Abstract
Glioblastoma is the most deadly and frequently occurring primary malignant tumor of the central nervous system. Genomic studies have shown that mutated oncogenes and tumor suppressor genes in glioblastoma mainly occur in three pathways: the RTK/Ras/PI3K signaling, the p53 and the Rb pathways. In this review, we summarize the modulatory effects of genetic aberrations in these three pathways to drugs targeting these specific pathways. We also provide an overview of the preclinical efforts made to identify genetic biomarkers of response and resistance. Knowledge of biomarkers will finally promote patient stratification in clinical trials, a prerequisite for trial design in the era of precision medicine.
KEYWORDS : brain tumor, genetic biomarkers, glioblastoma, personalized medicine, resistance, small-molecule kinase inhibitors
Practice points.
Three major pathways are deregulated in glioblastoma: the RTK/Ras/PI3K signaling, the p53 and the Rb tumor suppressor pathways.
These pathways are clinically attractive to target due to their frequent hyper- or inactivation.
Many drugs targeting these pathways appear to respond in only a subset of patients, and therefore the effect of a certain experimental drug will be diluted within an unselected study population.
Preclinical models that faithfully represent the tumor characteristics as observed in the patient must be applied to identify clinically relevant biomarkers and to filter out compounds that will likely not be effective for the patient.
Clinical trials need to be stratified according to the molecular predictor for response.
The aim of precision medicine is to tailor cancer treatment to the specific tumor characteristics of each individual patient. Glioblastomas carry a heterogeneous set of molecular aberrations driving tumor progression [1–3]. Integrative analyses of mutational and copy number data identified three major deregulated pathways in glioblastoma: the RTK/Ras/PI3K signaling, the p53 and the Rb pathway [1,3]. An average of 80–90% of glioblastomas contains at least one aberration in each pathway, highlighting their significance as possible therapeutic targets [1,3]. In addition to the increased insights into cancer biology, a large array of compounds has been developed that target different signaling pathways. However, many compounds do not pass preclinical testing and even fewer fulfill the required endpoints in clinical trials. An underlying reason may be attributed to the large patient-to-patient variability of glioblastoma. Many drugs appear to respond in only a subset of patients, and therefore the effect of a certain experimental drug will be diluted within an unselected study population [4]. As a result it will be difficult to detect a clinically relevant treatment effect that is restricted to a subpopulation with a specific predictive marker (for examples, see [4]). To avoid this problem, clinical trials need to be stratified according to the molecular predictor for response.
Appropriately, based on gene expression data The Cancer Genome Atlas (TCGA) consortium proposed glioblastoma to be divided into the following four subtypes: proneural, neural, classical and mesenchymal [2]. A later study by the TCGA looked at the extent of promoter methylation and identified a hypermethylated group of tumors which correlated with the presence of IDH1 mutations, which they named the glioma-CpG island methylator phenotype (G-CIMP) [5]. Interestingly, this intertumor heterogeneity can be preserved in the lab for in vitro experiments using glioma stem-like cells (GSC) [6]. In respect to the variety of glioma models available, patient-derived glioma cultures and orthotopic xenografts of patient-derived material are gaining more research focus since they appear to mimic the clinical responses more faithfully than established cell lines [7,8]. Since the discovery that serum-free neurobasal medium supplemented with basic FGF and EGF faithfully preserves the molecular aberrations of patient-derived glioblastomas, this culture method is increasingly being implemented in drug development research [7,8].
In this review, we provide an overview of the preclinical studies performed in the last decade on glioblastoma, investigating the value of genetic aberrations for the prediction of response to small-molecule therapeutics specifically targeting the RTK/Ras/PI3K, p53 or Rb pathway.
RTK/Ras/PI3K pathway
Approximately 90% of glioblastomas contain one or more genetic aberrations in the RTK/Ras/PI3K pathway [1,3], making it one of the most intensely studied pathways in drug development. Deregulation of this pathway affects several hallmarks of cancer [9], such as sustaining proliferative signaling, evading growth suppressors, activating invasion and resisting cell death.
EGFR inhibition
RTKs are altered in approximately 70% of glioblastomas, of which EGFR is most frequently mutated [3]. Approximately half of all glioblastomas contain amplifications of the EGFR gene and half of these contain in-frame deletions [3,10]. EGFRvIII is the predominant in-frame deletion, which arises due to the deletion of exons 2–7, leading to a constitutively active mutant receptor [11]. Missense mutations are usually mutually exclusive with the presence of in-frame deletions and occur in more than 20% of glioblastomas. These missense mutations are predominantly located in the extracellular domain of EGFR [12,13].
Due to the frequently occurring mutations in EGFR, it is a popular drug target. The clinical experience with EGFR inhibitors for glioblastoma was recently reviewed by Reardon and colleagues [14]. First-generation EGFR inhibitors compete with ATP-binding in the catalytic tyrosine kinase domain and bind reversibly to EGFR (such as erlotinib, gefitinib, lapatinib, PKI-166 and vandetanib). On the other side, second-generation EGFR inhibitors bind irreversibly to EGFR (such as NT113, neratinib and dacomitinib).
• EGFR aberrations in the context of EGFR inhibition
EGFR amplification has been shown to sensitize glioma stem-like cells (GSC) to erlotinib [15], dacomitinib [16] and lapatinib [12] (Figure 1). However, interestingly, the choice of the tumor model appears to play an important role in determining the eventual drug response. In the study performed by Eimer and colleagues the same clones cultured in serum-supplemented medium (SSM) were more resistant to erlotinib compared with GSCs [15]. There are various possible explanations underlying this observation, such as a failure to retain the increased EGFR copy number in SSM [17], the abundance of growth factors facilitating RTK switching [18] or perhaps coactivation of other RTKs [19]. Furthermore, a study performed in a different model, using orthotopic xenografts also did not observe sensitivity to erlotinib in amplified wild-type EGFR even in the presence of wild-type phosphatase and tensin homolog (PTEN) [20].
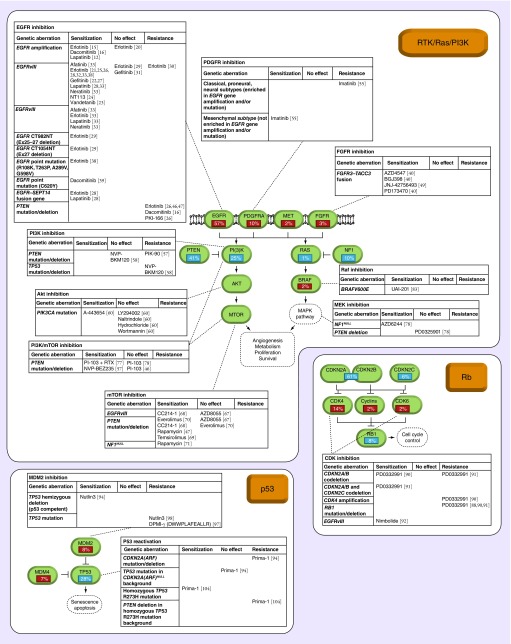
Figure 1. . Overview of the genetic aberrations conferring sensitivity or resistance to compounds targeting the RTK/Ras/PI3K, p53 or Rb pathway in glioblastoma.
The frequency of genetic amplifications (red), mutations and deletions (blue) are indicated in percentages. For each target molecule, a summary of the genetic biomarkers and its status of predicting sensitivity or resistance is given.
Besides EGFR amplification, several studies have found EGFRvIII to sensitize glioblastoma cell lines in vitro to erlotinib [21], gefitinib [22], vandetanib [23] and NT113 [24] compared with wild-type EGFR. This finding also appears to hold in orthotopic xenograft models of glioblastoma cell lines for erlotinib [25]. Especially cell lines with PTEN and EGFRvIII expression are more sensitive to erlotinib [26] and gefitinib [27] compared with wild-type EGFR. Recently it has also been reported in a glioblastoma stem-like cell culture carrying an EGFRvIII construct that it conveys an increased sensitivity to erlotinib and lapatinib compared with wild-type EGFR [28]. In addition, EGFRvIII sensitizes orthotopic xenografts of glioblastoma cell lines and patient-derived glioblastoma to vandetanib [23] and NT113 [24]. However other studies find no sensitization by EGFRvIII to erlotinib [29,30] or gefitinib [31]. Nevertheless, collectively there is more compelling evidence indicating that EGFRvIII sensitizes glioblastoma cells to EGFR inhibitors. A recent report, by Nathanson and colleagues [32] supports this by showing that the presence of more EGFRvIII extrachromosomal DNA promotes sensitivity to erlotinib [32].
Other types of EGFR in-frame deletions include EGFRvII and the recently discovered ‘EGFRvII-extended’. In EGFRvII, exons 14–15 are deleted. Whereas in the recently discovered EGFRvII-extended, exons 14–16 are deleted [33]. EGFRvII is present in around 10% of the EGFR mutations in GBM. Recently, Francis and colleagues have shown that EGFRvII is oncogenic and sensitive to a panel of EGFR inhibitors (Figure 1) [33].
In contrast to the frequently occurring EGFRvIII, recurrent in-frame C-terminal deletions are relatively rare [3,34,35]. Therefore it is not surprising that the functional consequences of these mutations have not yet been studied extensively. Cho and colleagues have performed one of the few studies looking into the functional consequence of C-terminal deletions [29]. They demonstrate that the EGFR C-terminal deletions CT982NT (exons 25–27 deletion) and CT1054NT (exon 27 deletion) are oncogenic in murine Ba/F3 pro-B lymphocytes and sensitize them to erlotinib [29].
Besides amplification or focal exon deletions of EGFR, single base mutations in EGFR can have profound effects on the response to EGFR inhibitors. EGFR mutations in glioblastoma appear to confer EGFR oncogene addiction [36,37]. Using murine Ba/F3 pro-B lymphocytes as a model system to investigate the oncogenic effect of different EGFR mutations, Lee and colleagues demonstrate that the EGFR ectodomain mutations (R108K, T263P, A289V, G598V) found in glioblastoma were more sensitive to erlotinib than the EGFR wild-type Ba/F3 cells [38]. Afterward Greenall and colleagues investigated the functional effects of additional ectodomain mutants and found that the glioma-specific EGFR C620Y mutation confers sensitivity to the pan-ERBB inhibitor, dacomitinib [39]. Interestingly, EGFR dependence cannot be exploited equally effective by all EGFR inhibitors. EGFR inhibitors can either bind to the conformationally active EGFR kinase domain (type I inhibitors, such as erlotinib) [36] or conformationally inactive EGFR kinase domain (type II inhibitors, such as lapatinib) [37]. In glioblastoma, activating point mutations occur predominantly in the extracellular domain of EGFR and confer sensitivity to EGFR type II inhibitors in contrast to lung cancer, where kinase domain mutations are predominant which confer sensitivity to EGFR type I inhibitors [12]. It is clear that drug response correlations identified in other types of cancer may not directly be extrapolated to glioblastoma and this should be taken into account when designing trials based on molecular markers independent of tumor histology (i.e., basket trials).
In contrast to the long-known EGFR point mutations, EGFR fusion genes were only recently found to play a role in glioblastoma, offering unique possibilities to investigate fusion oncogene dependency [3,28,40–45]. EGFR has been identified as one of the most frequently rearranged gene, fusing with genes such as SEPT14 (±4% of glioblastomas) [3,28], PSPH (±2%) [28] and SEC61G (±2%) [3]. Frattini and colleagues functionally investigated the EGFR–SEPT14 fusion gene and found that it sensitizes GSCs to erlotinib and lapatinib [28]. This report highlights the potential clinical relevance of fusion oncogenes in glioblastoma.
• PTEN mutations in the context of EGFR inhibition
PTEN acts downstream of EGFR and is an important repressor of PI3K–Akt–mTOR pathway activity. Loss of PTEN appears to confer resistance of glioblastoma cell lines in vitro to EGFR inhibitors such as erlotinib [26,46,47], dacomitinib [16] and PKI-166 [26]. However, resistance due to PTEN loss can be overcome by simultaneously inhibiting EGFR and mTOR [47].
FGFR inhibition
FGF signaling has been linked to proliferation, cellular survival and angiogenesis within a wide range of tumors and therefore the development of FGFR inhibitors is actively being pursued [48]. Although only 3% of glioblastomas contain aberrant FGFR [3,49], there is a renewed interest in FGFR inhibition in glioma due to the discovery of FGFR fusion genes [40,41,50,51]. Inhibition of fusion gene products is attractive due to the remarkable success of imatinib targeting the BCR–ABL1 fusion gene in chronic myeloid leukemia (CML) [52].
• FGFR fusion genes in the context of FGFR inhibition
The FGFR–TACC fusion gene was one of the first recurrent fusion genes to be discovered in glioblastoma [40]. Singh and colleagues discovered FGFR3–TACC1 and FGFR1−TACC3 (±3%) fusion genes and found that these drive gliomagenesis when introduced into astrocytes [40]. Recently, Di Stefano and colleagues showed that FGFR fusion genes occur mutually exclusive with EGFR amplification, EGFRvIII and IDH1/2 mutations whereas they appear to co-occur with CDK4 amplification [49]. Interestingly, GSCs transduced with these fusion genes are highly sensitive to FGFR inhibitors [40,49], suggesting a similar fusion gene dependence as in BCR–ABL-driven CML.
PDGFR inhibition
PDGFR is amplified or mutated in approximately 10% of glioblastomas. Glioblastomas of the proneural subtype are mainly driven by PDGFR signaling and a third of these tumors contain a PDGFRA amplification [2,53]. Apart from a sporadic gene fusion between VEGFRII and PDGFRA [43], PDGFRA is not known to be involved in recurrent gene fusions. In contrast, PDGFRA does contain a frequent in-frame deletion of 243 bp of exons 8–9, resulting in the constitutively active PDGFRA Δ8, 9 [43,54]. Cells carrying this deletion appear to be sensitive to PDGFR inhibitors [43], however it is yet unknown whether PDGFRAΔ8, 9 confers more sensitivity to these drugs compared with its wild-type counterpart.
• EGFR mutations in the context of PDGFR inhibition
Hägerstrand and colleagues studied 11 GSCs and subdivided them according to the expression of a set of genes [55]. A subset of glioblastomas, termed as ‘type A tumors’, contained more classical, neural and/or proneural tumors which were enriched for EGFR mutations. In contrast, ‘type B tumors’ were more related to the mesenchymal subtype. They observed that type A cultures were more resistant whereas type B tumors were more sensitive to imatinib. Possibly, type A cultures were more resistant to imatinib due to a decreased dependence on PDGFR signaling in the setting of increased coactivated mutant EGFR.
PI3K inhibition
PI3K is an intensely studied drug target for which inhibitors are being developed and tested within clinical trials for the treatment of glioblastoma (for review, see [56]). PI3Ks are divided into class I–III, of which class I is the most studied in glioblastoma. PIK3CA, PIK3CB and PIK3CD encode the class IA catalytic isoforms: p110α, p110β and p110δ, respectively. These isoforms heterodimerize with any of the regulatory subunits p85α, p85β, p55γ (encoded by PIK3R1, PIK3R2, PIK3R3, respectively). PI3K is mutated in 25% of glioblastomas, with 18% affecting the p110α and/or p85α subunits and 7% affecting other PI3K family genes [3].
• PTEN & TP53 mutations in the context of PI3K inhibition
Since PTEN is a known modulator of the PI3K–Akt–mTOR pathway, it is relevant to know whether PTEN mutations also modulate the response to PI3K inhibitors. Chen and colleagues found that PTEN-mutated glioma cell lines tend to be more resistant to single treatment with PI3K inhibition or radiation [57]. However, PI3K inhibition combined with radiation could decrease cell survival to a level comparable with the response of combinatorial treated PTEN wild-type glioma cell lines. So PI3K inhibitors are able to sensitize gliomas to radiation-induced cytotoxicity, only in the absence of PTEN wild-type expression [57].
Koul and colleagues tested NVP-BKM120 (a pan class I PI3K inhibitor) on a panel of 21 established glioma cell lines differing in PTEN and TP53 mutation status [58]. No relation was found between PTEN status and drug sensitivity, however there was a clear relation with the TP53 mutation status. TP53 wild-type cells were more sensitive and underwent apoptotic cell death, whereas p53 mutant cells underwent a mitotic catastrophe cell death [58].
Akt inhibition
Mutations in RTKs, PI3K and PTEN result in downstream hyperphosphorylation of Akt, leading to an increase in cell proliferation and this supports the rationale to inhibit Akt [59]. The development of catalytic and allosteric inhibitors provides the tools to target Akt.
• PIK3CA mutation in the context of Akt inhibition
Gallia and colleagues used a PIK3CA isogenic cell culture knockout system, in which one cell line contained mutant PIK3CA and as a consequence had hyperactivated Akt, whereas the other cell line contained wild-type PIK3CA [60]. Several PI3K/Akt inhibitors were screened for selective inhibition of cells with the mutant phenotype and they identified A-443654 as a selective PIK3CA mutant inhibitor. Due to previously reported metabolic toxicity problems in mice [61], Gallia and colleagues delivered A-443654 locally with polymers in a xenograft mouse model [60]. Polymers containing 30% A-443654 were well tolerated, but could only prolong survival for a few more days. Unfortunately, Akt inhibitors have shown limited single-agent efficacy in clinical trials, therefore, future research is focused more on combination therapies.
mTOR inhibition
mTOR is a master regulator of cell growth and is an essential node downstream of PI3K hyperactivation. As a consequence, mTOR has been the focus of numerous drug development programs for the past two decades. mTOR comprises mTORC1 and mTORC2 (for review, see [62,63]). It operates downstream of Akt and regulates functions in growth signaling pathways. First-generation mTOR inhibitors (such as rapamycin, temsirolimus, everolimus and sirolimus) inhibit only mTORC1 and not mTORC2. Whereas second-generation mTOR inhibitors compete for the ATP-binding catalytic site and inhibit both mTORC1 and mTORC2. Dual inhibition of mTORC1 and mTORC2 may increase the efficacy of mTOR inhibitors since compelling evidence points toward involvement of mTORC2 in treatment resistance [64], and gliomagenesis [65,66].
• EGFRvIII in the context of mTOR inhibition
EGFRvIII is constitutively activated and consequently leads to constitutive mTOR activation. There are contradictory reports about the predictive role of EGFRvIII for dual mTORC1/2 dual inhibitors. Luchman and colleagues studied GSCs differing in EGFRvIII status and did not report any difference in cell viability at a concentration of 2 uM of AZD8055 [67]. Whereas Gini and colleagues tested U87-cell lines differing in EGFRvIII status and reported that EGFRvIII mutant cells were more sensitive to CC214–1 in a dose-dependent manner compared with U87-EGFRvIII-negative cells [68].
• PTEN mutations in the context of mTOR inhibition
As with EGFRvIII, PTEN mutations lead to hyperactivated mTOR signaling. The predictive value of PTEN mutations for mTOR inhibitors has been debated. Several studies have found PTEN mutant glioma cell lines to be more sensitive for rapamycin analogs (such as everolimus and temsirolimus) [47,69,70] and dual mTORC1/2 inhibitors [68] compared with PTEN wild-type glioma cell lines. Weiler and colleagues concluded that they did not find any correlation between PTEN status and sensitivity to temsirolimus [69]. However a closer evaluation between the PTEN status and the provided IC50 value actually appears to reveal a strong tendency for PTEN mutants to be more sensitive to temsirolimus [69]. Yang and colleagues studied the efficacy of everolimus in glioma cell lines in parallel to a panel of patient-derived orthotopic xenografts [70]. Interestingly, the established PTEN mutant glioma cell lines were sensitive to mTOR inhibition, whereas xenografts of patient-derived PTEN mutant cells did not respond to everolimus [70]. They hypothesized that this discrepancy could be due to adaptive changes taking place with long-term culturing of tumor cells in vitro. A study using only GSCs (which better preserves the parental tumor [7]), supports this hypothesis, since it did not find any predictive value of PTEN mutant status for response to a dual mTORC1/2 inhibitor [67].
• NF1 deletions in the context of mTOR inhibition
Similar to PTEN, NF1 functions as a tumor suppressor gene. NF1 inactivation is enriched in the more malignant, mesenchymal subtype of glioblastoma [2], and leads to disinhibition of the Ras pathway. McGillicuddy and colleagues reported that in a subset of glioblastomas NF1 protein is degraded as a consequence of PKC hyperactivation, whereas a separate subset of glioblastomas inactivated NF1 through mutations [71]. Both mechanisms result in loss of NF1 function. Interestingly, they found that NF1-allele proficient tumors, in which PKC was hyperactivated, were sensitive to PKC inhibitors. Conversely, NF1 -/- tumors were insensitive to PKC inhibitors but were sensitive to mTOR inhibitors [71]. This study highlights that the precise mechanism by which NF1 function is inactivated, determines the selection strategy of the appropriately targeted therapy.
PI3K/mTOR dual inhibition
Active mTORC1 suppresses the PI3K–Akt pathway. Unfortunately, rapamycin analogs abrogate this negative feedback and activate the insulin receptor pathway which may diminish the potential therapeutic benefits of mTORC1 inhibition [72–74]. PI3K/mTOR dual inhibitors can counteract the derepression of the PI3K–Akt pathway and are therefore more effective than mTORC1 inhibitors as a single agent [75,76].
• PTEN mutations in the context of PI3K/mTOR dual inhibition
Studies investigating PI3K/mTOR dual inhibitors do not report drug resistance of PTEN mutants [46,57,77,78]. There was even a tendency of PTEN mutant glioma cell lines to be more sensitive to dual inhibition of PI3K and mTOR [57,77].
Raf inhibition
Specific inhibitors of mutated BRAF have produced impressive responses in patients with melanoma [79,80]. Therefore the treatment of BRAF-mutated glioblastomas may elicit similar spectacular responses. In contrast to the high incidence in melanomas, the BRAF V600E mutation is only detected in a few percent of glioblastomas [3]. BRAF V600E is more prevalent in pediatric patients and lower grade gliomas [81]. In the rare case of a BRAF V600E mutation, BRAF inhibition with vemurafenib may be clinically attractive as evidenced by a recent case report [82].
• BRAF V600E mutation in the context of BRAF inhibition
Vemurafenib is a BRAF mutant-specific inhibitor that has been effective in the treatment of BRAF V600E mutant melanomas [79]. Ahn and colleagues investigated the efficacy of UAI-201, a different BRAF mutant-specific inhibitor [83]. Similar to melanomas, glioma cell lines with the BRAF V600E mutation responded better to a BRAF mutant-specific inhibitor compared with BRAF wild-type cells [83].
MEK inhibition
Activated MAPK is an independent prognostic marker for poor overall survival in glioblastoma and since MAPK is activated by MEK, this suggests MEK hyperactivation to be implicated in more aggressive glioblastomas [84]. In support of this study, Brennan and colleagues report that the aggressive mesenchymal glioblastoma subtype, show an increased activation of MEK [3]. Therefore MEK inhibition may impede clinical progression of glioblastoma and may contribute to prolonged survival.
• NF1 deletions in the context of MEK inhibition
MEK inhibition within NF1-deficient cells has proven to be effective within AML [85]. Similarly a subset of NF1-deficient glioblastomas is highly sensitive to MEK inhibitors, whereas PTEN deficiency does not seem to modulate sensitivity to MEK inhibitors [78].
Rb pathway
Approximately 80% of glioblastomas contain one or more genetic aberrations in the Rb pathway [1,3]. Rb pathway deregulation leads to the sustainment of proliferative signaling, often through deletion or mutation of RB1 and the cyclin-dependent kinase inhibitors (CDKN2A, CDKN2C), and amplification of CDK4 or CDK6 [1,3].
CDK4/6 inhibition
Numerous studies indicated that the function of CDK4 and CDK6 is redundant in most mammalian cells, whereas glioblastomas appear to be more dependent on CDK4 and CDK6 for cell cycling compared with normal cells [86–88]. As a consequence CDK inhibitors may selectively impair tumor cells, while leaving normal cells intact. However, the first-generation CDK inhibitors were not very selective and inhibited multiple CDKs, such as CDK1/2/3/7 and CDK9. Second-generation CDK inhibitors are more selective for CDK4 and CDK6 and show more promising clinical responses (for review, see [89]).
• RB1 & CDKN2A/B/C mutation/deletion in the context of CDK4/6 inhibition
PD0332991 is a selective CDK4/6 inhibitor and is the most extensively investigated drug in relation to the Rb pathway. Homozygous deletion/mutation of RB1 [88,90,91] and CDK4 [90] amplification contribute to resistance to PD0332991. In contrast, p16INK4A (cyclin-dependent kinase inhibitor 2A, CDKN2A), p15INK4B (CDKN2B) and p18INK4C(CDKN2C) codeletion predicts sensitivity to PD0332991 [91]. The predictive status of CDKN2A/B codeletion without CDKN2C probably also predicts sensitivity, however reports have been contradictory since it has been linked to both tumor sensitization [90] and tumor resistance [91].
• EGFRvIII in the context of CDK4/6 inhibition
Nimbolide is a cytotoxic component extracted from the leaf of Azadirachta Indica and inhibits multiple proteins, including CDK4 and CDK6. Intriguingly, Karkare and colleagues report that EGFRvIII mutant cells are especially sensitive to this natural extract [92]. Nevertheless, the drug is known to also inhibit Akt, MAPK and the JAK-STAT3 pathway, thus the tumor suppressive effect may also be due to the inhibition of proteins other than CDK4/6.
p53 pathway
Approximately 80–90% of glioblastomas contain one or more genetic aberrations in the p53 pathway [1,13]. p53 pathway deregulation leads to resistance of cell death and evasion of growth suppression, often through amplification of MDM2/4 or inactivation of TP53 [1,3].
MDM2 inhibition
MDM2 inhibits p53 transcriptional activity and stimulates its nuclear export and degradation. Roughly half of human cancers, contain either deleted or mutated TP53 [93]. In the remaining cancers p53 protein function is often diminished due to interacting inhibiting proteins, such as MDM2 [93]. Reactivation of p53 by MDM2 interference of the MDM2–p53 interaction is therefore an interesting approach for inducing p53-mediated apoptosis.
• TP53 mutations in the context of MDM2 inhibition
The mode of TP53 inactivation has a major influence on the response to MDM2 inhibition. First, hemizygous TP53 deletion (p53 competent tumors) will be discussed. Shchors and colleagues have dissected the different mechanisms involved in the loss of p53 function [94]. They used a genetically engineered mouse model (GEMM) with Gfap-HRasV12 crossed into a p53ERTAM background, resulting in the three mouse strains Gfap-HRasV12; Tp53KI/KI, Gfap-HRasV12; Tp53+/KI and Gfap-HRasV12; Tp53+/+. The knocked-in Tp53ERTAM is functional only in the presence of 4-hydroxytamoxifen (4-OHT). As a result Tp53KI/KI mice are functionally p53null in the absence of 4-OHT, but change into p53wt upon systemic administration of tamoxifen (TAM), which is metabolized in vivo to 4-OHT. Gfap-HRasV12; Tp53+/KI and Gfap-HRasV12; Tp53+/+ GEMMs modeled the evolution of gliomas in which Ras pathway activation precedes p53 pathway inactivation. Shchors and colleagues show that functional p53 is present in Gfap-HRasV12;Tp53+/KI tumor cells and could induce p53-mediated apoptosis by irradiation. Since p53 and its downstream apoptosis pathway were intact, the likely explanation for p53 inactivity in Gfap-HRasV12; TP53+/KI and Gfap-HRasV12; Tp53+/+ tumors must lie in the upstream signal to activate p53 in response to oncogenic signaling. Normally oncogenic signaling, such as by Myc [95] and Ras [96], leads to activation of p19ARF (p14ARF in humans) and reactivates p53 function by suppression of MDM2. Indeed, MDM2 expression was high in these tumors and exposure to the MDM2 inhibitor (nutlin3) induced significant cell death [94]. This indicates that the upstream Ras oncogene-sensing p19ARF/Mdm2 (p14ARF/HDM2 in humans) pathway is dysfunctional, eventually resulting in the impaired reactivation of p53. Thus p53 competent tumors (containing at least one TP53wt allele) are likely to inactivate the upstream activating p53 pathway during tumor formation and can therefore benefit from MDM2 inhibition.
Besides TP53 deletions, TP53 can be genetically inactivated by point mutations. Studies using small-molecule inhibitors or peptides specifically blocking the MDM2–p53 interaction, reveal that TP53 mutant glioblastoma cells are resistant to MDM2 inhibition. This is because only p53 wild-type cells are able to undergo p53-dependent apoptosis [97,98].
p53 reactivation
TP53 is mutated (considering missense, truncating and in-frame mutations) in 20% of glioblastomas [3]. Missense mutations can confer loss of the tumor suppressor function and can lead to an oncogenic gain of function (for review, see [99]). For example, patients with germline mutant TP53 have an earlier onset of cancer than patients with mutations resulting in loss of p53 protein expression [100]. Understandably, restoring wild-type p53 function is an attractive therapeutic approach. Several synthetic peptides and small molecules have been developed to restore p53 wild-type activity from mutant p53 [101–103].
• TP53 mutation in the context of p53 reactivation
Gfap-HRasV12;Tp53KI/KI mice are able to model tumors in which sporadic Tp53 loss precedes or coincides Ras activation. These mouse models probably behave similarly to human glioblastomas that directly inactivate TP53 (via homozygous deletion or mutation) while retaining CDKN2A [94]. Shchors and colleagues [94] found that p53 reactivation with 4-OHT induced massive cell death in these GEMMs. Similarly, human glioma cell lines containing mutated TP53 with wild-type p14ARF (i.e., CDNK2A(ARF)) benefitted from p53 reactivation with the drug prima-1 [104], whereas the other combinations of mutations of TP53 and CDKN2A did not [94]. Interestingly, the expression of PTEN in the presence of mutant TP53 induces an even increased sensitivity to p53 reactivation [104].
Conclusion & future perspective
In this review, we provide an overview of the preclinical efforts to identify genetic biomarkers that predict response to small-molecule compounds specifically targeting the RTK/Ras/PI3K, p53 or Rb pathway (Figure 1). The application of targeted therapy with small-molecule kinase inhibitors has yet to lead to clinical breakthroughs in the treatment of glioblastoma (for review, see [105]). Even though the neuro-oncology research community has made great progress, the translation of preclinical findings into clinical trials and to patient benefit is lagging behind. This disconnect is also known as the translational gap [106]. The underlying reasons can be attributed to flawed preclinical models and flawed clinical trial designs.
For several decades, established glioblastoma cell lines were a popular tool to investigate the in vitro response to different compounds. However, recent research demonstrates that these models bear little resemblance to the original tumor of patients [6,7]. In contrast, in different types of cancer several reports indicate that response rates of patient-derived primary cultures and xenografts correlate well with that of patients (for review, see [107]). Next to the use of established cell lines, 2D ex vivo culture models may not accurately reflect the tumor pathophysiology as observed in the patient. For example, drug sensitivity profiles of 2D glioma cultures do not readily correlate with those of 3D cultured glioma cells [108].
Another factor influencing the lack of correlation between preclinical and clinical efficacy, is that the appropriate concentration of the small-molecule compounds may not reach and sustain therapeutic levels within the glioma tissue or (peri)tumoral area. This may be due to unfavorable pharmacodynamics resulting in inefficient penetration of the blood–brain barrier in the (peri)tumoral area. In addition, drug concentrations that show decrease of tumor cell viability in preclinical models may not be achievable in the patient due to drug toxicity. Even when the drug does effectively reach the tumor, intratumor heterogeneity may impede effective homogeneous tumor cell death.
Intratumor heterogeneity may complicate the translation of in vitro drug responses to in vivo patient responses, because the presence of biomarkers linked with specific drug responses may vary within the tumor [109–112]. Indeed, recent studies demonstrate that different subclones within the same tumor may have different in vitro drug sensitivity profiles [113,114]. To circumvent the problem of intratumor heterogeneity, it may be beneficial to identify targetable driver events that are uniformly expressed in the tumor, such as IDH1/2 mutations or FGFR-TACC fusion genes. This may delay or prevent the outgrowth of subclones without these specific driver events. However, a recent publication showed a case in which an IDH1 mutation was lost during tumor progression [115]. In this respect it remains to be determined whether biomarkers generally used to stratify patients, may also hold for predicting response in the setting of intratumor heterogeneity. A yet unanswered question is whether the presence of different genetic biomarkers within a specific tumor leads to a differential in vivo sensitivity of the subclones to the drug in question.
Besides intratumor hetereogeneity, also interpatient variability (i.e., intertumor heterogeneity) plays an important role in clinical assessment of drug efficacy. Many clinical trials still test the experimental drug in question in an unselected patient population. Usually only a subset of patients respond to experimental drugs and consequently the effect of the drug will be diluted [4]. Therefore clinical trials should be stratified with validated biomarkers that are predictive for the drug under study. However, predictive biomarkers of drug response identified in other types of cancer may not directly be extrapolated to glioblastoma. For example, inhibition of BRAF V600E in melanoma induces dramatic responses, whereas colon cancer patients harboring exactly the same BRAF V600E mutation show only a very limited response to this drug [116]. Thus the context in which the mutation occurs has a major influence on the value of predictive biomarkers and cannot immediately be translated to the treatment of other types of cancer.
Besides biomarker-guided patient stratification, the vast array of inhibitors which are already available should be incorporated into ‘smart drug schemes’. Many drug dimensions should be considered when designing smart drug schemes, such as: implementing synergistic drug combinations; implementing synergistic sequential drug schemes in order to minimize drug toxicity and maximize drug synergy [117]; implementing drug holidays in order to resensitize tumors to targeted therapy [32].
Finally, it is imperative to incorporate preclinical evidence into transformative clinical trial designs, such as multicenter umbrella trials or basket trials [4,118]. In an umbrella trial patients with a specific type of cancer are matched to a specific treatment arm based on their molecular profile of their cancer. In contrast, in basket trials the effect of a targeted drug is tested in patients whose cancer contains a specific molecular aberration irrespective of their type of cancer. Besides these larger scale trials, Phase 0 trials should also be pursued more. In these studies the experimental drug is administered before tumor resection which allows the investigation of drug-target effects and the assessment of pharmacokinetic–pharmacodynamic relationships in humans. For example, Vivanco and colleagues have applied this approach in order to identify that lapatinib does not reach sufficient intratumor concentrations in glioblastoma patients [12].
In conclusion, targeted therapies still provide hope for the treatment of patients with glioblastoma. Especially drugs targeting the RTK/Ras/PI3K, p53 or Rb pathway are attractive due to the frequent aberrations found in these pathways. However, preclinical models that faithfully represent the tumor characteristics as observed in the patient must be applied to filter out compounds that will likely not be effective in the patient. After compounds have successfully passed the appropriate filters of preclinical models, they should be tested in synergistic drug combination schemes in well-designed, biomarker-guided clinical trials.
Acknowledgements
The authors thank Wichor Bramer for assistance in composing relevant search terms. We would also like to thank Lotte Berghauser Pont and Zineb Belcaid for critical proofreading.
Footnotes
Financial & competing interests disclosure
This work was supported by the Foundation STOPBraintumors.org. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The first comprehensive sequencing study on glioblastoma that has defined the core mutated pathways.
- 2.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sleijfer S, Bogaerts J, Siu LL. Designing transformative clinical trials in the cancer genome era. J. Clin. Oncol. 2013;31(15):1834–1841. doi: 10.1200/JCO.2012.45.3639. [DOI] [PubMed] [Google Scholar]
- 5.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balvers RK, Kleijn A, Kloezeman JJ, et al. Serum-free culture success of glial tumors is related to specific molecular profiles and expression of extracellular matrix-associated gene modules. Neuro Oncol. 2013;15(12):1684–1695. doi: 10.1093/neuonc/not116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]; •• Describes the protocol for culturing GSCs.
- 8.Pollard SM, Yoshikawa K, Clarke ID, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4(6):568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Kastenhuber ER, Huse JT, Berman SH, et al. Quantitative assessment of intragenic receptor tyrosine kinase deletions in primary glioblastomas: their prevalence and molecular correlates. Acta Neuropathol. 2014;127(5):747–759. doi: 10.1007/s00401-013-1217-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang HS, Nagane M, Klingbeil CK, et al. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J. Biol. Chem. 1997;272(5):2927–2935. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- 12.Vivanco I, Ian Robins H, Rohle D, et al. Differential sensitivity of glioma- versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov. 2012;2(5):458–471. doi: 10.1158/2159-8290.CD-11-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes how the spectrum of mutations differs between gliomas and lung cancer and how this affects drug sensitivity.
- 13.Brennan CW, Verhaak RG, Mckenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reardon DA, Wen PY, Mellinghoff IK. Targeted molecular therapies against epidermal growth factor receptor: past experiences and challenges. Neuro Oncol. 2014;16(Suppl. 8):viii7–viii13. doi: 10.1093/neuonc/nou232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eimer S, Dugay F, Airiau K, et al. Cyclopamine cooperates with EGFR inhibition to deplete stem-like cancer cells in glioblastoma-derived spheroid cultures. Neuro Oncol. 2012;14(12):1441–1451. doi: 10.1093/neuonc/nos266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zahonero C, Aguilera P, Ramirez-Castillejo C, et al. Preclinical test of dacomitinib, an irreversible EGFR inhibitor, confirms its effectiveness for glioblastoma. Mol. Cancer Ther. 2015;14(7):1548–1558. doi: 10.1158/1535-7163.MCT-14-0736. [DOI] [PubMed] [Google Scholar]
- 17.Pandita A, Aldape KD, Zadeh G, Guha A, James CD. Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes Chromosomes Cancer. 2004;39(1):29–36. doi: 10.1002/gcc.10300. [DOI] [PubMed] [Google Scholar]
- 18.Akhavan D, Pourzia AL, Nourian AA, et al. De-repression of PDGFR(beta) transcription promotes acquired resistance to EGFR tyrosine kinase inhibitors in glioblastoma patients. Cancer Discov. 2013;3(5):534–547. doi: 10.1158/2159-8290.CD-12-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stommel JM, Kimmelman AC, Ying H, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318(5848):287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 20.Sarkaria JN, Yang L, Grogan PT, et al. Identification of molecular characteristics correlated with glioblastoma sensitivity to EGFR kinase inhibition through use of an intracranial xenograft test panel. Mol. Cancer Ther. 2007;6(3):1167–1174. doi: 10.1158/1535-7163.MCT-06-0691. [DOI] [PubMed] [Google Scholar]
- 21.Efferth T, Ramirez T, Gebhart E, Halatsch ME. Combination treatment of glioblastoma multiforme cell lines with the anti-malarial artesunate and the epidermal growth factor receptor tyrosine kinase inhibitor OSI-774. Biochem. Pharmacol. 2004;67(9):1689–1700. doi: 10.1016/j.bcp.2003.12.035. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Taich ZJ, Goyal A, et al. Epigenetic suppression of EGFR signaling in G-CIMP+ glioblastomas. Oncotarget. 2014;5(17):7342–7356. doi: 10.18632/oncotarget.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yiin JJ, Hu B, Schornack PA, et al. ZD6474, a Multitargeted inhibitor for receptor tyrosine kinases, suppresses growth of gliomas expressing an epidermal growth factor receptor mutant, EGFRvIII, in the brain. Mol. Cancer Ther. 2010;9(4):929–941. doi: 10.1158/1535-7163.MCT-09-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida Y, Ozawa T, Yao TW, et al. NT113, a pan-ERBB Inhibitor with high brain penetrance, inhibits the growth of glioblastoma xenografts with EGFR amplification. Mol. Cancer Ther. 2014;13(12):2919–2929. doi: 10.1158/1535-7163.MCT-14-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lal B, Goodwin CR, Sang Y, et al. EGFRvIII and c-Met pathway inhibitors synergize against PTEN-null/EGFRvIII+ glioblastoma xenografts. Mol. Cancer Ther. 2009;8(7):1751–1760. doi: 10.1158/1535-7163.MCT-09-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N. Engl. J. Med. 2005;353(19):2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 27.Cemeus C, Zhao TT, Barrett GM, Lorimer IA, Dimitroulakos J. Lovastatin enhances gefitinib activity in glioblastoma cells irrespective of EGFRvIII and PTEN status. J. Neurooncol. 2008;90(1):9–17. doi: 10.1007/s11060-008-9627-0. [DOI] [PubMed] [Google Scholar]
- 28.Frattini V, Trifonov V, Chan JM, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat. Genet. 2013;45(10):1141–1149. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho J, Pastorino S, Zeng Q, et al. Glioblastoma-derived epidermal growth factor receptor carboxyl-terminal deletion mutants are transforming and are sensitive to EGFR-directed therapies. Cancer Res. 2011;71(24):7587–7596. doi: 10.1158/0008-5472.CAN-11-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulte A, Liffers K, Kathagen A, et al. Erlotinib resistance in EGFR-amplified glioblastoma cells is associated with upregulation of EGFRvIII and PI3Kp110delta. Neuro Oncol. 2013;15(10):1289–1301. doi: 10.1093/neuonc/not093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lo HW, Cao X, Zhu H, Ali-Osman F. Constitutively activated STAT3 frequently coexpresses with epidermal growth factor receptor in high-grade gliomas and targeting STAT3 sensitizes them to Iressa and alkylators. Clin. Cancer Res. 2008;14(19):6042–6054. doi: 10.1158/1078-0432.CCR-07-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nathanson DA, Gini B, Mottahedeh J, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343(6166):72–76. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes how glioma cells can shed mutant EGFR from extrachromosomal DNA as a way of drug resistance to EGFR inhibitors.
- 33.Francis JM, Zhang CZ, Maire CL, et al. EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discov. 2014;4(8):956–971. doi: 10.1158/2159-8290.CD-13-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ekstrand AJ, Sugawa N, James CD, Collins VP. Amplified and rearranged epidermal growth factor receptor genes in human glioblastomas reveal deletions of sequences encoding portions of the N- and/or C-terminal tails. Proc. Natl Acad. Sci. USA. 1992;89(10):4309–4313. doi: 10.1073/pnas.89.10.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60(5):1383–1387. [PubMed] [Google Scholar]
- 36.Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J. Biol. Chem. 2002;277(48):46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- 37.Wood ER, Truesdale AT, Mcdonald OB, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64(18):6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 38.Lee JC, Vivanco I, Beroukhim R, et al. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med. 2006;3(12):2264–2273. doi: 10.1371/journal.pmed.0030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greenall SA, Donoghue JF, Gottardo NG, Johns TG, Adams TE. Glioma-specific Domain IV EGFR cysteine mutations promote ligand-induced covalent receptor dimerization and display enhanced sensitivity to dacomitinib in vivo . Oncogene. 2015;34(13):1658–1666. doi: 10.1038/onc.2014.106. [DOI] [PubMed] [Google Scholar]
- 40.Singh D, Chan JM, Zoppoli P, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337(6099):1231–1235. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker BC, Annala MJ, Cogdell DE, et al. The tumorigenic FGFR3–TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J. Clin. Invest. 2013;123(2):855–865. doi: 10.1172/JCI67144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng S, Fu J, Vegesna R, et al. A survey of intragenic breakpoints in glioblastoma identifies a distinct subset associated with poor survival. Genes Dev. 2013;27(13):1462–1472. doi: 10.1101/gad.213686.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ozawa T, Brennan CW, Wang L, et al. PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes Dev. 2010;24(19):2205–2218. doi: 10.1101/gad.1972310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Charest A, Lane K, McMahon K, et al. Fusion of FIG to the receptor tyrosine kinase ROS in a glioblastoma with an interstitial del(6)(q21q21) Genes Chromosomes Cancer. 2003;37(1):58–71. doi: 10.1002/gcc.10207. [DOI] [PubMed] [Google Scholar]
- 45.Charest A, Wilker EW, Mclaughlin ME, et al. ROS fusion tyrosine kinase activates a SH2 domain-containing phosphatase-2/phosphatidylinositol 3-kinase/mammalian target of rapamycin signaling axis to form glioblastoma in mice. Cancer Res. 2006;66(15):7473–7481. doi: 10.1158/0008-5472.CAN-06-1193. [DOI] [PubMed] [Google Scholar]
- 46.Fan QW, Cheng CK, Nicolaides TP, et al. A dual phosphoinositide-3-kinase alpha/mTOR inhibitor cooperates with blockade of epidermal growth factor receptor in PTEN-mutant glioma. Cancer Res. 2007;67(17):7960–7965. doi: 10.1158/0008-5472.CAN-07-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang MY, Lu KV, Zhu S, et al. Mammalian target of rapamycin inhibition promotes response to epidermal growth factor receptor kinase inhibitors in PTEN-deficient and PTEN-intact glioblastoma cells. Cancer Res. 2006;66(16):7864–7869. doi: 10.1158/0008-5472.CAN-04-4392. [DOI] [PubMed] [Google Scholar]
- 48.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat. Rev. Cancer. 2010;10(2):116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 49.Di Stefano AL, Fucci A, Frattini V, et al. Detection, characterization and inhibition of FGFR–TACC fusions in IDH wild-type glioma. Clin. Cancer Res. 2015;21(14):3307–3317. doi: 10.1158/1078-0432.CCR-14-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parker BC, Engels M, Annala M, Zhang W. Emergence of FGFR family gene fusions as therapeutic targets in a wide spectrum of solid tumours. J. Pathol. 2014;232(1):4–15. doi: 10.1002/path.4297. [DOI] [PubMed] [Google Scholar]
- 51.Parker BC, Zhang W. Fusion genes in solid tumors: an emerging target for cancer diagnosis and treatment. Chin. J. Cancer. 2013;32(11):594–603. doi: 10.5732/cjc.013.10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Druker BJ, Sawyers CL, Kantarjian H, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N. Engl. J. Med. 2001;344(14):1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 53.Brennan C, Momota H, Hambardzumyan D, et al. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS ONE. 2009;4(11):e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clarke ID, Dirks PB. A human brain tumor-derived PDGFR-alpha deletion mutant is transforming. Oncogene. 2003;22(5):722–733. doi: 10.1038/sj.onc.1206160. [DOI] [PubMed] [Google Scholar]
- 55.Hagerstrand D, He X, Lindh MB, et al. Identification of a SOX2-dependent subset of tumor-and sphere-forming glioblastoma cells with a distinct tyrosine kinase inhibitor sensitivity profile. Neuro Oncol. 2011;13(11):1178–1191. doi: 10.1093/neuonc/nor113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wen PY, Lee EQ, Reardon DA, Ligon KL, Alfred Yung WK. Current clinical development of PI3K pathway inhibitors in glioblastoma. Neuro Oncol. 2012;14(7):819–829. doi: 10.1093/neuonc/nos117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen JS, Zhou LJ, Entin-Meer M, et al. Characterization of structurally distinct, isoform-selective phosphoinositide 3′-kinase inhibitors in combination with radiation in the treatment of glioblastoma. Mol. Cancer Ther. 2008;7(4):841–850. doi: 10.1158/1535-7163.MCT-07-0393. [DOI] [PubMed] [Google Scholar]
- 58.Koul D, Fu J, Shen R, et al. Antitumor activity of NVP-BKM120 – a selective pan class I PI3 kinase inhibitor showed differential forms of cell death based on p53 status of glioma cells. Clin. Cancer Res. 2012;18(1):184–195. doi: 10.1158/1078-0432.CCR-11-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akbani R, Ng PK, Werner HM, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat Commun. 2014;5:3887. doi: 10.1038/ncomms4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gallia GL, Tyler BM, Hann CL, et al. Inhibition of Akt inhibits growth of glioblastoma and glioblastoma stem-like cells. Mol. Cancer Ther. 2009;8(2):386–393. doi: 10.1158/1535-7163.MCT-08-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo Y, Shoemaker AR, Liu X, et al. Potent and selective inhibitors of Akt kinases slow the progress of tumors in vivo. Mol. Cancer Ther. 2005;4(6):977–986. doi: 10.1158/1535-7163.MCT-05-0005. [DOI] [PubMed] [Google Scholar]
- 62.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12(1):9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 63.Guertin DA, Sabatini DM. The pharmacology of mTOR inhibition. Sci. Signal. 2009;2(67):pe24. doi: 10.1126/scisignal.267pe24. [DOI] [PubMed] [Google Scholar]
- 64.Tanaka K, Babic I, Nathanson D, et al. Oncogenic EGFR signaling activates an mTORC2-NF-(kappa)B pathway that promotes chemotherapy resistance. Cancer Discov. 2011;1(6):524–538. doi: 10.1158/2159-8290.CD-11-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Masri J, Bernath A, Martin J, et al. mTORC2 activity is elevated in gliomas and promotes growth and cell motility via overexpression of rictor. Cancer Res. 2007;67(24):11712–11720. doi: 10.1158/0008-5472.CAN-07-2223. [DOI] [PubMed] [Google Scholar]
- 66.Bashir T, Cloninger C, Artinian N, et al. Conditional astroglial Rictor overexpression induces malignant glioma in mice. PLoS ONE. 2012;7(10):e47741. doi: 10.1371/journal.pone.0047741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luchman HA, Stechishin OD, Nguyen SA, Lun XQ, Cairncross JG, Weiss S. Dual mTORC1/2 blockade inhibits glioblastoma brain tumor initiating cells in vitro and in vivo and synergizes with temozolomide to increase orthotopic xenograft survival. Clin. Cancer Res. 2014;20(22):5756–5767. doi: 10.1158/1078-0432.CCR-13-3389. [DOI] [PubMed] [Google Scholar]
- 68.Gini B, Zanca C, Guo D, et al. The mTOR kinase inhibitors, CC214–1 and CC214–2, preferentially block the growth of EGFRvIII-activated glioblastomas. Clin. Cancer Res. 2013;19(20):5722–5732. doi: 10.1158/1078-0432.CCR-13-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weiler M, Pfenning PN, Thiepold AL, et al. Suppression of proinvasive RGS4 by mTOR inhibition optimizes glioma treatment. Oncogene. 2013;32(9):1099–1109. doi: 10.1038/onc.2012.137. [DOI] [PubMed] [Google Scholar]
- 70.Yang L, Clarke MJ, Carlson BL, et al. PTEN loss does not predict for response to RAD001 (everolimus) in a glioblastoma orthotopic xenograft test panel. Clin. Cancer Res. 2008;14(12):3993–4001. doi: 10.1158/1078-0432.CCR-07-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mcgillicuddy LT, Fromm JA, Hollstein PE, et al. Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis. Cancer Cell. 2009;16(1):44–54. doi: 10.1016/j.ccr.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun SY, Rosenberg LM, Wang X, et al. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005;65(16):7052–7058. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- 73.O'reilly KE, Rojo F, She QB, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66(3):1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shi Y, Yan H, Frost P, Gera J, Lichtenstein A. Mammalian target of rapamycin inhibitors activate the AKT kinase in multiple myeloma cells by up-regulating the insulin-like growth factor receptor/insulin receptor substrate-1/phosphatidylinositol 3-kinase cascade. Mol. Cancer Ther. 2005;4(10):1533–1540. doi: 10.1158/1535-7163.MCT-05-0068. [DOI] [PubMed] [Google Scholar]
- 75.Maira SM, Stauffer F, Brueggen J, et al. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol. Cancer Ther. 2008;7(7):1851–1863. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- 76.Serra V, Markman B, Scaltriti M, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008;68(19):8022–8030. doi: 10.1158/0008-5472.CAN-08-1385. [DOI] [PubMed] [Google Scholar]
- 77.Liu TJ, Koul D, Lafortune T, et al. NVP-BEZ235, a novel dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor, elicits multifaceted antitumor activities in human gliomas. Mol. Cancer Ther. 2009;8(8):2204–2210. doi: 10.1158/1535-7163.MCT-09-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.See WL, Tan IL, Mukherjee J, Nicolaides T, Pieper RO. Sensitivity of glioblastomas to clinically available MEK inhibitors is defined by neurofibromin 1 deficiency. Cancer Res. 2012;72(13):3350–3359. doi: 10.1158/0008-5472.CAN-12-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hauschild A, Grob JJ, Demidov LV, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, Phase 3 randomised controlled trial. Lancet. 2012;380(9839):358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 81.Schindler G, Capper D, Meyer J, et al. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011;121(3):397–405. doi: 10.1007/s00401-011-0802-6. [DOI] [PubMed] [Google Scholar]
- 82.Robinson GW, Orr BA, Gajjar A. Complete clinical regression of a BRAF V600E-mutant pediatric glioblastoma multiforme after BRAF inhibitor therapy. BMC Cancer. 2014;14:258. doi: 10.1186/1471-2407-14-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ahn JH, Lee YW, Ahn SK, Lee M. Oncogenic BRAF inhibitor UAI-201 induces cell cycle arrest and autophagy in BRAF mutant glioma cells. Life Sci. 2014;104(1–2):38–46. doi: 10.1016/j.lfs.2014.03.026. [DOI] [PubMed] [Google Scholar]
- 84.Patil CG, Nuno M, Elramsisy A, et al. High levels of phosphorylated MAP kinase are associated with poor survival among patients with glioblastoma during the temozolomide era. Neuro Oncol. 2013;15(1):104–111. doi: 10.1093/neuonc/nos272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lauchle JO, Kim D, Le DT, et al. Response and resistance to MEK inhibition in leukaemias initiated by hyperactive Ras. Nature. 2009;461(7262):411–414. doi: 10.1038/nature08279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Malumbres M, Sotillo R, Santamaria D, et al. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell. 2004;118(4):493–504. doi: 10.1016/j.cell.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 87.Barriere C, Santamaria D, Cerqueira A, et al. Mice thrive without Cdk4 and Cdk2. Mol. Oncol. 2007;1(1):72–83. doi: 10.1016/j.molonc.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Michaud K, Solomon DA, Oermann E, et al. Pharmacologic inhibition of cyclin-dependent kinases 4 and 6 arrests the growth of glioblastoma multiforme intracranial xenografts. Cancer Res. 2010;70(8):3228–3238. doi: 10.1158/0008-5472.CAN-09-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat. Rev. Cancer. 2011;11(8):558–572. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 90.Cen L, Carlson BL, Schroeder MA, et al. P16-Cdk4-Rb axis controls sensitivity to a cyclin-dependent kinase inhibitor PD0332991 in glioblastoma xenograft cells. Neuro Oncol. 2012;14(7):870–881. doi: 10.1093/neuonc/nos114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wiedemeyer WR, Dunn IF, Quayle SN, et al. Pattern of retinoblastoma pathway inactivation dictates response to CDK4/6 inhibition in GBM. Proc. Natl Acad. Sci. USA. 2010;107(25):11501–11506. doi: 10.1073/pnas.1001613107. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Provides a comprehensive overview of how the different modes of Rb pathway inactivation affect drug sensitivity.
- 92.Karkare S, Chhipa RR, Anderson J, et al. Direct inhibition of retinoblastoma phosphorylation by nimbolide causes cell-cycle arrest and suppresses glioblastoma growth. Clin. Cancer Res. 2014;20(1):199–212. doi: 10.1158/1078-0432.CCR-13-0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 94.Shchors K, Persson AI, Rostker F, et al. Using a preclinical mouse model of high-grade astrocytoma to optimize p53 restoration therapy. Proc. Natl Acad. Sci. USA. 2013;110(16):E1480–E1489. doi: 10.1073/pnas.1219142110. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Provides a comprehensive overview of how the different modes of p53 pathway inactivation affect drug sensitivity.
- 95.Murphy DJ, Junttila MR, Pouyet L, et al. Distinct thresholds govern Myc's biological output in vivo. Cancer Cell. 2008;14(6):447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Junttila MR, Karnezis AN, Garcia D, et al. Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature. 2010;468(7323):567–571. doi: 10.1038/nature09526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu M, Li C, Pazgier M, et al. D-peptide inhibitors of the p53-MDM2 interaction for targeted molecular therapy of malignant neoplasms. Proc. Natl Acad. Sci. USA. 2010;107(32):14321–14326. doi: 10.1073/pnas.1008930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Villalonga-Planells R, Coll-Mulet L, Martinez-Soler F, et al. Activation of p53 by nutlin-3a induces apoptosis and cellular senescence in human glioblastoma multiforme. PLoS ONE. 2011;6(4):e18588. doi: 10.1371/journal.pone.0018588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25(3):304–317. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zerdoumi Y, Aury-Landas J, Bonaiti-Pellie C, et al. Drastic effect of germline TP53 missense mutations in Li-Fraumeni patients. Hum. Mutat. 2013;34(3):453–461. doi: 10.1002/humu.22254. [DOI] [PubMed] [Google Scholar]
- 101.Friedler A, Hansson LO, Veprintsev DB, et al. A peptide that binds and stabilizes p53 core domain: chaperone strategy for rescue of oncogenic mutants. Proc. Natl Acad. Sci. USA. 2002;99(2):937–942. doi: 10.1073/pnas.241629998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bykov VJ, Issaeva N, Shilov A, et al. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat. Med. 2002;8(3):282–288. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- 103.Yu X, Vazquez A, Levine AJ, Carpizo DR. Allele-specific p53 mutant reactivation. Cancer Cell. 2012;21(5):614–625. doi: 10.1016/j.ccr.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huang X, Zhang Y, Tang Y, et al. A novel PTEN/Mutant p53/c-Myc/Bcl-XL axis mediates context-dependent oncogenic effects of PTEN with implications for cancer prognosis and therapy. Neoplasia. 2013;15(8):952–965. doi: 10.1593/neo.13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.De Witt Hamer PC. Small molecule kinase inhibitors in glioblastoma: a systematic review of clinical studies. Neuro Oncol. 2010;12(3):304–316. doi: 10.1093/neuonc/nop068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ma C, Zhao G, Cruz MH, Siden A, Yakisich JS. Translational gap in glioma research. Anticancer Agents Med. Chem. 2014;14(8):1110–1120. doi: 10.2174/1871520614666140825110907. [DOI] [PubMed] [Google Scholar]
- 107.Hidalgo M, Amant F, Biankin AV, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014;4(9):998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fernandez-Fuente G, Mollinedo P, Grande L, Vazquez-Barquero A, Fernandez-Luna JL. Culture dimensionality influences the resistance of glioblastoma stem-like cells to multikinase inhibitors. Mol. Cancer Ther. 2014;13(6):1664–1672. doi: 10.1158/1535-7163.MCT-13-0854. [DOI] [PubMed] [Google Scholar]
- 109.Sottoriva A, Spiteri I, Piccirillo SG, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl Acad. Sci. USA. 2013;110(10):4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Shows that the gene expression-based molecular subtypes of glioblastoma can occur heterogeneously within the same tumor.
- 110.Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Piccirillo SG, Spiteri I, Sottoriva A, et al. Contributions to drug resistance in glioblastoma derived from malignant cells in the sub-ependymal zone. Cancer Res. 2015;75(1):194–202. doi: 10.1158/0008-5472.CAN-13-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Szerlip NJ, Pedraza A, Chakravarty D, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc. Natl Acad. Sci. USA. 2012;109(8):3041–3046. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Soeda A, Hara A, Kunisada T, Yoshimura S, Iwama T, Park DM. The evidence of glioblastoma heterogeneity. Sci. Rep. 2015;5:7979. doi: 10.1038/srep07979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Meyer M, Reimand J, Lan X, et al. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc. Natl Acad. Sci. USA. 2015;112(3):851–856. doi: 10.1073/pnas.1320611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Favero F, Mcgranahan N, Salm M, et al. Glioblastoma adaptation traced through decline of an IDH1 clonal driver and macro-evolution of a double-minute chromosome. Ann. Oncol. 2015;26(5):880–887. doi: 10.1093/annonc/mdv127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483(7387):100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 117.Lee MJ, Ye AS, Gardino AK, et al. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell. 2012;149(4):780–794. doi: 10.1016/j.cell.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Verweij J, De Jonge M, Eskens F, Sleijfer S. Moving molecular targeted drug therapy towards personalized medicine: issues related to clinical trial design. Mol. Oncol. 2012;6(2):196–203. doi: 10.1016/j.molonc.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]