Abstract
Human papillomaviruses (HPVs) co-evolve slowly with the human host and each HPV genotype displays epithelial tropisms. We assessed the evolution of intra HPV genotype variants within samples, and their association to anogenital site, cervical cytology and HIV status. Variability in the L1 gene of 35 HPV genotypes was characterized phylogenetically using maximum likelihood, and portrayed by phenotype. Up to a thousand unique variants were identified within individual samples. In-depth analyses of the most prevalent genotypes, HPV16, HPV18 and HPV52, revealed that the high diversity was dominated by a few abundant variants. This suggests high intra-host mutation rates. Clades of HPV16, HPV18 and HPV52 were associated to anatomical site and HIV co-infection. Particularly, we observed that one HPV16 clade was specific to vaginal cells and one HPV52 clade was specific to anal cells. One major HPV52 clade, present in several samples, was strongly associated with cervical neoplasia. Overall, our data suggest that tissue tropism and HIV immunosuppression are strong shapers of HPV evolution.
Keywords: HPV variability, HPV phylogenetics, Anogenital, Tissue tropism, HIV
Graphical abstract
Highlights
-
•
>1000 unique intra HPV type variants were identified within individual samples.
-
•
Distinctly different HPV variants are present in anal and vaginal samples.
-
•
Lineage specific HPV variant load is associated to HIV immunosuppression.
-
•
HPV variant evolution was associated with cervical cytology outcome, implying an influence of evolution on carcinogenesis.
1. Introduction
Papillomaviruses are extremely common and may cause anogenital warts and lesions in a range of different mammals [1]. In contrast to the relatively short evolutionary history and high mutation rate of the human immunodeficiency virus (HIV), the genetic sequence of HPVs and other papillomaviruses suggests a long history of host-linked evolution and adaptive radiation events [2]. HPVs may in rare cases cause cancers, however, most infections are cleared within less than two years, given a functional immune system [3]. Different HPV types display preference for different tissues, known as tropism [4]. Today, more than 300 papillomavirus types are fully sequenced of which 206 are classified as human associated genotypes [5]. Approximately sixty HPV types are known to infect the mucosal epithelia, mostly in the anogenital region, and at least 13 of these have oncogenic potential [6]. Persistent infection with one of these oncogenic HPVs is the etiological agent for several anogenital cancers and is aggravated by immunosuppression [7] and viral evolution [8], [9]. HPV DNA has been detected in cancers of different anogenital sites, with rates of cervix (98%), anus (88%), vagina (74%), penis (33%) and vulva (29%) [8], [10].
HPV genotypes are taxonomically classified by the > 10% genetic divergence of their respective L1 open reading frame (ORF) gene, encoding the viral capsid. HPV sub-types differ by 2–10% and variants display genetic differences in L1 by less than 2% [11]. In the context of our study, we have also used the term variant in all instances where unique sequences of a genotype are identified. HPV16 and HPV18 are the most common oncogenic genotypes worldwide [12], although type distribution of specific HPV genotypes is dependent on geographic location [6]. Notably, HPV52 shows increased prevalence in Asia and some parts of Africa. Certain geographic regions such as Zambia, Zimbabwe and China have recorded HPV58 and HPV52 as the most common types [13], [14], [15]. Developing countries make up 85% of the global burden of cervical cancer [16]. Still, there is paucity of data on the repertoire of oncogenic HPVs circulating in these regions [17]. A worldwide study on genotype variant level confirmed their dependence on geographic location [8]. Based on the risk of developing invasive cervical cancer, individual HPV16 variants show up to ten-fold differences [18]. For HPV52, a 7-fold difference between variants has been shown [19]. In Zimbabwe, where HIV prevalence is 14.5% among 15–49 year olds, cervical cancer makes up 34.6% of all cancers in black women [20], emphasizing the need for studies elucidating factors and conditions favouring HPV related carcinogenesis in the context of immunosuppression. Diversity of HPV is likely a distinctive feature of HIV co-infected individuals [21].
Next-generation sequencing (NGS) technology generates high-resolution data allowing in-depth characterization of HPV genetic variability [22], [23], [24], [25], [26] and associations between evolution and carcinogenesis. The aim of this study was to detect HPV intra-genotype variants in cervico-vaginal and anal swabs provided by women reporting for routine cervical cancer screening in Zimbabwe. HPV intra-genotype variation may elucidate evolution shaped by tissue tropism, cervical neoplasm and HIV status.
2. Materials and methods
2.1. Study population and sample size
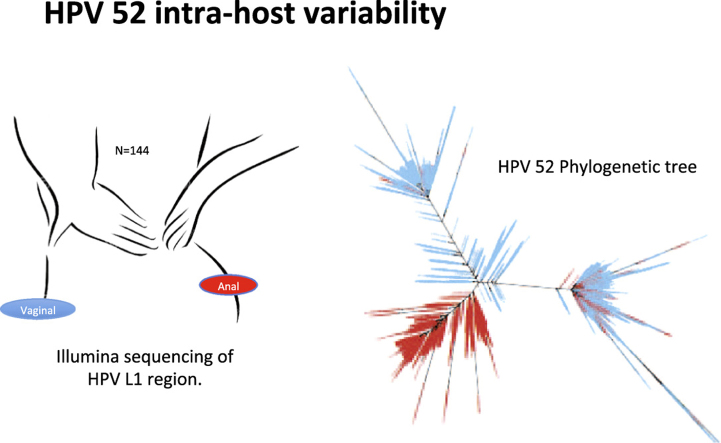
This cross-sectional study included women visiting a Visual Inspection with Acetic-acid (VIA) clinic within Parirenyatwa hospital in Harare, Zimbabwe. All participants were women from the general population, reporting for routine cervical cancer screening. A detailed description of the study population (N = 144) is reported previously [15].
2.2. Ethical approval
Ethical approval was obtained from The Joint Parirenyatwa hospital and College of Health Sciences Research Ethics Committee (reference: JREC210/14) and Medical Research Council of Zimbabwe (reference: MRCZ/A/1911). Written informed consent, in English or Shona, was obtained from the women who were ≥18 years, sexually active and had no history of a total abdominal hysterectomy.
2.3. Specimen collection
Enrolment period was from February to April 2015. A research nurse administered a structured questionnaire to capture demographics and survey data. On recruitment, HIV testing and counselling were offered to participants who did not have documented HIV status. For HPV investigation, two swabs were requested from each woman, one self-collected vaginal swab (VS) and one clinician-collected anal swab (CCAS). The women collected VS in a toilet within the clinic facility after the nurse explained the procedure and CCAS was collected in the examination room. The nurse gently inserted the swab into the anal canal until the shaft could not move further and rotated it for 10–30 s. All swabs were Dacron™ tipped with a firm plastic shaft and were immediately broken into a cryotube soon after collection and were stored in 500 μL lysis buffer from bioMerieux (containing guanadine thiocyanate) at −80 °C until analysed. After both swabs were collected, the research nurse inserted a speculum. A cytobrush was used to collect cells from the transformation zone of the cervix. A monolayer smear was made on a frosted glass slide and cytospray was used immediately to fix the slide. Lastly, VIA was then performed. Acetic acid was used to wipe the cervix. White precipitation was recorded as positive and a pink translucent colour was negative. All participants positive for VIA were immediately referred to colposcopy for treatment with loop electrosurgical excision procedure (LEEP) or cryotherapy.
2.4. Laboratory methods
DNA was extracted using conventional ammonium acetate method and HPV detection and genotyping by NGS on the MiSeq platform was performed as described previously [15]. The HPV amplicon sequencing was performed at the Norwegian HPV Reference Laboratory. The detection and genotyping of HPV was done by amplification of a 450 bp L1 gene fragment covering in all high risk HPV genotypes and a large fraction of low risk HPV genotypes using the PGMY09/11 primers, followed by sequencing using the MiSeq platform [27]. PGMY amplicons with Illumina-tailed adaptors were generated in 20 μL volumes using Phusion Master Mix (Thermo Fischer Scientific, MA), 0.1 μM of each primer in the PGMY09/11 sets and 5 μL sample DNA template under the following thermocycling conditions: 1 cycle of 98 °C for 30 s, 40 cycles of 98 °C for 10 s, 56 °C for 30 s, and 72 °C for 15 s, before a final extension at 72 °C for 10 min. Amplicons were cleaned up using modified PERFORMA® DTR V3 96-Well Short Plates and Quick- StepTM2 96-Well PCR Purification Kit (Edge Biosystems, MD) protocols. Amplicons were diluted 1:100 before being used as templates in indexing PCR in 20 μL volumes using Phusion Master Mix, 0.5 μM each index and 1 μL template under the following conditions: 1 cycle of 98 °C for 2 min, 12 cycles of 98 °C for 20 s, 65 °C for 30 s, and 72 °C for 30 s, and a final extension of 72 °C for 5 min. The resulting amplicon libraries were pooled using 5 μL from each library and cleaned up using 0.8× AMPure XP (Agencourt Beckman Coulter, CA) and 1 μL was run on the Bioanalyzer (Agilent Technologies, CA) for quality control and quantitation using the High Sensitivity DNA Analysis kit. The libraries containing PGMY amplicons were sequenced on the MiSeq platform (Illumina, CA) using V3 chemistry and 2 × 300 bp reads.
2.5. Cervical cytology
Cervical smears were collected on frosted glass slides, and were stained using the Papanicolaou procedure. The Bethesda system was used for classification [28]. A cytotechnologist signed out smears negative for intraepithelial lesion or malignancy (NILM). Smears with a lesion of any level were reviewed and signed out by a pathologist, who also reviewed 10% of all negative slides. Specimens without endocervical cells were deemed as unsatisfactory for evaluation. ‘Low-grade’ signified any result that was either low-grade squamous intraepithelial lesion (LGSIL) or atypical squamous cells of undetermined significance (ASC-US). ‘High-grade’ represented any result that was either high-grade squamous intra-epithelial lesions (HGSIL) or atypical squamous cells, cannot exclude HGSIL (ASC-H). ‘Unsatisfactory’ results signified that the slide was not satisfactory for evaluation i.e. had no distinct endothelial cells, was poorly stained and/or smear was poorly collected.
2.6. HIV testing
A serial algorithm was followed. Alere Determine™ HIV-1/2 rapid test (Alere North America, LLC, Orlando, FL) was used for screening and First Response™ HIV 1–2.0 Rapid test (Premier Medical Corporation Ltd., Kachigam, India) for confirming positive results. Both tests used finger-prick blood, a dried blood spot was collected and stored in case a tiebreaker test was required. Discordant results were tested using a GS HIV Combo Ag/Ab Enzyme Immuno-Assay kit (Bio-Rad Laboratories, California) using blood.
2.7. Phylogenetic analyses
An automated pipeline for HPV phylogeny was designed that can handle hundreds of thousand sequences generated. It was designed to reduce inaccuracies due to PCR artefacts and sequencing errors [29] to a minimum by only considering variants appearing multiple times in multiple samples and using stringent filtering settings.
Adaptors, primers, and low quality bases were removed from the raw sequence reads by Nesoni clip (version 0.115) [30]. Quality scores less than 20 and sequences shorter than 50 bp were removed. The PCR forward and reverse reads were merged using Pear version 0.9.6 [31] with a minimum possible length of 100. The merged sequences were clustered using Swarm [32] (v.2.0). Default parameter (d = 1) was used, which allows grouping of amplicons that only differ with maximum one nucleotide. The abundance level is activated and the built-in OUT refinement are deactivated. The seed sequence from each cluster was assigned to HPV types using BLASTN [33] (v.2.2.31+) and a self-combined reference file containing 1297 papillomavirus genome sequences with length of 5000 bp or more. The HPV reference types were included. Clustal Omega [34] (v.1.2.1) was used to do a multiple sequence alignment (MSA) for each HPV type. Unique sequences were excluded as they might be due to PCR artefacts or sequencing errors. The resulting (MSA) has been trimmed, removing positions in the start and in the end of the alignment containing more than 90 percent gaps. In addition, sequences containing more than 10 percent gaps were removed to increase the quality of the alignment. A sample was deemed HPV positive for a genotype if it had >99 sequence counts assigned to the genotype. However, all sequences from all samples were included in the phylogenetic analyses.
The phylogenetic trees were constructed using RAxML [35] (v. 8.2.0), a Maximum Likelihood method. The sequences were not mapped to a reference tree; however, the trees were built from the sequences available to get the unsupervised relationship between the sequences. RAxML produced a reduced file, containing one representative of each sequence. The model GTRCAT was used with the option f –a, to turn on rapid bootstrapping and search for the best-scoring tree in one program run. We have used four multiple runs to search for the best tree (-#). Both the bootstrap rapid random number of seed (-x) and the parsimony random number of seed (-p) was set to 000. Trees are presented in ‘Fan’ mode to enable a clear representation of the phylogenetic variability. Since only parts of L1 were sequenced and the designation of HPV variants would only be indicative, we did not attempt to name clades according to defined subtypes [36], [37], [38], [39], [40]. To visualize the bipartition file produced by RAxML we used FigTree [41], and coloured the trees according to sample name, anatomical site, HIV status and cytology result. In order to annotate the clades, we retrieved the genomes representing the sublineages of HPV16, HPV18 and HPV52 from Papillomavirus Episteme (PaVE) (https://pave.niaid.nih.gov/#home) [42]. These were aligned with the five most abundant sequences from each clade and a Neighbour Joining phylogeny identified sublineages using MAFFT v7 and 1000 bootstraps [43], [44]. However, not all sublineages could be resolved with the about 400 basepair long alignments.
2.8. Statistical analyses of the trees (HPV 16, 18 and 52)
Statistical analyses of HPV16, HPV18 and HPV52 trees were done in R studio (version 1.0.136) using the phylogenetic R packages APE (version 4.0) [45] and phytools (0.5–64)[46]. The basal node for each clade was determined by plotting “unrooted” trees, visual inspection of the trees and evaluation of number of variants (Nnode). HPV16 had in total 4273 branches and the tree where split into X and Y clade. HPV18 had in total 2401 branches and the tree where split into X and Y clade at variant number 2572. HPV52 had in total 4726 branches and the tree where split into X, Y and Z clade at variant number 5982 and 6019. The clades are shown in Fig. 4, Fig. 5, Fig. 6. Name of branches and their corresponding clade were merged with identification number of individual and samples, cervical cytology result, HIV status, sampling site and sequence depth. Pearson's chi-squared tests were used to attempt rejection of the null hypotheses that cytology, HIV and site of sampling were independent of clade. The tests were done for all variants and variants with more than 10 read pairs to monitor the influence of potential sequence artefacts. Nucleotide diversity was assessed using msa R package [47] to read the alignments into R. The nucleotide counts per position in the alignments was assessed and presented in percentage. The major allele (nucleotides present in more than 50% of the sequences) were removed for visualization purposes and the remaining nucleotide percentage were plotted as stacked bar charts for all samples and per site, cytology and HIV status. PopGenome version 2.2.4 [48]was used for calculation of nucleotide diversity and assessing neutrality and Tajima's D.
Fig. 4.
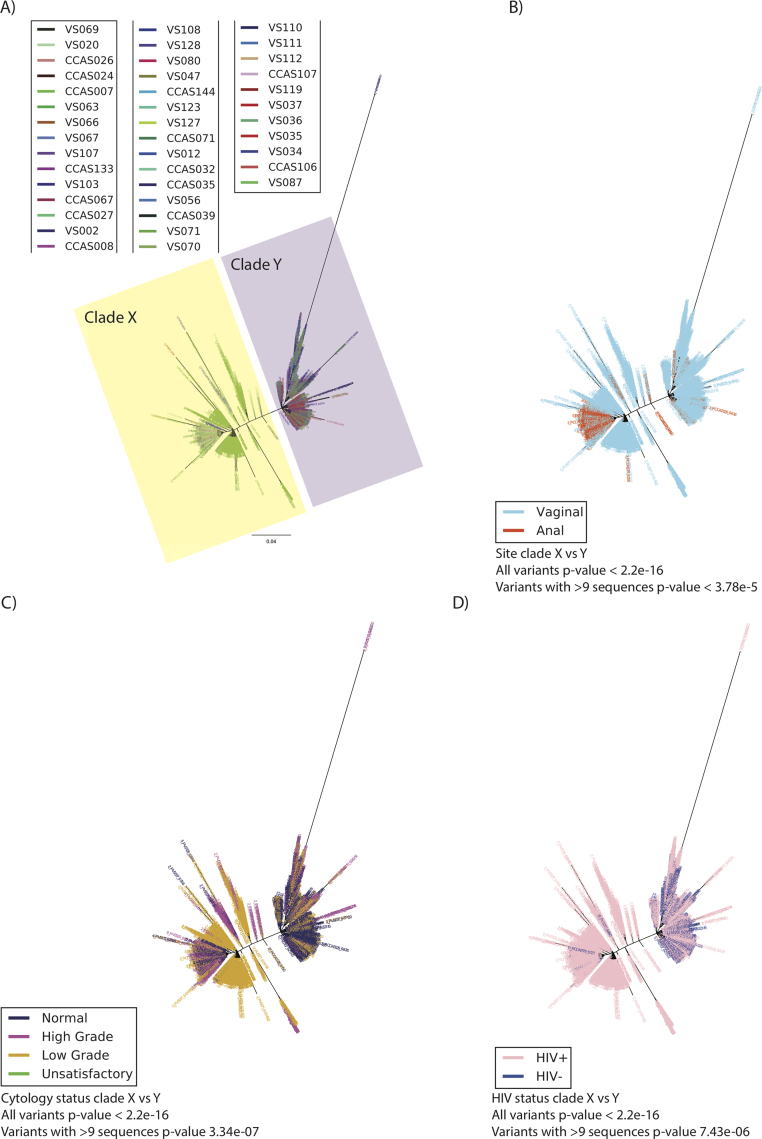
HPV 16 phylogenetic tree was constructed from 38 samples from 34 women and a total of 4275 variants using maximum likelihood (RAxML). The best tree was created from four multiple runs and 10,000 boot-straps and presented in ‘Fan’ mode. Two clades (X and Y) are observed. The tree was colour based on four variables; A) sample, B) site (vaginal or anal), C) cervical cytology outcome and D) HIV status. Cytology outcome were encoded according to Bethesda classification; Negative = Negative = Negative for intra-epithelial lesions and malignancy (NILM), High grade = High = High grade squamous intra-epithelial lesions (HGSIL) and Atypical squamous cells, cannot exclude HGSIL (ASC-H), Low grade = Low‐grade = Low-grade squamous intraepithelial lesion (LGSIL) and Atypical squamous cells of undetermined significance (ASC-US) and Unsatisfactory = slide = slide was not satisfactory for evaluation.
Fig. 5.
The HPV 18 phylogenetic tree was constructed from a total of 2403 variants of 41 samples from 37 women using maximum likelihood (RAxML). The best tree was created from four multiple runs and 10,000 bootstraps and presented in ‘Fan’ mode. Two clades (X and Y) are observed. The tree was color based on four variables; A) sample, B) site (vaginal or anal), C) cervical cytology outcome and D) HIV status. Cytology outcome were encoded according to Bethesda classification; Negative = Negative for intra-epithelial lesions and malignancy (NILM), High grade = High grade squamous intra-epithelial lesions (HGSIL) and Atypical squamous cells, cannot exclude HGSIL (ASC-H), Low grade = Low-grade squamous intraepithelial lesion (LGSIL) and Atypical squamous cells of undetermined significance (ASC-US) and Unsatisfactory = slide was not satisfactory for evaluation.
Fig. 6.
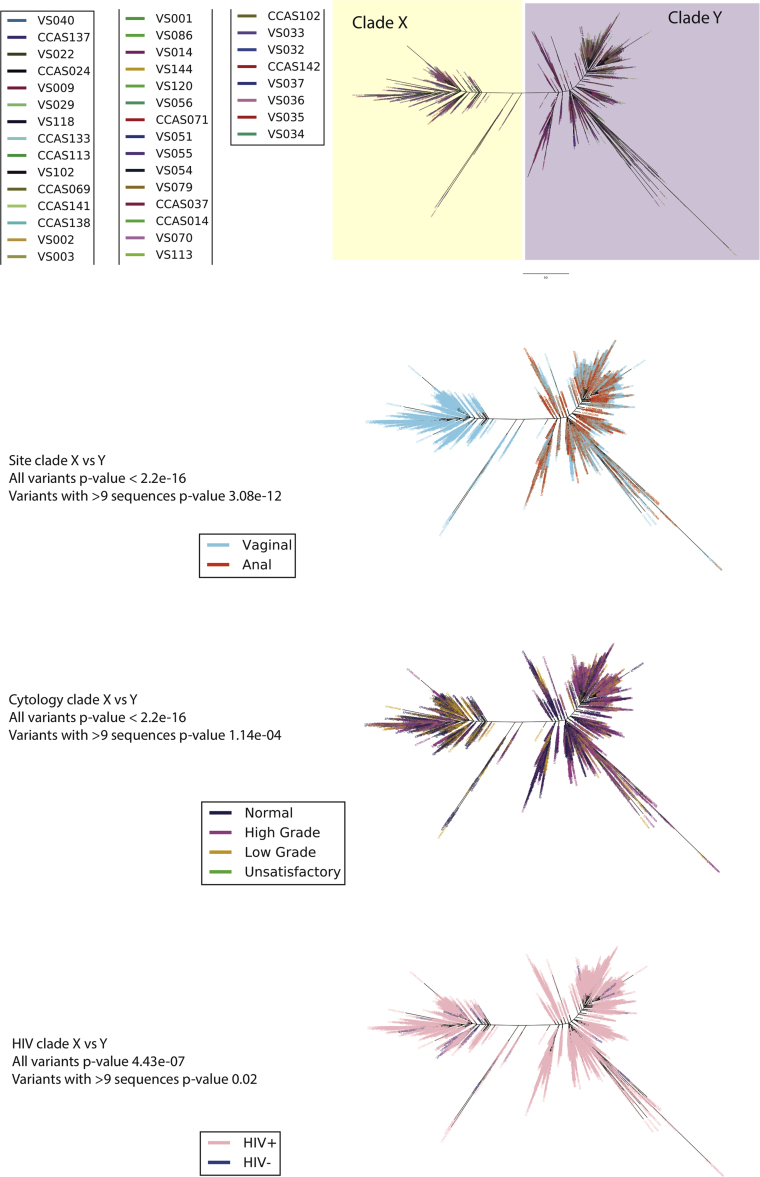
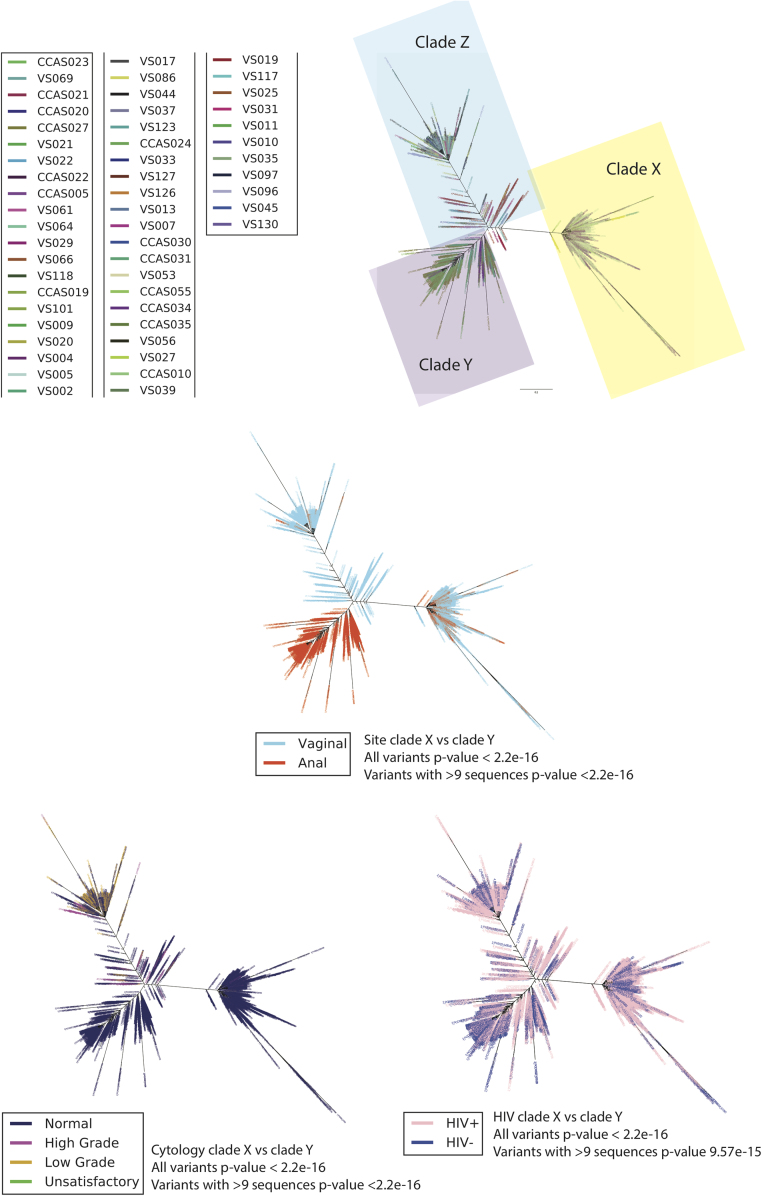
The HPV 52 phylogenetic tree was constructed from a total of 4728 variants of 53 samples from 44 women using maximum likelihood (RAxML). The best tree was created from four multiple runs and 10,000 bootstraps and presented in ‘Fan’ mode. Three distinct clades were observed, Clades X, Y and Z. The tree was color based on four variables; A) sample, B) site (vaginal or anal), C) cervical cytology outcome and D) HIV status. Cytology outcome were encoded according to Bethesda classification; Negative = Negative for intra-epithelial lesions and malignancy (NILM), High grade = High-grade squamous intra-epithelial lesions (HGSIL) and Atypical squamous cells, cannot exclude HGSIL (ASC-H), Low grade = Low-grade squamous intraepithelial lesion (LGSIL) and Atypical squamous cells of undetermined significance (ASC-US) and Unsatisfactory = slide was not satisfactory for evaluation.
2.9. Protein analyses (HPV 16, 18 and 52)
Sequences were aligned in fasta format and were translated to protein sequences in MEGA (v 7.0.21) after removing gaps from the aligned data. All the amino acid sites with more than 20% variation were taken into further analysis. The number of amino acids was counted for variants with 10 or more read pairs. In addition, amino acid counts that were less than 10, at a specific amino acid site, were excluded from the analysis. Pearson's chi-squared test was used to test if amino acid variation was dependent on sampling site, HIV and cytology status. Statistical analysis was done in R studio (Version 0.99.903). The significance threshold was set at p < 0.05.
All sequences are deposited to the Sequence Read Archive (SRA) (BioProject PRJNA393628, study accession SRP111438), the multiple sequence alignment is available at Mendeley data and custom R-scripts are available from the authors upon request.
3. Results
HPV L1 variants were identified in vaginal and anal swabs collected simultaneously from 144 women screened in Zimbabwe (Fig. 1), a cohort described in detail by Dube Mandishora and colleagues [15]. High prevalence of vaginal HPV (72%), anal HPV (48%) and HIV (49%) infection characterized the cohort. Thirty percent had cervical cytological findings suggestive for pre-malignancy or malignancy findings, by the Bethesda classification; 18 women were diagnosed high-grade and four had invasive cancer. Twelve women had low-grade lesions. Overall, HPV16, HPV18 and HPV52 were most prevalent (24%, 26% and 31% respectively), and were selected for in-depth analyses (Table 1). In the context of this study, we use the term variants for sequences differing from the consensus at least one position and are displayed as nodes on the trees. All nodes within a short genetic distance were termed clades. Thirty-five HPV genotype specific phylogenetic trees were constructed from all genetic variants (Fig. 4, Fig. 5, Fig. 6, Supplementary Figs. S1-S32 and Table S2). Each variant embodies at least two identical sequences and each node represents a variant within a sample. The trees consist of clades comprising similar nodes. The distributions of variants in each individual tree were analysed according to: i) individual sample, ii) collection site, iii) HIV status and iv) cervical cytological diagnosis.
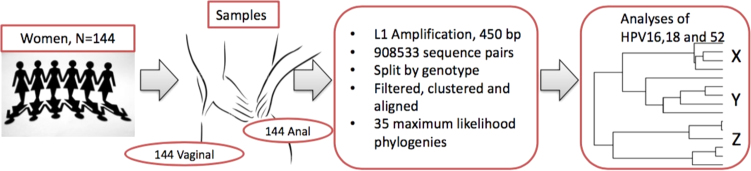
Fig. 1.
Summary of phylogenetic tree construction from 144 women who each provided duplicate samples (anal + vaginal swabs). The L1 region of HPV was sequenced using next generation sequencing on the Illumina MiSeq platform. The sequence pairs generated were filtered, clustered and aligned to produce 35 phylogenetic trees. HPV 16, 18 and 52 were further analysed for in-depth statistical description of the distribution of clades by site of HPV infection, cervical cytology result and HIV statuses of the women.
Table 1.
Summary of sequences and variables used for construction and in-depth analyses of HPV16, HPV18 and HPV52 phylogenetic trees. Data depicting total number of samples and sequence counts stratified by anatomical site, HIV status and cytology results. N represents the total number of women positive for a particular HPV genotype. These numbers were further grouped according to vaginal and anal sites, HIV status and cervical cytology. Cytology was reported according to Bethesda classification, with the following combinations used; Normal cervical cytology signified a Pap smear that was negative for intra-epithelial lesions and malignancy (NILM). ‘Low-grade’ signified any result that was either low-grade squamous intraepithelial lesion (LGSIL) or atypical squamous cells of undetermined significance (ASC-US). ‘High-grade’ represented any result that was either high-grade squamous intra-epithelial lesions (HGSIL) or atypical squamous cells, cannot exclude HGSIL (ASC-H). ‘Unsatisfactory’ results signified that the slide was not satisfactory for evaluation.
| HPV types & clades | HPV positive women (N) | HPV positive samples | Variant counts | Variants /samples and site |
Sequences/samples and HIV status |
Sequences/samples and cervical cytology status |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Anal | Vaginal | Positive | Negative | Normal | Low grade | High grade | ||||
| 16 | 34 | 38 | 4275 | |||||||
| 16× | 13 | 1377 | 0/0 | 1377/13 | 1333/8 | 44/5 | 486/9 | 596/1 | 295/3 | |
| 16Y | 27 | 2898 | 1247/12 | 1651/15 | 2869/20 | 29/7 | 722/11 | 543/7 | 1631/7 | |
| 18 | 37 | 41 | 2403 | |||||||
| 18× | 25 | 1363 | 211/11 | 1152/14 | 1353/16 | 10/9 | 150a/16 | 917/3 | 294/6 | |
| 18Y | 22 | 1040 | 30/4 | 1010/18 | 741/12 | 299/10 | 561a/14 | 381/3 | 94/4 | |
| 52 | 44 | 53 | 4728 | |||||||
| 52× | 15 | 1530 | 319/10 | 1211/5 | 1156/10 | 374/5 | 1506/11 | 5/2 | 19/2 | |
| 52Y | 7 | 1488 | 1487/6 | 1/1 | 872/6 | 616/1 | 1488/7 | 0/0 | 0/0 | |
| 52Z | 39 | 1710 | 46/2 | 1664/37 | 1277/20 | 433/19 | 1002/27 | 561/4 | 144/6 | |
Four samples had unsatisfactory cervical cytology.
3.1. HPV variability
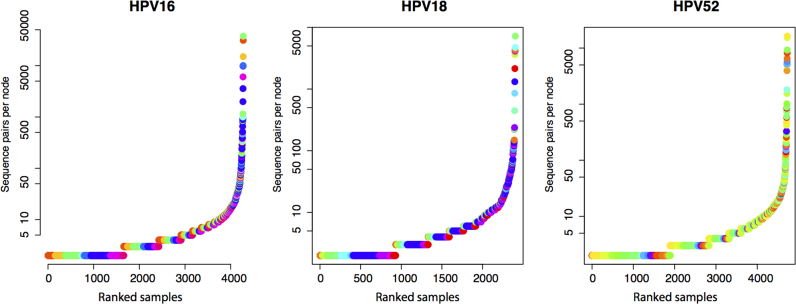
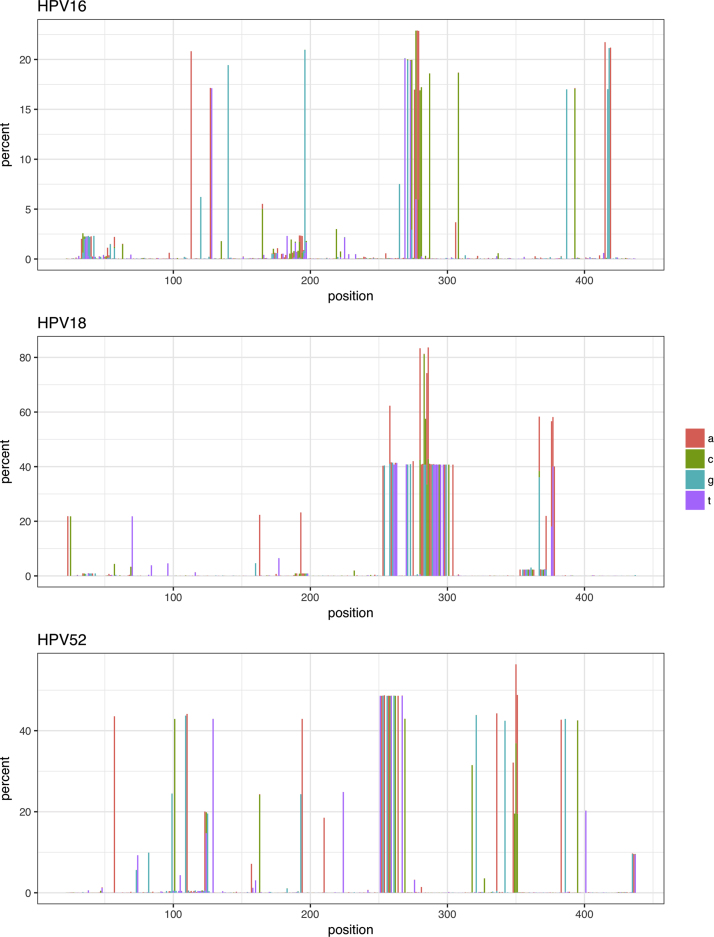
We obtained 153,335 sequence pairs from 38 HPV16-positive samples from 34 women. These HPV16 sequence pairs were assembled to about 450 bp fragments, (aligning to about 6600–7010 position of the HPV16 genome) which included 4275 unique variants (Fig. 2A). The four most abundant variants accounted for 49% of the sequences. Most variants were represented by only a few sequences. This distribution of variants can also be seen in the individual samples (Supplementary Fig. S33A). The alignment consisted of 333 polyallelic sites. The nucleotide variability consists of up to about 20% of the sequences at certain alignment positions or as very minor (<5%) alleles (Fig. 3A). Tajima's D were calculated to −0.70.
Fig. 2.
Distribution of sequences per variant. In HPV genotypes A) HPV16, B) HPV18 and c) HPV52 for all samples included in the trees. Each sample is represented by a unique colour.
Fig. 3.
Percentage of the minor allele throughout the amplified region of L1 for HPV 16, 18 and 52. Each nucleotide is represented by a unique colour.
We obtained a total of 44,988 sequence pairs (Supplementary Table S1 and S3) from 41 HPV18-positive samples from 37 women. The 450 bp fragments (aligning to 6570–6990 position of the HPV18 genome) represented 2403 unique variants. The four most sequence abundant variants were constructed from 37% of the sequences (Fig. 2B and Supplementary Fig. S33B). 325 polyallelic positions were identified in the alignment of which a large fraction was located in the 275–300 positions (Fig. 3B). Tajima's D were calculated to be − 2.32.
We obtained a total of 118,150 HPV52 sequence pairs from 53 samples and 44 women. The 450 bp fragments (aligning to 6630–7000 position) represented 4728 unique variants. The four most sequence abundant variants contained 33% of the sequences (Fig. 2C and Supplementary Fig. S2) indicating a more uniform distribution of sequences in each variant compared to HPV16. The alignment contained 309 polyallelic positions and more than 30 sites had minor variant frequencies of about 40% (Fig. 3C). Tajima's D were calculated to be −1.37.
The more nucleotide variability was observed in vaginal compared to anal samples (Supplementary Fig. S35). More variability was also observed in HIV+ samples for HPV16 and HPV18 (Supplementary Fig. S36). We could not observe any difference in variability in samples with different cytology status (Supplementary Fig. S37).
3.2. HPV16
Two distinct clades were observed in the HPV16 phylogenetic tree, here named Clade X related to the A3 sublineage (1377 variants, representing 34,139 sequences, from 13 samples) and Clade Y related to the unresolved (using 408 basepair of L1) C1/D1/D2 sublineage (2898 variants, representing 119,196 sequences, from 27 samples) (Table 1, Fig. 4, Supplementary Table S1 and S3). Each individual sample harbours multiple variants belonging to one or both clades. Minor nucleotide differences between neighbouring variants in the tree displayed a ‘fishbone’ pattern, indicative of a close phylogenetic relationship between the variants (Fig. 4A).
Unexpectedly, when stratifying our results by anatomical site, Clade X was exclusively of vaginal origin (blue in colour) whereas samples from both anal and vaginal sites were represented in Clade Y (both blue and red in colour) (Fig. 4B). Given the 4275 unique variants in the two clades of the HPV 16 tree, the probability that Clade X should harbour samples exclusively of vaginal origin at random is very small (Chi-square test, p < 2.2 × 10−16). Similarly, this association was also highly significant when only considering abundant variants with >9 sequences (p = 3.082 × 10−12). Stratifying clades by cervical cytology, clades X and Y both comprised normal, high-grade and low-grade lesions (Fig. 4C); Clade Y comprised statistically significantly more variants from samples with normal and high-grade lesions. Thus, there was a significant association between the clade and cervical cytology result (p < 2.2 × 10–16 and p = 0.0001144 for the abundant variants). 84% of the HPV16 variants emanated from HIV positive women (Fig. 4D) and we found that HIV status and clades were significantly associated (p = 4.434 × 10−7) (Supplementary Table S4).
We analysed non-synonymous variation to assess impact of variability on the protein sequence of the viral capsid. Non-synonymous variation in L1 amino acid position 353 (Table 2), results in proline and threonine residues in our dataset. The amino acid variation was associated to cytology status (Supplementary Table S5).
Table 2.
The number of amino acids variants and phenotype for selected non-synonymous positions. A) HPV16, position 353 shows number of variants with Pro/Thr variation. B) HPV18, show position 323 with variants Val/Ile/- and position 347 with variants Phe/Ser. C) HPV52 show position 380 with variants Lys/Thr, position 383 with variants Asp/Ser, position 473 with variants Asp/Glu/- and position 478 with variants Asp/Glu/-. The extent of variation of amino acid is stratified by anatomical site, HIV status and cervical cytology outcome (grouped into high-grade, low-grade and normal). (-) Denotes gap in the alignment.
| A) | ||||
|---|---|---|---|---|
|
HPV16 | ||||
| Phenotype | Amino acid position | 353 |
||
| Amino acids | Pro | Thr | p-value | |
| Site | CCAS | 134 | 61 | 0.2681 |
| VS | 407 | 149 | ||
| HIV status | HIV+ | 514 | 199 | 1 |
| HIV- | 27 | 11 | ||
| Cervical cytology | High-grade | 253 | 65 | < 2.2 × 10−16 |
| Low-grade | 206 | 36 | ||
| Normal | 80 | 109 | ||
| B) | ||||||||
|---|---|---|---|---|---|---|---|---|
|
HPV18 | ||||||||
| Phenotype | Amino acid position | 323 |
347 |
|||||
| Amino acid | Val | Ile | - | p-value | Phe | Ser | p-value | |
| Site | CCAS | 9 | 34 | 9 | 5.76 × 10−6 | 0 | 45 | 2.92 × 10−4 |
| VS | 260 | 183 | 17 | 110 | 331 | |||
| HIV status | HIV+ | 60 | 9 | 1 | 1.69 × 10−8 | 2 | 67 | 4.62 × 10−5 |
| HIV- | 209 | 208 | 25 | 108 | 309 | |||
| Cervical cytology | High-grade | 12 | 41 | 0 | <2.2 × 10–16 | 0 | 53 | <2.2 × 10–16 |
| Low-grade | 173 | 138 | 2 | 108 | 201 | |||
| Normal | 80 | 37 | 24 | 1 | 118 | |||
|
C) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
HPV52 | |||||||||||||||
| Phenotype | Amino acid position | 380 |
383 |
473 |
478 |
||||||||||
| Amino acid | Lys | Thr | p-value | Asp | Ser | p-value | Asp | Glu | - | p-value | Asp | Glu | - | p-value | |
| Site | CCAS | 172 | 89 | 0.0757 | 90 | 175 | 0.009 | 166 | 82 | 24 | 0.018 | 0 | 240 | 33 | <2.2 × 10–16 |
| VS | 224 | 158 | 172 | 215 | 202 | 158 | 44 | 114 | 247 | 44 | |||||
| HIV status | HIV+ | 126 | 38 | 5.19 × 10−6 | 42 | 125 | 6.693 × 10−6 | 119 | 38 | 13 | 1.31 × 10−5 | 32 | 126 | 13 | 0.1792 |
| HIV- | 270 | 209 | 220 | 265 | 249 | 202 | 55 | 82 | 361 | 64 | |||||
| Cervical cytology | High-grade | 19 | 5 | 3.03 × 10−10 | 9 | 15 | 5.353 × 10−10 | 18 | 5 | 1 | 6.03 × 10−10 | 5 | 18 | 1 | 5.562 × 10–14 |
| Low-grade | 67 | 3 | 3 | 67 | 61 | 4 | 1 | 34 | 31 | 1 | |||||
| Normal | 309 | 239 | 250 | 307 | 288 | 231 | 66 | 74 | 438 | 75 | |||||
3.3. HPV18
Two distinct clades were observed in the HPV18 phylogenetic tree, named Clade X related to the B1 sublineage (20,987 sequences in 1363 variants from 25 samples) and Clade Y related to the A sublineages (24,001 sequences in 1040 variants from 22 samples) (Fig. 5 and Table 1). Clade X was further divided into three sub-clades. Two of these sub-clades were from one sample while the third sub-clade was represented by multiple samples (Fig. 5A). Clade Y was further clustered into two sub-clades.
Clade X was of both vaginal and anal origin (blue and red colour), though two sub-clades emanating from a single sample were of vaginal origin (Fig. 5B). >97% of Clade Y's variants were of vaginal origin. The intermediate variants were of both anal and vaginal origin. There was significant association between the site of infection and the clade (p < 2.2 × 10−16 and p = 3.781 × 10−5 for the abundant variants). One of the sub-clades in Clade X was predominantly from samples with high-grade lesions although normal and low-grade lesion samples were also represented (Fig. 5C). The two other sub clades in Clade X were made up of sequences from a low-grade lesion sample. > 50% of the variants in Clade Y were from samples with normal cervical cytology results. The longest branch in this clade was from a high-grade lesion sample. The intermediate clades, emanating from Clade X, were made up of samples with normal, high-grade and low-grade lesions. There was significant association between the clade derived and the cervical cytology outcome for all clades (p = <2.2 × 10−16 and p = 3.335 × 10−7 for the abundant variants). In total, 87% of the variants were from HIV infected women. Clade Y had 299 variants from 10 samples from HIV negative women. For all the clades, there was significant association between the clade derived and the HIV status of the women (p = <2.2 × 10−16 and p = 7.428 × 10−6 for the abundant variants) (Fig. 5D) (Supplementary Table S4).
Non-synonymous substitutions were analysed in L1 amino acid positions 323 and 347. In position 323 both isoleucine and valine were encoded. In position 347, phenylalanine and serine were encoded. Each of these amino acid codons was significantly associated with anatomical site, HIV and cytology statuses (Table 2 and Supplementary Table S5).
3.4. HPV52
Three distinct clades were observed in the phylogenetic tree, named Clade X related to the D1 sublineage, clade Y related to the B2 sublineage and clade Z related to the A1/A2 sublineage (Table 1, Supplementary Table S1 and S3 and Fig. 6). Clades Y and Z showed multiple variants with small nucleotide differences portrayed in a characteristic ‘fishbone’ pattern (Fig. 6A). All clades were each represented by at least four different samples. Notably, Clade Y was exclusively of anal origin (red in colour) and >97% of variants in Clade Z was of vaginal origin (blue in colour, with a few red variants) whilst Clade X originated from both (20%) anal and (80%) vaginal samples (both blue and red in colour) (Fig. 6B). Similar to HPV16, there was a highly significant association between the clade derived and the site of HPV infection (p < 2.2 × 10−16 for all and only the most abundant variants). Variants in Clade Y, represented by seven women, were exclusively from samples with normal cervical cytology results (Fig. 6C). >99% of variants in Clade X was from samples with normal cervical cytology results, while variants in Clade Z were represented by 59% normal, 32% high grade and 8% low-grade results. There was significant association on the clade derived and the cervical cytology result (p < 2.2 × 10−16). Clade X, Y, Z harboured both HIV positive and negative samples. There was significant association between the derived clade and the HIV status of the women for all the variants (p < 2.2 × 10−16 and p = 9.574 × 10−15 for the most abundant variants) (Fig. 6D) (Supplementary Table S4).
Substitutions were analysed in four different L1 amino acid positions; 380, 383, 473 and 478. The variably encoded amino acids were threonine and lysine for position 380, serine and aspartic acid for position 383, whilst for positions 473 and 478 the variably encoded amino acids were glutamic acid and aspartic acid (Supplementary Table S5). The amino acid variation at positions 380 was associated with HIV and cytology status, 383 and 473 had significant association with sample type, HIV and cytology status. At position 478 the variation was associated with sample type and cytology status (Table 2).
3.5. Summary of the remaining 32 HPV genotypes
High-risk genotypes 31, 35, 39, 45, 51, 52, 56, 58 and 59, low-risk types 6, 11, 32, 40, 42, 44, 61, 70, 71, 72, 81, 84, 86, 89, 91 and 114, probable high-risk types 26, 53, 66, 68, 73 and 82 were assessed (Supplementary Figs. S1-S32 and Tables S2). The total number of samples for each genotype ranged from 1 to 21. Genotypes with at least five samples were HPV genotypes 35, 40, 71, 86 and 91. HPV35 displayed a characteristic ‘fishbone’ phylogenetic tree with three distinct clades and numerous intermediate variants. HPV40 and HPV70 were clearly split into three different clades, whereas each of HPV genotypes 39, 66, 68 and 72 split into two different clades. The HPV91 phylogenetic tree was drawn from two samples with a total variant count of 57. Trees for HPV genotypes 42, 54, 73, 82 and 89 were not resolved, as shown by the ‘dandelion’ shape of the phylogenetic tree. Diverse and numerous variants were observed in HPV genotypes 11, 32, 44, 45, 53, 58, 61, 71, 81, 83, 84 and 91 phylogenetic trees, shown by characteristic ‘fishbone’ patterns. Variants in HPV genotypes 31, 32, 35, 40, 58, 59, 69, 71, 72, 73, 81 and 83 were of vaginal origin. HPV84 was exclusively of anal origin and HPV11, HPV45 and HPV70 were typically of anal origin with a few branches of vaginal origin. In terms of cervical cytological diagnosis, HPV69 and HPV86 were noticeably from low-grade lesions, whilst genotypes 26, 32, 33, 42 and 72 were from samples with high-grade lesions. Genotypes 35, 40, 66, 70, 71, 83 and 84 were from normal cytology. Based on HIV status, phylogenetic trees for HPV types 31, 35, 58, 66 and 69 were visibly from HIV negative samples. Nineteen HPV genotypes were observably from HIV positive samples; these included HPV11 and HPV33.
4. Discussion
To our knowledge this is the first study to associate HPV intra-host and intra-genotype variants to anogenital site of origin, cervical cytology outcome and HIV co-infection. Microbial genetic diversity has in the pre-omics era been vastly underestimated [49], [50], [51]. Complex communities consisting of microbial species, types and variants beyond the abundant types, can be assessed with high resolution using NGS [22]. We find high intra-HPV genotype variability within the conserved L1 gene. This genetic variability is found both within samples and between individuals and are considerably higher than the known human mutation rate of one mutation in every 30 million base pairs [52]. Each infection is dominated by a few abundant variants, challenging the longstanding fallacy of low intra-host viral genotype variability [53].
Emerging evidence of high amount of intra-genotype variability has been shown in HPV6 and HPV11 [19], as well as intra-host diversity in HPV16 [54] and in rare genotypes such as HPV53, HPV66 and HPV70 [55]. Zhang and colleagues recently sequenced the E6-E7- L1 regions of HPV52 and found that many variants emanated from positive selection mutations [56], [57]. The lack of natural selection may be explained by insufficient time to purge variants. It is becoming clear that these variants may play a role in carcinogenesis [55] and the accumulation of intra-lesion HPV diversity has been suggested to “be a hidden factor contributing to different outcomes after infections by the same HPV type” [54], including the resultant cancer [18].
The phylogenetic trees illustrated that multiple variants were identified within each participant and sample. Two plausible, but not mutually exclusive, explanations for this phenomenon are 1) the women are infected by different variants (co-infected), and 2) different HPV variants are generated within the host [58]. The up to 4700 different and closely related variants per genotype and up to thousands within the same sample identified here support the latter. The HPV genes are replicated by the host replication machinery [59] suggesting that a very low human autosomal–like mutation rate would be operating. However, polymerases activated in infected cells are not always high-fidelity enzymes and may result in errors [58]. Also APOBEC deaminases are known to participate in generating driver-mutations in HPV-associated cancers [60]. The recruitment of APOBEC enzymes and the error-prone processes during DNA repair may explain the high mutation rate [61], [62]. Mirabello and colleagues have recently confirmed the existence of intra-individual diversity in HPV16 genomes [63]. They attribute this diversity to APOBEC. New variants generated by replication errors may not be infectious due to protein malfunctions, reduced fitness for uptake in the cell basal layer or epitope changes. Indeed, variants of HPV16, HPV18 and HPV52 that changed the protein sequence of the capsid protein encoded by L1 were found. These observations, in light of the slow evolution of HPV at the population level, suggest that strong selection pressures are at play in each infection cycle [64], [65], [66]. This is in line with the negative Tajima's D index shown here for HPV16, HPV18 and HPV52, indicating population expansion after a recent bottleneck. Observation of having distinct HPV16 and HPV52 variants in anal and vaginal sites, is best explained by an association to tissue tropism [67]. Molecular changes, particularly non-synonymous changes, may explain biases in viral survival in different tissues. Like HIV, the HPV virus may take up different virulence and selection strategies during an infection. The processes of carcinogenesis in the anal cells is slightly different from that of the vaginal mucosa [68]. A weakness of the study is the lack of anal cytology results that might have shed light on the process of carcinogenesis in anal versus cervical cells. Cancers from these two sites are known to be histologically similar and mostly start from the squamous-columnar transformation zone [69]. We report a difference in HPV variant distribution between the sites of infection, suggesting the presence of other factors that may affect the natural history of HPV. Nicolas-Parraga and colleagues [8] are one of few who have identified HPV16 (mono-infection) variants in invasive cancers from five anogenital sites; cervix, vulva, vagina, penis and anus. Although their sample set is made up of individuals with cancer and our sample set is from women reporting for routine screening, it is still important to note their inferences. With the limited diversity resolution given by Sanger sequencing, they identified different HPV16 variants from the various anogenital regions. They ruled out the possibility of tissue tropism playing a significant role, contradicting our study where we consider tissue tropism to be a contributing factor to the HPV evolution. Our study only relies on parts of the L1 gene to explain the tissue tropism and did not involve HPV from cancer tissue. Future studies that include other HPV genes, participants’ immunologic profiles and other anogenital microbiome may give a more detailed outlook on the anogenital tissue tropism. The vaginal and anal origin sub-clades of HPV16 and HPV52 showed intra individual on-going and independent evolution. This supports the notion that the virus is thriving differently in the two different anogenital sites and therefore different tropism is likely to generate different diversity spectrum.
Unique to our study is the significant association between HIV status and HPV clade, observed for all the three most prevalent HPV genotypes. This suggests that immunosuppression differs in importance for clearance and persistence of each clade. To our knowledge these clade specific differences have not been described earlier, although many studies show that HPV persistence is associated with the immune status of the individual [13], [70], [71], [72].
HPV16, HPV18 and HPV52 all showed associations between clade, amino acids sequence and the cytological diagnosis. This is first of all evident in the HPV52 tree, where Clade Z is mostly made up of low and high-grade lesions. There was also distinct representation of low-grade lesions on Clade X of HPV18. Emerging information about sub-clade differences in risk of cervical cancer progression [6], [8], [39], [73] is in compliance with our result.
The cross-sectional design of the study with no follow-up data available in the cohort prevents the possibility to assess progression or regression of lesions relative to HPV variants. We recommend prospective case control studies to be carried out to fully relate these variants to HPV persistence and carcinogenesis. This study benefits from high sequence depth that makes it possible to uncover low frequency variation, therefore our total number of variants may seem extreme. The limitation of our study is that these variants may also arise to some degree from errors during the NGS protocol. Although we have reduced sequencing errors to a minimum and presented variation with two thresholds (more than two and nine sequence pairs), errors may inflate variations to some degree. Another limitation of our study was not being able to separate multiple and mono-infections of HPV, considering that HPV genotypes co-exist. One or several co-infecting types may drive some of the associations described here.
Deep sequencing a segment of the HPV L1 gene of 35 different HPV types in anal and vaginal swabs from 144 Zimbabwean women revealed tens of thousands genetic variants. Thousands of closely related variants within the same sample suggest intra-host variability, which is surprising given the overall slow rate of HPV evolution at the population level. Highly significant associations between related sequences organized in clades and anatomical site, HIV status and cytology were identified. These results suggest that tissue tropisms and host immune status may drive HPV evolution in different directions and potentially influence rates of HPV diversification.
Acknowledgments
This work was supported by the Letten foundation grant. We acknowledge the recruiting nurse, Ms. Mary Mucheche for her contribution in sample and data collection. The authors thank Dr. Linda Vos for contributing to the bioinformatics analyses and Ms. Ellen Myrvang and Dr. Roger Meisal for the sequence production and Elina Vinberg for coordination of the study. Authors also thank Prof. Joel Palefsky and Ms. Maria DaCosta for the technical support and training on DNA extraction. Ms. Fiona Mtisi and Mr. Kidson Mataruka assisted with sample sorting and DNA extraction at UZCHS-CTU laboratory, Zimbabwe. The sequencing was performed at Akershus University Hospital, Norway. The co-authors would like to acknowledge Ignacio Bravo and the second reviewer for the constructive criticism during the review process.
Acknowledgments
Author contributions
RSDM, OHA and TBR designed the specific objectives of the study, interpreted the results, and wrote the paper. RSDM, KD, ZMC, NC and BSP coordinated the broad concept of the study, sample and data collection. DNA extraction was coordinated by NC and ZMC and performed by RSDM. IKC and SL coordinated the deep sequencing. KSG and TBR performed bioinformatics and biostatistical analyses. SL performed the amino acid substitution analyses. MN and ZMC interpreted the clinical outlook of the results. BSP provided funding for the study. All authors discussed the results and critically reviewed the manuscript.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.pvr.2018.04.006.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- 1.zur Hausen H. Papillomaviruses in the causation of human cancers - a brief historical account. Virology. 2009;384:260–265. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 2.Pimenoff V.N., Oliveira D., Mendes C., Bravo I.G. Transmission between archaic and modern human ancestors during the evolution of the oncogenic human papillomavirus 16. Mol. Biol. Evol. 2017;34:4–19. doi: 10.1093/molbev/msw214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franco E.L., Villa L.L., Sobrinho J.P., Prado J.M., Rousseau M.-C., Désy M., Rohan T.E. Epidemiology of acquisition and clearance of cervical human papillomavirus infection in women from a high-risk area for cervical cancer. J. Infect. Dis. 1999;180:1415–1423. doi: 10.1086/315086. [DOI] [PubMed] [Google Scholar]
- 4.Egawa N., Egawa K., Griffin H., Doorbar J. Human papillomaviruses; epithelial tropisms, and the development of neoplasia. Viruses. 2015;7:3863–3890. doi: 10.3390/v7072802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.HPV reference center Karolinska, Reference clones at International HPV Reference Center, 2016. http://www.hpvcenter.se/html/refclones.html?%3C?Php%20echo%20time( );%20?%3E (Accessed 4 June 2017).
- 6.Burk R.D., Harari A., Chen Z. Human papillomavirus genome variants. Virology. 2013;445:232–243. doi: 10.1016/j.virol.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palefsky J.M., Holly E.A., Ralston M.L., Da Costa M., Greenblatt R.M. Prevalence and risk factors for anal human papillomavirus infection in human immunodeficiency virus (HIV)-positive and high-risk HIV-negative women. J. Infect. Dis. 2001;183:383–391. doi: 10.1086/318071. [DOI] [PubMed] [Google Scholar]
- 8.Nicolás‐Párraga S., Gandini C., Pimenoff V.N., Alemany L., de Sanjosé S., Xavier Bosch F., Bravo I.G. HPV16 variants distribution in invasive cancers of the cervix, vulva, vagina, penis, and anus. Cancer Med. 2016;5:2909–2919. doi: 10.1002/cam4.870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L., Liao H., Yang B., Geffre C.P., Zhang A., Zhou A., Cao H., Wang J., Zhang Z., Zheng W. Variants of human papillomavirus type 16 predispose toward persistent infection. Int. J. Clin. Exp. Pathol. 2015;8:8453–8459. [PMC free article] [PubMed] [Google Scholar]
- 10.Burd E.M. Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 2003;16:1–17. doi: 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernard H.-U., Burk R.D., Chen Z., van Doorslaer K., zur Hausen H., de Villiers E.-M. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401:70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clifford G., Gallus S., Herrero R., Muñoz N., Snijders P., Vaccarella S., Anh P., Ferreccio C., Hieu N., Matos E., Molano M., Rajkumar R., Ronco G., de Sanjosé S., Shin H., Sukvirach S., Thomas J., Tunsakul S., Meijer C., Franceschi S. Worldwide distribution of human papillomavirus types in cytologically normal women in the International Agency for Research on Cancer HPV prevalence surveys: a pooled analysis. Lancet. 2005;366:991–998. doi: 10.1016/S0140-6736(05)67069-9. [DOI] [PubMed] [Google Scholar]
- 13.Sahasrabuddhe V.V., Mwanahamuntu M.H., Vermund S.H., Huh W.K., Lyon M.D., Stringer J.S.A., Parham G.P. Prevalence and distribution of HPV genotypes among HIV-infected women in Zambia. Br. J. Cancer. 2007;96:1480–1483. doi: 10.1038/sj.bjc.6603737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan P.K.S., Luk A.C.S., Park J.-S., Smith-McCune K.K., Palefsky J.M., Konno R., Giovannelli L., Coutlée F., Hibbitts S., Chu T.-Y., Settheetham-Ishida W., Picconi M.A., Ferrera A., Marco F.D., Woo Y.-L., Raiol T., Piña-Sánchez P., Cheung J.L.K., Bae J.-H., Chirenje M.Z., Magure T., Moscicki A.-B., Fiander A.N., Stefano R.D., Cheung T.-H., Yu M.M.Y., Tsui S.K.W., Pim D., Banks L. Identification of human papillomavirus type 58 lineages and the distribution worldwide. J. Infect. Dis. 2011;203:1565–1573. doi: 10.1093/infdis/jir157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dube Mandishora R.S., Christiansen I.K., Chin’ombe N., Duri K., Ngara B., Rounge T.B., Meisal R., Ambur O.H., Palefsky J.M., Stray-Pedersen B., Chirenje Z.M. Genotypic diversity of anogenital human papillomavirus in women attending cervical cancer screening in Harare, Zimbabwe. J. Med. Virol. 2017 doi: 10.1002/jmv.24825. [DOI] [PubMed] [Google Scholar]
- 16.IARC, GLOBOCAN Cancer Fact Sheets: Cervical Cancer. 〈http://globocan.iarc.fr/old/FactSheets/cancers/cervix-new.asp〉 (Accessed 11 March 2017).
- 17.Bruni L., Diaz M., Castellsagué M., Ferrer E., Bosch F.X., de Sanjosé S. Cervical human papillomavirus prevalence in 5 continents: meta-analysis of 1 million women with normal cytological findings. J. Infect. Dis. 2010;202:1789–1799. doi: 10.1086/657321. [DOI] [PubMed] [Google Scholar]
- 18.Mirabello L., Yeager M., Cullen M., Boland J.F., Chen Z., Wentzensen N., Zhang X., Yu K., Yang Q., Mitchell J., Roberson D., Bass S., Xiao Y., Burdett L., Raine-Bennett T., Lorey T., Castle P.E., Burk R.D., Schiffman M. HPV16 Sublineage associations with histology-specific cancer risk using HPV whole-genome sequences in 3200 women. J. Natl. Cancer Inst. 2016;108 doi: 10.1093/jnci/djw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burk R.D., Chen Z., Harari A., Smith B.C., Kocjan B.J., Maver P.J., Poljak M. Classification and nomenclature system for human Alphapapillomavirus variants: general features, nucleotide landmarks and assignment of HPV6 and HPV11 isolates to variant lineages. Acta Dermatovenerol. Alp. Panon. Adriat. 2011;20:113–123. [PMC free article] [PubMed] [Google Scholar]
- 20.Zimbabwe Cancer Registry, Pattern of Cancer in Zimbabwe, 2013.
- 21.Siqueira J.D., Alves B.M., Prellwitz I.M., Furtado C., Meyrelles Â.R., Machado E.S., Seuánez H.N., Soares M.A., Soares E.A. Identification of novel human papillomavirus lineages and sublineages in HIV/HPV-coinfected pregnant women by next-generation sequencing. Virology. 2016;493:202–208. doi: 10.1016/j.virol.2016.03.027. [DOI] [PubMed] [Google Scholar]
- 22.Arroyo L.S., Smelov V., Bzhalava D., Eklund C., Hultin E., Dillner J. Next generation sequencing for human papillomavirus genotyping. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2013;58:437–442. doi: 10.1016/j.jcv.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 23.Au C.H., Wa A., Ho D.N., Chan T.L., Ma E.S.K. Clinical evaluation of panel testing by next-generation sequencing (NGS) for gene mutations in myeloid neoplasms. Diagn. Pathol. 2016;11 doi: 10.1186/s13000-016-0456-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barzon L., Militello V., Lavezzo E., Franchin E., Peta E., Squarzon L., Trevisan M., Pagni S., Dal Bello F., Toppo S., Palù G. Human papillomavirus genotyping by 454 next generation sequencing technology. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2011;52:93–97. doi: 10.1016/j.jcv.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Yi X., Zou J., Xu J., Liu T., Liu T., Hua S., Xi F., Nie X., Ye L., Luo Y., Xu L., Du H., Wu R., Yang L., Liu R., Yang B., Wang J., Belinson J.L. Development and validation of a new HPV genotyping assay based on next-generation sequencing. Am. J. Clin. Pathol. 2014;141:796–804. doi: 10.1309/AJCP9P2KJSXEKCJB. [DOI] [PubMed] [Google Scholar]
- 26.Shen-Gunther J., Wang Y., Lai Z., Poage G.M., Perez L., Huang T.H.M. Deep sequencing of HPV E6/E7 genes reveals loss of genotypic diversity and gain of clonal dominance in high-grade intraepithelial lesions of the cervix. BMC Genom. 2017;18 doi: 10.1186/s12864-017-3612-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gravitt P.E., Peyton C.L., Alessi T.Q., Wheeler C.M., Coutlée F., Hildesheim A., Schiffman M.H., Scott D.R., Apple R.J. Improved amplification of genital human papillomaviruses. J. Clin. Microbiol. 2000;38:357–361. doi: 10.1128/jcm.38.1.357-361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nayar R., Wilbur D. Springer; 2015. The Bethesda System for Reporting Cervical Cytology: Definitions, Criteria, and Explanatory Notes. [Google Scholar]
- 29.Schirmer M., Ijaz U.Z., D’Amore R., Hall N., Sloan W.T., Quince C. Insight into biases and sequencing errors for amplicon sequencing with the Illumina MiSeq platform. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gku1341. (e37–e37) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nesoni: High Throughput Sequencing Analysis Tools, Victorian Bioinformatics Consortium. 〈https://github.com/Victorian-Bioinformatics-Consortium/nesoni〉, 2017.
- 31.PEAR - Paired-end read merger, (n.d.). 〈http://sco.h-its.org/exelixis/web/software/pear/index.html〉 (Accessed 1 May 2017).
- 32.Mahé F., Rognes T., Quince C., de Vargas C., Dunthorn M. Swarm: robust and fast clustering method for amplicon-based studies. Peer J. 2014;2 doi: 10.7717/peerj.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christiam Camacho, George Coulouris, Vahram Avagyan, Ning Ma, Jason Papadopoulos, Kevin Bealer, ThomasL Madden, Blast, (n.d.). http://download.springer.com/static/pdf/101/art%253A10.1186%252F1471-2105-10-421.pdf?OriginUrl=http%3A%2F%2Fbmcbioinformatics.biomedcentral.com%2Farticle%2F10.1186%2F1471-2105-10-421&token2=exp=1473082023~acl=%2Fstatic%2Fpdf%2F101%2Fart%25253A10.1186%25252F1471-2105-10-421.pdf*~hmac=b0c83e0c3a83afdd472d4fddb006f2c96f4146ac8ac6b3887cdbac0439bdf54a (Accessed 5 September 2016). [DOI] [PMC free article] [PubMed]
- 34.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J.D., Higgins D.G. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cornet I., Gheit T., Franceschi S., Vignat J., Burk R.D., Sylla B.S., Tommasino M., Clifford G.M. the I.H.V.S. group, human papillomavirus type 16 genetic variants: phylogeny and classification based on E6 and LCR. J. Virol. 2012;86:6855–6861. doi: 10.1128/JVI.00483-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King A.J., Sonsma J.A., Vriend H.J., van der Sande M.A.B., Feltkamp M.C., Boot H.J., Koopmans M.P.G. Genetic diversity in the major capsid L1 protein of HPV-16 and HPV-18 in the Netherlands. PLoS One. 2016;11 doi: 10.1371/journal.pone.0152782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ntova C.K., Kottaridi C., Chranioti A., Spathis A., Kassanos D., Paraskevaidis E., Karakitsos P. Genetic variability and phylogeny of high risk HPV type 16, 18, 31, 33 and 45 L1 gene in greek women. Int. J. Mol. Sci. 2011;13:1–17. doi: 10.3390/ijms13010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sichero L., Ferreira S., Trottier H., Duarte-Franco E., Ferenczy A., Franco E.L., Villa L.L. High grade cervical lesions are caused preferentially by non-European variants of HPVs 16 and 18. Int. J. Cancer. 2007;120:1763–1768. doi: 10.1002/ijc.22481. [DOI] [PubMed] [Google Scholar]
- 40.Tornesello M.L., Duraturo M.L., Giorgi-Rossi P., Sansone M., Piccoli R., Buonaguro L., Buonaguro F.M. Human papillomavirus (HPV) genotypes and HPV16 variants in human immunodeficiency virus-positive Italian women. J. Gen. Virol. 2008;89:1380–1389. doi: 10.1099/vir.0.83553-0. [DOI] [PubMed] [Google Scholar]
- 41.FigTree, (n.d.). 〈http://tree.bio.ed.ac.uk/software/figtree/〉 (Accessed 5 September 2016).
- 42.PaVE: Papilloma Virus Genome Database, (n.d.). 〈https://pave.niaid.nih.gov/#home〉 (Accessed 14 March 2018).
- 43.Kuraku S., Zmasek C.M., Nishimura O., Katoh K. A leaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013;41:W22–W28. doi: 10.1093/nar/gkt389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2017 doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paradis E., Claude J., Strimmer K. APE: analyses of Phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 46.Revell L.J. phytools: an R package for phylogenetic comparative biology (and other things) Methods Ecol. Evol. 2012;3:217–223. [Google Scholar]
- 47.Bodenhofer U., Bonatesta E., Horejš-Kainrath C., Hochreiter S. msa: an R package for multiple sequence alignment. Bioinforma. Oxf. Engl. 2015;31:3997–3999. doi: 10.1093/bioinformatics/btv494. [DOI] [PubMed] [Google Scholar]
- 48.Pfeifer B., Wittelsbürger U., Ramos-Onsins S.E., Lercher M.J. PopGenome: an efficient swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 2014;31:1929–1936. doi: 10.1093/molbev/msu136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woese C.R., Fox G.E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stein J.L., Marsh T.L., Wu K.Y., Shizuya H., DeLong E.F. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J. Bacteriol. 1996;178:591–599. doi: 10.1128/jb.178.3.591-599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torsvik V., Goksøyr J., Daae F.L. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dolgin E. Human mutation rate revealed. Nat. News. 2009 [Google Scholar]
- 53.Stewart A.C., Eriksson A.M., Manos M.M., Muñoz N., Bosch F.X., Peto J., Wheeler C.M. Intratype variation in 12 human papillomavirus types: a worldwide perspective. J. Virol. 1996;70:3127–3136. doi: 10.1128/jvi.70.5.3127-3136.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Oliveira C.M., Bravo I.G., Santiago e Souza N.C., Genta M.L.N.D., Fregnani J.H.T.G., Tacla M., Carvalho J.P., Longatto-Filho A., Levi J.E. High-level of viral genomic diversity in cervical cancers: a Brazilian study on human papillomavirus type 16. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2015;34:44–51. doi: 10.1016/j.meegid.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 55.Gurgel A.P.A.D., Chagas B.S., do Amaral C.M.M., Albuquerque E.M.B., Serra I.G.S.S., da J., Silva Neto C., Muniz M.T.C., de Freitas A.C. Prevalence and Genetic Variability in Capsid L1 Gene of Rare Human Papillomaviruses (HPV) Found in Cervical Lesions of Women from North-East Brazil. BioMed. Res. Int. 2013;2013:1–7. doi: 10.1155/2013/546354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y., Cao M., Wang M., Ding X., Jing Y., Chen Z., Ma T., Chen H. Genetic variability in E6, E7, and L1 genes of human papillomavirus genotype 52 from Southwest China. Gene. 2016;585:110–118. doi: 10.1016/j.gene.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 57.Zhang C., Park J.-S., Grce M., Hibbitts S., Palefsky J.M., Konno R., Smith-McCune K.K., Giovannelli L., Chu T.-Y., Picconi M.A., Piña-Sánchez P., Settheetham-Ishida W., Coutlée F., De Marco F., Woo Y.-L., Ho W.C.S., Wong M.C.S., Chirenje M.Z., Magure T., Moscicki A.-B., Sabol I., Fiander A.N., Chen Z., Chan M.C.W., Cheung T.-H., Burk R.D., Chan P.K.S. Geographical distribution and risk association of human papillomavirus genotype 52–variant lineages. J. Infect. Dis. 2014;210:1600–1604. doi: 10.1093/infdis/jiu310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loeb L.A., Monnat R.J. DNA polymerases and human disease. Nat. Rev. Genet. 2008;9:594–604. doi: 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- 59.Conger K.L., Liu J.-S., Kuo S.-R., Chow L.T., Wang T.S.-F. Human papillomavirus DNA replication interactions between the viral E1 protein and two subunits of human DNA polymerase α/primase. J. Biol. Chem. 1999;274:2696–2705. doi: 10.1074/jbc.274.5.2696. [DOI] [PubMed] [Google Scholar]
- 60.Henderson S., Chakravarthy A., Su X., Boshoff C., Fenton T.R. APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep. 2014;7:1833–1841. doi: 10.1016/j.celrep.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 61.Chen J., Furano A.V. Breaking bad: the mutagenic effect of DNA repair. DNA Repair. 2015;32:43–51. doi: 10.1016/j.dnarep.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kukimoto I., Mori S., Aoyama S., Wakae K., Muramatsu M., Kondo K. Hypermutation in the E2 gene of human papillomavirus type 16 in cervical intraepithelial neoplasia. J. Med. Virol. 2015;87:1754–1760. doi: 10.1002/jmv.24215. [DOI] [PubMed] [Google Scholar]
- 63.Mirabello L., Yeager M., Yu K., Clifford G.M., Xiao Y., Zhu B., Cullen M., Boland J.F., Wentzensen N., Nelson C.W., Raine-Bennett T., Chen Z., Bass S., Song L., Yang Q., Steinberg M., Burdett L., Dean M., Roberson D., Mitchell J., Lorey T., Franceschi S., Castle P.E., Walker J., Zuna R., Kreimer A.R., Beachler D.C., Hildesheim A., Gonzalez P., Porras C., Burk R.D., Schiffman M. HPV16 E7 genetic conservation is critical to carcinogenesis. Cell. 2017;170:1164–1174. doi: 10.1016/j.cell.2017.08.001. (e6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Firth C., Kitchen A., Shapiro B., Suchard M.A., Holmes E.C., Rambaut A. Using time-structured data to estimate evolutionary rates of double-stranded DNA viruses. Mol. Biol. Evol. 2010;27:2038–2051. doi: 10.1093/molbev/msq088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Doorslaer K. Evolution of the papillomaviridae. Virology. 2013;445:11–20. doi: 10.1016/j.virol.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 66.Bravo I.G., Félez-Sánchez M. Papillomaviruses. Evol. Med. Public Health. 2015;2015:32–51. doi: 10.1093/emph/eov003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.D’Souza G., Burk R.D., Zhong Y., Minkoff H., Massad L.S., Xue X., Watts D.H., Anastos K., Palefsky J.M., Levine A.M., Colie C., Castle P.E., Strickler H.D. Cervicovaginal HPV infection Before and After hysterectomy: evidence of different tissue tropism for oncogenic and non-oncogenic HPV types in a cohort of HIV-positive and HIV-negative women. Int. J. Cancer J. Int. Cancer. 2012;131:1472–1478. doi: 10.1002/ijc.27363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scurry J., Wells M. Viruses in anogenital cancer. Epithel. Cell Biol. 1992;1:138–145. [PubMed] [Google Scholar]
- 69.Darragh T.M., Winkler B. Anal cancer and cervical cancer screening: key differences. Cancer Cytopathol. 2011;119:5–19. doi: 10.1002/cncy.20126. [DOI] [PubMed] [Google Scholar]
- 70.Adler D., Wallace M., Bennie T., Abar B., Sadeghi R., Meiring T., Williamson A.-L., Bekker L.-G. High risk human papillomavirus persistence among HIV-infected young women in South Africa. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2015;33:219–221. doi: 10.1016/j.ijid.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baay M.F.D., Kjetland E.F., Ndhlovu P.D., Deschoolmeester V., Mduluza T., Gomo E., Friis H., Midzi N., Gwanzura L., Mason P.R., Vermorken J.B., Gundersen S.G. Human papillomavirus in a rural community in Zimbabwe: the impact of HIV co-infection on HPV genotype distribution. J. Med. Virol. 2004;73:481–485. doi: 10.1002/jmv.20115. [DOI] [PubMed] [Google Scholar]
- 72.Chin-Hong P.V., Palefsky J.M. Human papillomavirus anogenital disease in HIV-infected individuals. Dermatol. Ther. 2005;18:67–76. doi: 10.1111/j.1529-8019.2005.05009.x. [DOI] [PubMed] [Google Scholar]
- 73.Hang D., Yin Y., Han J., Jiang J., Ma H., Xie S., Feng X., Zhang K., Hu Z., Shen H., Clifford G.M., Dai M., Li N. Analysis of human papillomavirus 16 variants and risk for cervical cancer in Chinese population. Virology. 2016;488:156–161. doi: 10.1016/j.virol.2015.11.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material