Abstract
Background
Human leukocyte antigen (HLA) DQ2 and DQ8 are important risk factors for some autoimmune diseases such as celiac disease (CD), but their possible role in other diseases and health conditions is not fully explored.
Objectives
The objective of this article is to examine the distribution of HLA DQ2 and HLA DQ8 in an adult general population, and their association with health indicators (diseases, symptoms and biomarkers).
Methods
In this cross-sectional, population-based study, 2293 individuals were screened for HLA DQ2 and DQ8; CD-associated alleles (DQA*0201*03*05/DQB*02*0301/0304*0302/0305) and DQB1*02 homozygosity were determined for screen-positive participants. The National Patient Registry provided diagnosis information.
Results
A total of 47.7% (1093/2293) individuals were positive for DQ2 and/or DQ8: 31.2% (716/2293) only DQ2, 11.9% (273/2293) only DQ8, 4.1% (93/2293) both DQ2 and DQ8. Among nine individuals diagnosed with CD, 89.9% (8/9) had DQ2.5cis, 22.2% (2/9) DQ8 and 22.2% (2/9) DQ2.2 (two both DQ2 and DQ8). HLA DQ2.5 was associated with higher thyroid-stimulating hormone levels, while DQ2/DQ8-positive participants had significantly lower prevalence of irritable bowel syndrome (IBS). DQ2/DQ8 were strongly associated with CD, but no other registry-based diagnoses.
Conclusion
In this general Danish population, 47.7% were HLA DQ2/DQ8 positive and thus potentially at risk for CD. All individuals with CD were DQ2/DQ8 positive; the majority DQ2.5. Surprisingly, DQ2/DQ8-positivity was associated with lower IBS prevalence.
Keywords: Celiac disease, HLA DQ2, HLA DQ8, epidemiology
Key summary
- Summarize the established knowledge on this subject:
- Celiac disease is a lifelong autoimmune disease caused by a harmful immune response to gluten proteins in genetically susceptible individuals.
- Human leukocyte antigen (HLA) DQ2 and HLA DQ8 are important risk factors for some autoimmune diseases such as celiac disease; however, their possible role in other diseases and health conditions is not fully explored.
- What are the significant and/or new findings of this study?
- Our study showed that 47.7% of individuals were HLA DQ2/DQ8 positive and thus potentially at risk for celiac disease in this general Danish population.
- We found that all individuals with celiac disease were HLA DQ2/DQ8 positive; the majority DQ2.5.
- Furthermore, HLA DQ2/DQ8 positivity was associated with lower prevalence of irritable bowel syndrome, but not with other diagnoses/diseases.
Introduction
Celiac disease (CD) is a lifelong autoimmune disease caused by a harmful immune response to gluten proteins. Ingestion of gluten-containing grains, such as wheat, rye and barley, results in inflammation of the small intestine and villous atrophy in genetically susceptible individuals.1,2 Like other autoimmune diseases, CD has a strong human leukocyte antigen (HLA) association.2 The physiological role of HLA is to present antigenic peptides to T lymphocytes. HLA class II genes are encoded in the major histocompatibility complex on the short arm of chromosome 6 and include the principal factors for genetic susceptibility for CD.3 An essential step in CD pathogenesis is the HLA DQ-mediated presentation of gluten peptides to CD4+ T lymphocytes: Deamidated gluten peptides (deaminated by the enzyme transglutaminase 2) bind to HLA-DQ molecules and are presented to gluten-specific CD4+ T cells stimulating the immune response leading to CD.2,4 However, CD is a multifactorial and polygenic disorder, thus HLA is necessary but not sufficient for the development of CD.2 Approximately 30%–40% of Western populations carry HLA DQ2 or DQ8, while only 1% is affected by CD.3
About 90% of individuals with CD carry HLA-DQ2.5. HLA-DQ2.5 can be expressed in cis configuration, where the alleles are located on the same chromosome (DQA1*05:01–DQB1*02:01) or in trans configuration, where the alleles are located on opposite chromosomes (DQA1*05:05–DQB1*03:01/DQA1*02:01–DQB1*02:02).5 While individuals with CD who do not express HLA-DQ2.5 usually carry either HLA-DQ2.2 (DQA1*02:01–DQB1*02:02) or HLA-DQ8 (DQA1*03–DQB1*03:02),2 very few express HLA DQ7.5 (DQA1*05:05–DQB1*03:01), DQ2.3 (DQA1*03–DQB1*02) or DQ8.5 (DQA1*05–DQB1*03:02).2,6,7 The differences in risk of CD between the haplotypes are linked to the binding of gluten peptides and following T-cell response.5 Studies have shown that DQB1*02 homozygosity is associated with increased risk of CD,2,8 and that individuals homozygous for DQ2.5 and DQ8 have increased risk of CD.2
Although HLA DQ2 and DQ8 are known to be associated with autoimmune diseases such as CD,2,9–11 knowledge is limited about the association of HLA DQ2 and DQ8 with other diseases and health indicators.
The distribution of HLA DQ2 and DQ8 in a general Danish population has not previously been reported, although the distribution of HLA-DR in Danes was studied in 1981.12–14 We have conducted a screening for CD in a general Danish population.15 In a further analysis of this cohort we aimed to map the distribution of HLA DQ2 and HLA DQ8 haplotypes and examine their association with CD and other diseases, symptoms and biomarkers.
Material and methods
Study population
This study was based on the five-year follow-up of the Health2006 cohort and designed as a cross-sectional, population-based study. A detailed description of the Health2006 cohort profile has been published elsewhere.16 The participants invited to the Health2006 cohort were drawn as a random sample from the background population aged 18–69 years, living in 11 municipalities in the Southwestern part of the greater Copenhagen area. All eligible participants in the Health2006 cohort were invited to a five-year follow-up examination including essentially the same study protocol15 with the addition of screening for HLA DQ2/DQ8 known to be associated with CD, and screening for CD by measurements of CD-related antibodies: immunoglobulin (Ig)A against tissue transglutaminase (TTG), IgA and IgG against deamidated gliadin peptide, and for those with IgA deficiency: IgG TTG. The screening for CD has been described elsewhere.15,17 A total of 3405 participants were eligible for invitation and 2308 individuals (54.2% female) were re-examined. The mean age at the five-year-follow-up examination was 55.7 years (range: 24–76 years). The study included: structured self-administered questionnaires on general health, symptoms and diagnoses of disease, lifestyle, socioeconomic variables among other issues; physical examination and collection of blood samples.
Screening for HLA DQ2 and HLA DQ8
A total of 2293 individuals (15 not included because of missing blood sample or for technical reasons) were screened for HLA DQ2/DQ8 by the Celiac Gene Screen test, and alleles associated with CD were determined for 1175 HLA DQ2/DQ8-positive participants with the Celiac Gene Alleles test (BioDiagene, Palermo, Italy, distributed by Thermo Fisher Scientific, Allerød, Denmark) identifying by a sequence-specific primer-polymerase chain reaction (SSP-PCR) the HLA alleles DQA1*0201, *03, *05 and DQB1*02, *0301/0304, *0302/0305, as well as DQB1*02 homozygosity.
Covariates
Allergy
Serum samples were screened for specific immunoglobulin E (sIgE) to a panel of inhalant allergens using Phadiatop® (ImmunoDiagnostics, Allerød, Denmark) and a panel of food allergens by using food mix Fx5. Participants were considered Phadiotop positive and Fx5 positive when ≥0.35 kU/l.
Skin prick test reactivity against a panel of 10 common inhalant allergens was performed and atopy was defined as a positive reaction (≥3 mm) to at least one of 10 allergens. Groups of allergens (pollens, animals, or house dust mites) were also considered. Methods for allergy testing have been described elsewhere.18
Irritable bowel syndrome (IBS)
Definition of IBS was based on a symptom cluster of frequent self-reported gastrointestinal symptoms during the past 12 months: “Subjects stating that they often experience both abdominal pain and distension and additionally, either borborygmi or altering stool consistency, or both.19,20 This definition is based on valid statistical criteria in population-based data.19
Biomarkers
At follow-up, fasting blood samples were analyzed for glucose and glycated hemoglobin (HBA1c), and hemoglobin and anti-cyclic citrullinated peptide (anti-CCP) were measured. In addition, at baseline serum levels of 25-hydroxyvitamin D3 (25-OH-D3), anti-thyroperoxidase (anti-TPO), parathyroid hormone (PTH), thyroid-stimulating hormone (TSH) and free thyroxine (FT4) were measured.
Registry-based diagnoses
Our cohort was linked to data from the National Patient Registry by use of the unique personal identification number. The National Patient Registry holds information on diagnose codes (International Classification of Disease, eighth revision (ICD-8) and tenth revision (ICD-10)) registered from 1977,21 and for this study we obtained diagnoses registered until end of 2014. We selected 10 diagnosis groups for further analyses; see Table 1 and Supplementary Table S1.
Table 1.
Results of biochemical analyses, symptoms and diagnoses stratified by HLA DQ2 and HLA DQ8 status among participants in the Health2006 five-year follow-up study.
| All HLA-negative participants | All HLA DQ2- and/or DQ8-positive participantsa | p valueb | HLA DQ2-positive participantsa | HLA DQ8-positive participantsa | HLA DQ2- and DQ8-positive participantsa | p valuec | |
|---|---|---|---|---|---|---|---|
| Number of participants | 1200 | 1093 | 716 | 273 | 93 | ||
| Median (5/95%) | Median (5/95%) | Median (5/95%) | Median (5/95%) | Median (5/95%) | |||
| Biomarkers | |||||||
| Hemoglobin (mmol/l) | 8.8 (7.7/10.0) | 8.8 (7.6/9.9) | 0.311s | 8.8 (7.6/10.0) | 8.7 (7.5/9.9) | 8.7 (7.5/9.9) | 0.492t |
| Vitamin D (nmol/l) | 70.0 (28.1/122.8) | 71.8 (26.2/120.1) | 0.599s | 71.6 (26.9/120.1) | 75.5 (23.5/127.1) | 68.7 (27.1/118.3) | 0.463t |
| Plasma glucose (mmol/l) | 5.3 (4.4/6.8) | 5.2 (4.4/6.7) | 0.683s | 5.3 (4.4/6.7) | 5.2 (4.4/6.6) | 5.2 (4.4/6.4) | 0.668t |
| HbA1c (%) | 5.5 (4.9/6.2) | 5.5 (4.9/6.1) | 0.222s | 5.5 (5.0/6.1) | 5.5 (4.9/6.3) | 5.4 (4.9/6.1) | 0.199t |
| TPO (U/ml) | 11.0 (8.0/166.0) | 12.0 (8.0/118.0) | 0.129s | 12.0 (8.0/171.0) | 12.0 (8.0/92.0) | 11.0 (8.0/48.0) | 0.088t |
| TSH (mU/l) | 1.4 (0.5/3.6) | 1.4 (0.5/3.6) | 0.250s | 1.4 (0.6/3.7) | 1.4 (0.5/3.0) | 1.5 (0.4/4.2) | 0.281t |
| FT4 (pmol/l) | 12.3 (9.9/15.0) | 12.3 (9.9/15.1) | 0.487s | 12.2 (9.8/14.8) | 12.3 (10.1/15.8) | 12.1 (10.0/15.6) | 0.185t |
| Anti-CCP (U/ml) | 1.9 (0.7/4.0) | 1.9 (0.7/3.9) | 0.967s | 1.9 (0.7/3.8) | 2.0 (0.8/3.9) | 2.1 (0.6/3.7) | 0.889t |
| % (n/total N) | % (n/total N) | % (n/total N) | % (n/total N) | % (n/total N) | |||
| Symptoms/diagnoses | |||||||
| Diabetes mellitus Type 1d | 0.6 (7/1200) | 0.5 (5/1093) | 0.677u | 0.3 (2/716) | 1.1 (3/273) | 0.0 (0/93) | 0.383v |
| Cardiovascular diseasee | 7.4 (89/1200) | 8.1 (89/1093) | 0.516u | 8.8 (63/716) | 5.9 (16/273) | 9.7 (9/93) | 0.354u |
| Inflammatory bowel diseasef | 0.9 (11/1200) | 0.8 (9/1093) | 0.811u | 0.7 (5/716) | 1.1 (3/273) | 1.1 (1/93) | 0.923v |
| Asthmag | 4.2 (50/1200) | 3.8 (41/1093) | 0.611u | 3.8 (27/716) | 3.3 (9/273) | 4.3 (4/93) | 0.910v |
| Chronic obstructive pulmonary diseaseh | 2.8 (33/1200) | 1.9 (21/1093) | 0.191u | 2.2 (16/716) | 1.8 (5/273) | 0.0 (0/93) | 0.391v |
| Liver diseasei | 0.8 (9/1200) | 0.6 (7/1093) | 0.753u | 0.7 (5/716) | 0.4 (1/273) | 1.1 (1/93) | 0.833v |
| Gastrointestinal cancerj | 1.1 (13/1200) | 1.1 (12/1093) | 0.973u | 1.0 (7/716) | 1.5 (4/273) | 1.1 (1/93) | 0.845v |
| Lymphomak | 0.3 (4/1200) | 0.6 (7/1093) | 0.288u | 0.4 (3/716) | 1.1 (3/273) | 1.1 (1/93) | 0.202v |
| Celiac diseasel | 0.0 (0/1200) | 0.8 (9/1093) | 0.000v | 1.0 (7/716) | 0.0 (0/273) | 2.2 (2/93) | 0.000v |
| Any autoimmune diseasem | 6.3 (76/1200) | 7.5 (82/1093) | 0.270u | 6.8 (49/716) | 8.1 (22/273) | 9.7 (9/93) | 0.456u |
| Irritable bowel syndromen | 4.6 (55/1197) | 2.6 (28/1086) | 0.010u | 3.1 (22/711) | 1.9 (5/271) | 0.0 (0/93) | 0.017v |
| Allergy | |||||||
| Phadiatop > 0.35 | 30.9 (370/1198) | 29.0 (317/1092) | 0.333u | 29.9 (214/715) | 24.5 (67/273) | 31.2 (29/93) | 0.224u |
| Food mix, Fx5 > 0.35 | 4.1 (49/1198) | 4.1 (45/1092) | 0.971u | 5.2 (37/715) | 1.5 (4/273) | 4.3 (4/93) | 0.050v |
| Atopyo | 32.9 (391/1188) | 30.7 (331/1080) | 0.248u | 31.0 (220/710) | 27.1 (72/266) | 34.4 (32/93) | 0.270u |
| Skin prick test, positive: | |||||||
| Pollenp | 27.4 (325/1188) | 26.3 (284/1080) | 0.569u | 26.6 (189/710) | 22.6 (60/266) | 32.3 (30/93) | 0.256u |
| Animal danderq | 14.4 (171/1188) | 14.3 (154/1080) | 0.927u | 14.8 (105/710) | 10.5 (28/266) | 18.3 (17/93) | 0.217u |
| Dust mitesr | 12.0 (142/1188) | 9.4 (101/1080) | 0.046u | 9.7 (69/710) | 9.0 (24/266) | 5.4 (5/93) | 0.101u |
The participants were screened for HLA DQ8 and HLA DQ2 with the Celiac Gene Screen test, and the alleles associated with CD were identified by Celiac Gene Alleles test (BioDiagene, Palermo, Italy, distributed by Thermo Fisher Scientific, Allerød, Denmark).
P value for the difference between all HLA DQ2- and DQ8-positive versus -negative, p value ≥ 0.05 in bold.
P value for the difference between the four groups: all HLA DQ2- and DQ8-negative, HLA DQ2-positive, HLA DQ8-positive, and both HLA DQ8- and DQ2-positive, p value ≥ 0.05 in bold.
Diabetes mellitus Type 1: ICD-8: 249, ICD-10: E10.
Including: ischemic heart disease (ICD-8: 410–414, ICD-10: I20–I25) and stroke (ICD-8: 431–434, 436, ICD-10: I60–I69).
Including: Crohn’s disease (ICD-8: 563.01, ICD-10: K50) and ulcerative colitis (ICD-8: 563.19, ICD-10: K51).
Asthma: ICD-8: 493, ICD-10: J45, J46.
Chronic obstructive pulmonary disease: ICD-8: 491–492 and ICD-10: J42–J44.
Liver disease: ICD-8: 570–573 and ICD-10: K70–K77.
Gastrointestinal cancer: ICD-8: 140–154 and ICD-10: C00–C18, C20–C21.
Lymphoma: ICD-8: 200-202 and 275.59 and ICD-10: C81–86, C88 and C915.
Celiac disease: ICD-8: 269.00 and ICD-10: K90.0.
aIncluding thyrotoxicosis(ICD-8: 242.00, ICD-10: E05.0), type 1 diabetes (ICD-8: 249, ICD-10: E10), multiple sclerosis (ICD-8: 340, ICD-10: G35), iridocyclitis (ICD-8: 364, ICD-10: H20), Crohn’s disease (ICD-8: 563.01, ICD-10: K50), ulcerative colitis (ICD-8: 563.19, ICD-10: K51), psoriasis vulgaris (ICD-8: 696.09–10, 696.19, ICD-10: L40), seropositive rheumatoid arthritis (ICD-8: 712.19, 712.39, 712.59, ICD-10: M05–M06), polymyalgia rheumatica (ICD-8: 446.30–31, 446.39, ICD-10: M31.5–6, M35.3), pernicious anemia (ICD-8: 281.0, ICD-10: D51.0), autoimmune hemolytic anemia (ICD-8: 283.90–91, ICD-10: D59.1), idiopathic thrombocytopenic purpura (ICD-8: 446.49, ICD-10: D69.3), autoimmune thyroiditis (ICD-8: 245.03, ICD-10: E06.3), primary adrenocortical insufficiency (ICD-8: 255.1, ICD-10: E27.1), Guillain-Barré syndrome (ICD-8: 354, ICD-10: G61.0), autoimmune hepatitis (ICD-8: 571.93, ICD-10: K75.4), primary biliary cirrhosis (ICD-8: 571.90, ICD-10: K74.3), celiac disease (ICD-8: 269.00, ICD-10: K90.0), pemphigus (ICD-8: 694, ICD-10: L10), pemphigoid (ICD-8: 694.05, ICD-10: L12), alopecia areata (ICD-8: 704.00, ICD-10: L63), vitiligo (ICD-8: 709.01, ICD-10: L80), juvenile arthritis (ICD-8: 712.09, ICD-10: M08), Wegener’s granulomatosis (ICD-8: 446.29, ICD-10: M31.3), dermatopolymyositis (ICD-8: 716, ICD-10: M33), myasthenia gravis (ICD-8: 733.09, ICD-10: G70.0), systemic sclerosis (ICD-8: 734.0, ICD-10: M34), systemic lupus erythematosus (ICD-8: 734.19, ICD-10: M32), Sjögren’s syndrome (ICD-8: 734.90, ICD-10: M35.0), and nkylosing spondylitis (ICD-8: 712.49, ICD-10: M45).
Irritable bowel syndrome is defined as a cluster of frequent self-reported gastrointestinal symptoms during the past 12 months: “Subjects stating that they often experience both abdominal pain and distension and additionally, either borborygmi or altering stool consistency, or both.”
Atopy defined as a positive reaction to at least one of 10 allergens: birch, grass (Phleum pratense) mugwort, horse, cat, dog, two house dust mites (Dermatophagoides pteronyssinis and D. farinae), and two molds (Cladosporium herbarum and Alternaria alternata).
Pollen, at least one positive of birch, grass (P. pratense) or mugwort.
Animal dander, at least one positive for horse, cat or dog.
Dust mites, at least one positive for one of two house dust mites: D. pteronyssinis and D. farina.
Wilcoxon test.
Kruskal–Wallis test.
Chi-square test.
Fisher’s exact test.
Anti-CCP: anti-cyclic citrullinated peptide; CD: celiac disease; FT4: free thyroxine; HbA1c: hemoglobin A1c; HLA: human leukocyte antigen; ICD-8: International Statistical Classification of Diseases and Related Health Problems, eighth revision; ICD-10: International Statistical Classification of Diseases and Related Health Problems, 10th revision; TPO: thyroperoxidase; TSH: thyroid-stimulating hormone.
Statistical analysis
To investigate the association between variables, different tests were used: Wilcoxon test, Kruskal–Wallis tests and independent samples t test for continuous variables; and Chi-square test and Fisher’s exact test for categorical data. Complete case analyses were performed, thus the total number of observations in the analyses differs because of missing data. P values < 0.05 (two sided) were considered significant. Statistical analyses were performed with software package SAS 9.4 (SAS Institute, NC, USA).
Ethics
Informed written consent was obtained from all participants prior to participation. This study was approved by the Ethics Committee of the Capital Region of Denmark (H-3-2011-081, September 6, 2011), and we followed the recommendations of the 1975 Declaration of Helsinki.
Results
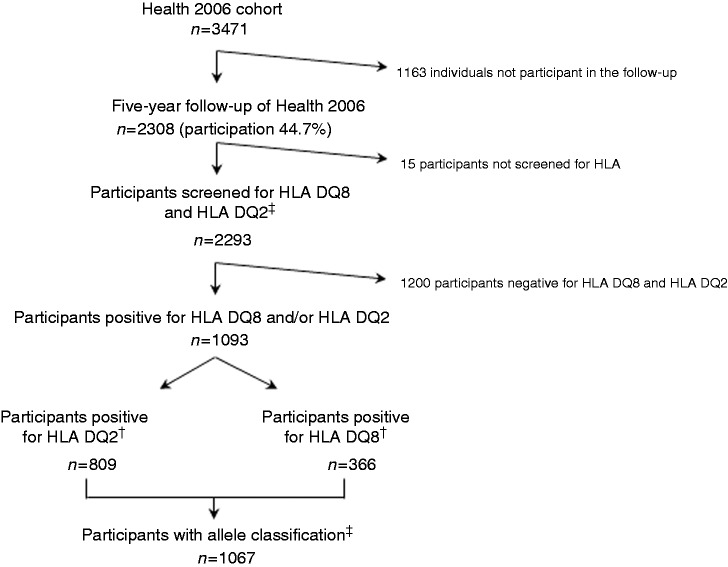
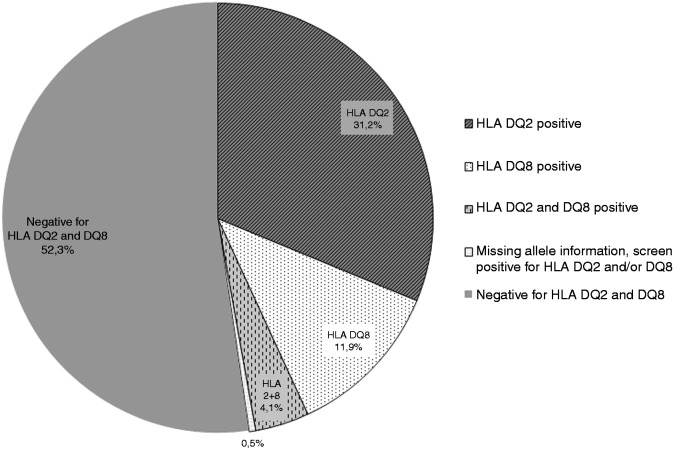
Figure 1 illustrates the flow of participants. A total of 2293 individuals were screened for HLA DQ2 and HLA DQ8 and 1093 (48%) were positive. Among the HLA DQ2- and HLA DQ8-positive participants, 1082 (99%) were allele classified. We found that 31% were DQ2 positive, 12% were DQ8 positive and 4% were both DQ2 and DQ8 positive (Figure 2).
Figure 1.
Overview of study and flow of participants in the cohort.
*The participants were screened for HLA DQ8 and HLA DQ2 with the Celiac Gene Screen test, and the alleles associated with celiac disease were identified by the Celiac Gene Alleles test (BioDiagene, Palermo, Italy, distributed by Thermo Fisher Scientific, Allerød, Denmark).
†Among the HLA DQ2 and HLA DQ8 positive individuals, 93 participants were both DQ2 and DQ8 positive.
‡Eleven participants positive for HLA DQ2 and/or HLA DQ8 were not allele classified.
HLA: human leukocyte antigen.
Figure 2.
Prevalence of celiac disease predisposition in a Danish general population.
HLA: human leukocyte antigen.
Table 2 shows the characteristics of the HLA DQ2/DQ8-positive and -negative participants, and the separate groups of HLA DQ2-positive, HLA DQ8-positive and both HLA DQ2- and DQ8-positive. There were no significant differences between the HLA DQ2/DQ8-positive and -negative participants, nor between the four groups: all HLA DQ2/DQ8-negative participants, HLA DQ2-positive, HLA DQ8-positive, and both HLA DQ2- and DQ8-positive.
Table 2.
Characteristics of the participants positive and negative for HLA DQ2 and HLA DQ8.
| All HLA DQ2- and DQ8-negative participants | All HLA DQ2-and DQ8-positive participantsa | p valueb | HLA DQ2-positive participants a | HLA DQ8-positive participantsa | HLA DQ2- and DQ8-positive participants a | p valuec | |
|---|---|---|---|---|---|---|---|
| Number of participants | 1200 | 1093 | 716 | 273 | 93 | ||
| % (n/total N) | % (n/total N) | % (n/total N) | % (n/total N) | % (n/total N) | |||
| Sex | 0.564d | 0.803d | |||||
| Male | 46.6 (559/1200) | 45.4 (496/1093) | 46.4 (332/716) | 44.0 (120/273) | 43.0 (40/93) | ||
| Female | 53.4 (641/1200) | 54.6 (597/1093) | 53.6 (384/716) | 56.0 (153/273) | 57.0 (53/93) | ||
| Age at examination | 0.064d | 0.433d | |||||
| 24–44 years | 22.9 (275/1200) | 19.2 (210/1093) | 19.1 (137/716) | 18.7 (51/273) | 20.4 (19/93) | ||
| 45–59 years | 37.9 (455/1200) | 41.5 (453/1093) | 41.5 (297/716) | 42.1 (115/273) | 39.8 (37/93) | ||
| ≥ 60 years | 39.2 (470/1200) | 39.3 (430/1093) | 39.4 (282/716) | 39.2 (107/273) | 39.8 (37/93) | ||
| Education | 0.289d | 0.460d | |||||
| ≤ 13 years of school/no vocational training | 7.6 (90/1185) | 7.4 (80/1089) | 7.2 (51/712) | 7.0 (19/273) | 10.8 (10/93) | ||
| Short vocational training (<3 years) | 57.7 (684/1185) | 55.3 (602/1089) | 55.9 (398/712) | 55.0 (150/273) | 51.6 (48/93) | ||
| Medium length vocational training (3–4 years) | 22.8 (270/1185) | 26.3 (286/1089) | 26.0 (185/712) | 28.6 (78/273) | 22.6 (21/93) | ||
| Long education vocational training(>4 years) | 11.9 (141/1185) | 11.1 (121/1089) | 11.0 (78/712) | 9.5 (26/273) | 15.1 (14/93) | ||
| Smoking | 0.569 d | 0.640d | |||||
| Current smoker | 15.7 (187/1192) | 15.9 (173/1086) | 16.5 (117/711) | 15.1 (41/271) | 15.1 (14/93) | ||
| Past smoker | 40.4 (482/1192) | 38.3 (416/1086) | 38.0 (270/711) | 36.2 (98/271) | 45.2 (42/93) | ||
| Never smoker | 43.9 (523/1192) | 45.8 (497/1086) | 45.6 (324/711) | 48.7 (132/271) | 39.8 (37/93) | ||
| Median (5%/95%) | Median (5%/95%) | Median (5%/95%) | Median (5%/95%) | Median (5%/95%) | |||
| Alcohol consumption: Units per week in the past 12 months | 5.5 (0.0/25.0) | 6.0 (0.0/28.0) | 0.102e | 6.0 (0.0/28.0) | 7.0 (0.0/28.0) | 5.0 (0.0/25.0) | 0.329g |
| Mean (95% CI) | Mean (95% CI) | Mean (95% CI) | Mean (95% CI) | Mean (95% CI) | |||
| Body mass index | 26.1 (25.9–26.4) | 25.8 (25.6–26.1) | 0.131f | 25.8 (25.5–26.1) | 25.9 (25.4–26.5) | 25.6 (24.7–26.5) | 0.302g |
The participants were screened for HLA DQ8 and HLA DQ2 with the Celiac Gene Screen test, and the alleles associated with CD were identified by the Celiac Gene Alleles test (BioDiagene, Palermo, Italy, distributed by Thermo Fisher Scientific, Allerød, Denmark).
P value for the difference between all HLA DQ2- and DQ8-positive versus negative.
P value for the difference between the four groups: all HLA DQ2- and DQ8-negative, HLA DQ2-positive, HLA DQ8-positive, and both HLA DQ8- and DQ2-positive.
Chi-square test.
Wilcoxon two-sample test.
Independent samples t test.
Kruskal–Wallis test.
CD: celiac disease; HLA: human leukocyte antigen.
With respect to biomarkers, there were no significant differences when comparing the HLA DQ2/DQ8-positive and -negative participants, or the four groups (described above) (Table 1). However, when selecting all HLA DQ2.5-positive participants, a high-risk group with respect to CD, we found a significant difference in TSH level between the HLA DQ2/DQ8-negative and the HLA DQ2.5-positive participants. No significant differences were found for the other biomarkers with respect to DQ2.5. When tested separately, the haplotypes DQ2.2, DQ8 and DQ2.3 had no significant difference compared with the DQ2/8 negative (Table 3).
Table 3.
Results of biochemical analyses, symptoms and diagnoses stratified by HLA haplotypes DQ2.5, DQ2.2, DQ2.3 and DQ8 among participants in the Health2006 five-year follow-up.
| All HLA-negative participantsa | All participants with DQ2.5b | p value negative vs DQ2.5 | All participants with DQ2.2c | p value negative vs DQ2.2 | All participants with DQ8d | p value negative vs DQ8 | All participants with DQ2.3 e | p value negative vs DQ2.3 | |
|---|---|---|---|---|---|---|---|---|---|
| Number of participants | 1200 | 594 | 223 | 361 | 29 | ||||
| Median (5/95%) | Median (5/95%) | Median (5/95%) | Median (5/95%) | Median (5/95%) | |||||
| Biomarkers | |||||||||
| Hemoglobin (mmol/l) | 8.8 (7.7/10.0) | 8.8 (7.6/10.0) | 0.620u | 8.8 (7.9/9.9) | 0.830u | 8.7 (7.5/9.9) | 0.123u | 8.8 (7.5/10.0) | 0.739u |
| Vitamin D (nmol/l) | 70.0 (28.1/122.8) | 69.8 (25.2/118.8) | 0.593u | 75.0 (31.2/121.7) | 0.193u | 73.5 (25.8/122.3) | 0.359u | 79.9 (34.2/127.7) | 0.831u |
| Plasma glucose (mmol/l) | 5.3 (4.4/6.8) | 5.3 (4.4/7.0) | 0.946u | 5.3 (4.5/6.4) | 0.689u | 5.3 (4.4/7.0) | 0.258u | 5.1 (4.4/6.3) | 0.075u |
| HbA1c (%) | 5.5 (4.9/6.2) | 5.5 (5.0/6.2) | 0.639u | 5.4 (4.9/6.0) | 0.106u | 5.5 (5.0/6.2) | 0.051u | 5.5 (4.9/6.0) | 0.578u |
| TPO (U/ml) | 11.0 (8.0/166.0) | 12.0 (8.0/170.0) | 0.282u | 12.0 (8.0/91.0) | 0.582u | 12.0 (8.0/86.0) | 0.437u | 11.0 (10.0/285.0) | 0.258u |
| TSH (mU/l) | 1.4 (0.5/3.6) | 1.5 (0.6/4.1) | 0.046u | 1.5 (0.5/3.3) | 0.698u | 1.4 (0.5/3.5) | 0.894u | 1.5 (0.7/5.4) | 0.224u |
| FT4 (pmol/l) | 12.3 (9.9/15.0) | 12.2 (9.8/14.8) | 0.109u | 12.3 (10.0/15.1) | 0.792u | 12.3 (10.1/15.8) | 0.535u | 12.2 (10.0/14.1) | 0.987u |
| Anti-CCP (U/ml) | 1.9 (0.7/4.0) | 2.0 (0.7/4.0) | 0.568u | 1.9 (0.6/3.7) | 0.159u | 2.0 (0.7/3.9) | 0.700u | 1.6 (0.7/3.4) | 0.166u |
| % (n/total N) | % (n/total N) | % (n/total N) | % (n/total N) | % (n/total N) | |||||
| Symptoms/diagnoses | |||||||||
| Diabetes mellitus Type 1f | 0.6 (7/1200) | 0.3 (2/594) | 0.726x | 0.0 (0/223) | 0.605x | 0.8 (3/361) | 0.706x | 0.0 (0/29) | 1.000x |
| Cardiovascular diseaseg | 7.4 (89/1200) | 9.4 (56/594) | 0.141w | 6.7 (15/223) | 0.716w | 6.9 (25/361) | 0.753w | 17.2 (5/29) | 0.064x |
| Inflammatory bowel diseaseh | 0.9 (11/1200) | 0.3 (2/594) | 0.241x | 1.8 (4/223) | 0.274x | 1.1 (4/361) | 0.759x | 0.0 (0/29) | 1.000x |
| Asthmai | 4.2 (50/1200) | 4.2 (25/594) | 0.967w | 3.6 (8/223) | 0.854x | 3.3 (12/361) | 0.472w | 6.9 (2/29) | 0.350x |
| Chronic obstructive pulmonary diseasej | 2.8 (33/1200) | 2.2 (13/594) | 0.479w | 1.8 (4/223) | 0.410w | 1.4 (5/361) | 0.140w | 0.0 (0/29) | 1.000x |
| Liver diseasek | 0.8 (9/1200) | 0.7 (4/594) | 1.000x | 0.9 (2/223) | 0.686x | 0.6 (2/361) | 1.000x | 3.5 (1/29) | 0.213x |
| Gastrointestinal cancerl | 1.1 (13/1200) | 1.4 (8/594) | 0.625w | 0.0 (0/223) | 0.241x | 1.4 (5/361) | 0.583x | 0.0 (0/29) | 1.000x |
| Lymphomam | 0.3 (4/1200) | 0.3 (2/594) | 1.000x | 0.9 (2/223) | 0.239x | 1.1 (4/361) | 0.089x | 0.0 (0/29) | 1.000x |
| Celiac diseasen | 0.0 (0/1200) | 1.4 (8/594) | 0.000x | 0.9 (2/223) | 0.025x | 0.6 (2/361) | 0.053x | 0.0 (0/29) | - |
| Any autoimmune diseaseo | 6.3 (76/1200) | 7.9 (47/594) | 0.213w | 5.8 (13/223) | 0.775w | 8.6 (31/361) | 0.137w | 0.0 (0/29) | 0.253x |
| Irritable bowel syndromep | 4.6 (55/1197) | 2.7 (16/590) | 0.055w | 3.2 (7/220) | 0.346w | 1.4 (5/359) | 0.006w | 6.9 (2/29) | 0.394x |
| Allergy | |||||||||
| Phadiatop > 0.35 | 30.9 (370/1198) | 28.5 (169/593) | 0.300w | 64.9 (144/222) | 0.211w | 73.4 (265/361) | 0.118w | 72.4 (21/29) | 0.704w |
| Food mix, Fx5 > 0.35 | 4.1 (49/1198) | 5.1 (30/593) | 0.347w | 94.1 (209/222) | 0.237w | 98.1 (354/361) | 0.054w | 100.0 (29/29) | 0.266w |
| Atopyq | 32.9 (391/1188) | 30.3 (179/590) | 0.274w | 33.9 (75/221) | 0.766w | 29.4 (104/354) | 0.211w | 24.1 (7/29) | 0.320w |
| Skin prick test, positive: | |||||||||
| Pollenr | 27.4 (325/1188) | 26.3 (155/590) | 0.627w | 30.3 (67/221) | 0.367w | 25.4 (90/354) | 0.472w | 17.2 (5/29) | 0.226w |
| Animal danders | 14.4 (171/1188) | 13.7 (81/590) | 0.705w | 17.2 (38/221) | 0.282w | 12.7 (45/354) | 0.424w | 24.1 (7/29) | 0.177x |
| Dust mitest | 12.0 (142/1188) | 8.6 (51/590) | 0.035w | 10.9 (24/221) | 0.644w | 8.2 (29/354) | 0.048w | 10.3 (3/29) | 1.000x |
The participants were screened for HLA DQ8 and HLA DQ2 with the Celiac Gene Screen test, and the alleles associated with CD were identified by Celiac Gene Alleles test (BioDiagene, Palermo, Italy, distributed by Thermo Fisher Scientific, Allerød, Denmark).
DQ2.5cis: DQB1*02, DQA1*05 and DQ2.5trans: encoded by DQA1*05-DQB1*03:01 and DQA1*02:01-DQB1*02.
DQ2.2: DQB1*02, DQA1*0201.
DQ8: DQA1*03, DQB1*0302/0305.
DQ2.3: DQB1*02, DQA1*03.
Diabetes mellitus Type 1: ICD-8: 249, ICD-10: E10.
Including: ischemic heart disease (ICD-8: 410–414, ICD-10: I20–I25) and stroke (ICD-8: 431–434, 436, ICD-10: I60–I69).
Including: Crohn’s disease (ICD-8: 563.01, ICD-10: K50) and ulcerative colitis (ICD-8: 563.19, ICD-10: K51).
Asthma: ICD-8: 493, ICD-10: J45, J46.
Chronic obstructive pulmonary disease: ICD-8: 491–492 and ICD-10: J42–J44.
Liver disease: ICD-8: 570–573 and ICD-10: K70–K77.
Gastrointestinal cancer: ICD-8: 140–154 and ICD-10: C00–C18, C20–C21.
Lymphoma: ICD-8: 200–202 and 275.59 and ICD-10: C81–86, C88 and C915.
Celiac disease, ICD-8: 269.00 and ICD-10: K90.0.
aIncluding thyrotoxicosis (ICD-8: 242.00, ICD-10: E05.0), Type 1 diabetes (ICD-8: 249, ICD-10: E10), multiple sclerosis (ICD-8: 340, ICD-10: G35), iridocyclitis (ICD-8: 364, ICD-10: H20), Crohn’s disease (ICD-8: 563.01, ICD-10: K50), ulcerative colitis (ICD-8: 563.19, ICD-10: K51), psoriasis vulgaris (ICD-8: 696.09–10, 696.19, ICD-10: L40), seropositive rheumatoid arthritis (ICD-8: 712.19, 712.39, 712.59, ICD-10: M05–M06), polymyalgia rheumatica (ICD-8: 446.30–31, 446.39, ICD-10: M31.5–6, M35.3), pernicious anemia (ICD-8: 281.0, ICD-10: D51.0), autoimmune hemolytic anemia (ICD-8: 283.90–91, ICD-10: D59.1), idiopathic thrombocytopenic purpura (ICD-8: 446.49, ICD-10: D69.3), autoimmune thyroiditis (ICD-8: 245.03, ICD-10: E06.3), primary adrenocortical insufficiency (ICD-8: 255.1, ICD-10: E27.1), Guillain-Barré syndrome (ICD-8: 354, ICD-10: G61.0), autoimmune hepatitis (ICD-8: 571.93, ICD-10: K75.4), primary biliary cirrhosis (ICD-8: 571.90, ICD-10: K74.3), celiac disease (ICD-8: 269.00, ICD-10: K90.0), pemphigus (ICD-8: 694, ICD-10: L10), pemphigoid (ICD-8: 694.05, ICD-10: L12), alopecia areata (ICD-8: 704.00, ICD-10: L63), vitiligo (ICD-8: 709.01, ICD-10: L80), juvenile arthritis (ICD-8: 712.09, ICD-10: M08), Wegener’s granulomatosis (ICD-8: 446.29, ICD-10: M31.3), dermatopolymyositis (ICD-8: 716, ICD-10: M33), myasthenia gravis (ICD-8: 733.09, ICD-10: G70.0), systemic sclerosis (ICD-8: 734.0, ICD-10: M34), systemic lupus erythematosus (ICD-8: 734.19, ICD-10: M32), Sjögren’s syndrome (ICD-8: 734.90, ICD-10: M35.0), and nkylosing spondylitis (ICD-8: 712.49, ICD-10: M45).
Irritable bowel syndrome is defined as a cluster of frequent self-reported gastrointestinal symptoms during the past 12 months: “Subjects stating that they often experience both abdominal pain and distension and additionally, either borborygmi or altering stool consistency, or both.”
Atopy defined as a positive reaction to at least one of 10 allergens: birch, grass (Phleum pratense) mugwort, horse, cat, dog, two house dust mites (Dermatophagoides pteronyssinis and D. farinae), and two molds (Cladosporium herbarum and Alternaria alternata).
Pollen, at least one positive of birch, grass (P. pratense) or mugwort.
Animal dander, at least one positive for horse, cat or dog.
Dust mites, at least one positive for one of two house dust mites: D. pteronyssinis and D. farina.
Wilcoxon test.
Chi-square test.
Fisher’s exact test (p values ≥ 0.05 in bold).
Anti-CCP: anti-cyclic citrullinated peptide; CD: celiac disease; FT4: free thyroxine; HbA1c: hemoglobin A1c; HLA: human leukocyte antigen; ICD-8: International Statistical Classification of Diseases and Related Health Problems, eighth revision; ICD-10: International Statistical Classification of Diseases and Related Health Problems, 10th revision; TPO: thyroperoxidase; TSH: thyroid-stimulating hormone.
We also compared the different groups on allergy parameters, IgE tests and skin prick test. When comparing the HLA DQ2/DQ8-positive with the HLA-negative participants, we found that the negative had a significantly higher number of positive skin prick tests for dust mites; however, there were no other significant differences with respect to allergy (Table 1). The same results were found when comparing the HLA-negative participants with all HLA DQ2.5-positive and all HLA DQ8-positive separately, but the haplotypes DQ2.2 and DQ2.3 did not have the same low number of positive skin prick tests for dust mites (Table 3). However, when comparing the four groups (described above), we found no significant differences among dust mite allergy, although there were significant differences among the groups when screened for serum-specific IgE to food allergens by food mix (Fx5) (Table 1). This difference was not found in the analysis where haplotypes were compared separately (Table 3).
We found no significant associations with respect to different diagnosis groups from the National Patient Registry, apart from a significantly higher prevalence of CD among HLA DQ2- and HLA DQ8-positive individuals as expected (Table 1).
Looking at IBS, we found significant differences between HLA DQ2/DQ8-negative and -positive, as well as the four groups (described above), showing that HLA DQ2/DQ8-positive participants had a significantly lower frequency of IBS (Table 1). This difference did not remain significant in the separate analyses of HLA DQ2.5-, DQ2.2- and DQ2.3-positive participants (Table 3).
All analyses were also conducted separately for individuals without CD, but the exclusion of CD did not change the results (data not shown).
Table 4 shows the distribution of HLA DQ2 and DQ8 haplotypes among the participants in this general population, and among the nine individuals with CD. We found HLA DQ2.5 to be the most frequent of the HLA DQ2 and DQ8 haplotypes: 25% among the total population and 89% among the individuals with CD, followed by DQ8 haplotype (16%/22%), DQ2.2 haplotype (10%/22%) and DQ2.3 haplotype (1%/none). Table 5 shows the allele distribution for alleles associated with CD in this cohort.
Table 4.
HLA DQ2 and HLA DQ8 haplotype distribution in a general Danish population.
| All (n = 2293) | Individuals homozygotic for DQB1*02 | Individuals with celiac diseasea (n = 9) | Individuals homozygotic for DQB1*02 | |
|---|---|---|---|---|
| % (n/n total) | % (n/n total) | |||
| DQ2- and DQ8-negative | 52.3 (1200/2293) | – | ||
| DQ2.5cis | 20.9 (480/2293) | 27 homozygotic for DQB1*02 | 66.7 (6/9) | Two homozygotic for DQB1*02 |
| DQ8 | 11.7 (268/2293) | – | ||
| DQ2.2 | 7.2 (165/2293) | 16 homozygotic for DQB1*02 | – | |
| DQ2.5cis and DQ8 | 2.5 (58/2293) | 11.1 (1/9) | ||
| DQ2.2 and DQ8 | 1.2 (28/2293) | 11.1 (1/9) | ||
| DQ2.5cis and DQ2.2 | 1.2 (27/2293) | 27 homozygotic for DQB1*02 | 11.1 (1/9) | One homozygotic for DQB1*02 |
| DQ2.5trans | 1.0 (22/2293) | – | ||
| DQ2.3 | 0.5 (12/2293) | 4 homozygotic for DQB1*02 | – | |
| DQ2.5cis and DQ2.3 | 0.3 (7/2293) | 7 homozygotic for DQB1*02 | – | |
| DQ2.3 and DQ8 | 0.3 (7/2293) | – | ||
| DQ8.5 | 0.2 (5/2293) | – | ||
| DQ2.2 and DQ2.3 | 0.1 (3/2293) | 3 homozygotic for DQB1*02 | – | |
| Missing allele information | 0.5 (11/2293) | 0.0 (0/9) |
Celiac disease cases: eight found by screening (published earlier12) and one with known celiac disease from the National Patient Registry.
HLA: human leukocyte antigen.
Table 5.
Allele distribution in a general Danish population.
| Allele | % (n/total N) |
|---|---|
| DQB1*02 | 35.3 (809/2293) |
| DQA1*05 | 30.6 (702/2293) |
| DQA1*03 | 20.6 (473/2293) |
| DQB1*0302/0305 | 16.0 (366/2293) |
| DQA1*0201 | 14.8 (340/2293) |
| DQB1*0301/0304 | 10.6 (244/2293) |
Discussion
In this general Danish population, 47.7% were positive for either HLA DQ2 or DQ8 or both. All individuals with CD were HLA DQ2 and/or HLA DQ8 positive, the majority HLA DQ2.5 being positive. We compared the different HLA DQ2 and HLA DQ8 groups with respect to biomarkers, diagnoses, symptoms and allergy test results and found that HLA DQ2.5 was associated with higher TSH levels, while HLA DQ2/DQ8-positive participants had significantly lower frequencies of IBS. There was no statistically significant association between HLA DQ2/DQ8 and diagnoses other than CD.
Surprisingly, we found that this general Danish population had a higher prevalence of HLA DQ2/DQ8 than reported in other studies,3,22–24 although the prevalence in Denmark of CD has been reported to be lower than other European countries even though it is increasing.25 The lower prevalence of CD in Denmark may be due to differences in exposure to environmental risk factors or underdiagnosis of CD. Our study extends the results from other studies since DQ2.5 (both cis and trans), DQ2.2 and DQ2.3 all were measured. Other studies often reported only DQ2.5cis thereby possibly estimating a lower prevalence of individuals having HLA DQ2-risk alleles. This explanation was supported by a study by DiGiacomo et al.,24 who found that 29% had DQ2.5/8, but the figure rose to 39% when counting the total HLA DQ2/8. When counting only HLA DQ2.5cis and DQ8 the percentages in our study were: DQ2.5cis: 25% (572/2293), DQ8: 16% (361/2293), and 2.5cis and/or DQ8: 41% (933/2293), comparable to the 22% among the control group in a Swedish study for DQ2.5,26 but higher for DQ8 than numbers from Mexico and Brazil.27,28 Compared with the prevalence table of different populations in Alarida et al.,29 our numbers are higher for DQ2 (DQ2:31%/DQ2.5 25% vs 23%), but comparable for DQ8 (DQ8 16% vs 28.3%), and the Scandinavian numbers from the table (11% DQ2 and 15% DQ8) are lower than ours. Our HLA DQ2/8 prevalence is also higher than the one found in a screening study from Latvia comparable to our study.30
Our findings suggest that measuring total HLA DQ2/8 risk alleles as compared with only DQ2.5cis and DQ8 identifies more individuals at risk of CD, which could have important implications for clinical practice as HLA DQ risk assessment sometimes is used to exclude a diagnosis of CD. This was also demonstrated in a Danish study of individuals with celiac disease in which seven individuals were HLA2.5trans or DQ2.2 positive.31 In the present study, however, all of the nine individuals diagnosed with CD had either DQ2.5cis or DQ8.
As it is known that CD and diabetes Type 1 share links to HLA,9,10,32 we had expected an association between Type 1 diabetes and HLA DQ2/DQ8, but we found no significant difference among the groups with respect to diabetes. This might be due to a small proportion of individuals with diabetes Type 1 in the cohort, caused by selection for the study population and low prevalence of diabetes Type 1, as well as the fact that susceptible HLA antigens for diabetes Type 1 are more numerous than for CD, giving less power. We found no association between HLA DQ2/DQ8 positivity and autoimmune diseases,9–11 which might be due to a lack of power; however, twin studies have shown that CD is more highly genetically determined than other autoimmune diseases.12 Furthermore, we found an association between TSH and DQ2.5 that has not been previously known, and not found for the other thyroid markers (TPO, FT4). A reason could be that TSH is a more sensitive marker for hypothyroidism, as well as the fact that TSH is a quantitative parameter measured in all participants, giving more power to detect differences.
In line with a study from Italy,24 we found that the proportion of IBS was lower among HLA DQ2/8-positive individuals. However, the Italian study also found that IBD and liver diseases were more prevalent among HLADQ2/8-positive individuals, which we could not confirm in this present study. Other studies have found IBS symptoms to be more prevalent in patients with CD33,34; however, none of the nine individuals with CD in this present study had IBS according to our IBS definition.
We found a significant difference between HLA DQ2/8-positive and -negative individuals for dust mite allergy. HLA has earlier been associated with allergy, playing a role in antigen/allergen presentation35,36 and IgE dysregulation37; however, the association is not clear and different alleles and allergies have been tested. Few studies have associated HLA DQ2/DQ8 with allergy. One found an association between DQ8 and hevein-specific IgE immune response in individuals with latex allergy,38 while others found DQ2 to be associated with olive pollen39 and the allele DQB1*0301 to be associated with grass pollen allergy.36 Thus, the association of HLA DQ2/8 with allergy remains unclear.
The population-based design and the screening of all participants both for CD antibodies and for HLA DQ2/DQ8 are major strengths of our study. However, the relatively low prevalence of CD limits the possibilities of comparing participants with and without CD. Our study is limited to the HLAs known to be associated with CD and we were therefore not able to describe other HLA alleles, and since the test is an SSP-PCR for specific alleles, we cannot be sure about the typing with respect to homozygosity and heterozygosity on the alleles other than DQB1*02. Selection bias is not likely as we have no reason to believe that HLA genotype is associated with participation in the study. The linkage to the National Patient Registry is a strength that gives us the possibility of accurate information about diagnoses. However, there is a risk of misclassification in the registries that can influence the frequencies of diseases. There is also a risk of misclassification of IBS that was based on the self-reported questionnaire for IBS symptoms, and we had no information on duration and severity. Also, the differences between the IBS definition used and the Rome criteria could lead to misclassification of IBS and inconsistent results between studies.
In conclusion, 48% of individuals were positive for either HLA DQ2 or HLA DQ8 or both in this general Danish population and thus potentially at risk for CD. All individuals with CD were HLA DQ2 and/or DQ8 positive, and the majority HLA DQ2.5. We found that HLA DQ2.5 was associated with higher TSH levels, while HLA DQ2- and/or HLA DQ8-positive participants had a significantly lower prevalence of IBS. HLA DQ2/DQ8 was strongly associated with CD, but no other registry-based diagnoses.
Supplemental Material
Supplemental material, Supplementary table for The distribution of HLA DQ2 and DQ8 haplotypes and their association with health indicators in a general Danish population by Line Lund Kårhus, Betina H Thuesen, Tea Skaaby, Jüri J Rumessen and Allan Linneberg in United European Gastroenterology Journal
Acknowledgements
We would like to thank all staff members at the Research Centre for Prevention and Health for contributing to the data collection and health examinations. We would also like to thank Thermo Fisher Scientific, Allerød, Denmark; Lone Søgaard and Bjarne Kristiensen for their help with the HLA analyses and interpretation of the results; and Pia Schytte Hansen for performing the HLA screening.
Declaration of conflicting interests
None declared.
Funding
This work was supported by the Tryg Foundation (7-11-0213), Dansk Cøliaki Forening (the Danish Celiac Disease Patient Organization), and Thermo Fisher Scientific, Allerød, Denmark. Thermo Fisher Scientific performed the HLA screening.
Informed consent
Informed written consent was obtained from all participants prior to participation.
Ethics approval
This study was approved by the Ethics Committee of the Capital Region of Denmark (H-3-2011-081, September 6, 2011), and we followed the recommendations of the 1975 Declaration of Helsinki.
References
- 1.Lebwohl B, Ludvigsson JF, Green PH. Celiac disease and non-celiac gluten sensitivity. BMJ 2015; 351: h4347–h4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sollid LM. The roles of MHC class II genes and post-translational modification in celiac disease. Immunogenetics 2017; 69: 605–616. [DOI] [PubMed] [Google Scholar]
- 3.Husby S, Koletzko S, Korponay-Szabó IR, et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J Pediatr Gastroenterol Nutr 2012; 54: 136–160. [DOI] [PubMed] [Google Scholar]
- 4.Sollid LM, Iversen R, Steinsbø Ø, et al. Small bowel, celiac disease and adaptive immunity. Dig Dis 2015; 33: 115–121. [DOI] [PubMed] [Google Scholar]
- 5.Sollid LM, Qiao SW, Anderson RP, et al. Nomenclature and listing of celiac disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules. Immunogenetics 2012; 64: 455–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodd M, Kim CY, Lundin KE, et al. T-cell response to gluten in patients with HLA-DQ2.2 reveals requirement of peptide-MHC stability in celiac disease. Gastroenterology 2012; 142: 552–561. [DOI] [PubMed] [Google Scholar]
- 7.Tollefsen S, Hotta K, Chen X, et al. Structural and functional studies of trans-encoded HLA-DQ2.3 (DQA1*03:01/DQB1*02:01) protein molecule. J Biol Chem 2012; 287: 13611–13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lionetti E, Castellaneta S, Francavilla R, et al. Introduction of gluten, HLA status, and the risk of celiac disease in children. N Engl J Med 2014; 371: 1295–1303. [DOI] [PubMed] [Google Scholar]
- 9.Denham JM, Hill ID. Celiac disease and autoimmunity: Review and controversies. Curr Allergy Asthma Rep 2013; 13: 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahaly GJ, Schuppan D. Celiac disease and endocrine autoimmunity. Dig Dis 2015; 33: 155–161. [DOI] [PubMed] [Google Scholar]
- 11.Kokaraki G, Daniilidis M, Yiangou M, et al. Major histocompatibility complex class II (DRB1*, DQA1*, and DQB1*) and DRB1*04 subtypes’ associations of Hashimoto’s thyroiditis in a Greek population. Tissue Antigens 2009; 73: 199–205. [DOI] [PubMed] [Google Scholar]
- 12.Bogdanos DP, Smyk DS, Rigopoulou EI, et al. Twin studies in autoimmune disease: Genetics, gender and environment. J Autoimmun 2012; 38: J156–J169. [DOI] [PubMed] [Google Scholar]
- 13.Jakobsen BK, Morling N, Platz P, et al. HLA-DR phenotype and HLA-B,DR haplotype frequencies in 704 unrelated Danes. Tissue Antigens 1981; 18: 270–275. [DOI] [PubMed] [Google Scholar]
- 14.Madsen M, Graugaard B, Lamm LU, et al. HLA-DR genes and antigens in the Danish population. A study of 500 unrelated Danes. Tissue Antigens 1981; 18: 258–269. [DOI] [PubMed] [Google Scholar]
- 15.Horwitz A, Skaaby T, Kårhus LL, et al. Screening for celiac disease in Danish adults. Scand J Gastroenterol 2015; 50: 824–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thuesen BH, Cerqueira C, Aadahl M, et al. Cohort profile: The Health2006 cohort, research centre for prevention and health. Int J Epidemiol 2014; 43: 568–575. [DOI] [PubMed] [Google Scholar]
- 17.Kårhus LL, Thuesen BH, Rumessen JJ, et al. Symptoms and biomarkers associated with celiac disease: Evaluation of a population-based screening program in adults. Eur J Gastroenterol Hepatol 2016; 28: 1298–1304. [DOI] [PubMed] [Google Scholar]
- 18.Thuesen BH, Heede NG, Tang L, et al. No association between vitamin D and atopy, asthma, lung function or atopic dermatitis: A prospective study in adults. Allergy 2015; 70: 1501–1504. [DOI] [PubMed] [Google Scholar]
- 19.Kay L, Jorgensen T. Redefining abdominal syndromes. Results of a population-based study. Scand J Gastroenterol 1996; 31: 469–475. [DOI] [PubMed] [Google Scholar]
- 20.Heinsvig Poulsen C, Falgaard Eplov L, Hjorthøj C, et al. Gastrointestinal symptoms related to the irritable bowel syndrome—A longitudinal population-based register study. Scand J Gastroenterol 2016; 51: 420–426. [DOI] [PubMed] [Google Scholar]
- 21.Nickelsen TN. Data validity and coverage in the Danish National Health Registry. A literature review [article in Danish]. Ugeskr Laeger 2001; 164: 33–37. [PubMed] [Google Scholar]
- 22.Berzina L, Shtauvere-Brameus A, Ludvigsson J, et al. Newborn screening for high-risk human leukocyte antigen markers associated with insulin-dependent diabetes mellitus: The ABIS study. Ann N Y Acad Sci 2002; 958: 312–316. [DOI] [PubMed] [Google Scholar]
- 23.Catassi C, Gatti S, Lionetti E. World perspective and celiac disease epidemiology. Dig Dis 2015; 33: 141–146. [DOI] [PubMed] [Google Scholar]
- 24.DiGiacomo D, Santonicola A, Zingone F, et al. Human leukocyte antigen DQ2/8 prevalence in non-celiac patients with gastrointestinal diseases. World J Gastroenterol 2013; 19: 2507–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grode L, Bech BH, Jensen TM, et al. Prevalence, incidence, and autoimmune comorbidities of celiac disease: A nation-wide, population-based study in Denmark from 1977 to 2016. Eur J Gastroenterol Hepatol 2018; 30: 83–91. [DOI] [PubMed] [Google Scholar]
- 26.Ploski R, Ascher H, Sollid LM. HLA genotypes and the increased incidence of coeliac disease in Sweden. Scand J Gastroenterol 1996; 31: 1092–1097. [DOI] [PubMed] [Google Scholar]
- 27.Cecilio LA, Bonatto MW. The prevalence of HLA DQ2 and DQ8 in patients with celiac disease, in family and in general population. Arq Bras Cir Dig 2015; 28: 183–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mejía-León ME, Ruiz-Dyck KM, Calderón de la Barca AM. HLA-DQ genetic risk gradient for type 1 diabetes and celiac disease in northwestern Mexico. Rev Gastroenterol Mex 2015; 80: 135–143. [DOI] [PubMed] [Google Scholar]
- 29.Alarida K, Harown J, Di Pierro MR, et al. HLA-DQ2 and -DQ8 genotypes in celiac and healthy Libyan children. Dig Liver Dis 2010; 42: 425–427. [DOI] [PubMed] [Google Scholar]
- 30.Leja M, Shums Z, Nikitina-Zake L, et al. Prevalence estimation of celiac disease in the general adult population of Latvia using serology and HLA genotyping. United European Gastroenterol J 2015; 3: 190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lund F, Hermansen MN, Pedersen MF, et al. Mapping of HLA- DQ haplotypes in a group of Danish patients with celiac disease. Scand J Clin Lab Invest 2015; 75: 519–522. [DOI] [PubMed] [Google Scholar]
- 32.Adlercreutz EH, Svensson J, Hansen D, et al. Prevalence of celiac disease autoimmunity in children with type 1 diabetes: Regional variations across the Oresund strait between Denmark and southernmost Sweden. Pediatr Diabetes 2015; 16: 504–509. [DOI] [PubMed] [Google Scholar]
- 33.Sainsbury A, Sanders DS, Ford AC. Prevalence of irritable bowel syndrome-type symptoms in patients with celiac disease: A meta-analysis. Clin Gastroenterol Hepatol 2013; 11: 359–365. [DOI] [PubMed] [Google Scholar]
- 34.Silvester JA, Graff LA, Rigaux L, et al. Symptoms of functional intestinal disorders are common in patients with celiac disease following transition to a gluten-free diet. Dig Dis Sci 2017; 62: 2449–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blumenthal MN. Genetics of asthma and allergy. Allergy Asthma Proc 2000; 21: 55–59. [DOI] [PubMed] [Google Scholar]
- 36.Boehncke WH, Loeliger C, Kuehnl P, et al. Identification of HLA-DR and -DQ alleles conferring susceptibility to pollen allergy and pollen associated food allergy. Clin Exp Allergy 1998; 28: 434–441. [DOI] [PubMed] [Google Scholar]
- 37.Granada M, Wilk JB, Tuzova M, et al. A genome-wide association study of plasma total IgE concentrations in the Framingham Heart Study. J Allergy Clin Immunol 2012; 129: 840–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rihs HP, Chen Z, Ruëff F, et al. HLA-DQ8 and the HLA-DQ8-DR4 haplotype are positively associated with the hevein-specific IgE immune response in health care workers with latex allergy. J Allergy Clin Immunol 2002; 110: 507–514. [DOI] [PubMed] [Google Scholar]
- 39.Quiralte J, Llanes E, Barral P, et al. Ole e 2 and Ole e 10: New clinical aspects and genetic restrictions in olive pollen allergy. Allergy 2005; 60: 360–365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplementary table for The distribution of HLA DQ2 and DQ8 haplotypes and their association with health indicators in a general Danish population by Line Lund Kårhus, Betina H Thuesen, Tea Skaaby, Jüri J Rumessen and Allan Linneberg in United European Gastroenterology Journal