Abstract
Brown adipose tissue (BAT) plays an important role in regulation of energy expenditure while adapting to a cold environment. BAT thermogenesis depends on uncoupling protein 1 (UCP1), which is expressed in the inner mitochondrial membranes of BAT. Gene expression profiles induced by cold exposure in BAT have been studied, but the metabolomic biological pathway that contributes to the activation of thermogenesis in BAT remains unclear. In this study, we comprehensively compared the relative levels of metabolites between the BAT of rats kept at room temperature (22 °C) and of those exposed to a cold temperature (4 °C) for 48 h using capillary electrophoresis (CE) time-of-flight mass spectrometry (TOFMS) and liquid chromatography (LC)-TOFMS. We identified 218 metabolites (137 cations and 81 anions) by CE-TOFMS and detected 81 metabolites (47 positive and 34 negative) by LC-TOFMS in BAT. We found that cold exposure highly influenced the BAT metabolome. We showed that the cold environment lead to lower levels of glycolysis and gluconeogenesis intermediates and higher levels of the tricarboxylic acid (TCA) cycle metabolites, fatty acids, and acyl-carnitine metabolites than control conditions in the BAT of rats. These results indicate that glycolysis and β-oxidation of fatty acids in BAT are positive biological pathways that contribute to the activation of thermogenesis by cold exposure, thereby facilitating the generation of heat by UCP1. These data provide useful information for understanding the basal metabolic functions of BAT thermogenesis in rats in response to cold exposure.
Keywords: Brown adipose tissue, Metabolomics, Cold exposure
Highlights
-
•
We found that cold exposure highly influenced the brown adipose tissue (BAT) metabolome using CE-TOFMS and LC-TOFMS.
1. Introduction
Adipose tissue is a major metabolic organ and plays a key role in energy homeostasis. Based on cell morphology and tissue function, there are two types of adipose tissue, white and brown adipose tissue (WAT and BAT, respectively) in mammals. The physiological roles of these adipose tissues are different: WAT is highly adapted to store excess energy in the form of triglycerides, whereas BAT dissipates energy to produce heat by converting glucose and fatty acids to the resulting proton-motive force. BAT thermogenesis depends on uncoupling protein 1 (UCP1), which is specifically expressed in brown fat mitochondria and responsible for the unique metabolic function of BAT. UCP1 is known to dissipate the proton gradient across the inner mitochondrial membrane, thus uncoupling the electron transfer system from adenosine triphosphate (ATP) synthesis, which in turn causes the energy to be dissipated as heat [[1], [2], [3]]. Thus, BAT is a crucial tissue for regulation of energy expenditure for adaptation to a cold environment.
To understand the mechanisms of BAT thermogenesis, we previously reported the gene expression profiles involved in energy metabolism in BAT compared to WAT in rats [[4], [5], [6]]. The transcript levels of the proteins involved in the transport and catabolism of glucose and fatty acids in BAT were elevated in response to 48 h of exposure to 4 °C. Particularly, UCP1 was expressed only in BAT, and cold exposure elevated its transcript level [4]. In addition, we reported the transcript levels of various genes involved in the activation of thermogenesis in BAT through exposure to the cold using microarray analysis and revealed that cold exposure leads to transcriptional upregulation of thermogenic genes such as sarcomeric mitochondrial creatine kinase and myoglobin expressions in BAT [7]. Consistent with our reports, a previous study demonstrated that expression of UCP1 mRNA was increased in the BAT of mice in response to 24 h of exposure to the cold compared with room temperature controls [8]. Moreover, fatty acids are one of the major energy sources in the BAT of rats in response to cold exposure [1]. The intracellular fatty acid-binding proteins (FABPs) bind long-chain fatty acids and act as fatty acid transport proteins [9]. FABPs are known to exist in eight isoforms in mammals. We previously found that the transcript level of the heart-type fatty acid-binding protein 3 (FABP3) was elevated 100-fold in BAT of rats exposed to the cold compared to those kept at room temperature [4,10,11]. Although gene expression profiles are useful to understand the mechanisms underlying the activation of thermogenesis in BAT, the products of individual protein expression on several transduction pathways remains unclear.
Integration of omics data generated by the different high-throughput technologies including genomics, transcriptomics, proteomics, and metabolomics has widely expanded the understanding of the cellular mechanisms in the systems biology field. Metabolome analysis has been recognized as a useful approach to analyze chemical processes involving low molecular weight metabolites and helped to understand pathological conditions occurring in an organism [12]. Although gene expression profiles induced by exposure to the cold in BAT are well studied [4,7,11], metabolomic biological pathway contributions to the activation of thermogenesis in BAT has not been clearly elucidated. Recently, Lu et al. showed that acute cold exposure induced many significant metabolic changes in BAT and increased lipid metabolites such as diglyceride, monoglyceride, and fatty acid levels in the BAT of mice during the early phase of cold exposure (up to 6 h) [13]. They also demonstrated that glycolysis and pentose phosphate pathway metabolites had significant changes in gene expression in BAT after 4 h of cold exposure. Moreover, it was reported that loss of the mechanistic target of the rapamycin complex 1 (mTORC1) affected glucose, lipid, and oxidative metabolism in the BAT of mice following chronic cold exposure (2 weeks) using metabolomics [14]. However, little is known about the many metabolic statuses including those of glucose, fatty acid metabolism, and tricarboxylic acid (TCA) cycle involved in BAT thermogenesis of rats in response to cold exposure for 48 h compared with the gene expression profiles. Therefore, in this study, we utilized metabolomic approaches to focus on the comprehensive metabolic changes between the BAT of rats kept at room temperature (22 °C) and those exposed to a cold temperature (4 °C) for 48 h using capillary electrophoresis (CE) time-of-flight mass spectrometry (TOFMS) and liquid chromatography (LC)-TOFMS.
2. Materials and methods
2.1. Animals and tissue sampling
Specific pathogen-free male Wistar rats (5 weeks) from Japan SLC Inc. (Hamamatsu, Japan) were used according to ethical guidelines for Animal Experiments of Tokushima University with approval of the Institutional Animal Care and Use Committee of Tokushima University (Approval number: T28–29). In this experiment, six rats were randomly assigned to two subgroups (n = 3 in each group) as follows: (1) rats kept at room temperature (22 °C) for 48 h; (2) rats kept at 4 °C for 48 h. Prior to the experiment, rats were housed in groups at room temperature for one week under a 12-h light/dark cycle and provided a normal diet and water ad libitum. All rats were euthanized by cervical dislocation. An interscapular BAT was isolated from three rats in each group, and then the samples were combined to avoid differences between individuals. BAT samples were stored at −80 °C until metabolome analysis.
2.2. Quantitative real-time polymerase chain reaction (PCR)
Total RNA was isolated from BAT using an RNeasy Lipid Mini Kit (Qiagen, Valencia, CA, USA) and cDNA was synthesized from 500 ng total RNA using PrimeScript RT Master Mix (Perfect Real Time, TaKaRa Bio, Otsu, Japan). Quantitative real-time PCR was performed using StepOne Plus (Applied Biosystems, Foster City, CA, USA). Template cDNA (5 ng/μl) was mixed with Fast SYBR® Green Master Mix (Thermo Fisher Scientific, Waltham, MA, USA), distilled water, and primers (final concentration 500 nM). The reaction was performed at 95 °C for 20 s, followed by 40 cycles at 95 °C for 3 s and 60 °C for 30 s. The following primer sets were used: UCP1 forward, 5′-GTACCCAGCTGTGCAATGAC-3′, and UCP1 reverse, 5′-GATGACGTTCCAGGATCCGA-3′; FABP3 forward, 5′-ACCAAGCCGACCACAATCAT-3′, and FABP3 reverse, 5′-TCCCACTTCTGCACATGGAC-3′; RPLP0 forward, 5′-ATTGGCTACCCGACTGTTGC-3′, and RPLP0 reverse, 5′-CCGCAAATGCAGATGGATCG-3′. The relative mRNA levels of the various genes were normalized to that of ribosomal protein lateral stalk subunit P0 (RPLP0) mRNA as an internal control.
2.3. Sample preparation
Metabolite extraction and metabolome analysis were conducted at Human Metabolome Technologies Inc. (HMT, Tsuruoka, Yamagata, Japan) as follows. For CE-TOFMS analysis, approximately 200 mg of each frozen BAT sample was plunged into 6 ml of 50% acetonitrile/Milli-Q water containing 10 μM of an internal standard solution (H3304-1002, HMT) at 0 °C. The tissue was homogenized using a homogenizer (Micro Smash MS100R, Tomy Digital Biology Co., Ltd., Tokyo, Japan) at 1500 rpm for 120 s 5 times on ice and then the homogenate was centrifuged at 2300 ×g at 4 °C for 5 min. After that, 800 μl of the upper aqueous layer was centrifugally filtered through a Millipore 5-kDa cutoff filter (UltrafreeMC-PLHCC, HMT) at 9100 ×g at 4 °C for 120 min to remove macromolecules. The filtrate was centrifugally concentrated and re-suspended in 50 μl Milli-Q water for CE-TOFMS analysis at HMT.
For LC-TOFMS analysis, approximately 50 mg of each frozen BAT sample was plunged into 500 μl of 1% formic acid/acetonitrile containing 20 μM of an internal standard solution (H3304–1002, HMT) at 0 °C. The tissue was homogenized using a homogenizer (Micro Smash MS100R) at 1500 rpm for 120 s 3 times on ice. The mixture was homogenized again after adding 167 μl Milli-Q water, and then the homogenate was centrifuged at 2300 ×g at 4 °C for 5 min. The supernatant was then mixed with 500 μl of 1% formic acid/acetonitrile and 167 μl Milli-Q water, and the solution was centrifugally filtered through 3-kDa cutoff filter (NANOCEP 3 K OMEGA, PALL Corporation, Michigan, USA) at 9100 ×g at 4 °C for 120 min to remove macromolecules, and then phospholipids were removed using a Hybrid SPE phospholipid cartridge (55261-U, Supelco, Bellefonte, PA, USA). The filtrate was desiccated and then re-suspended in 100 μl of 50% isopropanol/Milli-Q water for LC-TOFMS analysis at HMT.
2.4. CE-TOFMS measurements
Metabolome analysis was conducted using a Dual Scan package at HMT using CE-TOFMS and LC-TOFMS for ionic and non-ionic metabolites, respectively, based on previously described methods [15,16]. The CE-TOFMS analysis were carried out using an Agilent CE system (Agilent Technologies, Palo Alto, CA, USA) equipped with an Agilent 6210 TOFMS (Agilent Technologies) at a service facility at HMT. Cationic metabolites were analyzed with a fused silica capillary (50 μm internal diameter × 80 cm total length) with cationic electrophoresis buffer (H3301-1001, HMT) as the electrolyte. The sample solution was injected at a pressure of 50 mbar for 10 s. The applied voltage was set at 27 kV. Electrospray ionization-mass spectrometry (ESI-MS) was conducted in the positive-ion mode and the capillary voltages were set at 4000 V. The spectrometer was scanned from m/z 50 to 1000. Anionic metabolites were analyzed with a fused silica capillary (50 μm × 80 cm) with anionic electrophoresis buffer (H3302-1021, HMT) as the electrolyte. The sample solution was injected at a pressure of 50 mbar for 25 s. The applied voltage was set at 30 kV. ESI-MS was conducted in the negative-ion mode and the capillary voltages were set at 3500 V. The spectrometer was scanned from m/z 50 to 1000.
2.5. LC-TOFMS measurements
The LC-TOFMS analysis were carried out using an Agilent 1200 series RRLC system SL (Agilent Technologies) equipped with an Agilent 6230 TOFMS (Agilent Technologies) at a service facility at HMT. The system was run in gradient mode using an octadecylsilane column (2 × 50 mm, 2 μm) set at 40 °C. Solvent A was 0.1% formic acid containing H2O and solvent B was 0.1% formic acid and 2 mM ammonium hydrogen carbonate containing isopropanol:acetonitrile:H2O at 60:30:5; the flow rate was 0.3 ml/min. The gradient was set as follows: 1% B (0–0.5 min), increasing linearly to 100% B (from 13.5 min) and to 100% B at 20 min. MS analysis was carried out in both positive and negative-ion ESI modes of detection. The operating parameters were: drying gas (N2) flow rate, 10 L/min; drying gas temperature, 350 °C; nebulizer pressure, 40 psi; capillary voltage, 3500 V. The mass scanning range was m/z 100–1700.
2.6. Metabolome data analysis
Peaks were extracted using MasterHands, automatic integration software ver.2.17.1.11 (Keio University, Tsuruoka, Yamagata, Japan) to obtain peak information including the m/z; migration time (MT) and retention time (RT) for CE-TOFMS and LC-TOFMS measurements, respectively; and peak area. Signal peaks corresponding to isotopomers, adduct ions, and other product ions of known metabolites were excluded. The remaining peaks were annotated with putative metabolites from the HMT metabolite database based on their MT/RT and m/z values as determined by TOFMS. The tolerance range for the peak annotation was configured at ±0.5 mm for MT/RT and ±10 ppm for m/z. In addition, the areas of the annotated peaks were normalized based on internal standard levels, and then the resultant relative area values were further normalized by sample amounts to obtain a relative level for each metabolite. Detected metabolites were plotted on metabolic pathway maps using VANTED (Visualization and Analysis of Networks containing Experimental Data) software.
2.7. Statistical analysis
Quantitative real-time PCR was performed at least three times to confirm the reproducibility of the results. The results are shown as the mean ± SD of three independent rats in each group. The Student's t-test was used for statistical analysis. Statistical significance was set at P < 0.05. GraphPad Prism 6.00 software (GraphPad Software, Inc., La Jolla, CA) was used for data analysis.
3. Results and discussion
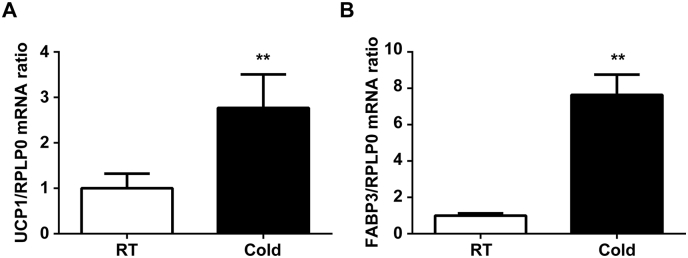
BAT plays a key role in thermogenesis in small rodents and newborn humans [1]. We examined the expression of two genes, UCP1 and FABP3, by quantitative real-time PCR to determine whether there was a significant difference in BAT from rats exposed to cold for 48 h, as we reported previously [4,10,11]. In agreement with our previous studies [4,10,11], both UCP1 and FABP3 mRNA levels were significantly increased in BAT of rats exposed to a cold temperature compared with those kept at room temperature (UCP1: 2.8-fold, **P < 0.01; FABP3: 7.6-fold, **P < 0.01, Fig. 1A and B, respectively). Therefore, we decided to use these BAT samples in the subsequent experiments.
Fig. 1.
Expression of UCP1 and FABP3 mRNAs in BAT of cold-exposed rats. Rats were maintained at 22 °C (room temperature: RT) or 4 °C (cold temperature: Cold) for 48 h. UCP1 and FABP3 mRNAs were analyzed by quantitative real-time PCR. Data are mean ± SD of three independent rats in each group. **P < 0.01, compared with room temperature.
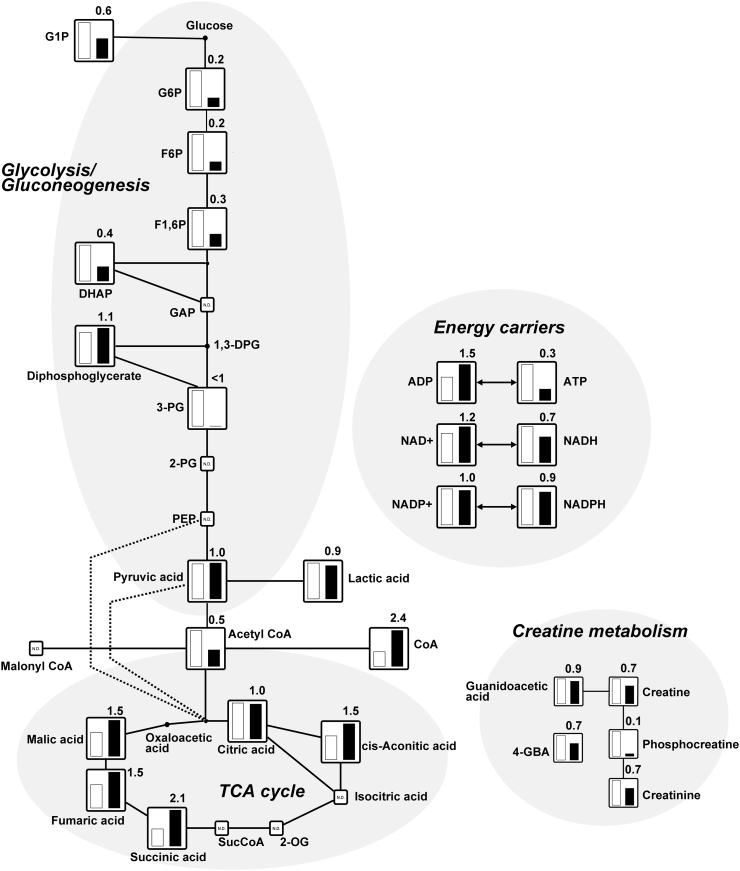
To focus on the comprehensive metabolic changes on BAT samples isolated from rats maintained at room temperature and exposed to a cold temperature for 48 h, we performed metabolomics analyses using CE-TOFMS and LC-TOFMS. In our analysis, CE-TOFMS revealed 218 metabolites (137 cations and 81 anions), and LC-TOFMS detected 81 metabolites (47 positive and 34 negative). We comprehensively compared the relative levels of metabolites between the BAT of rats kept at room temperature and those exposed to a cold temperature. Fig. 2 shows that the relative levels of metabolites in the glycolysis/gluconeogenesis, TCA cycle, energy carriers, and creatine metabolism were illustrated on a metabolic pathway map. Interestingly, our results show that the response of BAT to cold exposure was reduced in six key metabolites for glycolysis/gluconeogenesis compared to the BAT of rats kept at room temperature. Particularly, we observed that glucose 6-phosphate (G6P), fructose 6-phosphate (F6P), and fructose 1,6-diphosphate (F1,6P) were reduced to 0.2-fold, 0.2-fold, and 0.3-fold, respectively, in the BAT of rats exposed to the cold as compared to BAT of rats kept at room temperature. The level of glycolytic intermediate dihydroxyacetone-phosphate (DHAP) was also reduced to 0.4-fold in response to cold exposure. Glucose is imported into the cell by glucose transporter (GLUT). In our previous study, we showed that the transcript levels of GLUT isoforms, including GLUT1, GLUT3, and GLUT4, were significantly elevated in the BAT of rats exposed to the cold compared to those maintained at room temperature [4]. In addition, the transcript levels of hexokinase (HK) isozymes, which are enzymes of glucose metabolism that phosphorylate glucose to G6P, were also increased in response to cold exposure. These results suggest that BAT utilizes more glucose as an energy source in rats exposed to a cold temperature than those kept at room temperature. Cold exposure likely increases the regulation of glucose metabolism in BAT. Therefore, cold exposure is associated with increased glucose uptake in BAT to act as an energy source in heat production. Although the relative levels of intermediates of glucose metabolism in BAT exposed to cold were relatively lower than those in the BAT of rats kept at room temperature, pyruvic acid which is the end-product of glycolysis was not altered by cold exposure. In addition, lactic acid, which is derived from the conversion of pyruvic acid produced by glycolysis, was also not altered by cold exposure. Acetyl CoA produced either through glucose oxidation or fatty acid β-oxidation is used at the initial step of the TCA cycle in mitochondria. We observed that acetyl CoA levels reduced to 0.5-fold in response to cold exposure, suggesting that acetyl CoA is used to promote cellular respiration.
Fig. 2.
Metabolites related to glycolysis/gluconeogenesis, TCA cycle, energy carriers, and creatine metabolism (see Supplementary Table 1). The relative levels of the annotated metabolites are represented as bar graphs (open column, BAT of rats kept at room temperature: closed column, the BAT of rats exposed to the cold temperature). N.D. indicates “not detected.” Height of column with large quantity is set to 100%, and that for the smaller quantity is shown proportionally. Numbers on the columns represent the relative levels of metabolites from the BAT of rats exposed to the cold compared to those from the BAT of rats kept at room temperature. <1 indicates that the value for the BAT of rats exposed to cold is below the detection limit. The abbreviations are as follows: G1P, glucose 1-phosphate; G6P, glucose 6-phosphate; F6P, fructose 6-phosphate; F1,6P, fructose 1,6-diphosphate; DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde 3-phosphate; 1,3-DPG, 1,3-diphosphoglycerate; 3-PG, 3-phosphoglycerate; 2-PG, 2-phosphoglycerate; PEP, phosphoenolpyruvate; ADP, adenosine diphosphate; ATP, adenosine triphosphate; NAD, nicotinamide adenine dinucleotide; NADH, nicotinamide adenine dinucleotide phosphate; NADP, nicotinamide adenine dinucleotide phosphate; NADPH, reduced nicotinamide adenine dinucleotide phosphate; and 4-GBA, 4-guanidinobutyric acid.
Metabolites related to glycolysis/gluconeogenesis, TCA cycle, energy carriers, and creatine metabolism (see Supplementary Table 1). The relative levels of the annotated metabolites are represented as bar graphs (open column, BAT of rats kept at room temperature: closed column, the BAT of rats exposed to the cold temperature). N.D. indicates “not detected.” Height of column with large quantity is set to 100%, and that for the smaller quantity is shown proportionally. Numbers on the columns represent the relative levels of metabolites from the BAT of rats exposed to the cold compared to those from the BAT of rats kept at room temperature. <1 indicates that the value for the BAT of rats exposed to cold is below the detection limit. The abbreviations are as follows: G1P, glucose 1-phosphate; G6P, glucose 6-phosphate; F6P, fructose 6-phosphate; F1,6P, fructose 1,6-diphosphate; DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde 3-phosphate; 1,3-DPG, 1,3-diphosphoglycerate; 3-PG, 3-phosphoglycerate; 2-PG, 2-phosphoglycerate; PEP, phosphoenolpyruvate; ADP, adenosine diphosphate; ATP, adenosine triphosphate; NAD, nicotinamide adenine dinucleotide; NADH, nicotinamide adenine dinucleotide phosphate; NADP, nicotinamide adenine dinucleotide phosphate; NADPH, reduced nicotinamide adenine dinucleotide phosphate; and 4-GBA, 4-guanidinobutyric acid.
The TCA cycle is a central route for energy metabolism. The cycle is composed of a series of biochemical reactions occurring in the mitochondrial matrix [17]. We observed elevated the relative levels of metabolites of the TCA cycle, including cis-Aconitic acid, succinic acid, fumaric acid, and malic acid, which were higher in the BAT of rats exposed to the cold compared to those kept at room temperature (1.5-fold, 2.1-fold, 1.5-fold, and 1.5-fold, respectively). In our previous study, three subunits (α, β, and γ) of NAD+-dependent isocitrate dehydrogenase [EC 1.1.1.41], which catalyzes the conversion of isocitric acid to 2-oxoglutaric acid in the TCA cycle, were expressed most significantly in the BAT of rats [18], whereas both isocitric acid and 2-oxoglutaric acid were not detected in this study. These results suggest that the pathways of glycolysis and β-oxidation of fatty acids are activated in BAT exposed to a cold temperature.
ATP, which is the main cellular energy source, is produced primarily from glycolysis and mitochondrial respiration. ATP degrades into adenosine diphosphate (ADP) and adenosine monophosphate (AMP) by hydrolysis. The relative levels of ADP (1.5-fold) and AMP (5.5-fold, data not shown) were higher in BAT exposed to the cold temperature compared to those kept at room temperature, while the ratio of ATP was markedly reduced to 0.3-fold. BAT has a relatively low capacity for ATP synthesis as compared to their respiratory potential [1]. These results suggest that most of the energy is dissipated as heat rather than being converted to ATP in BAT exposed to cold temperature. Nicotinamide adenine dinucleotide (NAD) is an important coenzyme in the cell. The oxidized form of the electron carrier nicotinamide adenine dinucleotide (NAD+) is reduced to NADH during glycolysis, pyruvate oxidation, the TCA cycle, and β-oxidation of fatty acids. Pyruvic acid conversion into lactic acid reoxidizes NADH into NAD+, which is needed for glycolytic pathway maintenance. The relative levels of NAD+ was slightly higher in BAT exposed to cold temperatures (1.2-fold), whereas the NADH ratio was lower (0.7-fold). Nicotinamide adenine dinucleotide phosphate (reduced form; NADPH) and NADP+ (oxidized form) were not altered by cold exposure. BAT likely has a higher capacity for NAD+ biosynthesis with cold exposure.
The creatine/phosphocreatine pathway also plays a central role in energy metabolism. Creatine and its associated enzyme creatine kinase [EC 2.7.3.2] facilitate the shuttling of high energy phosphates in the form of phosphocreatine between the sites of ATP generation, i.e. mitochondrial oxidative phosphorylation and glycolysis, and compartmentalized ATP consumption [19]. We previously reported that the transcript level of sarcomeric mitochondrial creatine kinase was markedly up-regulated by 12.5-fold in BAT exposed to a cold temperature compared with those kept at room temperature using microarray analysis and revealed that three creatine kinase isozymes (cytoplasmic brain type, cytoplasmic muscle type, and sarcomeric mitochondrial type) were upregulated in BAT exposed to the cold [7]. In this study, the relative levels of creatine were reduced to 0.7-fold in BAT exposed to cold. Moreover, the relative levels of phosphocreatine were markedly reduced to 0.1-fold, and creatinine was reduced to 0.7-fold. These results suggest that the lower levels of metabolites associated in creatine metabolism of BAT exposed to cold temperatures may be due to ATP depletion.
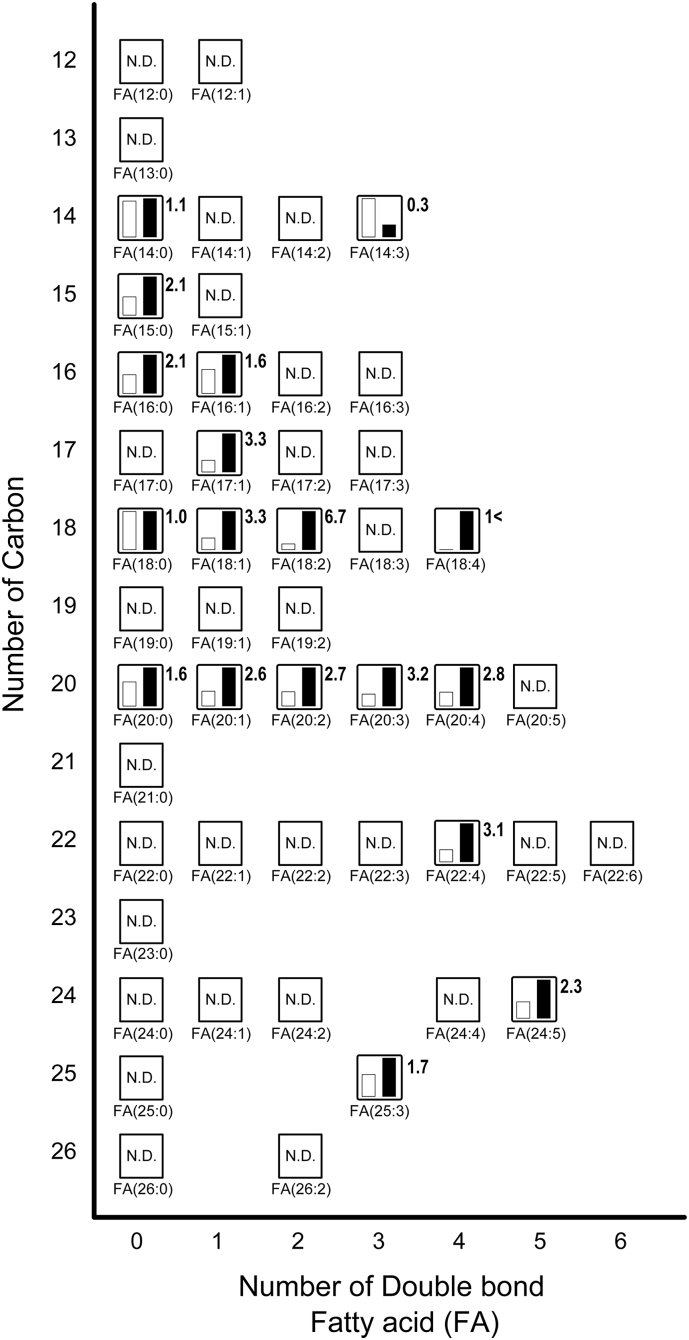
Next, we focused on the comparative relative levels of metabolites to fatty acids and acyl-carnitine in Fig. 3, Fig. 4, respectively. Fatty acids play an important role for thermogenesis in the BAT of rats in response to cold exposure [1]. In addition, fatty acids are known to activate UCP1 function in BAT [1]. Oxidizing fatty acids by uncoupling the proton gradient produces heat instead of ATP in mitochondria, thus contributing to thermogenesis [20]. In this study, we confirmed that the expression of UCP1 mRNA was increased by cold exposure (Fig. 1A). Free fatty acids are produced when triacylglycerols are converted enzymatically to unsaturated and saturated fatty acids. The released free fatty acids are transported to the mitochondria following their conversion to fatty acyl-CoAs by long chain acyl-CoA synthetase 1 [21]. As shown in Fig. 3, various long chain fatty acids in BAT were increased in response to cold exposure. Fatty acids derived from intracellular triacylglycerol hydrolysis are essential for BAT thermogenesis as the only activators of UCP1. The most induced fatty acid in BAT by cold exposure was FA(18:2) (known as linoleic acid), which was elevated by 6.7-fold compared to the BAT of rats kept at room temperature. In a group of saturated fatty acids, the levels of FA(15:0) (known as pentadecanoic acid) and FA(16:0) (known as palmitic acid) in BAT exposed to the cold were both increased by 2.1-fold. It is reported that BAT exhibited higher levels of saturated triacylglycerol compared to other tissues in mice such as the brain, liver, and kidneys through the mass spectrometry survey of lipid abundance [22]. Our results indicated that induced saturated fatty acids may be because of increased levels of saturated triacylglycerol in BAT exposed to cold temperatures. In addition, Lu et al. reported that BAT exhibited increased lipid metabolites including fatty acids levels and that saturated fatty acids were significantly increased rather than unsaturated fatty acids after only 2 h of cold exposure in BAT [13]. However, under our experimental conditions, the relative levels of unsaturated fatty acids in the BAT exposed to the cold exhibited a greater increase than those of saturated fatty acids. For example, the relative levels of FA(17:1), FA(18:1), FA(20:3), and FA(22:4) increased >3-fold in BAT exposed to cold temperatures. We observed that only FA(14:3) reduced to 0.3-fold while others increased in a group of identified unsaturated fatty acids after 48 h of cold exposure in BAT. In contrast, it was reported that acute cold exposure increased the number of down-regulated unsaturated fatty acids in BAT rather than those of up-regulated unsaturated fatty acids [13]. In our previous study, the transcript levels of FABP3, which is a member of the fatty acid binding protein family, was dramatically enhanced in response to cold exposure [4,10,11]. In this study, we confirmed that the expression of FABP3 mRNA was significantly increased by cold exposure (Fig. 1B). FABP3 reversibly binds to hydrophobic ligands such as saturated and unsaturated long-chain fatty acids with high affinity [23]. FABP3 contributes to the transport and delivery of fatty acids from the cytoplasm to the mitochondria, where β-oxidation occurs in BAT. These results suggest that FABPs are important molecules relying on fatty acid uptake for metabolism in BAT, and increasing intracellular fatty acids availability by cold exposure causes the activation of β-oxidation leading to the production of heat by UCP1 in mitochondria.
Fig. 3.
Metabolites related to fatty acids (see Supplementary Table 2). The relative levels of annotated metabolites are represented as bar graphs (open column, the BAT of rats kept at room temperature: closed column, the BAT of rats exposed to the cold temperature). N.D. indicates ‘not detected.’ Height of column with large quantity is set to 100%, and that for the smaller quantity is shown proportionally. Numbers beside the columns represent the relative levels of metabolites from the BAT of rats exposed to the cold to those from the BAT of rats kept at room temperature. 1< indicates that the value for the BAT of rats kept at room temperature is below the detection limit.
Metabolites related to fatty acids (see Supplementary Table 2). The relative levels of annotated metabolites are represented as bar graphs (open column, the BAT of rats kept at room temperature: closed column, the BAT of rats exposed to the cold temperature). N.D. indicates ‘not detected.’ Height of column with large quantity is set to 100%, and that for the smaller quantity is shown proportionally. Numbers beside the columns represent the relative levels of metabolites from the BAT of rats exposed to the cold to those from the BAT of rats kept at room temperature. 1< indicates that the value for the BAT of rats kept at room temperature is below the detection limit.
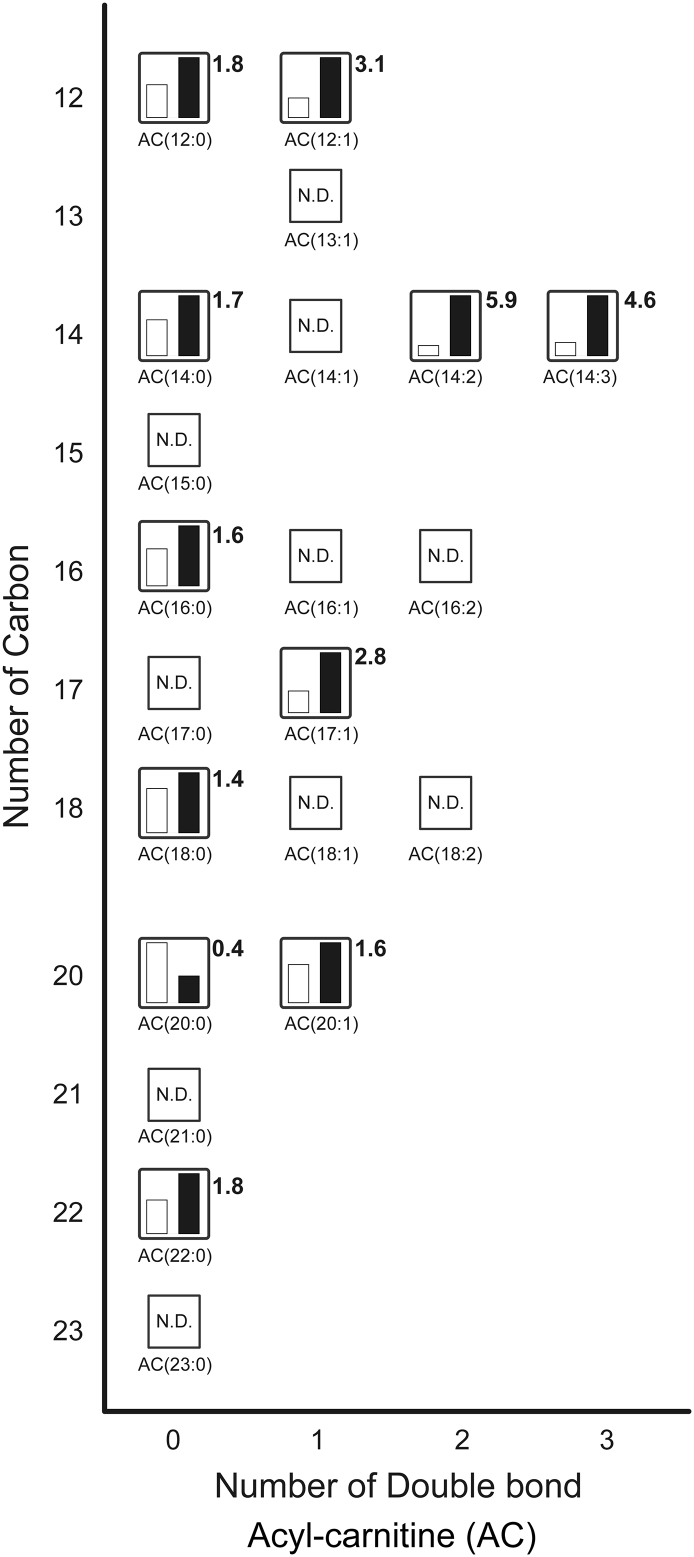
Fig. 4.
Metabolites related to acyl-carnitine (see Supplementary Table 3). The relative levels of annotated metabolites are represented as bar graphs (open column, the BAT of rats kept at room temperature: closed column, the BAT of rats exposed to the cold temperature). N.D. indicates ‘not detected.’ Height of column with large quantity is set to 100%, and that for the smaller quantity is shown proportionally. Numbers beside the columns represent the relative levels of metabolites from the BAT of rats exposed to the cold to those from the BAT of rats kept at room temperature.
Metabolites related to acyl-carnitine (see Supplementary Table 3). The relative levels of annotated metabolites are represented as bar graphs (open column, the BAT of rats kept at room temperature: closed column, the BAT of rats exposed to the cold temperature). N.D. indicates ‘not detected.’ Height of column with large quantity is set to 100%, and that for the smaller quantity is shown proportionally. Numbers beside the columns represent the relative levels of metabolites from the BAT of rats exposed to the cold to those from the BAT of rats kept at room temperature.
Fig. 4 shows acyl-carnitine (AC) metabolites as measured by LC-TOFMS. The relative levels of AC(12:1), AC(14:2), AC(14:3), and AC(17:1) were elevated by 3.1-fold, 5.9-fold, 4.6-fold, and 2.8-fold, respectively, in BAT of cold exposed rats. We observed that cold exposure reduced only AC(20:0) to 0.4-fold among the detected metabolites. Long chain acyl-carnitines act as important carriers during fatty acid β-oxidation. First, long chain fatty acids enter the mitochondria via carnitine palmitoyltransferase 1 (CPT1) activity. CPT1 is the mitochondrial enzyme that is located on the outer mitochondrial membrane and catalyzes the first and essential step in the β-oxidation of long chain fatty acids. Fatty acids are converted into acyl-CoA by acyl-CoA synthase. Then the acyl-CoA is converted into acyl-carnitine by CPT1. Acyl-carnitine cross the mitochondrial membrane through the carnitine transporter and is converted back into acyl-CoA by CPT2. This is followed by β-oxidation to supply energy. Acyl-carnitine results from incomplete fatty acid β-oxidation [20,24,25]. In our previous study, we found that the transcript levels of proteins involved in the import of acyl-CoA into the mitochondria, such as CPT1b, carnitine carrier, and CPT2, were markedly elevated in the BAT of rats exposed to the cold [4]. These elevated protein levels may contribute to the increase in the number of acyl-carnitines in mitochondria. β-oxidation in BAT is primarily induced by cold exposure. Taken together, these results suggest that accumulation of intracellular fatty acids and enhanced CPT1 activity increased the number of acyl-carnitines, resulting in UCP1 activation to produce heat in the BAT of cold exposed rats.
In this study, comprehensive metabolome analysis of BAT was performed using CE-TOFMS and LC-TOFMS. We found that many metabolites involved in glucose and fatty acid metabolism showed differences between the BAT of rats kept at room temperature and those exposed to cold temperatures. Particularly, the cold environment caused lower levels of glycolysis/gluconeogenesis intermediates and higher levels of the TCA cycle, fatty acid, and acyl-carnitine metabolites than normal conditions in the BAT of rats. Our results indicate that β-oxidation is a positive biological pathway that contributes to the activation of thermogenesis in BAT by cold exposure, thereby facilitating the generation of heat by UCP1. Although further studies are required to clearly identify their roles, these results provide useful information for understanding the basal metabolic functions in BAT thermogenesis of rats in response to exposure to cold.
The following are the supplementary data related to this article.
Metabolites shown in Fig. 2
Metabolites shown in Fig. 3
Metabolites shown in Fig. 4
Funding sources
This work was supported in part by JSPS KAKENHI Grant Number JP17K08274 (Grant-in-Aid for Scientific Research (C), YS) and JP17K17352 (Grant-in-Aid for Young Scientists (B), YH).
Conflict of interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Acknowledgment
This study was supported by the Support Center for Advanced Medical Sciences, Institute of Biomedical Sciences, Tokushima University Graduate School.
References
- 1.Nicholls D.G., Locke R.M. Thermogenic mechanisms in brown fat. Physiol. Rev. 1984;64:1–64. doi: 10.1152/physrev.1984.64.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Lowell B.B., Spiegelman B.M. Towards a molecular understanding of adaptive thermogenesis. Nature. 2000;404:652–660. doi: 10.1038/35007527. [DOI] [PubMed] [Google Scholar]
- 3.Ricquier D., Bouillaud F. The uncoupling protein homologues: UCP1, UCP2, UCP3, StUCP and AtUCP. Biochem. J. 2000;345(Pt 2):161–179. [PMC free article] [PubMed] [Google Scholar]
- 4.Daikoku T., Shinohara Y., Shima A., Yamazaki N., Terada H. Specific elevation of transcript levels of particular protein subtypes induced in brown adipose tissue by cold exposure. Biochim. Biophys. Acta. 2000;1457:263–272. doi: 10.1016/s0005-2728(00)00107-9. [DOI] [PubMed] [Google Scholar]
- 5.Kajimoto K., Daikoku T., Kita F., Yamazaki N., Kataoka M., Baba Y., Terada H., Shinohara Y. PCR-select subtraction for characterization of messages differentially expressed in brown compared with white adipose tissue. Mol. Genet. Metab. 2003;80:255–261. doi: 10.1016/j.ymgme.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Unami A., Shinohara Y., Kajimoto K., Baba Y. Comparison of gene expression profiles between white and brown adipose tissues of rat by microarray analysis. Biochem. Pharmacol. 2004;67:555–564. doi: 10.1016/j.bcp.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe M., Yamamoto T., Kakuhata R., Okada N., Kajimoto K., Yamazaki N., Kataoka M., Baba Y., Tamaki T., Shinohara Y. Synchronized changes in transcript levels of genes activating cold exposure-induced thermogenesis in brown adipose tissue of experimental animals. Biochim. Biophys. Acta. 2008;1777:104–112. doi: 10.1016/j.bbabio.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 8.Shore A.M., Karamitri A., Kemp P., Speakman J.R., Graham N.S., Lomax M.A. Cold-induced changes in gene expression in brown adipose tissue, white adipose tissue and liver. PLoS One. 2013;8 doi: 10.1371/journal.pone.0068933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Storch J., Thumser A.E. The fatty acid transport function of fatty acid-binding proteins. Biochim. Biophys. Acta. 2000;1486:28–44. doi: 10.1016/s1388-1981(00)00046-9. [DOI] [PubMed] [Google Scholar]
- 10.Daikoku T., Shinohara Y., Shima A., Yamazaki N., Terada H. Dramatic enhancement of the specific expression of the heart-type fatty acid binding protein in rat brown adipose tissue by cold exposure. FEBS Lett. 1997;410:383–386. doi: 10.1016/s0014-5793(97)00619-4. [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto T., Yamamoto A., Watanabe M., Kataoka M., Terada H., Shinohara Y. Quantitative evaluation of the effects of cold exposure of rats on the expression levels of ten FABP isoforms in brown adipose tissue. Biotechnol. Lett. 2011;33:237–242. doi: 10.1007/s10529-010-0444-0. [DOI] [PubMed] [Google Scholar]
- 12.Bujak R., Struck-Lewicka W., Markuszewski M.J., Kaliszan R. Metabolomics for laboratory diagnostics. J. Pharm. Biomed. Anal. 2015;113:108–120. doi: 10.1016/j.jpba.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 13.Lu X., Solmonson A., Lodi A., Nowinski S.M., Sentandreu E., Riley C.L., Mills E.M., Tiziani S. The early metabolomic response of adipose tissue during acute cold exposure in mice. Sci. Rep. 2017;7:3455. doi: 10.1038/s41598-017-03108-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Labbe S.M., Mouchiroud M., Caron A., Secco B., Freinkman E., Lamoureux G., Gelinas Y., Lecomte R., Bosse Y., Chimin P., Festuccia W.T., Rochard D., Laplante M. mTORC1 is required for brown adipose tissue recruitment and metabolic adaptation to cold. Sci. Rep. 2016;6 doi: 10.1038/srep37223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohashi Y., Hirayama A., Ishikawa T., Nakamura S., Shimizu K., Ueno Y., Tomita M., Soga T. Depiction of metabolome changes in histidine-starved Escherichia coli by CE-TOFMS. Mol. BioSyst. 2008;4:135–147. doi: 10.1039/b714176a. [DOI] [PubMed] [Google Scholar]
- 16.Ooga T., Sato H., Nagashima A., Sasaki K., Tomita M., Soga T., Ohashi Y. Metabolomic anatomy of an animal model revealing homeostatic imbalances in dyslipidaemia. Mol. BioSyst. 2011;7:1217–1223. doi: 10.1039/c0mb00141d. [DOI] [PubMed] [Google Scholar]
- 17.Anderson N.M., Mucka P., Kern J.G., Feng H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell. Jul 26 2017 doi: 10.1007/s13238-017-0451-1. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shinohara Y., Daikoku T., Kajimoto K., Shima A., Yamazaki N., Terada H. Expression of NAD(+)-dependent isocitrate dehydrogenase in brown adipose tissue. Biochem. Biophys. Res. Commun. 2001;281:634–638. doi: 10.1006/bbrc.2001.4351. [DOI] [PubMed] [Google Scholar]
- 19.Kitzenberg D., Colgan S.P., Glover L.E. Creatine kinase in ischemic and inflammatory disorders. Clin. Transl. Med. 2016;5:31. doi: 10.1186/s40169-016-0114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 21.Ellis J.M., Frahm J.L., Li L.O., Coleman R.A. Acyl-coenzyme A synthetases in metabolic control. Curr. Opin. Lipidol. 2010;21:212–217. doi: 10.1097/mol.0b013e32833884bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jain M., Ngoy S., Sheth S.A., Swanson R.A., Rhee E.P., Liao R., Clish C.B., Mootha V.K., Nilsson R. A systematic survey of lipids across mouse tissues. Am. J. Physiol. Endocrinol. Metab. 2014;306:E854–68. doi: 10.1152/ajpendo.00371.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furuhashi M., Hotamisligil G.S. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 2008;7:489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foster D.W. The role of the carnitine system in human metabolism. Ann. N. Y. Acad. Sci. 2004;1033:1–16. doi: 10.1196/annals.1320.001. [DOI] [PubMed] [Google Scholar]
- 25.Houten S.M., Wanders R.J. A general introduction to the biochemistry of mitochondrial fatty acid beta-oxidation. J. Inherit. Metab. Dis. 2010;33:469–477. doi: 10.1007/s10545-010-9061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Metabolites shown in Fig. 2
Metabolites shown in Fig. 3
Metabolites shown in Fig. 4