Abstract
Parkinson’s disease (PD) features nonmotor symptoms such as olfactory dysfunction referred to as hyposmia, an initial sign of disease progression. Metabolic dysfunction can contribute to neurodegenerative diseases, and various xenobiotics and endogenous compounds are also involved in the pathogenesis of PD. Although aerobic exercise was found to induce preservation or improvement in olfactory function in PD patients in a recent study, the exact underlying mechanism for this effect is not clear. We aimed to investigate the influence of an enriched environment (EE) on olfactory dysfunction especially via metabolic pathways related to detoxification enzymes. Eight-month-old transgenic (Tg) PD mice that overexpress human A53T α-synuclein (α-syn) were randomly allocated to an EE or standard conditions for 2 mo. The buried food test showed that EE group had significantly improved olfactory function compared to the control group. Reverse transcription polymerase chain reaction (PCR) and real-time quantitative PCR showed that expression of the detoxification enzymes––cytochrome P450 family 1 subfamily A member 2, paraoxonase 1, alcohol dehydrogenase 1, UDP glucuronosyltransferase family 2 member A1 complex locus, aldehyde oxidase homolog 2, and aldehyde glutathione peroxidase 6––was significantly increased in the olfactory bulb (OB) of the PD control group, but these enzymes were normalized in the EE group. Immunohistochemical staining of the OB showed that oxidative stress and nitrated α-syn were significantly increased in the control group but decreased in the EE group. In conclusion, we suggest that exposure to an EE decreases both oxidative stress and nitrated α-syn, resulting in normalized detoxification enzymes and amelioration of olfactory dysfunction.
Keywords: enriched environment, olfactory dysfunction, Parkinson’s disease, detoxification enzymes, oxidative stress
Introduction
An enriched environment (EE) has been used in various research studies of rearing animals for therapeutic effects through the complex combination of physical, cognitive, and social stimulation1,2. When animals are exposed to an EE, even the adult brain can undergo biochemical and histologic changes, consequently promoting brain function3. Biochemical changes including neurogenesis, axonal sprouting, and dendritic arborization are stimulated by an EE4,5. As a result, performance in various behaviors is improved by an EE, and previous studies have reported that an EE is a potential therapeutic strategy for recovery from brain damage6–9.
Parkinson’s disease (PD) is one of the most common adult neurodegenerative diseases. Hyposmia is an initial sign and one of the earliest symptoms of PD. As an olfactory dysfunction, hyposmia can be detected before motor symptoms are displayed in PD10–16. The loss of dopaminergic neurons in the substantia nigra is known as an essential discovery in PD, and Lewy bodies, eosinophilic cytoplasmic inclusions composed of abnormal aggregates of proteins such as α-synuclein (α-syn), have an important role in several neurodegenerative diseases because they can induce inflammatory reactions, mitochondrial dysfunction, and subsequent oxidative stress17. It is thought that numerous etiologies including genetic susceptibility and environmental exposure play an important role in the pathophysiology of PD18. Additionally, previous research has shown that metabolic dysfunction can contribute to neurodegenerative diseases17, and various xenobiotics and endogenous compounds are involved in the pathogenesis of PD19–23.
In previous studies using a mouse model of PD pathology by the administration of 1-methyl-4-phenyl-1,2,3,6-tetrahydrophyridine (MPTP), the therapeutic effect of an EE in behavioral recovery of motor function has been reported24,25. On the other hand, there is no research on the effect of an EE on olfactory dysfunction, which is a nonmotor symptom, in PD mice. Additionally, in a recent study, aerobic exercise induced the preservation or improvement of olfactory function in PD patients26. However, the exact mechanism underlying olfactory dysfunction in PD is not yet known, and it is not clear why exercise can lead to an improvement in olfactory function. Thus, this study aims to confirm the recovery of olfactory dysfunction by an EE in a PD transgenic (Tg) mouse model in which human A53T α-syn is overexpressed. Through this Tg mouse model of PD, we investigated the influence of EE on olfactory dysfunction, especially via the metabolic pathways related to detoxification enzymes.
Materials and Methods
Tg Mouse Model
The human α-syn (A53T) Tg line G2-3 (B6.Cg-Tg [Prnp-SNCA*A53T] 23 Mkle/J; Jackson Laboratories, stock no. 006823, Bar Harbor, ME, USA) was used to generate both wild-type (WT) and Tg mice. The Tg mice produced heterozygous offspring that overexpressed one copy of A53T mutant human α-syn. All animals were housed in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care and provided food and water ad libitum with alternating 12-h light/dark cycles according to animal protection regulations. The experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC 2017-0039).
Genotyping
Genotyping of mice was performed based on a protocol from Jackson Laboratories. Genomic DNA (gDNA) was extracted from a 2 mm piece of each mouse tail using the standard procedure of the prepGEM Tissue Kit (ZyGEM, Hamilton, New Zealand). The tissue from the mouse tail was incubated with 1 μL of prepGEM, 10 μL of Buffer Gold (ZyGEM, Hamilton, New Zealand), and 89 μL of autoclaved 3′ distilled water at 75 °C for 15 min and 95 °C for 5 min. The following primers were used for the polymerase chain reaction (PCR): transgene forward, 5′-TCATGAAAGGACTTTCAAAGGC-3′ transgene reverse, 5′-CCTCCCCCAGCCTAGACC-3′ (transgene = ∼500 bp); internal positive control forward, 5'-CTAGGCCACAGAATTGAAAGATCT-3′; internal positive control reverse, 5′-GTAGGTGGAAATTCTAGCATCC-3′ (internal positive control = 324 bp). Electrophoresis was performed by loading 10 μL of each PCR product on a 1.5% agarose gel (Medicago, Quebec, Canada).
EE
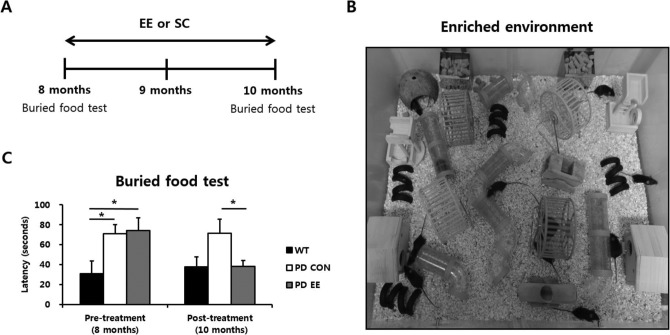
For the EE, mice were housed for 2 mo in a large cage (86 × 76 × 31 cm3) containing novel objects such as tunnels, shelters, toys, and running wheels for voluntary exercise and allowing for social interaction (12 to 15 mice/cage), whereas the control group mice were housed in standard cages (27 × 22.5 × 14 cm3) without social interaction (3 to 5 mice/cage). A schematic time line of this experiment from 8 to 10 mo of age is provided in Fig. 1A. An image of EE is provided in Fig. 1B.
Fig. 1.
Experimental design and effect of an enriched environment on olfactory dysfunction in Parkinson's disease. (A) Schematic timeline of the experiment in a mouse model of PD. (B) An image of EE. (C) Buried food test result. The latency time of finding food in PD control group (N = 14, 71.0 ± 9.2 sec, P < 0.05) and PD EE group (N = 11, 74.2 ± 12.8 sec, P < 0.05) significantly increased compared to WT group (N = 8, 30.7 ± 4.6 sec) at 8 mo of age. The result of latency time to find food at 10 mo of age showed that the PD EE group (37.9 ± 6.1 sec, P < 0.05) significantly decreased compared to PD control group (71.3 ± 14.2 sec). Abbreviations: PD = Parkinson's disease; EE = enriched environment; WT = wild type. *P < 0.05 is based on a one-way analysis of variance followed by a post hoc test.
Buried Food Test
The buried food test measures how quickly an overnight-fasted animal can find a small piece of familiar palatable food, such as cookies, cereal, chocolate chips, or food pellets, that is hidden underneath a layer of bedding27. The test was performed twice on mice at 8 and 10 mo of age. Before starting the test, the mice were fasted for 14 to 18 h to ensure motivation. The mice were placed individually in clean holding cages for 5 min, transferred to test cages for 2 min, and then returned to the holding cage, while a pellet was buried approximately 0.5 cm below the bedding in a random location to eliminate a learning component. After food placement, each mouse was placed in the center of the test cage and given 5 min to find the pellet, while the latency to sniff, dig up, and begin eating food was recorded using a stop watch28.
RNA Preparation
Total RNA was extracted from the mouse olfactory bulb (OB) using Trizol (Invitrogen Life Technologies, Carlsbad, CA, USA). After deoxyribonuclease (DNase) digestion and clean-up procedures, the RNA samples were quantified, aliquoted, and stored at −80 °C until further use. RNA purity was evaluated by the A260:A280 ratio and analyzed with an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA).
Reverse Transcription PCR (RT-PCR)
We performed RT-PCR for genes that were related to detoxification enzymes. For RT-PCR, the following reaction-specific primers were used: cytochrome P450 family 1 subfamily A member 2 (CYP1A2), forward 5′-GCTTCTCCATAGCCTCGGAC-3′ and reverse 5′-TTAGCCACCGATTCCACCAC-3′; paraoxonase 1 (PON1), forward 5′-ATGACGCAGAGAATCCTCCC-3′ and reverse 5′-TTTGTACACAGAGGCGACCG-3′; alcohol dehydrogenase 1 (ADH1), forward 5′-GACATAGAAGTCGCACCCCC-3′ and reverse 5′-CCAACGCTCTCAACAATGCC-3′; aldehyde oxidase homolog 2 (AOH2), forward 5′-CTCGGGGAGTCTGGGATGTT-3′ and reverse 5′-GTTTTTGGGTCATCTCTCGGG-3′; UDP glucuronosyltransferase family 2 member A1 complex locus (UGT2A1), forward 5′-CTAGGAATGAGTCTTGGTGGGA-3′ and reverse 5′-GGCCACAAGGACAGTCACATTA-3′; glutathione peroxidase 6 (GPX6), forward 5′-CAGAAGTTGTGGGGTTCCTGT-3′ and reverse 5′-TGCCAGTCACCCCTTTGTTG-3′; and mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH), forward 5′-CATCACTGCCACCCAGAAGACTG-3′ and reverse 5′-ATGCCAGTGAGCTTCCCGTTCAG-3′.
Real-time Quantitative PCR (RT-qPCR)
Complementary DNAs (cDNAs) were synthesized from sample RNAs with the ReverTra Ace® qPCR RT Master Mix with gDNA Remover (TOYOBO, Osaka, Japan). Then, 1 μL of cDNA in a total volume of 20 μL was used in the following reaction. RT-qPCR was performed in triplicate on a LightCycler 480 (Roche Applied Science, Mannheim, Germany) using the LightCycler 480 SYBR Green master mix (Roche Applied Science), and the thermocycler conditions were as follows: amplifications were performed starting with a 5-min template preincubation step at 95 °C, followed by 40 cycles at 95 °C for 20 sec, 62 °C for 20 sec, and 72 °C for 15 sec. The melting curve analysis began at 95 °C for 5 sec, followed by 1 min at 60 °C. The specificity of the produced amplification product was confirmed by the melting curve analysis and showed a distinct single sharp peak with the expected melting temperature (Tm) for all samples. The GAPDH gene was used as the internal control. The expression level of each gene of interest was obtained using the 2−ΔΔCt method.
Immunohistochemistry
Animals were euthanized and perfused with 4% paraformaldehyde. Harvested brain tissues were cryosectioned at a slice thickness of 16 μm along the sagittal plane, and immunohistochemistry staining was performed on 4 sections over a range of over 128 μm. The sections were stained with primary antibodies against induced nitric oxide (iNOS, 1:400, Abcam, Cambridge, UK) and α-syn (Nitrate Tyr 125, Nitrate Tyr 133, Fluorescein isothiocynate (FITC); 1:400, Novus, Littleton, CO, USA). Stained sections were then mounted on glass slides with fluorescent mounting medium containing 4,6-diamidino-2-phenylindole (Vectashield, Vector, Burlingame, CA, USA). Stained sections were analyzed using a fluorescent microscope (Axio Imager M2, Zeiss, Stockholm, Sweden). Images of nitrated α-syn and iNOS in glomerular cell layer of OB were evaluated using ZEN Imaging Software version 2.1 (Blue edition, Zeiss). Images of the area (mm2) of the glomerular cell layer of OB were obtained and layers were merged and converted to volume (mm3). Area of nitrate α-syn or iNOS was also obtained using ZEN Imaging Software (Blue edition, Zeiss), and layers were merged and converted to volume. The volume of nitrate α-syn or iNOS was calculated via the formula: percent density (%) = [α-syn or iNOS (mm3)/total volume of glomerular cell layer of OB (mm3)] × 100.
Statistical Analysis
Statistical analyses were performed using Statistical Package for Social Sciences software version 23.0 (IBM Corporation, Armonk, NY, USA). Data are expressed as the mean ± standard error of the mean. The interaction (group × time) of results in the buried food test was analyzed using a two-way analysis of variance (ANOVA). Then, 3 groups (WT, PD control, and PD EE) were compared using a one-way ANOVA in each period pretreatment (8 mo) and posttreatment (10 mo), respectively. Both results of RT-qPCR and immunohistochemistry were analyzed using a one-way ANOVA followed by a post hoc Bonferroni test to compare 3 groups. The results of RT-qPCR in different ages (10 and 13 mo) of PD were analyzed using an independent t test. Statistical significance was accepted when P < 0.05.
Results
EE Ameliorates Olfactory Dysfunction in PD
We first determined whether an EE could restore olfactory dysfunction using the buried food test at 8 and 10 mo of age in a mouse model of PD (Fig. 1C). A significant interaction (group × time) of buried food test was not detected in a two-way ANOVA (P = 0.132), revealing there was no time effect. To identify the main effect of treatment, a one-way ANOVA was used at 8 and 10 mo of age, separately. The result based on a one-way ANOVA showed that latency time of finding food in PD control group (N = 14, 71.0 ± 9.2 sec, P < 0.05) and PD EE group (N = 11, 74.2 ± 12.8 sec, P < 0.05) were significantly increased compared to that of WT group (N = 8, 30.7 ± 4.6 sec) at 8 mo of age. At 10 mo of age, the latency time of finding food in PD EE group (37.9 ± 6.1 sec, P < 0.05) was significantly decreased compared to PD control group (71.3 ± 14.2 sec). These results suggest that EE ameliorates olfactory dysfunction in PD.
Expression of Genes Related to Detoxification Enzymes in the OB According to RT-qPCR
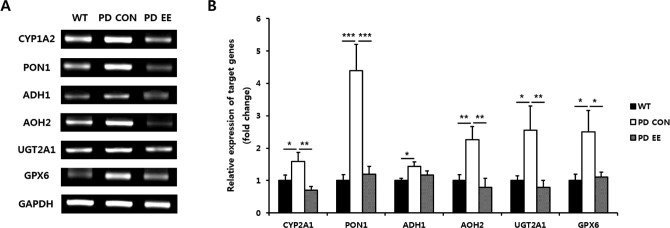
We performed RT-qPCR for quantitative analysis and confirmed expression patterns similar to the RT-PCR results (Fig. 2). The expression levels of genes in the OB of the PD control (N = 5) and PD EE group (N = 5) relative to the WT mice (N = 5) were compared at 10 mo of age. The expression of CYP1A2 (1.59-fold, P < 0.05), PON1 (4.39-fold, P < 0.001), ADH1 (1.43-fold, P < 0.05), AOH2 (2.26-fold, P < 0.01), UGT2A1 (2.55-fold, P < 0.05), and GPX6 (2.50-fold, P < 0.05) was significantly increased in the PD control group compared to the WT group. On the other hand, the expression of CYP1A2 (0.70-fold, P < 0.01), PON1 (1.20-fold, P < 0.001), ADH1 (1.17-fold), AOH2 (0.79-fold, P < 0.01), UGT2A1 (0.79-fold, P < 0.01), and GPX6 (1.10-fold, P < 0.05) was decreased in the PD EE group compared to the PD control group.
Fig. 2.
Expression of genes related with detoxification enzymes in the olfactory bulb. (A) RT-PCR analysis of 6 genes related to detoxification enzymes in the OB of PD mice at 10 mo of age: Cytochrome P450 family 1 subfamily A member 2 (CYP1A2), paraoxonase 1 (PON1), alcohol dehydrogenase 1 (ADH1), aldehyde oxidase homolog 2 (AOH2), UDP glucuronosyltransferase family 2 member A1 complex locus (UGT2A1), and aldehyde glutathione peroxidase 6 (GPX6). (B) Quantitative comparison of gene expression in the PD control (N = 5) and PD EE group (N = 5) relative to WT mice (N = 5) at 10 mo of age determined by RT-qPCR. The expression of CYP1A2 (1.59-fold, P < 0.05), PON1 (4.39-fold, P < 0.001), ADH1 (1.43-fold, P < 0.05), AOH2 (2.26-fold, P < 0.01), UGT2A1 (2.55-fold, P < 0.05), and GPX6 (2.50-fold, P < 0.05) was significantly increased in the PD control group compared to the WT group. On the other hand, the expression of CYP1A2 (0.70-fold, P < 0.01), PON1 (1.20-fold, P < 0.001), ADH1 (1.17-fold), AOH2 (0.79-fold, P < 0.01), UGT2A1 (0.79-fold, P < 0.01), and GPX6 (1.10-fold, P < 0.05) was decreased in the PD EE group compared to the PD control group. Abbreviations: RT-PCR = reverse transcription polymerase chain reaction; OB = olfactory bulb; PD = Parkinson's disease; EE = enriched environment; WT = wild type; RT-qPCR = real-time quantitative polymerase chain reaction. *P < 0.05, **P < 0.01, and ***P < 0.001 are based on a one-way analysis of variance followed by a post hoc test.
Expression of Genes Related to Detoxification Enzymes in Early and Late Stage of PD
We performed RT-qPCR to compare expression of genes related to detoxification enzymes at 10 and 13 mo in the OB of PD (Fig. 3). Gene expressions of early stage mice, 10 mo of age, of PD mice (N = 5) relative to the same age of WT mice (N = 5) and that of PD mice (N = 7) in late stage, 13 mo of age, relative to the same age of WT mice (N = 5) were compared. PD mice in late stage showed the decrease in CYP1A2 (0.78-fold), PON1 (0.28-fold, P < 0.01), ADH1 (0.75-fold), AOH2 (0.54-fold, P < 0.05), UGT2A1 (0.70-fold), and GPX6 (1.16-fold) compared to the PD mice in early stage.
Fig. 3.
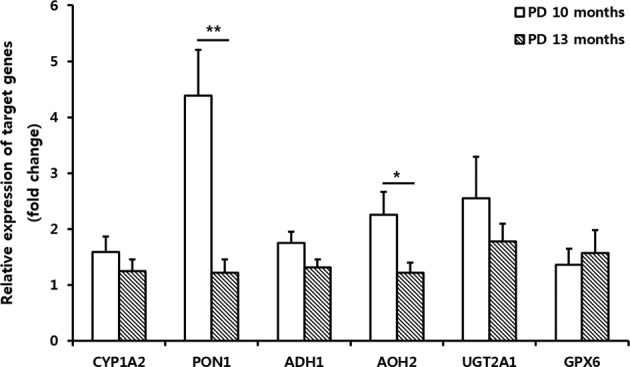
Expression of genes related with detoxification enzymes in early and late stage of PD. Quantitative comparison of gene expression of detoxification enzymes in the OB of PD mice relative to the WT mice was determined by RT-qPCR. Gene expressions of early stage mice, 10 mo of age, of PD mice (N = 5) relative to the same age of WT mice (N = 5) and that of PD mice (N = 7) in late stage, 13 mo of age, relative to the same age of WT mice (N = 5) were compared. PD mice in late stage showed the decrease in CYP1A2 (0.78-fold), PON1 (0.28 fold, P < 0.01), ADH1 (0.75-fold), AOH2 (0.54-fold, P < 0.05), UGT2A1 (0.70-fold), and GPX6 (1.16-fold) compared to the PD mice in early stage. Abbreviations: PD = Parkinson's disease; OB = olfactory bulb; WT = wild type; RT-qPCR = real-time quantitative polymerase chain reaction; CYP1A2 = cytochrome P450 family 1 subfamily A member 2; PON1 = paraoxonase 1; ADH1 = alcohol dehydrogenase 1; AOH2 = aldehyde oxidase homolog 2; UGT2A1 = UDP glucuronosyltransferase family 2 member A1 complex locus; GPX6 = aldehyde glutathione peroxidase 6. *P < 0.05 and **P < 0.01 are based on an independent t test.
EE Ameliorates Oxidative Stress in PD
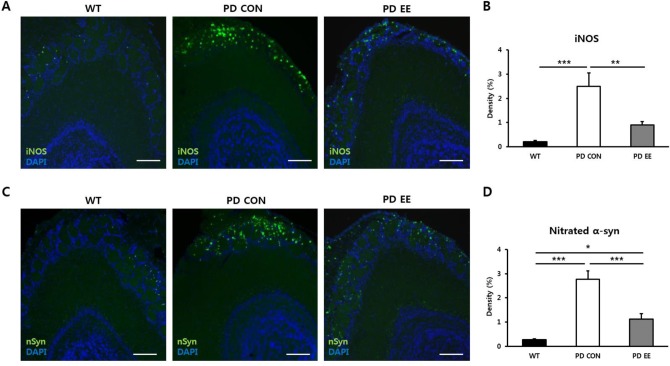
To evaluate the level of oxidative stress in the glomerular layer of the OB, we performed immunohistochemical staining of iNOS in 10-mo-old WT, PD control, and PD EE mice (N = 5 per group; Fig. 4A). The density of iNOS staining was significantly higher in the PD control group (2.5 ± 0.53%; P < 0.001) than in the WT group (0.2 ± 0.06%). However, the density of iNOS was significantly lower in the PD EE group (0.9 ± 0.13%; P < 0.01) than in the PD control group (Fig. 4B). These results suggest that EE ameliorates oxidative stress in PD.
Fig. 4.
An enriched environment ameliorates oxidative stress and nitrated α-syn in PD. (A) Images of the iNOS immunohistochemistry staining in the glomerular layer of the olfactory bulb. (B) The density of iNOS was significantly higher in PD control group (2.5 ± 0.53%, P < 0.001) than in WT group (0.2 ± 0.06%). However, the density of nitrated α-syn was significantly lower in PD EE group than in both PD control group and WT group based on a one-way ANOVA (1.1 ± 0.21%, P < 0.001, P < 0.05, respectively). Scale bars = 200 μm. Abbreviations: α-syn = human A53T α-synuclein; PD = Parkinson's disease; iNOS = induced nitric oxide; WT = wild type; EE = enriched environment; ANOVA = analysis of variance. *P < 0.05, **P < 0.01, and ***P < 0.001 are based on a one-way ANOVA followed by a post hoc test.
EE Ameliorates Nitrated α-Syn in PD
To evaluate the density of nitrated α-syn in the glomerular layer of the OB, we also performed immunohistochemical staining of nitrated α-syn in 10-mo-old WT, PD control, and PD EE mice (N = 5 per group; Fig. 4C). The density of nitrated α-syn staining was significantly higher in the PD control group (2.8 ± 0.33%; P < 0.001) than in the WT group (0.3 ± 0.04%). However, the density of nitrated α-syn staining was significantly lower in the PD EE group (1.1 ± 0.21%; P < 0.001, P < 0.05, respectively) than both in the PD control group and WT group (Fig. 4D). These results suggest that EE ameliorates nitrated α-syn level in PD.
Discussion
Although motor symptoms of PD are caused by degeneration of dopaminergic neurons in the substantia nigra in association with Lewy bodies, it has recently been shown that degeneration starts in other sites such as the glossopharyngeal and vagal nerves and olfactory structures16. Since Ansari and Jonson reported olfactory dysfunction in PD in 197510, many reports have described impairments of odor detection and discrimination in PD, and hyposmia that precedes motor symptoms by several years12,13,15 which can be an initial sign of PD14. In our study, a neurobehavioral test of olfactory function showed that EE ameliorated olfactory dysfunction in PD mice compared to the PD control group without exposure to an EE. Although a similar A53T α-syn mouse model, B6;C3-Tg(Prnp-SNCA*A53T)83Vle/J, of PD was previously reported to have olfactory dysfunction27, we report on the recovery of olfactory function in PD through an EE.
Previous studies have shown that CYP1A2 is related to detoxification of MPTP in PD29. MPTP is a neurotoxic compound that can lead to Parkinsonian features in humans and mice19,20,30 and is known as one of the best molecules to establish a PD animal model31. Moreover, PON1, which has a role in the hydrolysis of the toxic metabolites of pesticides, and has various roles in antioxidative function, anti-inflammatory ability, protection against cardiovascular disease via an inverse correlation with atherosclerosis, and metabolism of organophosphates32–34. Its effects on the hydrolyzation of organophosphates and antioxidant capacity suggest the putative relation of PON1 in PD pathogenesis33. However, its exact role should be elucidated, because many studies have shown inconsistent results between PON1 polymorphism and the risk of PD35–38.
As a representative enzyme for alcohol metabolism, ADH participates in the metabolism of aldehyde, retinol, steroid, and lipid peroxidation products as well as alcohols39,40 and also has a defensive function against alcohols and aldehydes without production of toxic radicals41. It is well known that chronic consumption of ethanol can induce neurologic disorders such as neurodegenerative and neurocognitive deficits through oxidative stress and mitochondrial dysfunction42. There have been several studies showing the relationship between ADH and PD risk43. Furthermore, AOH, one of the molybdoflavoenzymes, is another non-P450 phase I enzyme. It has broad specificity for various substrates, endogenous molecules, and xenobiotics44, as well as aldehydes, which have some toxicities related with neurodegenerative disease, cardiovascular diseases, stroke, and cancer45.
UGT, as key enzymes of phase II metabolism, has an important role in biotransformation and detoxification via a glucuronidation reaction and is the most important pathway of detoxification. These enzymes have numerous substrates such as xenobiotics, including chemical carcinogens and environmental substances, as well as endogenous metabolites46. UTG2A1 is expressed in the rat and mouse OB47, indicating that UTG2A1 as a “metabolic barrier” might have a protective role against exogenous toxins invading via the nasal cavity, which can be a direct route of entry for xenobiotics48. Likewise, GPX is an antioxidant enzyme that detoxifies hydroxyl radicals. Because oxidative stress is believed to be an important cause of nigral neuronal cell death, many human-based studies have been performed to elucidate the mechanism of PD.
In our study, these detoxification enzymes such as CYP1A2, PON1, ADH1, AOH2, UGT2A1, and GPX6 significantly increased in early stage of PD as a compensatory mechanism. This result showed that detoxification enzymes were increased as a compensatory mechanism for the increment of various toxic factors after the onset of disease. From 8 to 10 months of age, EE normalized these enzymes. However, most enzymes decreased at 13 mo of age compared to 10 mo of age in PD, suggesting that detoxification enzymes attempt to detoxify the toxic factors in early stage, but in the late stage of the disease, these detoxification enzymes finally decreased their activities, causing increased oxidative stress and exacerbating the disease at last in PD.
Histological analysis in the glomerular layer of the OB showed that oxidative stress and nitrated α-syn were significantly increased in the PD control group compared to the WT group, but those were significantly decreased in the PD EE group compared to the PD control group at 10 mo of age in PD. Oxidative stress contributes to pathogenic modification of α-syn such as nitration of tyrosine residues that have been observed in the brains of patients afflicted with PD49. The result of immunohistochemistry with an iNOS antibody was similar to that of nitrated α-syn. Oxidative stress in PD upregulated nitrated α-syn; however, since the mice were exposed to an EE, oxidative stress decreased and nitrated α-syn was reduced. Additionally, previous research reported that PD might occur by selective α-syn nitration50. Due to its nitration, α-syn becomes more difficult to degrade through proteolysis, which changes its properties as a synaptic protein. Therefore, nitration of α-syn regulated the mechanism, onset, and progression of synucleinopathy, which also depended on the formation of the nitrated α-syn form50. Thus, an EE alleviated olfactory dysfunction in a PD mouse model through decreased oxidative stress and nitrated α-syn in the OB.
Conclusion
In a Tg mouse model of PD that overexpressed human A53T α-syn, exposure to an EE reduced oxidative stress and nitrated α-syn, resulting in normalized detoxification enzymes and amelioration of olfactory dysfunction.
Footnotes
Authors’ Note: Soohyun Wi and Jang Woo Lee equally contributed to this study.
Ethical Approval: The experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC 2017-0039).
Statement of Human and Animal Rights: All animals were housed in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care and provided food and water ad libitum with alternating 12-h light/dark cycles according to animal protection regulations.
Statement of Informed Consent: There are no human subjects in this article and informed consent is not applicable.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by grants from the National Research Foundation (NRF-2014R1A2A1A11052042; 2015M3A9B40 67068) and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health and Welfare (HI16C1012, HI16C1013, and HI16C1176), Republic of South Korea.
References
- 1. Lee MY, Yu JH, Kim JY, Seo JH, Park ES, Kim CH, Kim H, Cho SR. Alteration of synaptic activity-regulating genes underlying functional improvement by long-term exposure to an enriched environment in the adult brain. Neurorehabil Neural Repair. 2013;27(6):561–574. [DOI] [PubMed] [Google Scholar]
- 2. Rosenzweig MR, Bennett EL, Hebert M, Morimoto H. Social grouping cannot account for cerebral effects of enriched environments. Brain Res. 1978;153(3):563–576. [DOI] [PubMed] [Google Scholar]
- 3. Will B, Galani R, Kelche C, Rosenzweig MR. Recovery from brain injury in animals: relative efficacy of environmental enrichment, physical exercise or formal training (1990-2002). Prog Neurobiol. 2004;72(3):167–182. [DOI] [PubMed] [Google Scholar]
- 4. Cummins RA, Walsh RN, Budtz-Olsen OE, Konstantinos T, Horsfall CR. Environmentally-induced changes in the brains of elderly rats. Nature. 1973;243(5409):516–518. [DOI] [PubMed] [Google Scholar]
- 5. Holloway RL., Jr Dendritic branching: some preliminary results of training and complexity in rat visual cortex. Brain Res. 1966;2(4):393–396. [DOI] [PubMed] [Google Scholar]
- 6. Kim MS, Yu JH, Kim CH, Choi JY, Seo JH, Lee MY, Yi CH, Choi TH, Ryu YH, Lee JE, et al. Environmental enrichment enhances synaptic plasticity by internalization of striatal dopamine transporters. J Cereb Blood Flow Metab. 2016;36(12):2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nithianantharajah J, Hannan AJ. Enriched environments, experience-dependent plasticity and disorders of the nervous system. Nat Rev Neurosci. 2006;7(9):697–709. [DOI] [PubMed] [Google Scholar]
- 8. Rosenzweig MR, Bennett EL. Psychobiology of plasticity: effects of training and experience on brain and behavior. Behav Brain Res. 1996;78(1):57–65. [DOI] [PubMed] [Google Scholar]
- 9. van Praag H, Kempermann G, Gage FH. Neural consequences of environmental enrichment. Nat Rev Neurosci. 2000;1(3):191–198. [DOI] [PubMed] [Google Scholar]
- 10. Ansari KA, Johnson A. Olfactory function in patients with Parkinson’s disease. J Chronic Dis. 1975;28(9):493–7. [DOI] [PubMed] [Google Scholar]
- 11. Ascherio A, Schwarzschild MA. The epidemiology of Parkinson’s disease: risk factors and prevention. Lancet Neurol. 2016;15(12):1257–1272. [DOI] [PubMed] [Google Scholar]
- 12. Delamarre A, Meissner WG. Epidemiology, environmental risk factors and genetics of Parkinson’s disease. Presse Med. 2017;46(2 Pt 1):175–181. [DOI] [PubMed] [Google Scholar]
- 13. Haehner A, Boesveldt S, Berendse HW, Mackay-Sim A, Fleischmann J, Silburn PA, Johnston AN, Mellick GD, Herting B, Reichmann H, et al. Prevalence of smell loss in Parkinson’s disease—a multicenter study. Parkinsonism Relat Disord. 2009;15(7):490–494. [DOI] [PubMed] [Google Scholar]
- 14. Haehner A, Hummel T, Hummel C, Sommer U, Junghanns S, Reichmann H. Olfactory loss may be a first sign of idiopathic Parkinson’s disease. Mov Disord. 2007;22(6):839–42. [DOI] [PubMed] [Google Scholar]
- 15. Ponsen MM, Stoffers D, Booij J, van Eck-Smit BL, Wolters E, Berendse HW. Idiopathic hyposmia as a preclinical sign of Parkinson’s disease. Ann Neurol. 2004;56(2):173–81. [DOI] [PubMed] [Google Scholar]
- 16. Qutubuddin AA, Chandan P, Carne W. Degenerative movement disorders of the central nervous system In: Cifu DX, editor. Braddom’s physical medicine and rehabilitation. Philadelphia (PA): Saunders Elsevier; 2015. p. 1017–1027. [Google Scholar]
- 17. Procaccini C, Santopaolo M, Faicchia D, Colamatteo A, Formisano L, de Candia P, Galgani M, De Rosa V, Matarese G. Role of metabolism in neurodegenerative disorders. Metabolism. 2016;65(9):1376–1390. [DOI] [PubMed] [Google Scholar]
- 18. Deng Y, Newman B, Dunne MP, Silburn PA, Mellick GD. Further evidence that interactions between CYP2D6 and pesticide exposure increase risk for Parkinson’s disease. Ann Neurol. 2004;55(6):897. [DOI] [PubMed] [Google Scholar]
- 19. Apte SN, Langston JW. Permanent neurological deficits due to lithium toxicity. Ann Neurol. 1983;13(4):453– 455. [DOI] [PubMed] [Google Scholar]
- 20. Kopin IJ, Markey SP. MPTP toxicity: implications for research in Parkinson’s disease. Annu Rev Neurosci. 1988;11:81–96. [DOI] [PubMed] [Google Scholar]
- 21. Williams AC, Ramsden DB. Autotoxicity, methylation and a road to the prevention of Parkinson’s disease. J Clin Neurosci. 2005;12(1):6–11. [DOI] [PubMed] [Google Scholar]
- 22. Maruyama W, Abe T, Tohgi H, Naoi M. An endogenous MPTP-like dopaminergic neurotoxin, N-methyl(R)salsolinol, in the cerebrospinal fluid decreases with progression of Parkinson’s disease. Neurosci Lett. 1999;262(1):13–16. [DOI] [PubMed] [Google Scholar]
- 23. Giannoccaro MP, La Morgia C, Rizzo G, Carelli V. Mitochondrial DNA and primary mitochondrial dysfunction in Parkinson’s disease. Mov Disord. 2017;32(3):346–363. [DOI] [PubMed] [Google Scholar]
- 24. Faherty CJ, Raviie Shepherd K, Herasimtschuk A, Smeyne RJ. Environmental enrichment in adulthood eliminates neuronal death in experimental Parkinsonism. Brain Res Mol Brain Res. 2005;134(1):170–179. [DOI] [PubMed] [Google Scholar]
- 25. Goldberg NR, Haack AK, Meshul CK. Enriched environment promotes similar neuronal and behavioral recovery in a young and aged mouse model of Parkinson’s disease. Neuroscience. 2011;172:443–452. [DOI] [PubMed] [Google Scholar]
- 26. Rosenfeldt AB, Dey T, Alberts JL. aerobic exercise preserves olfaction function in individuals with Parkinson’s disease. Parkinsons Dis. 2016;2016:9725089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Farrell KF, Krishnamachari S, Villanueva E, Lou H, Alerte TN, Peet E, Drolet RE, Perez RG. Non-motor Parkinsonian pathology in aging A53T alpha-synuclein mice is associated with progressive synucleinopathy and altered enzymatic function. J Neurochem. 2014;128(4):536–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang M, Crawley JN. Simple behavioral assessment of mouse olfaction. Curr Protoc Neurosci. 2009; Chapter 8:Unit 8.24. [DOI] [PMC free article] [PubMed]
- 29. Coleman T, Ellis SW, Martin IJ, Lennard MS, Tucker GT. 1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) is N-demethylated by cytochromes P450 2D6, 1A2 and 3A4—implications for susceptibility to Parkinson’s disease. J Pharmacol Exp Ther. 1996;277(2):685–690. [PubMed] [Google Scholar]
- 30. Sonsalla PK, Heikkila RE. The influence of dose and dosing interval on MPTP-induced dopaminergic neurotoxicity in mice. Eur J Pharmacol. 1986;129(3):339–345. [DOI] [PubMed] [Google Scholar]
- 31. Singh S, Singh K, Gupta SP, Patel DK, Singh VK, Singh RK, Singh MP. Effect of caffeine on the expression of cytochrome P450 1A2, adenosine A2A receptor and dopamine transporter in control and 1-methyl 4-phenyl-1,2,3,6-tetrahydropyridine treated mouse striatum. Brain Res. 2009;1283:115–126. [DOI] [PubMed] [Google Scholar]
- 32. Furlong CE, Marsillach J, Jarvik GP, Costa LG. Paraoxonases-1, -2 and -3: what are their functions? Chem Biol Interact. 2016;259(Pt B):51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Menini T, Gugliucci A. Paraoxonase 1 in neurological disorders. Redox Rep. 2014;19(2):49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Androutsopoulos VP, Kanavouras K, Tsatsakis AM. Role of paraoxonase 1 (PON1) in organophosphate metabolism: implications in neurodegenerative diseases. Toxicol Appl Pharmacol. 2011;256(3):418–424. [DOI] [PubMed] [Google Scholar]
- 35. Akhmedova SN, Yakimovsky AK, Schwartz EI. Paraoxonase 1 Met--Leu 54 polymorphism is associated with Parkinson’s disease. J Neurol Sci. 2001;184(2):179–182. [DOI] [PubMed] [Google Scholar]
- 36. Clarimon J, Eerola J, Hellstrom O, Tienari PJ, Singleton A. Paraoxonase 1 (PON1) gene polymorphisms and Parkinson’s disease in a Finnish population. Neurosci Lett. 2004;367(2):168–170. [DOI] [PubMed] [Google Scholar]
- 37. Carmine A, Buervenich S, Sydow O, Anvret M, Olson L. Further evidence for an association of the paraoxonase 1 (PON1) Met-54 allele with Parkinson’s disease. Mov Disord. 2002;17(4):764–766. [DOI] [PubMed] [Google Scholar]
- 38. Liu YL, Yang J, Zheng J, Liu DW, Liu T, Wang JM, Wang CN, Wang MW, Tian QB. Paraoxonase 1 polymorphisms L55M and Q192R were not risk factors for Parkinson’s disease: a HuGE review and meta-analysis. Gene. 2012;501(2):188–192. [DOI] [PubMed] [Google Scholar]
- 39. Duester G, Farres J, Felder MR, Holmes RS, Hoog JO, Pares X, Plapp BV, Yin SJ, Jornvall H. Recommended nomenclature for the vertebrate alcohol dehydrogenase gene family. Biochem Pharmacol. 1999;58(3):389–395. [DOI] [PubMed] [Google Scholar]
- 40. Westerlund M, Belin AC, Felder MR, Olson L, Galter D. High and complementary expression patterns of alcohol and aldehyde dehydrogenases in the gastrointestinal tract: implications for Parkinson’s disease. FEBS J. 2007;274(5):1212–1223. [DOI] [PubMed] [Google Scholar]
- 41. Hoog JO, Hedberg JJ, Stromberg P, Svensson S. Mammalian alcohol dehydrogenase—functional and structural implications. J Biomed Sci. 2001;8(1):71–76. [DOI] [PubMed] [Google Scholar]
- 42. Haorah J, Ramirez SH, Floreani N, Gorantla S, Morsey B, Persidsky Y. Mechanism of alcohol-induced oxidative stress and neuronal injury. Free Radic Biol Med. 2008;45(11):1542–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Anvret A, Ran C, Westerlund M, Gellhaar S, Lindqvist E, Pernold K, Lundstromer K, Duester G, Felder MR, Galter D, et al. Adh1 and Adh1/4 knockout mice as possible rodent models for presymptomatic Parkinson’s disease. Behav Brain Res. 2012;227(1):252–257. [DOI] [PubMed] [Google Scholar]
- 44. Garattini E, Fratelli M, Terao M. The mammalian aldehyde oxidase gene family. Hum Genomics. 2009;4(2):119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen CH, Ferreira JCB, Gross ER, Mochly-Rosen D. Targeting aldehyde dehydrogenase 2: new therapeutic opportunities. Physiol Rev. 2014;94(1):1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rowland A, Miners JO, Mackenzie PI. The UDP-glucuronosyltransferases: their role in drug metabolism and detoxification. Int J Biochem Cell Biol. 2013;45(6):1121–1132. [DOI] [PubMed] [Google Scholar]
- 47. Heydel J, Leclerc S, Bernard P, Pelczar H, Gradinaru D, Magdalou J, Minn A, Artur Y, Goudonnet H. Rat olfactory bulb and epithelium UDP-glucuronosyltransferase 2A1 (UGT2A1) expression: in situ mRNA localization and quantitative analysis. Brain Res Mol Brain Res. 2001;90(1):83–92. [DOI] [PubMed] [Google Scholar]
- 48. Ouzzine M, Gulberti S, Ramalanjaona N, Magdalou J, Fournel-Gigleux S. The UDP-glucuronosyltransferases of the blood-brain barrier: their role in drug metabolism and detoxication. Front Cell Neurosci. 2014;8:349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wong YC, Krainc D. α-Synuclein toxicity in neurodegeneration: mechanism and therapeutic strategies. Nat Med. 2017;23(2):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Giasson BI, Duda JE, Murray IV, Chen Q, Souza JM, Hurtig HI, Ischiropoulos H, Trojanowski JQ, Lee VM. Oxidative damage linked to neurodegeneration by selective alpha-synuclein nitration in synucleinopathy lesions. Science. 2000;290(5493):985–989. [DOI] [PubMed] [Google Scholar]