Abstract
Objective:
To test a free-breathing MRI protocol for anatomical and functional assessment during lung cancer radiotherapy by assessing two non-Cartesian acquisition schemes based on T1 weighted 3D gradient recall echo sequence: (i) stack-of stars (StarVIBE) and (ii) spiral (SpiralVIBE) trajectories.
Methods:
MR images on five healthy volunteers were acquired on a wide bore 3T scanner (MAGNETOM Skyra, Siemens Healthcare, Erlangen, Germany). Anatomical image quality was assessed on: (1) free breathing (StarVIBE), (2) the standard clinical sequence (volumetric interpolated breath-hold examination, VIBE) acquired in a 20 second (s) compliant breath-hold and (3) 20 s non-compliant breath-hold. For functional assessment, StarVIBE and the current standard breath-hold time-resolved angiography with stochastic trajectories (TWIST) sequence were run as multiphase acquisitions to replicate dynamic contrast enhancement (DCE) in one healthy volunteer. The potential application of the SpiralVIBE sequence for lung parenchymal imaging was assessed on one healthy volunteer. Ten patients with lung cancer were subsequently imaged with the StarVIBE and SpiralVIBE sequences for anatomical and structural assessment. For functional assessment, free-breathing StarVIBE DCE protocol was compared with breath-hold TWIST sequences on four prior lung cancer patients with similar tumour locations. Image quality was evaluated independently and blinded to sequence information by an experienced thoracic radiologist.
Results:
For anatomical assessment, the compliant breath-hold VIBE sequence was better than free-breathing StarVIBE. However, in the presence of a non-compliant breath-hold, StarVIBE was superior. For functional assessment, StarVIBE outperformed the standard sequence and was shown to provide robust DCE data in the presence of motion. The ultrashort echo of the SpiralVIBE sequence enabled visualisation of lung parenchyma.
Conclusion:
The two non-Cartesian acquisition sequences, StarVIBE and SpiralVIBE, provide a free-breathing imaging protocol of the lung with sufficient image quality to permit anatomical, structural and functional assessment during radiotherapy.
Advances in knowledge:
Novel application of non-Cartesian MRI sequences for lung cancer imaging for radiotherapy. Illustration of SpiralVIBE UTE sequence as a promising sequence for lung structural imaging during lung radiotherapy.
Introduction
MRI is being increasingly used in radiotherapy planning for its capability to acquire morphological and functional data during a single scan. However, MRI in lung cancer is challenging due to the low proton density of lung tissue, magnetic susceptibility from microscopic heterogeneity and respiratory motion. Hardware and software development has overcome some of these problems. A number of different protocols and sequences have been proposed for imaging the lung.1–3 T2 weighted (T2W) half-Fourier single-shot turbo spin echo (HASTE) sequence can be used to visualise pathological changes.4 Pulmonary nodules and mediastinal disease are best assessed with a T1 weighted (T1W) 3D gradient recall echo (GRE) sequence with breath-hold.3 A major issue with lung imaging is the effect of respiratory motion. To minimise impact of respiratory motion, respiratory gating or a breath-hold manoeuvre is performed during image acquisition.
Breath-hold techniques are challenging for patients undergoing radiotherapy for lung cancer. These patients usually have compromised pulmonary function and are, therefore, unable to maintain a breath-hold for the required length of time. Breath-hold non-compliance results in poorer image quality. Conventional sequences suffer from image quality degradation due to the way signal data is sampled. K-space is the raw data space where digitised MR signal is stored during acquisition. Since the data is reconstructed using Fourier transform, data in the middle of k-space contains the bulk of the signal contrast for the image and the edge contains data about image resolution (edges and boundaries).5 Conventional MRI sequences acquire k-space data in a line by line (Cartesian) manner, this makes them sensitive to motion as the data is acquired contiguously.5 If the object moves during acquisition (i.e. respiratory motion) this disturbs the phase encoding scheme resulting in artefacts, most evident in the phase encoding direction. Respiratory motion also impacts on functional imaging, where accuracy relies on consistent spatial positioning over time. For optimal assessment of tissue vasculature properties from dynamic contrast enhanced (DCE) imaging, contrast uptake is monitored over several minutes. Motion during these time frames can cause inter-frame misalignment which can affect functional assessment.6
An alternative method of overcoming sensitivity to motion is to change the way k-space is acquired and filled.7 Non-Cartesian acquisition schemes provide a different sampling geometry to manage motion. Two such acquisition schemes are radial and spiral techniques. Radial sampling is based on acquiring k-space data along radial spokes. Each radial projection passes through the centre of k-space, therefore, the signal is heavily averaged. A stack-of-stars technique is performed for in-plane sampling and Cartesian sampling is performed in the slice direction.8 Due to the overlap of spokes in the centre, the distribution of k-space data along individual spokes is averaged out. Although robust in minimising aliasing, ghosting and motion artefacts, radial trajectories are prone to blurring and radial streak artefacts inherent to the sampling scheme. The oversampling on the k-space results in increased acquisition time compared with conventional imaging.5 Due to its lower motion sensitivity radial stack-of-stars acquisition allows a free-breathing T1W sequence that can aid in accurate tumour volume identification and delineation.8 However, with the higher sampling requirement of k-space data, the stack-of-stars technique is limited by temporal resolution. Whilst a conventional T1W GRE sequence allows the entire thorax to be imaged in a 20 second (s) breath-hold, a radial GRE sequence requires a longer acquisition time to acquire the equivalent anatomical coverage. Without the breath-hold constraint it is possible to increase the spatial resolution compared to the Cartesian breath-hold scan. This, of course, increases the acquisition time even further.
Magnetic susceptibility in lung tissue causes signal loss due to extremely short T2* relaxation time of lung tissue, this limits the ability of MRI sequences with conventional echo time (TE) to acquire images of the lung parenchyma. To monitor changes within the tumour and also healthy lung, high resolution CT is used to acquire images of lung parenchyma to assess radiological changes within the lung and correspond this to histological outcomes.9 To acquire lung parenchyma images with MRI, sequences need to accommodate fast signal acquisition following excitation before signal decay. Ultrashort echo time (UTE) imaging has be shown to be ideal for imaging lung parenchyma and evaluating lung microstructure at the alveolar level.10,11 Most 3D UTE sequences combine short non-selective RF pulses with a 3D centre out radial trajectory. Scan time can be reduced by using a more efficient stack-of-spirals trajectory. Qian and Boada12 showed that a short effective TE can be realised with the stack-of-spirals trajectory, if the duration between excitation and spiral readout is minimised for each through-plane phase-encoding step separately.
Non-Cartesian trajectories and, in particular, radial and spiral k-space sampling techniques have potential applications in imaging for lung cancer in radiotherapy. In order to utilise the superior soft tissue contrast of MRI for target volume delineation for lung radiotherapy, management of respiratory motion is a prerequisite. Radial sampling provides a potential solution for a free-breathing T1W GRE sequence for anatomical delineation of lung tumour volume as well as a free-breathing DCE sequence. The spiral sampling technique coupled with UTE imaging provides information on lung parenchyma, which is not visible with standard imaging sequence, under free-breathing conditions. The aim of this paper is to describe the application of the two non-Cartesian k-space acquisition techniques to minimise the impact of respiratory motion on image quality, during lung MRI acquired for radiotherapy planning. These sequences were compared with existing corresponding Cartesian sequences.
Methodology
Imaging technique
For non-Cartesian imaging, a T1W volumetric interpolated breath-hold examination (VIBE) with radial acquisition trajectory (StarVIBE) was assessed for anatomical and DCE assessment of tumour volume. For comparison with Cartesian sequence, a T1W VIBE sequence with DIXON fat suppression technique was utilised for anatomical imaging. T1 VIBE with DIXON is a 3D sequence with contiguous and spatially registered slices. DIXON is more robust in the presence of varying tissue interfaces and suffers less from paramagnetic susceptibility artefacts (which are abundant in the lungs) compared to chemical fat suppression. However, DIXON is limited to Cartesian k-space sampling with the VIBE sequence but may be performed with a 20 sec breath-hold. The sequence may be degraded by respiratory motion, therefore patient compliance to breath-hold is important. For comparison with a Cartesian DCE sequence, time-resolved angiography with stochastic trajectories (TWIST) was used with a series of multiple short (20 sec) breath-hold periods over a course of time. TWIST is a variant of high resolution breath-hold 3D flash angiography which allows assessment of tumour and lung perfusion with high resolution 3D imaging. TWIST compared to StarVIBE provides better temporal resolution (can acquire the entire thorax with the same acquisition time) but the integrity of the data sets is reliant on patient compliance to breath-hold. Images are acquired over seven respiratory phases for a total of thirty eight measurements.
Lung microstructure imaging was performed with a works-in-progress (WIP) sequence based on UTE imaging with a stack-of-spirals acquisition scheme (SpiralVIBE) encompassing the entire thorax. Different to the original technique of Qian and Boada,12 which uses a slab selective excitation, the WIP sequence used in this work utilises a 60 μs non-selective rectangular pulse to achieve effective TE times of 50 μs. Images were acquired in a coronal orientation to minimise the number of through-plane phase encoding steps.13 Furthermore, the sequence incorporates navigator scans for respiratory gating. The navigator uses a Cartesian readout in head-feet direction. Navigator scans and imaging scans use the same excitation pulse and TR to sustain steady state. The information gained with the navigator is used to obtain scans at inspiration during a free-breathing acquisition. MRI sequences and parameters are outlined in Table 1.
Table 1.
Sequence parameters
| Parameters | Anatomical/structural | Functional | |||||
|---|---|---|---|---|---|---|---|
| StarVIBE | TWIST | ||||||
| StarVIBE | VIBE | SpiralVIBE | T1 map (variable flip angle) | DCE | T1 map (variable flip angle) | DCE | |
| TR/TE (ms) | 7.46/2.46 | 5.79/2.46 | 2.97/0.05 | 5.00/1.33 | 5/1.33 | 4.09/1.29 | 1.98/0.88 |
| Frequency FOV (mm) | 400 | 380 | 480 | 340 | 340 | 380 | 380 |
| Average FOV in slice direction (mm)a | 300 | 320 | 288 | 72 | 72 | 240 | 240 |
| Acquisition orientation | Transverse | Transverse | Coronal | Coronal | Coronal | Coronal | Coronal |
| Flip angle | 9 | 5 | 2 & 15 | 9 | 2 & 15 | 10 | |
| In-plane resolution acquired (mm) | 1.25 × 1.25 | 1.2 × 1.2 | 1.5 × 1.5 | 1.77 × 1.77 | 1.77 × 1.77 | 2.0 × 2.0 | 1.5 × 1.5 |
| Base resolution | 320 | 320 | 320 | 192 | 192 | 192 | 256 |
| Slice thickness (mm) | 4 | 4 | 1.5 | 4 | 4 | 4 | 4 |
| Bandwidth (Hz/Px) | 820 | 400 | NA | 810 | 810 | 390 | 1500 |
| Acquisition time (s) | 220 | 14–20 (BH) | 295 | 12 | 12 per measurement | 18 (BH) | 20 BH per measurement |
| Radial view/spiral interleaves | Radial views: 600 | NA | Spiral interleaves: 504 | Radial views: 300 | Radial views: 300 | NA | NA |
DCE, dynamic contrast enhanced; FOV, field of view; NA, not applicable; TR/TE, repetition time/echo time.
aFOV in slice direction = (slice thickness × no. of slices), this is variable per patient depending on the number of slices acquired.
Data acquisition
All images were acquired on a 3T wide bore scanner (MAGNETOM Skyra, Siemens Healthcare, Erlangen, Germany) using a 32-channel spine coil and a 18-channel surface coil placed directly on subjects thorax. Patients were positioned in a supine radiotherapy treatment setup, with their arms raised above their head in a customised vacuum bag (BlueBAG™, Elekta, Stockholm, Sweden) for immobilisation and a flat wing board (MTWB09 Wingboard, CIVCO Medical Solutions, Orange City, IA). Healthy volunteers were positioned in a similar setup without customised vacuum bag.
For comparison with CT data for structural assessment, each patient’s 4DCT radiotherapy planning CT was used. MRI and CT scans were performed prior to the start of patient’s treatment within one week of each other. All patient CT scans were performed on the departmental Phillips Brilliance Big Bore 16 slice CT scanner (Phillips Medical Systems, Cleveland, OH). For all patients the CT image matrix was 512 × 512 mm, slice thickness of 2 mm with 2 mm slice increments.
Anatomical and structural assessment
Healthy volunteers
The applicability of the T1 StarVIBE sequences was assessed on five healthy volunteers. For comparison, the existing T1 W breath-hold VIBE sequence with and without compliant breath-hold (representative of patient non-compliance to breath-hold instruction) was also acquired for these volunteers. The SpiralVIBE sequence was assessed on one healthy volunteer with free-breathing instructions to setup parameters to optimise image quality.
Patient imaging
Images from ten consecutive lung cancer patients, enrolled in a prospective study were acquired. Image acquisition for StarVIBE and SpiralVIBE was performed with free-breathing instruction. For anatomical imaging, StarVIBE sequence was acquired in transverse plane to match CT imaging plane. SpiralVIBE was acquired in coronal plane.
Functional assessment
Healthy volunteer
To assess impact of respiratory motion on DCE imaging, one healthy volunteer was scanned with StarVIBE and breath-hold TWIST sequences. The volunteer was instructed to maintain a compliant breath-hold during TWIST imaging, for StarVIBE imaging shallow breathing instruction was given. Interframe misalignment between imaging phases was used as assessment of respiratory motion impact.
Patient images
StarVIBE DCE images were acquired using the same four consecutive lung cancer patients imaged for anatomical assessment. FOV for DCE waslimited to the tumour in the anteroposterior direction to maintain a fast temporal resolution. Intravenous administration of Gadobutrol 4.535 g (Gadovist 1.0, Bayer, Leverkusen, Germany) mmol/kg half-dose at 4 ml/ sec was given during DCE imaging. Direct comparison of image quality between breath-hold and free-breathing DCE MRI sequence was not possible with the same patients due to the administration of contrast for DCE imaging. For comparison with the free-breathing, StarVIBE DCE protocol data sets from four previous patients imaged with breath-hold TWIST protocol were included in the assessment. The previous patients were matched for tumour location where possible with all four patients imaged with the free-breathing StarVIBE protocol.
Semi quantitative assessment
The visibility of tumour boundaries and bronchi for patient imaging was scored according to a four-point scale developed in-house (Table 2). For healthy volunteers, assessment of the pulmonary trunk was used as a tumour surrogate for image quality analysis. Images were scored by an experienced thoracic radiologist and radiation oncologist. A score of two or less was considered to be clinically acceptable for tumour volume delineation and structural definition.
Table 2.
Tumour/surrogate and bronchi score categories
| Score | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Edge detection | Edge clearly defined | Structure edge slightly blurred, not impairing definition of boundary | Considerable blurring of structure edge impacting on accurate definition of boundary | Significant blurring of boundary edge, definition of structure boundary not achievable |
| Artefacts | No artefacts | Minimal artefact not impairing image quality | Considerable artefacts impacting evaluation of anatomical structures | Extreme artefacts obscuring delineation of structures |
| Image noise | Minimal noise | Minimal noise not impairing image quality | Considerable noise impacting evaluation of anatomical structures | Extreme noise obscuring definition of structures |
| Overall image quality | Very good image quality | Fair image quality not impairing delineation of structures | Impaired image quality that may lead to incorrect delineation | Anatomical structure not definable |
For comparison of functional imaging, misalignment of interframe images was assessed with image subtraction between frames. To assess the impact of respiratory motion on DCE imaging between free-breathing and breath-hold, in-plane misregistration between imaging phases on subtracted images were used.
Results
Anatomical assessment
Healthy volunteers
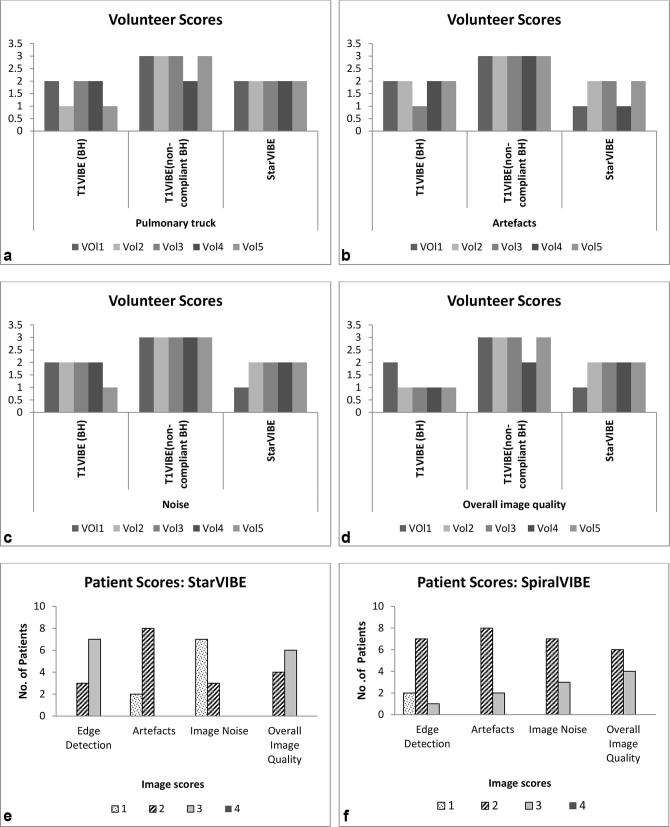
For each of the four image quality score criteria, both StarVIBE and compliant breath-hold T1 VIBE sequences scored better than non-compliant T1 VIBE for all five volunteers (Figure 1a–d). Sequence parameters for DCE StarVibe were initially copied from the anatomical StarVIBE sequence. However, based on the volunteer imaging, radial views of 600 for DCE StarVIBE resulted in an increase in acquisition time of 23 s. To maintain scan efficiency while limiting streak artefacts, the base resolution and radial views were reduced to 300 allow a 12-sec acquisition time per measurement. Significant signal saturation was present on the MR images anteriorly on the healthy volunteer UTE images. To overcome this, the surface coil was raised away from patient anatomy with sponges to physically normalise the signal.
Figure 1.
Healthy volunteer and patient anatomical scores.
Patient images
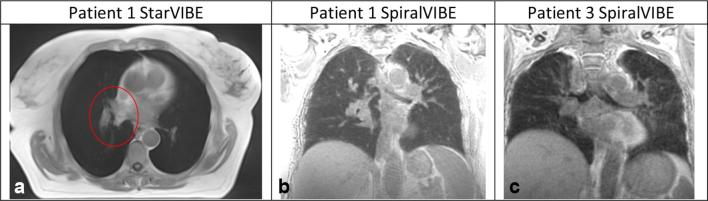
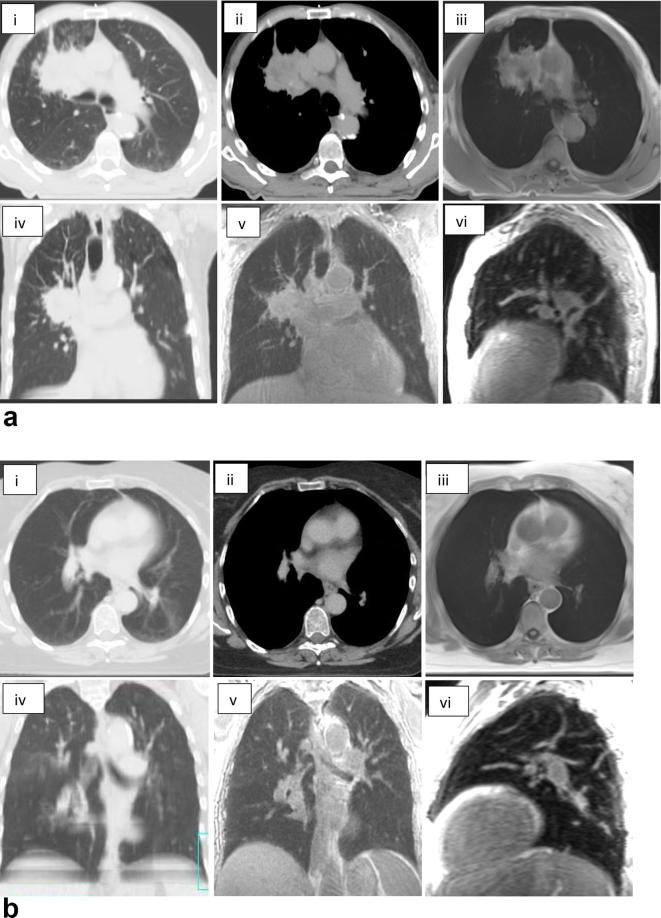
Figure 1e,f illustrate the image scores across the ten patients. All patient images scored two or less for image artefact and noise, seven patients had a score of three for edge detection. Figure 2 illustrates the blurred tumour edge for patient 1 for StarVIBE (a) and increased image noise for patients 1 and 3 for SpiralVIBE (b,c). There was more visual noise present in the SpiralVIBE sequence compared with CT, however, this did not impair overall image quality. Nine patients had a score of less than two for edge detection of tumour on the SpiralVIBE. Figure 3a and b(iii) illustrates the anatomical image quality acquired with the free-breathing non-Cartesian sequence StarVIBE. In comparison with the 4D planning CT (i-ii) the tumour boundary is slightly blurred, however, sufficient edge detail is present to define boundary. Signal from lung parenchyma is visible on SpiralVIBE images (v–vi).
Figure 2.
(a) Blurred anatomical detail to define tumour edge for patient 1 StarVIBE image defined by red ROI. (b) Grainy image, impacting on visualisation of bronchial structure, (c) Grainy image appearance, slight distortion of bronchial structures.
Figure 3.
(a): Images of a patient with non-small cell lung cancer (Patient 2), (b) Images of a patient with small cell lung cancer (Patient 1). (i) RT planning CT (lung window); (ii) RT planning CT (mediastinal window); (iii) Axial StarVIBE MRI; (iv) Radiotherapy planning CT coronal plane; (v) SpiralVIBE coronal plane (UTE); (vi) SpiralVIBE sagittal plane (UTE).
Functional
Healthy volunteer
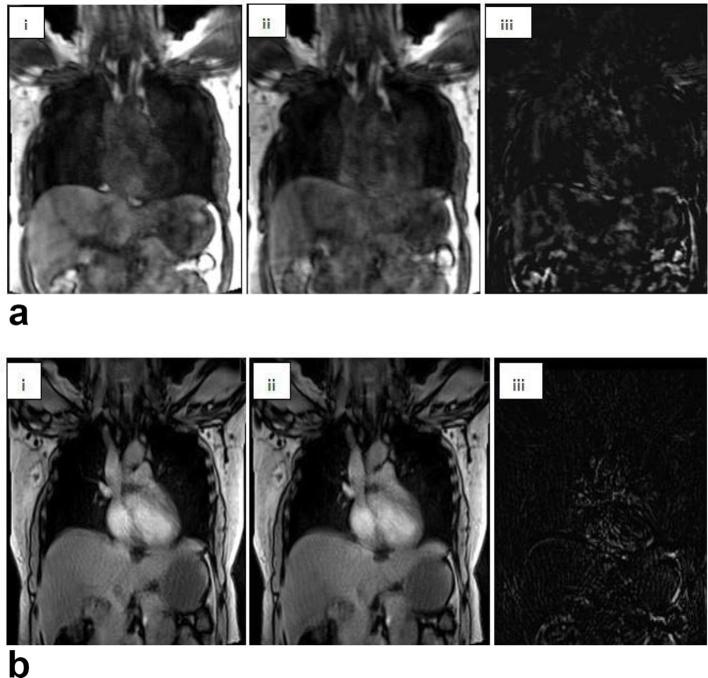
For inter-frame misalignment, a large variation between frames was noted on subtracted images for TWIST compared to StarVIBE sequences (Figure 4).
Figure 4.
(a): Breath-hold compliant TWIST image, (b): (i) Free-breathing StarVIBE; (ii) Same slice position different frames; (iii) Subtracted image between (i) and (ii).
Patient
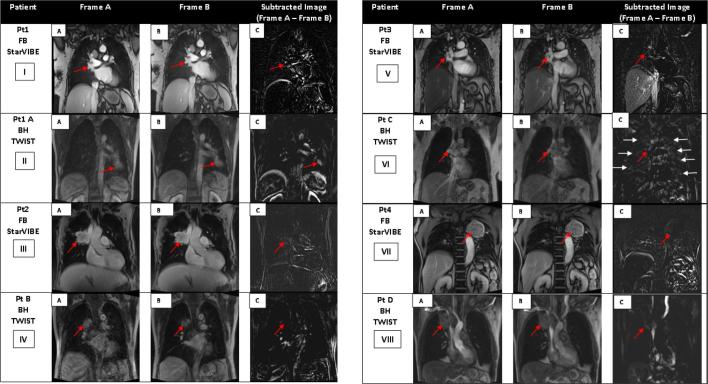
Inter-frame misalignment of patient data for free-breathing StarVIBE and breath-hold TWIST is shown in Figure 5; Phase 1 and 2 images were taken at the same slice position with a difference of no more than 5 s between phases. For patients imaged with free-breathing StarVIBE, although misalignment was noted near the liver dome and abdominal structures, alignment near the tumour volume was sufficient for a free-breathing sequence. For the breath-hold TWIST protocol only Patient B had minimal variation near the tumour volume (Figure 5iv iv,C). For the other breath-hold TWIST images, variation in signal near tumour position was seen for patient A, C and D (Figure 5).
Figure 5.
Coronal frames acquired during DCE acquisition at the same slice position during different phases, maximum of 5 s apart for free-breathing StarVIBE and breath-hold TWIST. Breath-hold TWIST images acquired during the same breath-hold phase. (a–b) Red arrows indicate position of tumour volume during the two phases. (c) Red arrow indicates tumour position on subtracted image, residual signal on subtracted image indicative of variation in tumour position between the two frames. Image vi (c) demonstrates residual signal within normal lung shown by white arrows. This is indicative of the non-compliance to breath-hold instruction during the breath-hold scan.
Discussion
This study demonstrates the potential application of two free-breathing non-Cartesian image sequences for cancer imaging in radiotherapy. The pre-contrast free-breathing T1W StarVIBE was shown to be robust in the presence of motion, however, slight blurring at the edges of tumour volume was evident. To optimise image quality, radial views were kept at 300 to minimise streak artefacts. Temporal resolution was reduced for StarVIBE compared to breath-hold T1 VIBE. For T1 VIBE, a 20 sec breath-hold was required to acquire the entire thorax, for StarVIBE this extended to approximately a 3 min free-breathing scan for the same FOV. Similar compromises had to be made for the DCE protocol. The breath-hold TWIST sequence allowed image acquisition of the entire thorax during a 5 min session with 20 sec breath-hold periods. To balance reduced temporal resolution with acquisition of sufficient data without prolonging scan times, the scan limit for the DCE study was limited to the region of tumour volume in the anteroposterior direction for the coronal acquisition planes. While motion correction function is available with most commercial software such as Tissue 4D (Siemens Healthcare, Erlangen, Germany) for perfusion analysis, motion correction only allows alignment of the dynamic images to the reference images. Motion correction did not improve the integrity of the image quality between imaging phases for the breath-hold TWIST sequence, it also did not improve artefacts within the acquired image. To maintain integrity of DCE data acquired as well as ensuring accurate post imaging analysis, StarVIBE demonstrated more efficient motion correction compared to breath-hold TWIST. With improved motion management on a T1 sequence, StarVIBE sequence can be used for delineation of tumour volume for lung.4 SpiralVIBE allowed the detection of signal from the lung parenchyma, which is not visible on MRI sequences with conventional TE. Results show that lung parenchyma imaging is possible with minimal respiratory artefacts during free-breathing acquisition. While both non-Cartesian sequences have longer acquisition times, breath-holds were avoided and this improved image quality in lung cancer patients with compromised breath-hold capacities ranging between15 to 20 sec for a single breath-hold.14,15 Minimum breath-hold durations recommended for most lung imaging sequence is20 sec.16
Other studies have demonstrated the effectiveness of StarVIBE in managing motion for abdominal and pelvic DCE imaging.17,18 DCE imaging in oncology allows for quantitative assessment of tumour status. It has been shown to be a non-invasive method to measure tumour properties based on the tumour microvasculature with potential for use as an imaging biomarker to assess treatment response.19,20 However, to ensure accurate analysis of tumour enhancement kinetics, data over a period of time needs to be acquired at high temporal resolution. In the presence of respiratory motion, tumour displacement and blurring can introduce significant errors in the measured parameters and pixel by pixel analysis. Therefore, appropriate motion management is necessary if DCE imaging is to be utilised in lung cancer radiotherapy as a potential biomarker to guide radiotherapy treatment and also to assess treatment response. The StarVIBE sequence provides a potential solution to improve quality of DCE data acquired for functional assessment of tumour volume. Apart from improving image integrity of functional imaging, free-breathing sequences are also of benefit for integrated MRI radiotherapy systems to facilitate online tracking of target volumes.21
Similarly, UTE imaging has been shown to enable visualisation of other lung disease within the lung parenchyma.10 Utilisation of UTE imaging in radiation therapy has mainly been for deriving substitute CT data for planning from the MRI data.22 However, UTE imaging can be utilised to assess lung architecture during the course of treatment and post treatment. T2* derived from UTE images can be utilised to differentiate between fibrosis and non-specific pneumonitis on CT.23 The SpiralVIBE sequence has further applications in assessing radiological changes that occur in the lung parenchyma during radiotherapy, particularly in relation to lung toxicities. The work presented in this paper is the first illustration of the free-breathing SpiralVIBE UTE for radiotherapy imaging and its potential application in monitoring changes during and post treatment. In the context of MRI integrated radiotherapy systems, UTE can allow visualisation of lung parenchyma to facilitate generation of pseudo CT for radiotherapy therapy or be used in lieu of the CT data.24 As SpiralVIBE is acquired during free breathing, it has the potential to provide equivalent information to current radiotherapy 3DCT images used for planning. This study is limited by the small number of healthy volunteers and patients. For functional image assessment, it would have been ideal to compare motion compensation between TWIST and StarVIBE sequence for the same patient; however, this is challenging due to contrast administration. All lung cancer patients undergoing MRI at Liverpool Hospital are now scanned using these free-breathing sequences.
Conclusion
The aim of this pilot study was to illustrate the application of a free-breathing non-Cartesian image acquisition for anatomical and structural imaging of lung for the purpose of radiotherapy imaging. Both non-Cartesian imaging sequences were able to achieve this with clinically acceptable image quality. MR imaging of lung cancer for the purposes of radiotherapy, where high spatial resolution is necessary, is feasible. The utilisation of non-Cartesian imaging sequence to manage respiratory motion should be considered where available to improve lung MRI image quality.
Acknowledgments
Acknowledgment
The SpiralVIBE sequence used here is a prototype research sequence provided by Siemens Healthcare.
Conflicts of Interest
Sonal Josan and Alto Stemmer are employees of Siemens Healthcare.
Contributor Information
Shivani Kumar, Email: shivani.kumar@sswahs.nsw.gov.au.
Robba Rai, Email: Robba.Rai@sswahs.nsw.gov.au.
Alto Stemmer, Email: alto.stemmer@siemens.com.
Sonal Josan, Email: sonal.josan@siemens.com.
Lois Holloway, Email: Lois.Holloway@sswahs.nsw.gov.au.
Shalini Vinod, Email: shalini.vinod@sswahs.nsw.gov.au.
Daniel Moses, Email: daniel.moses@gmail.com.
Gary Liney, Email: Gary.Liney@sswahs.nsw.gov.au.
References
- 1.Biederer J, Mirsadraee S, Beer M, Molinari F, Hintze C, Bauman G, et al. MRI of the lung (3/3)-current applications and future perspectives. Insights Imaging 2012; 3: 373–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cobben DC, de Boer HC, Tijssen RH, Rutten EG, van Vulpen M, Peerlings J, et al. Emerging Role of MRI for Radiation Treatment Planning in Lung Cancer. Technol Cancer Res Treat 2016; 15: NP47–NP60. [DOI] [PubMed] [Google Scholar]
- 3.Kumar S, Liney G, Rai R, Holloway L, Moses D, Vinod SK. Magnetic resonance imaging in lung: a review of its potential for radiotherapy. Br J Radiol 2016; 89: 20150431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar S, Rai R, Moses D, Choong C, Holloway L, Vinod SK, et al. MRI in radiotherapy for lung cancer: A free-breathing protocol at 3T. Pract Radiat Oncol 2017; 7: e175–e183. [DOI] [PubMed] [Google Scholar]
- 5.McRobbie DW, Moore EA, Graves MJ, Prince MR. MRI from Picture to Proton. Cambridge university press; 2007. [Google Scholar]
- 6.Hamy V, Dikaios N, Punwani S, Melbourne A, Latifoltojar A, Makanyanga J, et al. Respiratory motion correction in dynamic MRI using robust data decomposition registration - application to DCE-MRI. Med Image Anal 2014; 18: 301–13. [DOI] [PubMed] [Google Scholar]
- 7.Kierans A, Parikh N, Chandarana H. Recent Advances in MR Hardware and Software. Radiol Clin North Am 2015; 53: 599–610. [DOI] [PubMed] [Google Scholar]
- 8.Block KT, Chandarana H, Milla S, Bruno M, Mulholland T, Fatterpekar G, et al. Towards Routine Clinical Use of Radial Stack-of-Stars 3D Gradient-Echo Sequences for Reducing Motion Sensitivity. J Korean Soc Mag Resonan Med 2014; 18: 87–106. [Google Scholar]
- 9.Choi YW, Munden RF, Erasmus JJ, Park KJ, Chung WK, Jeon SC, et al. Effects of radiation therapy on the lung: radiologic appearances and differential diagnosis. Radiographics 2004; 24: 985–97. [DOI] [PubMed] [Google Scholar]
- 10.Dournes G, Grodzki D, Macey J, Girodet PO, Fayon M, Chateil JF, et al. Quiet Submillimeter MR Imaging of the Lung Is Feasible with a PETRA Sequence at 1.5 T. Radiology 2015; 276: 258–65. [DOI] [PubMed] [Google Scholar]
- 11.Johnson KM, Fain SB, Schiebler ML, Nagle S. Optimized 3D ultrashort echo time pulmonary MRI. Magn Reson Med 2013; 70: 1241–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qian Y, Boada FE. Acquisition-weighted stack of spirals for fast high-resolution three-dimensional ultra-short echo time MR imaging. Magn Reson Med 2008; 60: 135–45. [DOI] [PubMed] [Google Scholar]
- 13.Mugler III JP, Fielden WS, Meyer HC, Altes AT, Miller GW, Stemmer A. . Breath-hold UTE Lung Imaging using a Stack-of-Spirals Acquisition In: Proc Intl Soc Mag Reson Med; 2015. [Google Scholar]
- 14.Kumar SS, Holloway LC, Koh ES, Phan PD, Choong CH, Vinod S. Active breathing coordination to measure tumour motion in lung cancer patients: a feasibility study. Philadelphia, PA: Lippincott Williams & Wilkins; 2013. [Google Scholar]
- 15.Wong JW, Sharpe MB, Jaffray DA, Kini VR, Robertson JM, Stromberg JS, et al. The use of active breathing control (ABC) to reduce margin for breathing motion. Int J Radiat Oncol Biol Phys 1999; 44: 911–9. [DOI] [PubMed] [Google Scholar]
- 16.Biederer J, Beer M, Hirsch W, Wild J, Fabel M, Puderbach M. MRI of the lung (2/3). Why… when… how? Insights Imaging 2012; 3: 373–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandarana H, Block KT, Winfeld MJ, Lala SV, Mazori D, Giuffrida E, et al. Free-breathing contrast-enhanced T1-weighted gradient-echo imaging with radial k-space sampling for paediatric abdominopelvic MRI. Eur Radiol 2014; 24: 320–6. [DOI] [PubMed] [Google Scholar]
- 18.Reiner CS, Neville AM, Nazeer HK, Breault S, Dale BM, Merkle EM, et al. Contrast-enhanced free-breathing 3D T1-weighted gradient-echo sequence for hepatobiliary MRI in patients with breath-holding difficulties. Eur Radiol 2013; 23: 3087–93. [DOI] [PubMed] [Google Scholar]
- 19.Pauls S, Breining T, Muche R, Schmidt SA, Wunderlich A, Krüger S, et al. The role of dynamic, contrast-enhanced MRI in differentiating lung tumor subtypes. Clin Imaging 2011; 35: 259–65. [DOI] [PubMed] [Google Scholar]
- 20.Weller A, O'Brien MER, Ahmed M, Popat S, Bhosle J, McDonald F, et al. Mechanism and non-mechanism based imaging biomarkers for assessing biological response to treatment in non-small cell lung cancer. Eur J Cancer 2016; 59: 65–78. [DOI] [PubMed] [Google Scholar]
- 21.Bjerre T, Crijns S, af Rosenschöld PM, Aznar M, Specht L, Larsen R, et al. Three-dimensional MRI-linac intra-fraction guidance using multiple orthogonal cine-MRI planes. Phys Med Biol 2013; 58: 4943: 4943–50. [DOI] [PubMed] [Google Scholar]
- 22.Johansson A, Karlsson M, Nyholm T. CT substitute derived from MRI sequences with ultrashort echo time. Med Phys 2011; 38: 2708–14. [DOI] [PubMed] [Google Scholar]
- 23.Alex W, Sharon G, Sharon G, Veronica M, David C, David H. et al. 3D Ultrashort TE (UTE) MRI repeatability within the thorax and it's application to pulmonary fibrosis In: International Society for Magnetic Resonance in Medicine 23rd Annual Meeting; 2015. [Google Scholar]
- 24.Edmund JM, Nyholm T. A review of substitute CT generation for MRI-only radiation therapy. Radiat Oncol 2017; 12: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]