Abstract
The fruit fly Drosophila melanogaster, like most organisms, exhibits increased sleep amount and depth in young compared to mature animals. While the fly has emerged as a powerful model for studying sleep during development, qualitative behavioral features of sleep ontogeny and its genetic control are poorly understood. Here we find that, in addition to increased sleep time and intensity, young flies sleep with less place preference than mature adults, and, like mammals, exhibit more motor twitches during sleep. In addition, we show that ontogenetic changes in sleep amount, twitch, and place preference are preserved across sleep mutants with lesions in distinct molecular pathways. Our results demonstrate that sleep ontogeny is characterized by multifaceted behavioral changes, including quantitative and qualitative alterations to sleep as animals mature. Further, the preservation of sleep ontogenetic changes despite mutations that alter sleep time suggests independent genetic control mechanisms for sleep maturation.
Keywords: sleep, Drosophila, brain development, sleep ontogeny, behavioral genetics, sleep twitches
Statement of Significance
Our data combining high-resolution behavioral tracking with video monitoring in Drosophila provide novel insight into sleep during early development, revealing conserved features of sleep in young animals. Our findings support the idea that genes regulating sleep ontogeny are distinct from those that regulate overall sleep duration. Deepening our understanding of early life sleep is highly relevant towards studying the function for sleep in the developing brain.
Introduction
Nearly all species exhibit ontogenetic sleep changes, which most prominently include increased sleep amount in early life [1–3]. The ontogenetic hypothesis of sleep proposes that early life sleep facilitates ongoing brain maturation [1]. Consistent with this idea, sleep disturbances in humans during infancy and childhood can be deleterious to cognitive and emotional development and are highly prevalent across neurodevelopmental disorders [4–7].
Sleep in the fruit fly Drosophila melanogaster shares many features with mammalian sleep [8, 9] including ontogenetic changes, with increased sleep amount and intensity in early adult life [10–12]. We have previously shown that young adult flies sleep significantly more during the day and night than their mature counterparts, have more consolidated sleep, and are harder to arouse [12]. It is unknown whether other characteristics of sleep might undergo ontogenetic changes in Drosophila. For example, selection of a preferred resting location is a broadly conserved feature of sleep across species [13]. Several studies in mature flies have described stereotyped sleep place preferences, but it is unknown whether young flies also prefer to sleep in certain locations [14–17]. Additionally, work in young rodents has demonstrated increased motor twitch frequency during sleep episodes, which is thought to facilitate sensorimotor development [18]. Characterization of such qualitative changes in Drosophila sleep during early adult development could reveal additional conserved elements of early life sleep, enhancing the utility of this tractable model system for studying sleep ontogeny.
Most studies of sleep in Drosophila, including our prior work on sleep ontogeny, have utilized the single beam (SB) Drosophila activity monitoring (DAM) system, in which flies are individually housed in glass tubes with a single infrared (IR) beam in the center detecting movement [9, 10, 14]. Based on originally described behavioral criteria and later studies of arousability, the absence of beam breaks for a period of greater than or equal to 5 minutes constitutes sleep [10, 14, 19]. While this system is extremely useful for studying many aspects of sleep, it does not provide information about most small fly movements or location during sleep episodes, so behaviors such as twitching and sleep place preference cannot be evaluated. To investigate whether additional aspects of sleep behavior show ontogenetic change in flies, we used the multi-beam (MB) DAM system, which expands on the SB system by using 17 IR beams, providing higher resolution sleep-activity data as well location information [20]. Additionally, we performed high-resolution video monitoring to complement MB studies and evaluate more complex sleep behavior.
We have previously shown that high sleep amounts in young adult flies stem from reduced activity in wake-promoting dopaminergic projections to the sleep-promoting dorsal fan-shaped body (dFSB), resulting in increased dFSB activity to drive sleep [12]. As flies mature, dopaminergic projections become more active, inhibiting the dFSB and reducing sleep amount. Despite knowledge of involved circuitry, it remains unknown which genes regulate sleep ontogenetic change. Over the past several years, many genes have been discovered that control sleep duration in mature adults [8]. Given that these genes exert significant effects on mature adult sleep, it is possible that certain mutations may also interfere with sleep ontogenetic change. We thus examined sleep ontogeny in several short- or long-sleeping mutants with lesions in distinct molecular pathways; these include fumin (DAT transporter) insomniac (adapter for the Cul3 ubiquitin ligase), redeye (nAChR), shaker (K+ channel), sleepless (K+ channel regulator), taranis (cell cycle regulator), ADAR (RNA editing gene), and DTH (Drosophila tyrosine hydroxylase) [21–28].
Here, we report that ontogenetic changes from early to mature adulthood occur in multiple sleep behaviors in Drosophila, including sleep duration, motor twitches, and place preference. Further, we show that ontogenetic changes in sleep persist in all studied sleep mutants, suggesting that the genetic control of sleep ontogeny is distinct from the genetic control of overall sleep duration.
Materials and Methods
Fly stocks and husbandry
Flies were raised and maintained in bottles on standard molasses food at 25°C on a 12:12 hour light:dark cycle. Fly stocks were as follows: Iso31 [29], fumin [21], redeye [23], shaker [24] and sleepless [25] were gifts from Amita Sehgal, taranis [26] was a gift from Kyunghee Koh, DTH ple [2] BAC and DTHFS±Bac (“DA-deficient”) [28] were gifts from Jay Hirsh, UAS RNAi ADAR (VDRC #7764) was a gift from William Joiner, and insomniac [22] was a gift from Nicholas Stavropoulous. Fumin, redeye, sleepless, shaker, RNAi ADAR, and taranis were back-crossed >7 generations into Iso31. Insomniac was outcrossed to an isogenic w1118 strain as previously described [22]. Unless otherwise specified, all studied flies were males. For experiments in Supplementary Figure S1 involving changes in food type, flies were raised on the specified food type from eclosion until sleep monitoring.
MB analysis
For all ontogeny experiments, unless otherwise noted, day 1 males were compared to day 7–10 males of the same genotype [12]. We did not detect any differences in sleep between day 7–10 in our system (data not shown). Flies that were assayed as mature adults were initially collected as virgins within 4 hours of eclosion and housed in same sex vials in groups of 20 until loading at ZT6-9 (Zeitgeber Time) the day prior to data collection. Young adult flies were collected up to 4 hours after eclosion, and housed in same sex vials until loading along with mature adults. Individual flies were loaded into glass MB DAM tubes containing 5% sucrose and 2% agar unless otherwise specified, and placed into the MB DAM monitor (www.trikinetics.com). Sleep traces were created and total sleep amount analyzed using PySolo software [30]. Fly location heat maps were created using data from the “Dwell” function, which records fly position in each tube at each second over the course of a reading; data from all flies was averaged at each time point to give the average percentage of time spent in a given tube position. Location heat maps were scaled as such: a red heat map signal reflects the highest possible average percentage of time in a given position across all conditions in a given experiment. For example, if the greatest average amount of time spent in a given beam is 30/60 seconds across all conditions in a given experiment, the red signal would correspond to 50%. However, if the greatest average amount of time spent in a given beam is 60/60 seconds across all conditions in an experiment, the value of the red signal changes to correspond to 100%. Scaling in this manner facilitated comparison between groups. A blue signal indicates that 0% of time on average across all flies was spent in the given position. Sleep proportion heat maps were created using data from the “Moves” function, which only counts movement between beams rather than activity occurring within a beam. For each minute of analysis, the number of flies sleeping in a given beam was divided by the total number of sleeping flies (to be counted as “asleep”, each individual fly had to meet the previously defined criteria of 5 minutes or more of immobility). As such, sleep proportion heat maps are accounting for all instances of sleep occurring during a given minute (across all flies). A red signal in the sleep proportion heat map indicates that 100% of sleeping flies were located in the given beam, while blue indicates that 0% of sleeping flies were located in the given beam, or that no flies were sleeping during that minute. For example, in a given minute, eight flies may be sleeping, with 2/8 (25%) sleeping in beam 1 and 6/8 (75%) sleeping in beam 2. In the following minute, if one of the two sleeping flies in beam 1 wakes, the ratios and heat map signal change, with 1/7 (14%) in beam 1 and 6/7 (86%) in beam 2. Place preference index (PPI) for sleep and location heat maps was calculated as follows, where “time” refers to either minutes of sleep for sleep PPI or average number of seconds for PPI of location heat maps: [(time in 5 beams farthest from food) – (time in 5 beams closest to food) / (total time in all beams)]. A value of 1.0 corresponds to all sleep occurring in the beam farthest from the food, while a value of −1.0 corresponds to all sleep occurring in the beam closest to the food.
Video recording and analysis
To mimic MB recording conditions, individual flies were loaded into glass MB DAM tubes on the day prior to recording. Tubes were laid adjacent to one another, flat on a white surface for appropriate video contrast. The tubes were evenly spaced and illuminated to avoid variations in lighting. A Sony HDR CX405 was used to acquire videos of six flies simultaneously. For video experiments in Figure 2A, one-third of the DAM tube closest to the food was visible within frame for the video to optimize resolution and distinguish feeding/proboscis extensions from true inactivity. Our video recording approach prevented us from simultaneously monitoring in the MB system and directly correlating video and MB fly position. However, since the 17 IR beams in the MB system are evenly spaced at 3 mm apart and a fly body length is approximately 2–3 mm, we defined fly position and movement based on body lengths and made close comparisons to the MB system. In the MB monitoring system, it is likely that a fly within one body length of the food would register as being in the beam immediately adjacent to the food, while greater than one body length away from the food would likely register as the next beam. Likewise, if a fly moves more than one body length, this would likely register as inter-beam movement. Videos were scored in 1 minute bins, and fly behavior was categorized as “out of frame” (fly was not in the portion of the tube being recorded), “moving” (a change in location of greater than one body length), “still, away from food” (immobile and greater than one body length away from the food), “still, near food” (immobile and less than one body length away from the food) and “eating” (visible proboscis extensions contacting the food). Videos were scored at 5× speed in 1-minute bins; each minute was scored as one of the five described behaviors if the fly was engaged in that behavior for >30 seconds. Thirty minutes of recording was scored at four different circadian time points: ZT1, ZT3, ZT5, and ZT7. This video scoring method likely overestimates the errant registration of sleep when an animal is actually feeding: so a 3-minute “eating” episode followed by locomotion would not register as 5 continuous minutes of immobility in MB or appear as sleep in the heat maps. Even during prolonged feeding bouts, flies exhibit small movements that may register as activity, so the portion of time scored as “eating” may not entirely appear as immobility in the MB data.
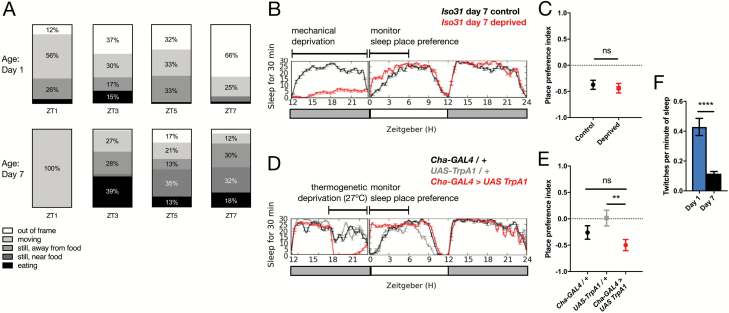
Figure 2.
Video analysis of sleep place preference and twitch in young adult flies. (A) Proportion of time spent on each behavior in a 30-minute window beginning at indicated ZT time, based on video recording (n = 12 flies for day 1, 12 flies for day 7). (B) Representative sleep traces of Iso31 day 7 controls (black) and mechanically deprived (red) flies. (C) Sleep place preference index during the first 6 hours following deprivation in control (black) versus mechanically deprived (red) Iso31 males (n = 48 per condition). (D) Representative sleep traces of Cha-GAL4 > UAS TrpA1 (red) and genetic controls (black/gray). (E) Sleep place preference index during the first 6 hours following exposure to 27°C in genetic controls (black/gray) versus Cha-GAL4>UAS-TrpA1 (red) (n = 32 for genetic controls and 64 for experimental genotype). (F) Twitches per minute of sleep in Iso31 males at day 1 versus day 7 (n = 20 for both ages). **p < 0.01, unpaired two-tailed Student’s t-test plus Welch’s correction (B, F), ordinary one-way ANOVA with multiple comparisons and Tukey’s test (E).
For video experiments in Figure 3E (twitch), one-third of the tube where we expected to observe the most sleep for the given genotype/age based on sleep heat maps was recorded. Twitches were scored at 10× speed using the following criteria: (1) occurring within a 5-minute (or longer) bout of behavioral quiescence; (2) at least one body length away from the food, or oriented away from the food; and (3) defined as clear bouts of activity in isolated appendages. Three hours of recording from ZT3 to ZT6 was scored per fly blind to experimental condition.
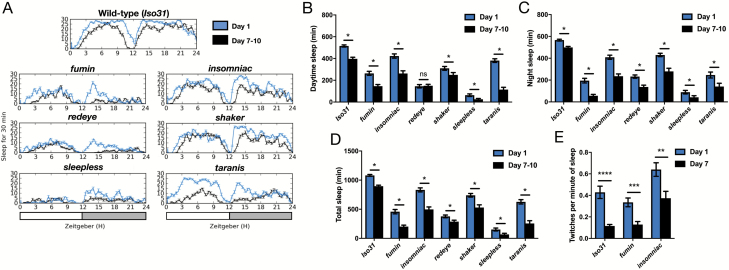
Figure 3.
Ontogenetic changes in sleep amount and twitch frequency are maintained in short-sleeping mutants. (A) Representative multi-beam sleep traces of Iso31 (wild-type) and short sleeping mutants fumin, redeye, sleepless, insomniac, shaker and taranis at day 1 (blue traces) and day 7–10 (black traces). (B–D) Quantification of day, night, and total sleep amounts in Iso31 and short sleeping mutants at day 1 versus day 7–10 post-eclosion (n for each genotype at day 1/day 7–10 is as follows: Iso31 66/67, fumin 24/24, insomniac 23/24, redeye 55/59, shaker 24/23, sleepless 38/39, taranis 38/38). (E) Twitches per minute of sleep from video recordings of Iso31, fumin, and insomniac day 1 versus day 7 males (n for each genotype at day 1/day 7–10 is as follows: Iso31 20/20, fumin 12/13, insomniac 17/13). ***p < 0.001, **p < 0.01, *p < 0.05; multiple Student’s t-tests with Holm-Sidak correction, α = 0.05 (B, C, D, E).
Arousal threshold experiments
Flies were placed in the same recording setup as in all video recording experiments. A stimulus was applied at four time points throughout the day: ZT1, ZT3, ZT5, and ZT7. We detected no differences in arousal within an experimental condition across the time points. The stimulus consisted of a 700 g weight dropped adjacent to recording apparatus from a height of 20 cm. “Low” intensity corresponds to two stimulus applications 1 second apart, and “medium” intensity corresponds to the stimulus being applied three times. A fly exhibiting activity within 10 seconds of stimulus application was scored as “aroused” by the stimulus.
Sleep deprivation experiments
Mechanical sleep deprivation was accomplished using a Trikinetics vortexer mounting plate, with shaking of monitors for 2 seconds randomly within every 20-second window for 12 hours during the night. Thermogenetic sleep deprivation was accomplished by raising the temperature to 27°C for the last 6 hours of the night; outside of this deprivation window, flies were monitored at 22°C.
Statistical analysis
Analysis was done using Prism (GraphPad Software). Unpaired Student’s t-test was used in Figure 1, B and D, Figure 2, C and F, Supplementary Figure S1, A, C and D, Supplementary Figure S2, B and C. ANOVA with Tukey’s test was used in Figure 2E, Supplementary Figure S1E, and Supplementary Figure S2A. ANOVA with Dunnett’s test was used in Supplementary Figure S3. Multiple Student’s t-tests with Holm-Sidak correction was used in Figure 3, B–E, Figure 4, B and C, Figure 5, B and C, and Figure 6, B–D and F–H. For significance: *p ≤ 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001. Each experiment was generated from a minimum of three independent biological replicates.
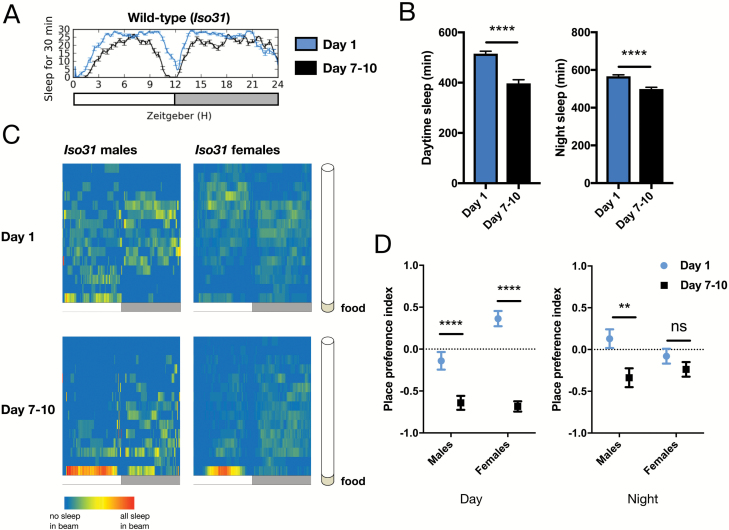
Figure 1.
Sleep place preference is diminished in young adult flies. (A) Representative sleep trace of Iso31 male wild-type flies monitored in the multi-beam system. Sleep amount is plotted in a rolling 30-minute window across the 24-hour day; white = day (ZT0–ZT12), gray = night (ZT12–ZT24). (B) Quantification of day and night sleep amounts (n = 66 for age day 1, n = 67 for age day 7–10). (C) Heat map showing the proportion of sleep occurring at each position of the multi-beam tube (shown along the y-axis) in Iso31 male and female wild-type flies at age day 1 (top) or age day 7–10 (bottom); sleep in each beam is plotted in 1-minute bins (x-axis). White = day (ZT0–ZT12), gray = night (ZT12–ZT24). Tube orientation is shown at right, with the beam closest to the food at the bottom of each heat map. Value for each 1-minute bin = (# of sleep episodes for that minute in given beam / all sleep episodes occurring in the given minute). Blue indicates that 0% of the sleep episodes during that minute occurred in the given beam, while red indicates that 100% of the sleep episodes during that minute occurred in the given beam. (D) Sleep place preference index from heat maps in (C). A value of 1.0 corresponds to all sleep occurring in the beam farthest from the food, while a value of −1.0 corresponds to all sleep occurring in the beam closest to the food. Graphs in this figure and all others are presented as means ± SEM. ****p < 0.0001, **p < 0.01, *p < 0.05; unpaired two-tailed Student’s t-test plus Welch’s correction.
Figure 4.
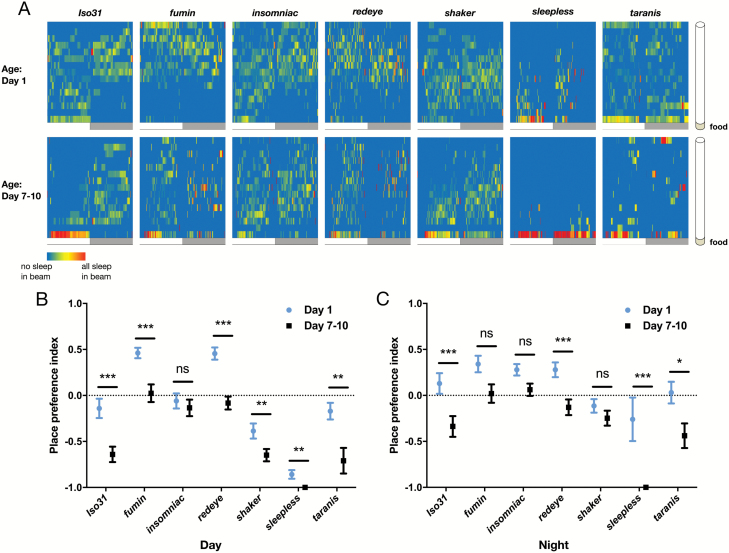
Location preferences of short-sleeping Drosophila mutants. (A) Representative heat maps showing the average percent of time spent in each beam for every 1-minute bin. Value for each 1-minute bin = percentage of time spent in beam during that minute, averaged over all flies. A red signal indicates that the highest average amount of time (across all genotypes/ages) was spent in that beam during that minute, while blue indicates that 0% of time on average was spent in the given beam during that minute. (B) Day and (C) night place preference indices from heat maps in (A). PPI = [(average time spent in 5 beams farthest from food) – (average time spent in 5 beams closest to food) / (total time in all beams)] (n for each genotype at day 1/day 7–10 is as follows: Iso31 66/67, fumin 24/24, insomniac 23/24, redeye 55/59, shaker 24/23, sleepless 38/39, taranis 38/38). *p < 0.05; multiple Student’s t-tests with Holm-Sidak correction, α = 0.05 (B and C).
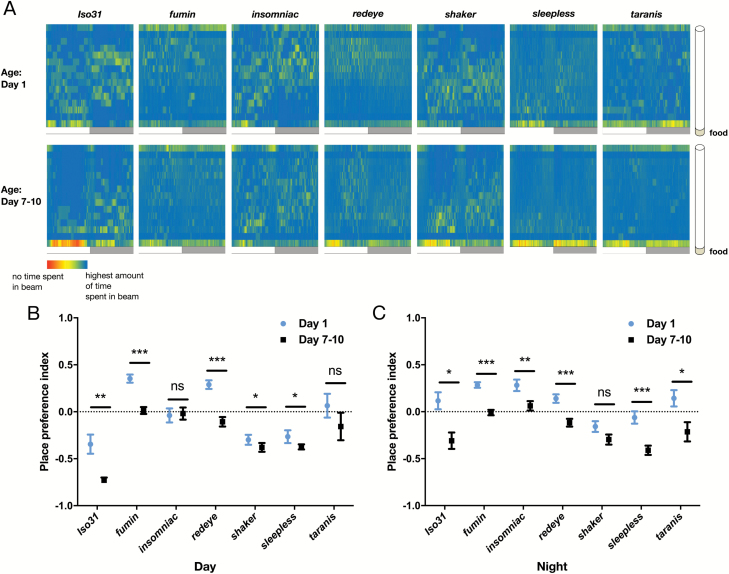
Figure 5.
Ontogenetic changes in sleep place preference persist in short-sleeping mutants. (A) Representative heat maps showing the proportion of sleep occurring in each beam in 1-minute bins. Value for each 1-minute bin = (# of sleep episodes for that minute in given beam / all sleep episodes occurring in the given minute). (B) Day and (C) night sleep place preference indices from heat maps in A (n for each genotype at day 1/day 7–10 is as follows: Iso31 66/67, fumin 24/24, insomniac 23/24, redeye 55/59, shaker 24/23, sleepless 38/39, taranis 38/38). ***p < 0.001, **p < 0.01; multiple Student’s t-tests with Holm-Sidak correction, α = 0.05 (B and C).
Figure 6.
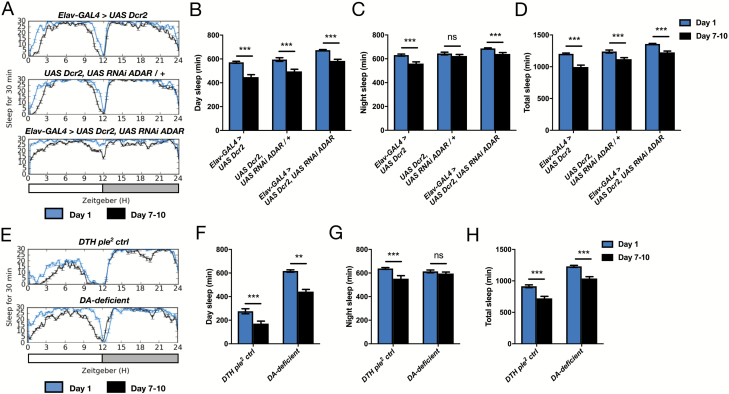
Ontogenetic changes in sleep amount persist in long sleepers. (A) Representative sleep traces of Elav-GAL4 > UAS Dcr2, UAS RNAi ADAR and genetic controls at day 1 (blue traces) and day 7–10 (black traces). (B–D) Quantification of day, night, and total sleep amounts in Elav-GAL4 > UAS Dcr2, UAS RNAi ADAR and genetic controls (n = 32 for all ages/genotypes). (E) Representative sleep traces of DTH ple [2] controls and DTHFS±Bac (“DA-deficient”) flies at day 1 (blue traces) and day 7–10 (black traces). (F–H) Quantification of day, night, and total sleep amounts in DTH ple [2] controls and DA-deficient flies (n = 24 for all ages/genotypes). ***p < 0.001, **p < 0.01; multiple Student’s t-tests with Holm-Sidak correction, α = 0.05 (B–D and F–H).
Results
Young flies show less preference for sleeping location
We first examined sleep ontogeny in wild-type Iso31 flies using the MB DAM system. Our previous work demonstrated robust ontogenetic changes in sleep amount and intensity when comparing flies on day 1 post-eclosion to day 7–10 in the SB DAM system [12], so here we also compared young (day 1) flies to mature (day 7–10). We found that ontogenetic changes in sleep amount were still present in the MB system, indicating that previously observed ontogenetic changes in the SB system were not a result of overestimating sleep amount in young flies (Figure 1, A and B). In the MB system, day 1 wild-type male flies slept an average of 1081 ± 16 minutes total throughout the day and night, while day 7–10 flies slept an average of 896 ± 17 minutes (Figure 1, A and B). Consistent with work in the SB system, wake activity was not different in young compared to mature adult flies monitored in the MB system (Supplementary Figure S1A).
Most animals prefer to sleep in defined locations [13] and previous work has shown that mature flies exhibit location preferences for their sleep episodes [14–17]. We wondered whether young flies also have a preferred sleeping location. We generated heat maps showing the proportion of sleep that occurred in each beam during each minute of monitoring (Figure 1C). We found a stark difference between where young (day 1) and mature (day 7–10) flies sleep: while mature flies in our experiments slept most in the beam closest to the food, consistent with previous reports [14–16] (Figure 1C, lower panels), young flies showed less preference regarding the location of their sleep episodes during the daytime, distributing themselves along the length of the tube (Figure 1C, upper panels). Further, night sleep episodes shifted towards the food side of the tube as flies matured (Figure 1C). Ontogenetic changes in sleep place preference were not specific to males: mature female flies preferred to sleep closer to the food, while young females showed less of a preference (Figure 1C, right panels). To quantify the change in sleep place preference we calculated a PPI for each condition, where −1.0 indicates sleep occurred closest to food and 1.0 indicates sleep occurred farthest from the food. We found that mature males and females displayed a significantly stronger food location preference during the daytime than did young males and females (Figure 1D). Males also exhibited a significant ontogenetic change in sleep PPI during the night, while females showed a similar trend (Figure 1D). We also observed that flies tended not to sleep two beams away from the food; this is consistent with the original observation that flies often turn away from the food, walk a few paces and then settle into a sleep bout [14]. We also confirmed that sleep place preferences are not dependent on nutritional value of the food source (Supplementary Figure S1, B–E).
In the SB system, prolonged feeding bouts or escape behaviors at either end of the DAM tube can erroneously register as sleep, as awake flies may not break the center beam when engaged in these behaviors [15, 16]. Despite the higher resolution, this confound between feeding and sleeping may also occur in the MB system. To determine whether the high amount of daytime sleep in this system may actually be feeding, we performed high-resolution video monitoring of flies in the MB DAM tubes, focusing on the one-third of the tube closest to the food. At this resolution, we could clearly distinguish proboscis extensions and feeding behaviors from rest. We defined position relative to the food using fly body lengths (see Methods). We analyzed 30 minutes of recording at four circadian time points in flies under 12:12: light:dark conditions: ZT1, ZT3, ZT5, and ZT7.
In mature flies, the majority of immobile episodes in the vicinity of the food consisted of true inactivity without feeding behavior, in keeping with previous work [15] (Figure 2A). These periods of inactivity were associated with increased arousal threshold compared to feeding flies near the food, confirming that the behavioral quiescence was indeed sleep (Supplementary Figure S2). Consistent with previous data, mature day 7 flies did not exhibit much sleep at ZT1 (Figure 2A, lower panel) [10]. We did find that ~40% of the time at ZT3, mature flies were actually feeding while immobile near the food (Figure 2A, lower panel). However, this only appeared to be true for a short period of time in the earlier part of the day, as at ZT5 and ZT7 respectively, mature flies only spent 13% and 18% of their time feeding but 35% and 32% of their time immobile near food (Figure 2A, lower panel, Supplementary Video S1). Moreover, our video analysis likely overestimates the errant registration of sleep when an animal is actually feeding (see Methods). In addition to the time spent immobile immediately adjacent to the food, flies spent an average of 23% of their time (across three timepoints, ZT3, ZT5, and ZT7) immobile in the video frame but not immediately next to the food. This indicates that they were often immobile in the one-third of the tube closest to the food, in keeping with our heat map data. We also noticed that for mature adults, very little time (an average of 6% across all time points) was spent in the two-thirds of the tube away from the food (“out of frame”). Overall, this video data corroborates our heat map analysis, confirming that mature flies are not merely eating all day, but genuinely prefer to sleep near the food.
In contrast to mature adults, video recording of young flies indicates lack of sleep place preference as well as greater amounts of immobility in general throughout the day (Figure 2A, upper panel). We observed a clear difference in the percent of time spent “moving” at ZT1 (56% compared to 100% for mature flies), consistent with previous observations that after lights on at ZT0, young flies have a shorter latency to sleep onset [12]. When scoring the videos, we noticed that awake, “moving” flies of either age generally walk back and forth across the full length of the tube and take less than 1 minute to do so, rather than just moving around on one side of the tube. Thus, if a fly was scored as “out of frame” for 1 minute or more, it is likely that the fly was immobile/sleeping. We thus considered “out of frame” as immobility away from the food, and saw that young flies spent much more time “out of frame” (an average of 36% across all time points, six times as much as mature flies). Video recordings of the entire tube confirmed that in addition to sleeping in the portion of the tube visible at higher resolution, young flies also slept in the two-thirds of the tube away from the food (Supplementary Video S2). Young flies did not only sleep away from the food, as the higher resolution recording showed that they spent an average of 20% of their time immobile within frame. Therefore, young flies have not developed a strong sleeping place preference. Importantly, mature flies do not simply eat more than young flies: mature flies were found to eat in more prolonged bouts (greater than 1 minute, scored as “eating”; Supplementary Video S3), while young flies ate in bouts of less than 30 seconds in between walking back and forth (Supplementary Video S4). Additionally, we have previously shown using the CAFÉ feeding assay [31] that young and mature flies eat the same amount over a 24-hour period [12]. In sum, our results demonstrate reduced sleep place preference in young adult flies.
To test whether the change in sleep place preference with maturation is a function of attenuated sleep drive, we examined whether sleep deprivation of mature adult flies also reduces sleep place preference. Wild-type mature flies (day 7) were mechanically sleep deprived for 12 hours during the night and sleep place preference during the subsequent period of rebound sleep was compared to non-deprived flies (Figure 2B). We detected no difference in sleep place preference between deprived and non-deprived flies during the rebound period (Figure 2C), despite increased sleep duration of sleep-deprived flies (Figure 2B). Because different types of wake-promoting stimuli result in distinct sleep rebound profiles [32], we next tested whether activation of wake-promoting neurons known to induce particularly large subsequent sleep rebound might yield a change to sleep place preference. Cha+ neurons were thermogenetically activated during the night, and we assessed subsequent sleep rebound the following day (Figure 2D). Once again, sleep-deprived animals in which Cha+ neurons had been activated did not display reduced sleep place preference in comparison to genetic controls also exposed to the elevated temperature, and may even display a slight increase in food side preference (Figure 2E). Additionally, we found that sleep depriving young, day 1 flies similarly did not alter sleep place preference (Supplementary Figure S2C). Together, these data suggest that the relative lack of sleep place preference in young flies is not simply due to their heightened sleep drive, and constitutes a distinct feature of sleep ontogeny.
Young flies exhibit more motor twitches during sleep
While analyzing video recordings we noticed that flies, like mammals, exhibit motor twitches during sleep [33]. Twitches are composed of clear bouts of activity in isolated appendages in between periods of behavioral quiescence [18]. It has been previously shown that mammals twitch more during sleep when young, which is thought to be critical for sensorimotor development [18, 34, 35]. We, therefore, investigated whether flies also undergo ontogenetic change in sleep twitch frequency. Using video recordings, we counted twitches during sleep episodes in day 1 versus day 7 wild-type flies. We found that like mammals, flies twitched significantly more when young (Figure 2F). We observed that these twitches consisted of stereotyped movements including leg and proboscis extensions (not near the food) as well as central thorax/abdominal muscle twitches resulting in full body contractions (Supplementary Videos S5–S8). Thus, another central feature of sleep ontogenetic change in mammals is also conserved in flies.
Short sleeping mutants exhibit ontogenetic changes in sleep amount
We next examined whether any of the known mutations that reduce sleep time in mature flies also interfere with sleep ontogeny. We investigated sleep ontogenetic change in several short sleeping mutants: fumin (fmn), insomniac (inc), redeye (rye), shaker (sh), sleepless (sss), and taranis (tara) [21–26]. Interestingly, we found that ontogenetic change in sleep amount persisted in all of these mutants, as day 1 flies invariably slept significantly more than day 7 flies of a given genotype (Figure 3, A–D). Even mutants with severely reduced sleep time as mature adults, such as sss or rye, still showed significantly increased sleep when young. Ontogenetic changes to both day and night sleep amount were significant for all mutants, except for rye, which only exhibited a change in night sleep (Figure 3, B and C). We next tested whether all mutants undergo a similar amount of sleep reduction from young adulthood to maturity compared to wild-type flies. We found that ontogenetic changes in sleep amount were particularly exaggerated in fmn and tara mutants, suggesting potential molecular pathways that may influence sleep maturation (Supplementary Figure S3). Overall, these results indicate that despite distinct molecular etiologies of a short sleeping phenotype, sleep ontogenetic change is still preserved across all studied short-sleeping mutants.
Are ontogenetic changes in sleep twitching similarly preserved in short-sleeping mutants? We focused on inc [1] mutants because of the less severe short sleep phenotype (higher chance of observing sleep episodes) and fmn mutants because of the exaggerated ontogenetic change in sleep duration. We found that, like wild-type flies, both inc [1] and fmn mutants twitched more when young (Figure 3E). Interestingly, we also observed that inc [1] flies twitched significantly more at both ages than did iso or fmn flies (Figure 3E), raising the interesting possibility that the inc [1] mutation may modulate overall sleep twitch amount.
Short sleeping mutants display ontogenetic changes in sleep place preference
Most studies of short sleeping mutants have focused on sleep and activity amounts, but it is not known whether these mutants have place preferences for their sleep or activity bouts. We first characterized overall location preferences of short sleeping mutants. Using the “Dwell” function in the MB system, which reports fly location for each second of recording, we created location heat maps showing the average percentage of time flies spent in each beam for each minute (Figure 4). The location heat map for Iso31 wild-type flies looks similar to the sleep proportion heat map (Figure 1C), suggesting that the majority of Dwell time consists of episodes that fit the criteria for sleep, and are thus included in the sleep proportion heat map. We also noticed that most of the short sleeping mutants tended to be located either in the beam closest to the food or farthest from the food, consistent with previously published MB data [36]. Based on our video scoring of flies moving within a tube, this likely reflects the tendency of awake, moving flies to walk back and forth, passing through the middle beams without much Dwell time while incurring Dwell time on either end as they turn within a beam. Further, we noticed that time spent in the beams closest to the food appeared greater at day 7–10 across all genotypes (Figure 4A). To quantify this change, we calculated an overall PPI (not limited to sleep) and found that ontogenetic changes in location preference occurred during both the day and night for the majority of mutants (Figure 4, B and C).
Since short-sleeping mutants spent more overall time near the food as mature adults, and given that ontogenetic changes in sleep amount were still present in these mutants, we tested whether they also develop sleep place preferences as they mature. We created sleep proportion heat maps for each mutant at day 1 and day 7 and quantified sleep PPI. Most short sleeping genotypes shifted the location of sleep episodes for at least part of the day/night towards the food side of the tube as they matured (Figure 5A). The difference in sleep PPI was significant during both the day and night in rye, sss and tara (Figure 5, B and C). Fmn and sh mutants displayed a significant shift in the same direction during either the day (fmn) or night (sh). Finally, inc [1] flies showed a similar but non-significant trend (Figure 5, B and C). Unlike wild-type flies, certain short-sleeping mutants (fmn and rye) preferred the side of the tube opposite from the food when young and shifted towards the food with maturation. Despite having a place preference for sleep even when young, these mutants still exhibited ontogenetic change in sleep place preference.
Long sleepers exhibit ontogenetic changes in sleep amount
Lastly, we sought to determine whether the genetic dissociation of overall sleep duration from sleep ontogenetic change also applies to mutations that increase overall sleep time. We investigated ontogenetic change in sleep amount in previously described genetic lesions that increase sleep: neuronal ADAR knockdown using Elav-GAL4 to express ADAR RNAi [27] and brain-specific deletion of DTH (“DA-deficient”) [28]. We found that although these lesions increase sleep time in mature flies compared to genetic controls, young flies of all genotypes still slept more than the respective mature flies (Figure 6). Therefore, all studied molecular lesions that alter overall sleep duration do not impinge on sleep ontogenetic change, suggesting unique genetic control of sleep ontogeny.
Discussion
Ontogenetic changes in sleep are highly conserved in Drosophila, with increased sleep time and intensity in young adulthood. However, developmental changes in other aspects of sleep have not previously been investigated. Further, despite the identification of several genes that regulate sleep in mature adults, prior work has not addressed the genetic regulation of sleep ontogeny. Here, we demonstrate that many aspects of sleep behavior undergo ontogenetic change in early adulthood, including sleep place preference and motor twitching. Additionally, we show that sleep ontogeny is not regulated by genes that control overall sleep amount, suggesting independent genetic control.
Consistent with our previous study in the SB DAM system [12], our results in the higher resolution MB system show that young adult flies sleep more than do mature adults. One drawback of the SB system is that it slightly overestimates sleep, as movement on either end of the tube that does not break the center beam will register as sleep [16, 20]. Thus, it was possible that past estimates of young fly sleep reflected a tendency to stay on one side of tube for prolonged periods of time, rather than actual sleep, but our data show this is not the case.
Other studies have reported sleep place preferences in mature flies [14–17]. Our MB and video data support the idea that mature flies prefer to sleep in the vicinity of the food source, consistent with other video-based measurements [14–16, 36]. Importantly, we show by video analysis in males that while a portion of sleep in the earlier part of the day (ZT3) may actually be feeding, this confound is not likely to account for sleep data later in the day, as we can visualize a high percentage of flies completely immobile near the food without feeding at ZT5 and ZT7. Previous video tracking data has also shown that in the second half of the day, flies sleep near the food [16]. Thus, we are confident that our MB data reflects a sleeping place preference in mature flies, which constitutes another sleep behavior that appears to be conserved in mice and humans [37, 38].
In contrast with mature flies, we found that young flies sleep with less preference for location. We wondered whether higher homeostatic sleep drive in young flies makes them less selective about where they sleep. However, our data show that sleep deprivation in mature flies does not similarly alter sleep place preference. While it is possible that a sleep-deprived mature fly does not mount the same degree of sleep drive as a young fly, brain regions involved in processing elevated sleep need show increased levels of activity in young flies and mature deprived flies [12, 39, 40], suggesting that these states are comparable. Thus, our results suggest that the lack of place preference in young flies is a unique feature of sleep ontogeny unrelated to elevated sleep drive.
Our video data also revealed that flies, like mammals, exhibit motor twitches during sleep that occur more frequently in young animals, including in short-sleeping mutants. We found that sleep twitches in both male and female flies are qualitatively similar to those observed in developing rodents, comprised of discrete, non-random jerky movements separated by periods of quiet rest [18, 33]. As in rodents, we also did not observe simultaneous twitches in different muscle groups. In young rodents, twitches allow peripheral muscle activity to trigger correlated responses in relevant areas of the somatosensory cortex [41]. Activity in the sensorimotor cortex is higher during active sleep (when twitching occurs) than during wake in young rodents, and sleep twitches are thought to facilitate sensorimotor development [33]. Phenotypic similarity between fly and mammalian twitching suggests that sleep twitches in developing flies may serve a similar purpose to that in mammals. While gross motor skills appear to be well-developed in young flies, behaviors that rely on more complex motor programs, such as courtship and aggression, exhibit ontogenetic changes which may stem in part from ongoing refinement and integration of sensorimotor systems [42, 43]. Indeed, we have previously shown that sleep deprivation in young adulthood leads to a persistent courtship deficit which stems from impaired growth of a courtship-relevant olfactory glomerulus [12]. In addition to this olfactory defect, it is also possible that early life sleep deprivation hinders twitch-dependent sensorimotor development, contributing to compromised courtship ability. Interestingly, motor twitches in young rats occur exclusively during active sleep, which is more frequent in young mammals and is intimately linked to brain development [33, 44]. Despite the lack of an identifiable active sleep analog in Drosophila, recent work showed that flies sleep in stages with varying levels of arousability, indicating that fly sleep may be more complex than previously appreciated [19, 45]. Further, work in honey bees demonstrated different sleep stages and spontaneous antennal movements during more active sleep [46]. It is thus possible that motor twitches in flies preferentially occur during a particular type of sleep that is more prevalent in early life. Even in the absence of clear active sleep in flies, the presence of sleep twitches and conservation of ontogenetic change in this behavior raises the possibility of a conserved function for twitching in relation to nervous system development.
What is the genetic basis of sleep ontogenetic changes? Here we examined whether known sleep regulatory genes also control features of sleep ontogeny. We found that all studied sleep mutants sleep significantly more as young adults, indicating that this basic aspect of sleep ontogeny is not regulated by any of these genes that exert dramatic effects on overall sleep amount. The presence of exaggerated ontogenetic sleep change in fmn mutants is consistent with our previous work showing that at the circuit level, increased sleep in young flies stems from hypoactivity of dopaminergic neurons [12]. Since short sleep in fmn mutants results from a compromised ability to clear wake-promoting dopamine from the synapse, young fmn flies likely have less dopamine to clear compared to mature fmn flies, allowing them to sleep more [12]. Intriguingly, sleep ontogenetic change persists in brain-specific dopamine-deficient flies, suggesting that low dopamine is not the only contributor to increased sleep in young flies. Importantly, analysis of all mutant genotypes focused on male flies, so it is still possible that certain mutations affect sleep ontogeny in females. However, since ontogenetic changes in sleep amount, daytime place preference, and sleep twitches are present in both males and females, we do not expect the genetic control of sleep ontogeny to be sex-specific. Notably, none of the studied short sleeping mutants achieve the high sleep amounts seen in young wild-type flies, indicating that the molecular lesions responsible for mature adult short sleep phenotypes still influence sleep at this earlier developmental stage. Similarly, young flies that are genetic long sleepers also sleep more than their respective controls at the same age. Yet, the persistence of ontogenetic change despite the presence of altered sleep amounts at both ages suggests an independent genetic control mechanism for sleep maturation. Building on the candidate-based approach described here, future work will utilize unbiased screens to identify sleep ontogeny regulatory genes.
Like wild-type flies, most studied short-sleeping mutants exhibit ontogenetic changes in sleep place preference, with sleep closer to the food as mature adults. Additionally, ontogenetic changes in sleep twitch frequency also occur in short sleeping fmn and inc [1] mutants. Thus, sleep ontogenetic change in these mutants is a multifaceted phenotype not limited to sleep amount, raising the possibility that an unknown master regulator controls several sleep ontogeny behaviors.
Collectively, our results indicate that sleep ontogenetic change encompasses multiple behaviors in Drosophila, including sleep time, place preference, and motor twitches. Our results are consistent with work in mammalian systems showing that sleep in early life is qualitatively different from sleep in adulthood, and likely serves an important function in the developing brain. Further, we show that ontogenetic changes persist across different molecular etiologies of short and long sleep, suggesting that the genetic control of sleep ontogeny is distinct from that of overall sleep amount. Future studies aimed at identifying the specific genes that regulate sleep ontogeny will lend insight into mechanisms controlling early life sleep and provide new avenues for investigating the role of early life sleep in brain development.
Supplementary material
Supplementary material is available at SLEEP online.
Funding
This work was funded by National Institutes of Health (NIH) grants K08 NS090461 (M.S.K.), T32 HL007953 and F31 NS105447 (L.C.D.), and a Burroughs Wellcome Career Award for Medical Scientists grant 100000861, March of Dimes Basil O’Connor Scholar Award grant 100000912, and Sloan Research Fellowship grant 100000879 (M.S.K.).
Acknowledgments
We thank David Garbe for experimental support. We thank David Raizen, Amita Sehgal, and members of the Kayser Lab for helpful input.
Notes
Conflict of interest statement. None declared.
References
- 1. Roffwarg HP, et al. Ontogenetic development of the human sleep-dream cycle. Science. 1966;152(3722):604–619. [DOI] [PubMed] [Google Scholar]
- 2. Kayser MS, et al. Sleep and development in genetically tractable model organisms. Genetics. 2016;203(1):21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kurth S, et al. Sleep and early cortical development. Curr Sleep Med Rep. 2015;1(1):64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gregory AM, et al. Parent-reported sleep problems during development and self-reported anxiety/depression, attention problems, and aggressive behavior later in life. Arch Pediatr Adolesc Med. 2008;162(4):330–335. [DOI] [PubMed] [Google Scholar]
- 5. Ednick M, et al. A review of the effects of sleep during the first year of life on cognitive, psychomotor, and temperament development. Sleep. 2009;32(11):1449–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. O’Brien LM. The neurocognitive effects of sleep disruption in children and adolescents. Child Adolesc Psychiatr Clin N Am. 2009;18(4):813–823. [DOI] [PubMed] [Google Scholar]
- 7. Kotagal S. Sleep in neurodevelopmental and neurodegenerative disorders. Semin Pediatr Neurol. 2015;22(2):126–129. [DOI] [PubMed] [Google Scholar]
- 8. Chakravarti L, et al. Unraveling the neurobiology of sleep and sleep disorders using Drosophila. Curr Top Dev Biol. 2017;121:253–285. [DOI] [PubMed] [Google Scholar]
- 9. Sehgal A, et al. Genetics of sleep and sleep disorders. Cell. 2011;146(2):194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shaw PJ, et al. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287(5459):1834–1837. [DOI] [PubMed] [Google Scholar]
- 11. Seugnet L, et al. Sleep deprivation during early-adult development results in long-lasting learning deficits in adult Drosophila. Sleep. 2011;34(2):137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kayser MS, et al. A critical period of sleep for development of courtship circuitry and behavior in Drosophila. Science. 2014;344(6181):269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Campbell SS, et al. Animal sleep: a review of sleep duration across phylogeny. Neurosci Biobehav Rev. 1984;8(3):269–300. [DOI] [PubMed] [Google Scholar]
- 14. Hendricks JC, et al. Rest in Drosophila is a sleep-like state. Neuron. 2000;25(1):129–138. [DOI] [PubMed] [Google Scholar]
- 15. Donelson NC, et al. High-resolution positional tracking for long-term analysis of Drosophila sleep and locomotion using the “tracker” program. PLoS One. 2012;7(5):e37250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zimmerman JE, et al. A video method to study Drosophila sleep. Sleep. 2008;31(11):1587–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rieger D, et al. The fruit fly Drosophila melanogaster favors dim light and times its activity peaks to early dawn and late dusk. J Biol Rhythms. 2007;22(5):387–399. [DOI] [PubMed] [Google Scholar]
- 18. Blumberg MS, et al. Spatiotemporal structure of REM sleep twitching reveals developmental origins of motor synergies. Curr Biol. 2013;23(21):2100–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van Alphen B, et al. A dynamic deep sleep stage in Drosophila. J Neurosci. 2013;33(16):6917–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garbe DS, et al. Context-specific comparison of sleep acquisition systems in Drosophila. Biol Open. 2015;4(11):1558–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kume K, et al. Dopamine is a regulator of arousal in the fruit fly. J Neurosci. 2005;25(32):7377–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stavropoulos N, et al. insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron. 2011;72(6):964–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shi M, et al. Identification of Redeye, a new sleep-regulating protein whose expression is modulated by sleep amount. Elife. 2014;3:e01473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cirelli C, et al. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434(7037):1087–1092. [DOI] [PubMed] [Google Scholar]
- 25. Koh K, et al. Identification of SLEEPLESS, a sleep-promoting factor. Science. 2008;321(5887):372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Afonso DJ, et al. TARANIS functions with cyclin A and Cdk1 in a novel arousal center to control sleep in Drosophila. Curr Biol. 2015;25(13):1717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robinson JE, et al. ADAR-mediated RNA editing suppresses sleep by acting as a brake on glutamatergic synaptic plasticity. Nat Commun. 2016;7:10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cichewicz K, et al. A new brain dopamine-deficient Drosophila and its pharmacological and genetic rescue. Genes Brain Behav. 2017;16(3):394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ryder E, et al. The drosdel deletion collection: a drosophila genomewide chromosomal deficiency resource. Genetics. 2007;177(1):615–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gilestro GF, et al. PySolo: a complete suite for sleep analysis in Drosophila. Bioinformatics. 2009;25(11):1466–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ja WW, et al. Prandiology of Drosophila and the CAFE assay. Proc Natl Acad Sci U S A. 2007;104(20):8253–8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seidner G, et al. Identification of Neurons with a privileged role in sleep homeostasis in Drosophila melanogaster. Curr Biol. 2015;25(22):2928–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blumberg MS. Developing sensorimotor systems in our sleep. Curr Dir Psychol Sci. 2015;24(1):32–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marcano-Reik AJ, et al. An abrupt developmental shift in callosal modulation of sleep-related spindle bursts coincides with the emergence of excitatory-inhibitory balance and a reduction of somatosensory cortical plasticity. Behav Neurosci. 2010;124(5):600–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tiriac A, et al. Rapid whisker movements in sleeping newborn rats. Curr Biol. 2012;22(21):2075–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cavanaugh DJ, et al. The Drosophila circadian clock gates sleep through time-of-day dependent modulation of sleep-promoting neurons. Sleep. 2016;39(2):345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sherwin CM. Preferences of individually housed TO strain laboratory mice for loose substrate or tubes for sleeping. Lab Anim. 1996;30(3):245–251. [DOI] [PubMed] [Google Scholar]
- 38. Spörrle M, et al. Sleeping in safe places: an experimental investigation of human sleeping place preferences from an evolutionary perspective. Evol Psychol. 2010;8(3):405–419. [PubMed] [Google Scholar]
- 39. Donlea JM, et al. Neuronal machinery of sleep homeostasis in Drosophila. Neuron. 2014;81(6):1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu S, et al. Sleep drive is encoded by neural plastic changes in a dedicated circuit. Cell. 2016;165(6):1347–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Khazipov R, et al. Early motor activity drives spindle bursts in the developing somatosensory cortex. Nature. 2004;432(7018):758–761. [DOI] [PubMed] [Google Scholar]
- 42. Long CE, et al. Relative male age, fertility, and competitive mating success in Drosophila melanogaster. Behav Genet. 1980;10(2):163–170. [DOI] [PubMed] [Google Scholar]
- 43. Hoffmann AA. The influence of age and experience with conspecifics on territorial behavior in Drosophila melanogaster. J Insect Behav. 1990;3(1):1–12. [Google Scholar]
- 44. Marks GA, et al. A functional role for REM sleep in brain maturation. Behav Brain Res. 1995;69(1–2):1–11. [DOI] [PubMed] [Google Scholar]
- 45. Yap MHW, et al. Oscillatory brain activity in spontaneous and induced sleep stages in flies. Nat Commun. 2017;8(1):1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sauer S, et al. The dynamics of sleep-like behaviour in honey bees. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2003;189(8):599–607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.