a much higher rate than CYP2A13. Molecular docking simulation studies showed that ligand-interaction energies (U values) for interaction of 2,5,2′,5′-TCB with CYP2A13 4EJH (NNK type), CYP2A13 2P85 (indole type), CYP2A13 3T3S (pilocarpine type), and CYP2A13 4EJG (nicotine type), were −22.2, −7.1, −2.9, and −5.0, respectively, and with CYP2A6 3T3R (pilocarpine type), CYP2A6 4EJJ (nicotine type), and CYP2A6 1Z10 (coumarin type) were 16.4, 62, and 39, respectively. These results suggest that 2,5,2′,5′-TCB more readily interacts with CYP2A13 than CYP2A6 in inducing type I binding spectra, having lower values of ligand-interaction energies, and that these interaction affinities are not consistent with the rates of 2,5,2′,5′-TCB oxidation by these P450 enzymes.
McGraw and Waller (2006) reported that 2,4,5,2′,5′-pentachlorobiphenyl is oxidized by CYP2A6 to a 4-hydroxylated metabolite at a rate of ∼0.5 nmol/min/nmol P450, with a Km value of 34 µM. The activity reported by them is comparable to the rate of metabolism of 2,5,2′,5′-TCB by CYP2A6 found in this study, suggesting that CYP2A6 is preferentially responsible for the metabolism of PCB congeners having chlorine at the 2 and 5 positions, and that it shows attenuated oxidation of PCB congeners when they have 2,4,5-chlorine atom substitution on one of the benzene rings. Schnellmann et al. (1983) found that 2,4,5,2′,4′,5′-hexachlorobiphenyl is not metabolized by human liver microsomes, although this congener has been shown to be oxidized by human CYP2B6 to a 3-hydroxyl product at a very low rate (0.0064 nmol/min/nmol P450) (Ariyoshi et al., 1995). In comparing three PCB congeners, Ghiasuddin et al. (1976) reported that 2,5,2′-trichlorobiphenyl is extensively metabolized by rat liver microsomes, followed by 2,5,2′,5′-TCB and then 2,4,5,2′,5′-pentachlorobiphenyl.
PCBs have been identified in air, soil, water, and other environmental samples and detected in human blood and tissue samples (McFarland and Clarke, 1989; Konishi et al., 2006; International Agency for Research on Cancer, 2015). Non-Aroclor or nonlegacy PCB contaminants have been determined in the environments of homes and cities and have accumulated in the bodies of exposed populations in various cities in the United States (Sun et al., 2006; Martinez et al., 2010; Hu et al., 2012; Koh et al., 2015). 2,4,3′,4′-TCB has been reported to be one of the most persistent TCB isomers in biologic samples and human milk and can be detected in food samples such as fish and other meats, eggs, and bovine milk (Akutsu et al., 2005; Konishi et al., 2006; Todaka et al., 2008). Yoshimura et al. (1973) reported that 2,4,3′,4′-TCB is metabolized in vivo in rats to 4-hydroxy-2,5,3′,4′-TCB, which is possibly formed through a 4,5-oxide followed by an NIH-shift of a chlorine atom from the 4 position (Koga et al., 1992), indicating the possible role of P450 enzymes in the reaction. However, little is known about the in vitro metabolism of 2,4,3′,4′-TCB in humans as well as in laboratory animals, probably due to its slow oxidation by P450 enzymes. In fact, our present studies showed that none of the oxygenated metabolites of 2,4,3′,4′-TCB were detected using the determined human and monkey P450 enzymes. We also found that 3,4,3′,4′-TCB, one of the coplanar PCB congeners present in human milk (Konishi et al., 2006), was not oxidized by human CYP1A1 and other P450s to oxidative metabolites, although rat CYP1A1 has been shown to catalyze 3,4,3′,4′-TCB to form 4- and 5-hydroxylated products at a molar ratio of 2.2:1 (Ishida et al., 1991). There are species-related differences in the metabolism of 3,4,5,3′,4′-pentachlorobiphenyl in rat and human CYP1A1, in that the former enzyme catalyzes 4-hydroxylation of this compound but human CYP1A1 does not (Yamazaki et al., 2011; Inui et al. 2014).
In conclusion, these studies showed that 2,5,2′,5′-TCB oxidation is catalyzed by CYP2A6 to form 4-hydroxy-2,5,2′,5′-TCB as a major metabolite and at least four minor products, including 3-hydroxy-2,5,2′,5′-TCB, in humans. CYP2A13 oxidized 2,5,2′,5′-TCB at a very slow rate compared with CYP2A6 and other human P450 enzymes, including CYP1A1, 1A2, 1B1, 2B6, 2D6, 2E1, 2C9, and 3A4, which did not oxidize 2,5,2′,5′-TCB. The orthologous monkey CYP2A24 (95% identity to CYP2A6) was found to oxidize 2,5,2′,5′-TCB to form 4- and 3-hydroxy-2,5,2′,5′-TCB at rates of 0.29 and 0.042 minute−1, respectively. Monkey CYP2A26 (93% identity to CYP2A6) catalyzed the oxidation of 2,5,2′,5′-TCB at lower rates than CYP2A24 but at much higher rates than CYP2A23 (94% identity to CYP2A13). Small amino acid changes in the SRS regions of human and monkey CYP2A enzymes are suggested to determine how these P450 enzymes catalyze the oxidation of 2,5,2′,5′-TCB. Ligand-interaction energies (U values) in molecular docking analysis were suggested to be useful markers to predict the molecular interaction of 12 chemicals (four TCBs, five known P450 2A substrates, and naphthalene, phenanthrene, and biphenyl) with human and monkey CYP2A enzymes.
Abbreviations
- b5
cytochrome b5
- GC-MS
gas chromatography–mass spectrometry
- HPB
4-hydroxy-1-(3-pyridyl)-1-butanone
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- NNK
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- P450
cytochrome P450
- PCB
polychlorinated biphenyl
- PDB
Protein Data Bank
- SRS
substrate-recognition site
- TCB
tetrachlorobiphenyl
Authorship Contributions
Participated in research design: Shimada, Kakimoto, Murayama, Takenaka.
Conducted experiments: Shimada, Kakimoto, Murayama.
Contributed new reagents or analytic tools: Koga, Uehara, Kim.
Performed data analysis: Shimada, Kakimoto, Takenaka, Murayama, Uehara, Yamazaki.
Wrote or contributed to the writing of the manuscript: Shimada, Yamazaki, Guengerich, Komori.
Footnotes
T.S., K.K., S.T., and M.K. were supported in part by grants from the Ministry of Education, Science, and Culture of Japan and the Ministry of Health and Welfare of Japan. N.K., S.U., H.Y., and F.P.G. were also supported partly by a Health and Labor Scientific Research Grant [Grant H27-food-designated-017], the Japan Society for the Promotion of Science Grant-in-Aid for Young Scientists B [Grant 15K18934] and Scientific Research [Grant 26460206], and by the United States Public Health Service [Grant R01 GM118122], respectively.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Akutsu K, Kuwabara K, Konishi Y, Matsumoto H, Murakami Y, Tanaka Y, Matsuda R, Hori S. (2005) [Congener-specific analysis of Pcbs in food samples by using GC/MS]. Shokuhin Eiseigaku Zasshi 46:99–108 in Japanese. [DOI] [PubMed] [Google Scholar]
- Ariyoshi N, Oguri K, Koga N, Yoshimura H, Funae Y. (1995) Metabolism of highly persistent PCB congener, 2,4,5,2′,4′,5′-hexachlorobiphenyl, by human CYP2B6. Biochem Biophys Res Commun 212:455–460. [DOI] [PubMed] [Google Scholar]
- Arnich N, Tard A, Leblanc JC, Le Bizec B, Narbonne JF, Maximilien R. (2009) Dietary intake of non-dioxin-like PCBs (NDL-PCBs) in France, impact of maximum levels in some foodstuffs. Regul Toxicol Pharmacol 54:287–293. [DOI] [PubMed] [Google Scholar]
- Borlakoglu JT, Haegele KD, Reich HJ, Dils RR, Wilkins JP. (1991) In vitro metabolism of [14C]4-chlorobiphenyl and [14C]2,2′,5,5′-tetrachlorobiphenyl by hepatic microsomes from rats and pigeons. Evidence against an obligatory arene oxide in aromatic hydroxylation reactions. Int J Biochem 23:1427–1437. [DOI] [PubMed] [Google Scholar]
- Brown RE, Jarvis KL, Hyland KJ. (1989) Protein measurement using bicinchoninic acid: elimination of interfering substances. Anal Biochem 180:136–139. [DOI] [PubMed] [Google Scholar]
- DeVore NM, Meneely KM, Bart AG, Stephens ES, Battaile KP, Scott EE. (2012) Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine. FEBS J 279:1621–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVore NM, Scott EE. (2012) Nicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone binding and access channel in human cytochrome P450 2A6 and 2A13 enzymes. J Biol Chem 287:26576–26585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVore NM, Smith BD, Wang JL, Lushington GH, Scott EE. (2009) Key residues controlling binding of diverse ligands to human cytochrome P450 2A enzymes. Drug Metab Dispos 37:1319–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto C, Yoda N, Uno Y, Iwasaki K, Umehara K, Kashiyama E, Yamazaki H. (2013) Comparison of p450 enzymes between cynomolgus monkeys and humans: p450 identities, protein contents, kinetic parameters, and potential for inhibitory profiles. Curr Drug Metab 14:239–252. [PubMed] [Google Scholar]
- Ghiasuddin SM, Menzer RE, Nelson JO. (1976) Metabolism of 2,5,2′ -trichloro-, 2,5,2′,5′ -tetrachloro-, and 2,4,5,2′,5′ -pentachlorobiphenyl in rat hepatic microsomal systems. Toxicol Appl Pharmacol 36:187–194. [DOI] [PubMed] [Google Scholar]
- Gotoh O. (1992) Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem 267:83–90. [PubMed] [Google Scholar]
- Guengerich FP.(2015) Human cytochrome P450 enzymes, in Cytochrome P450: Structure, Mechanism, and Biochemistry (Ortiz de Montellano PR. ed) 4th ed, pp 523–785, Kluwer Academic/Plenum Press, New York. [Google Scholar]
- Hanioka N, Saeki HK, Ishida C, Koga N, Yoshimura H. (1991) [Toxicological assessment of 2,5,2′,5′-tetrachlorobiphenyl and its major metabolite, 3-hydroxy-2,5,2′,5′-tetrachlorobiphenyl in rats]. Fukuoka Igaku Zasshi 82:191–196. [PubMed] [Google Scholar]
- He XY, Shen J, Hu WY, Ding X, Lu AY, Hong JY. (2004) Identification of Val117 and Arg372 as critical amino acid residues for the activity difference between human CYP2A6 and CYP2A13 in coumarin 7-hydroxylation. Arch Biochem Biophys 427:143–153. [DOI] [PubMed] [Google Scholar]
- Hu X, Adamcakova-Dodd A, Lehmler HJ, Hu D, Hornbuckle K, Thorne PS. (2012) Subchronic inhalation exposure study of an airborne polychlorinated biphenyl mixture resembling the Chicago ambient air congener profile. Environ Sci Technol 46:9653–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Agency for Research on Cancer (2015) Polychlorinated biphenyls and polybrominated biphenyls. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans 107:1–513 [PMC free article] [PubMed] [Google Scholar]
- Inui H, Itoh T, Yamamoto K, Ikushiro S, Sakaki T. (2014) Mammalian cytochrome P450-dependent metabolism of polychlorinated dibenzo-p-dioxins and coplanar polychlorinated biphenyls. Int J Mol Sci 15:14044–14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida C, Koga N, Hanioka N, Saeki HK, Yoshimura H. (1991) Metabolism in vitro of 3,4,3′,4′- and 2,5,2′,5′-tetrachlorobiphenyl by rat liver microsomes and highly purified cytochrome P-450. J Pharmacobiodyn 14:276–284. [DOI] [PubMed] [Google Scholar]
- Juvonen RO, Iwasaki M, Sueyoshi T, Negishi M. (1993) Structural alteration of mouse P450coh by mutation of glycine-207 to proline: spin equilibrium, enzyme kinetics, and heat sensitivity. Biochem J 294:31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto K, Nagayoshi H, Inazumi N, Tani A, Konishi Y, Kajimura K, Ohura T, Nakano T, Tang N, Hayakawa K, et al. (2015) Identification and characterization of oxidative metabolites of 1-chloropyrene. Chem Res Toxicol 28:1728–1736. [DOI] [PubMed] [Google Scholar]
- Koga N, Beppu M, Ishida C, Yoshimura H. (1989) Further studies on metabolism in vivo of 3,4,3′,4′-tetrachlorobiphenyl in rats: identification of minor metabolites in rat faeces. Xenobiotica 19:1307–1318. [DOI] [PubMed] [Google Scholar]
- Koga N, Kanamaru T, Kikuichi N, Oishi N, Kato S, Yoshimura H. (1998) Guinea pig liver cytochrome P450 responsible for 3-hydroxylation of 2,5,2′,5′-tetrachlorobiphenyl. Bull Environ Contam Toxicol 60:898–903. [DOI] [PubMed] [Google Scholar]
- Koga N, Kikuichi N, Kanamaru T, Ariyoshi N, Oguri K, Yoshimura H. (1996) Hamster liver cytochrome P450 (CYP2A8) as a 4-hydroxylase for 2,5,2′,5′-tetrachlorobiphenyl. Biochem Biophys Res Commun 225:685–688. [DOI] [PubMed] [Google Scholar]
- Koga N, Kikuichi-Nishimura N, Hara T, Harada N, Ishii Y, Yamada H, Oguri K, Yoshimura H. (1995a) Purification and characterization of a newly identified isoform of cytochrome P450 responsible for 3-hydroxylation of 2,5,2′,5′-tetrachlorobiphenyl in hamster liver. Arch Biochem Biophys 317:464–470. [DOI] [PubMed] [Google Scholar]
- Koga N, Kikuichi-Nishimura N, Yoshimura H. (1995b) Effect of cytochrome P450 inducers on liver microsomal metabolism of tetrachlorobiphenyls in rats, guinea pigs and hamsters. Biol Pharm Bull 18:705–710. [DOI] [PubMed] [Google Scholar]
- Koga N, Nishimura N, Kuroki H, Masuda Y, Yoshimura H. (1994) Metabolism of 3,5,3′,5′-tetrachlorobiphenyl by rat liver microsomes and purified P4501A1. Xenobiotica 24:775–783. [DOI] [PubMed] [Google Scholar]
- Koga N, Shin’yama A, Ishida C, Hanioka N, Yoshimura H. (1992) A new metabolite of 2,4,3′,4′-tetrachlorobiphenyl in rat feces. Chem Pharm Bull (Tokyo) 40:3338–3339. [DOI] [PubMed] [Google Scholar]
- Koh WX, Hornbuckle KC, Thorne PS. (2015) Human serum from urban and rural adolescents and their mothers shows exposure to polychlorinated biphenyls not found in commercial mixtures. Environ Sci Technol 49:8105–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi Y, Kitagawa M, Akutsu K, Tanaka Y. (2006) Surveillance of polychlorinated biphenyl congeneric patterns in human breast milk from 1973 to 2000 in Osaka, Japan. Environ Health Prev Med 11:38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunita N, Kashimoto T, Miyata H, Fukushima S, Hori S, Obana H. (1984) Causal agents of yusho. Am J Ind Med 5:45–58. [PubMed] [Google Scholar]
- Kuratsune M, Yoshimura T, Matsuzaka J, Yamaguchi A. (1972) Epidemiologic study on Yusho, a poisoning caused by ingestion of rice oil contaminated with a commercial brand of polychlorinated biphenyls. Environ Health Perspect 1:119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Norström K, Wang K, Hornbuckle KC. (2010) Polychlorinated biphenyls in the surficial sediment of Indiana Harbor and Ship Canal, Lake Michigan. Environ Int 36:849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsusue K, Ariyoshi N, Oguri K, Koga N, Yoshimura H. (1996) Involvement of cytochrome b5 in the metabolism of tetrachlorobiphenyls catalyzed by CYP2B1 and CYP1A1. Chemosphere 32:517–523. [DOI] [PubMed] [Google Scholar]
- McFarland VA, Clarke JU. (1989) Environmental occurrence, abundance, and potential toxicity of polychlorinated biphenyl congeners: considerations for a congener-specific analysis. Environ Health Perspect 81:225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw JE, Sr, Waller DP. (2006) Specific human CYP 450 isoform metabolism of a pentachlorobiphenyl (PCB-IUPAC# 101). Biochem Biophys Res Commun 344:129–133. [DOI] [PubMed] [Google Scholar]
- Mills SA, 3rd, Thal DI, Barney J. (2007) A summary of the 209 PCB congener nomenclature. Chemosphere 68:1603–1612. [DOI] [PubMed] [Google Scholar]
- Ohta C, Haraguchi K, Kato Y, Endo T, Koga N. (2009) [Metabolism of 2,2′,5,5′-tetrachlorobiphenyl (CB52) by rabbit liver microsomes]. Fukuoka Igaku Zasshi 100:200–209. [PubMed] [Google Scholar]
- Omura T, Sato R. (1964) The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem 239:2370–2378. [PubMed] [Google Scholar]
- Parikh A, Gillam EMJ, Guengerich FP. (1997) Drug metabolism by Escherichia coli expressing human cytochromes P450. Nat Biotechnol 15:784–788. [DOI] [PubMed] [Google Scholar]
- Preston BD, Miller JA, Miller EC. (1983) Non-arene oxide aromatic ring hydroxylation of 2,2′,5,5′-tetrachlorobiphenyl as the major metabolic pathway catalyzed by phenobarbital-induced rat liver microsomes. J Biol Chem 258:8304–8311. [PubMed] [Google Scholar]
- Safe S. (1993) Toxicology, structure-function relationship, and human and environmental health impacts of polychlorinated biphenyls: progress and problems. Environ Health Perspect 100:259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu P, Baba T, Guengerich FP. (1993) Expression of modified cytochrome P450 2C10 (2C9) in Escherichia coli, purification, and reconstitution of catalytic activity. Arch Biochem Biophys 306:443–450. [DOI] [PubMed] [Google Scholar]
- Sandhu P, Guo Z, Baba T, Martin MV, Tukey RH, Guengerich FP. (1994) Expression of modified human cytochrome P450 1A2 in Escherichia coli: stabilization, purification, spectral characterization, and catalytic activities of the enzyme. Arch Biochem Biophys 309:168–177. [DOI] [PubMed] [Google Scholar]
- Sansen S, Hsu MH, Stout CD, Johnson EF. (2007) Structural insight into the altered substrate specificity of human cytochrome P450 2A6 mutants. Arch Biochem Biophys 464:197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnellmann RG, Putnam CW, Sipes IG. (1983) Metabolism of 2,2′,3,3′,6,6′-hexachlorobiphenyl and 2,2′,4,4′,5,5′-hexachlorobiphenyl by human hepatic microsomes. Biochem Pharmacol 32:3233–3239. [DOI] [PubMed] [Google Scholar]
- Shimada T, Kim D, Murayama N, Tanaka K, Takenaka S, Nagy LD, Folkman LM, Foroozesh MK, Komori M, Yamazaki H, et al. (2013) Binding of diverse environmental chemicals with human cytochromes P450 2A13, 2A6, and 1B1 and enzyme inhibition. Chem Res Toxicol 26:517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Murayama N, Tanaka K, Takenaka S, Guengerich FP, Yamazaki H, Komori M. (2011) Spectral modification and catalytic inhibition of human cytochromes P450 1A1, 1A2, 1B1, 2A6, and 2A13 by four chemopreventive organoselenium compounds. Chem Res Toxicol 24:1327–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Takenaka S, Kakimoto K, Murayama N, Lim YR, Kim D, Foroozesh MK, Yamazaki H, Guengerich FP, Komori M. (2016a) Structure-function studies of naphthalene, phenanthrene, biphenyl, and their derivatives in interaction with and oxidation by cytochromes P450 2A13 and 2A6. Chem Res Toxicol 29:1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Takenaka S, Murayama N, Kramlinger VM, Kim JH, Kim D, Liu J, Foroozesh MK, Yamazaki H, Guengerich FP, et al. (2016b) Oxidation of pyrene, 1-hydroxypyrene, 1-nitropyrene and 1-acetylpyrene by human cytochrome P450 2A13. Xenobiotica 46:211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Takenaka S, Murayama N, Yamazaki H, Kim JH, Kim D, Yoshimoto FK, Guengerich FP, Komori M. (2015) Oxidation of acenaphthene and acenaphthylene by human cytochrome P450 enzymes. Chem Res Toxicol 28:268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Tanaka K, Takenaka S, Foroozesh MK, Murayama N, Yamazaki H, Guengerich FP, Komori M. (2009) Reverse type I binding spectra of human cytochrome P450 1B1 induced by flavonoid, stilbene, pyrene, naphthalene, phenanthrene, and biphenyl derivatives that inhibit catalytic activity: a structure-function relationship study. Chem Res Toxicol 22:1325–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Tanaka K, Takenaka S, Murayama N, Martin MV, Foroozesh MK, Yamazaki H, Guengerich FP, Komori M. (2010) Structure-function relationships of inhibition of human cytochromes P450 1A1, 1A2, 1B1, 2C9, and 3A4 by 33 flavonoid derivatives. Chem Res Toxicol 23:1921–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Tsumura F, Gillam EM, Guengerich FP, Inoue K. (2000) Roles of NADPH-P450 reductase in the O-deethylation of 7-ethoxycoumarin by recombinant human cytochrome P450 1B1 variants in Escherichia coli. Protein Expr Purif 20:73–80. [DOI] [PubMed] [Google Scholar]
- Smith BD, Sanders JL, Porubsky PR, Lushington GH, Stout CD, Scott EE. (2007) Structure of the human lung cytochrome P450 2A13. J Biol Chem 282:17306–17313. [DOI] [PubMed] [Google Scholar]
- Sun P, Basu I, Hites RA. (2006) Temporal trends of polychlorinated biphenyls in precipitation and air at chicago. Environ Sci Technol 40:1178–1183. [DOI] [PubMed] [Google Scholar]
- Todaka T, Hori T, Hirakawa H, Kajiwara J, Yasutake D, Onozuka D, Kato S, Sasaki S, Nakajima S, Saijo Y, et al. (2008) Congener-specific analysis of non-dioxin-like polychlorinated biphenyls in blood collected from 195 pregnant women in Sapporo City, Japan. Chemosphere 73:923–931. [DOI] [PubMed] [Google Scholar]
- Uehara S, Murayama N, Nakanishi Y, Nakamura C, Hashizume T, Zeldin DC, Yamazaki H, Uno Y. (2014) Immunochemical detection of cytochrome P450 enzymes in small intestine microsomes of male and female untreated juvenile cynomolgus monkeys. Xenobiotica 44:769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara S, Uno Y, Inoue T, Sasaki E, Yamazaki H. (2015) Substrate selectivities and catalytic activities of marmoset liver cytochrome P450 2A6 differed from those of human P450 2A6. Drug Metab Dispos 43:969–976. [DOI] [PubMed] [Google Scholar]
- Yakushiji T, Watanabe I, Kuwabara K, Tanaka R, Kashimoto T, Kunita N, Hara I. (1984) Postnatal transfer of PCBs from exposed mothers to their babies: influence of breast-feeding. Arch Environ Health 39:368–375. [DOI] [PubMed] [Google Scholar]
- Yakushiji T, Watanabe I, Kuwabara K, Yoshida S, Hori S, Fukushima S, Kashimoto T, Koyama K, Kunita N. (1979) Levels of organochlorine pesticides and polychlorinated biphenyls (PCBs) in mothers’ milk collected in Osaka Prefecture from 1969 to 1976. Arch Environ Contam Toxicol 8:59–66. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Nakamura M, Komatsu T, Ohyama K, Hatanaka N, Asahi S, Shimada N, Guengerich FP, Shimada T, Nakajima M, et al. (2002) Roles of NADPH-P450 reductase and apo- and holo-cytochrome b5 on xenobiotic oxidations catalyzed by 12 recombinant human cytochrome P450s expressed in membranes of Escherichia coli. Protein Expr Purif 24:329–337. [DOI] [PubMed] [Google Scholar]
- Yamazaki K, Suzuki M, Itoh T, Yamamoto K, Kanemitsu M, Matsumura C, Nakano T, Sakaki T, Fukami Y, Imaishi H, et al. (2011) Structural basis of species differences between human and experimental animal CYP1A1s in metabolism of 3,3′,4,4′,5-pentachlorobiphenyl. J Biochem 149:487–494. [DOI] [PubMed] [Google Scholar]
- Yano JK, Hsu MH, Griffin KJ, Stout CD, Johnson EF. (2005) Structures of human microsomal cytochrome P450 2A6 complexed with coumarin and methoxsalen. Nat Struct Mol Biol 12:822–823. [DOI] [PubMed] [Google Scholar]
- Yoshimura H, Yamamoto H, Saeki S. (1973) Metabolic studies on polychlorinated biphenyls. II. Metabolic fate of 2,4,3′,4′-tetrachlorobiphenyl in rats. Chem Pharm Bull (Tokyo) 21:2231–2236. [DOI] [PubMed] [Google Scholar]
- Yoshimura H, Yamamoto H, Yonezawa K. (1975) [Metabolic studies on polychlorinated biphenyls. VII. Metabolic fate of 2,5,2′,5′-tetrachlorobiphenyl in rats(author’s transl)]. Fukuoka Igaku Zasshi 66:555–562. [PubMed] [Google Scholar]