Abstract
The core LATS kinases of the Hippo tumor suppressor pathway phosphorylate and inhibit the downstream transcriptional co-activators YAP and TAZ, which are implicated in various cancers. Recent studies have identified various E3 ubiquitin ligases that negatively regulate the Hippo pathway via ubiquitination, yet few deubiquitinating enzymes (DUB) have been implicated. In this study, we report the DUB USP9X is an important regulator of the core kinases of this pathway. USP9X interacted strongly with LATS kinase and to a lesser extent with WW45, KIBRA, and Angiomotin, and LATS co-migrated exclusively with USP9X during gel filtration chromatography analysis. Knockdown of USP9X significantly downregulated and destabilized LATS and resulted in enhanced nuclear translocation of YAP and TAZ, accompanied with activation of their target genes. In the absence of USP9X, cells exhibited an epithelial-to-mesenchymal transition phenotype, acquired anchorage-independent growth in soft agar, and led to enlarged, disorganized, three-dimensional acini. YAP/TAZ target gene activation in response to USP9X knockdown was suppressed by knockdown of YAP, TAZ, and TEAD2. Deletion of USP9X in mouse embryonic fibroblasts resulted in significant downregulation of LATS. Furthermore, USP9X protein expression correlated positively with LATS but negatively with YAP/TAZ in pancreatic cancer tissues as well as pancreatic and breast cancer cell lines. Overall, these results strongly indicate that USP9X potentiates LATS kinase to suppress tumor growth.
Keywords: Hippo pathway, USP9X, LATS kinase, YAP, TAZ, TEAD
Introduction
The Hippo pathway is an evolutionarily conserved kinase cascade governing organ size and contact inhibition through regulation of cell proliferation and apoptosis during organ development and regeneration. Dysregulation of this signaling cascade contributes to tumorigenesis. The core components of the pathway consist of upstream regulators, a core kinase cassette, and downstream oncogenes. Once the core kinases are activated, LATS1/2 kinase phosphorylates downstream targets YAP and TAZ leading to their cytoplasmic retention and inactivation; otherwise, YAP and TAZ translocate into the nucleus through interaction with transcription factor TEAD to activate pro-proliferative and anti-apoptotic genes. YAP and TAZ have been shown to be fundamental for cancer initiation and growth of most solid tumors including liver, breast, lung and pancreatic cancers (1–6).
In most recent reports, key members of the Hippo pathway were revealed to be negatively regulated by ubiquitination. A set of E3 ubiquitin ligases were shown to destabilize LATS1 (7,8), LATS2 (9), WW45 (10) or AMOT (11), resulting in stimulation of YAP and TAZ activity. Ubiquitination and deubiquitination of proteins play an essential role in the regulation of homeostasis and are increasingly implicated in the onset and progression of various tumors. Nevertheless, little information regarding regulation of the Hippo pathway by deubiquitinating enzymes has been described.
The ubiquitin-specific protease 9X (USP9X/FAM) is a highly conserved member of the ubiquitin-specific proteases (USP) family of deubiquitinating enzymes (12). USP9X is known to play a crucial role in development and disease (13). Recently, more studies emerged implicating this deubiquitinating enzyme in initiation and progression of various cancers. In pancreatic ductal adenocarcinoma, USP9X was found to be the most commonly mutated gene inactivated in over 50% of tumors in a mouse mutagenesis screen of pancreatic cancer and its loss enhanced transformation of pancreatic cancer cells (14). Furthermore, elevated expression of USP9X has been linked to the poor prognosis of patients diagnosed with non-small lung carcinoma (15). Thus, USP9X may play a context-specific role in the progression of human cancers, acting either as a tumor suppressor or an oncoprotein. From our Hippo pathway proteomic data, USP9X stands out as one of the high confidence interactors.
In this study, we report the role of USP9X as a positive regulator of the Hippo pathway signaling. We reveal that USP9X interacts strongly with Hippo components LATS, moderately with WW45, to a lesser extent with KIBRA and Angiomotins. Only LATS and USP9X co-migrated strongly in the same fraction of cell lysates separated by FPLC. USP9X stabilizes each of these components and inhibits downstream activation of YAP/TAZ and TEAD. The expression of USP9X is correlated positively with LATS kinases and negatively with YAP/TAZ in pancreatic and breast cancer cells as well as pancreatic cancer patient tissues, indicating that USP9X synergize the Hippo components to mediate tumor suppressor function.
Materials and Methods
Cell Culture
Normal human pancreatic ductal epithelial (HPDE6) cell line was a kind gift from Dr. Ming-Sound Tsao (Princess Margaret Cancer Centre, UHN, Canada). HPDE6 cells were cultured in keratinocyte serum free (KSF) medium supplemented with epidermal growth factor and bovine pituitary extract (Gibco Life Technologies). All the rest of the cell lines were purchased from American Type Culture Collection (ATCC) (after year 2010) with cell authentication and mycoplasma tests. The cell authentication tests were last performed by DNA Sequencing Facility of the Institute of Molecular and Cell Biology, Singapore, using Promega GenePrint® 10 System in July 2017 and had >90% match with ATCC database. All the cell lines in the lab are routinely tested every 6 months for mycoplasma using Lonza MycoAlert mycoplasma detection kit and last mycoplasma tests were perform in July, 2017. The cells described in the experiments were used within 10 passages after thawing from the original vials. The three-dimensional culture of MCF10A mammary epithelial acini was performed exactly as outlined previously (16). Fibroblasts were isolated from day 14.5 mouse embryos following mating of Usp9xflox/flox females to Usp9xflox/Y males (17), and cultured as described (18). Animal studies have been conducted in accordance with an Institutional Animal Care and Use Committee (IACUC).
Plasmids
The cDNA of human USP9X was from NIH MCG 428. Flag-tagged and Myc-tagged versions of USP9X-wild-type (USP9X-WT) were constructed by PCR using the MCG clone as the template and cloned into pClneo vector (Promega). Myc-tagged enzyme defective mutant, USP9X-H1871A, was generated by introduction of histidine to alanine residue (1871) of the enzyme catalytic domain by PCR. The HA-tagged Ubiquitin plasmid was purchased from Addgene (#17608). Flag-tagged LATS2 fragments and HA-tagged USP9X fragments were constructed by PCR using pClneo-Flag-LATS2 and pClneo-HA-USP9X as a template, respectively. All the rest of plasmids were described previously (19–22). Cre cDNA was cloned into pLVXpuro from Clontech.
Antibodies and reagents
USP9X (#A301-350A) and LATS (#A300-479A) antibodies were purchased from Bethyl Laboratories Inc. YAP (#4912), phospho-YAP127 (#4911), TAZ (#4883), MST1 (#3682), MST2 (#3952) and Pan-TEAD (#13295) antibodies were from Cell Signaling Technology. KIBRA (#LS-C175353) antibody was purchased from LifeSpan BioSciences, WW45 (#sc-374366) antibody was from Santa Cruz Biotechnology and AMOT antibody was from Abnova as previously described (19,20). Fibronectin (#610077), vimentin (#550513) and E-cadherin (#610404) were from BD Biosciences. Anti-Flag antibody was from Sigma, anti-HA and anti-Myc antibodies were purchased from Roche. Immunohistochemistry analyses were completed using USP9X (#55054-1-AP) antibody purchased from Proteintech, LATS1/2 (#ab70565) from Abcam and YAP/TAZ (#8418) from Cell Signaling Technology. Western blot analyses were performed as described previously (23). Mouse anti-HA-agarose antibody was from Sigma. Anti c-Myc monoclonal agarose conjugate was purchased from Nacalai Tesque Inc. and E2 view red anti-Flag M2 affinity gel was from Sigma. MG-132 proteasome inhibitor was obtained from Calbiochem. Nuclear and cytoplasmic fractions were prepared using NE-PER Nuclear and Cytoplasmic Extraction reagents (#78833, Thermo Scientific).
Gel filtration analysis
1.2 mg of HEK293 cell lysate was fractionated using the Superose™ 6 HR 10/30 column (GE Healthcare), using the ÄKTA™ Purifier 10 FPLC system (GE Healthcare). Fractionation was performed in PBS at 4°C. Flow rate was 0.3 ml/min and fractions of 0.6 ml each were collected. The fractions were precipitated with trichloroacetic acid (TCA), resolved by SDS-PAGE and transferred to PVDF membrane (Millipore) for subsequent immunodetection.
Anchorage-independent growth in soft agar, Cell motility and Invasion assays (Transwell assays), Retrovirus generation, Infection, Immunoprecipitation and Mass Spectrometry and data analysis
Lentivirus Generation and Infection
293FT cell line and ViraPower Packaging Mix for lentivirus production were purchased from Thermo Fisher Scientific and lentiviruses were produced according to their instruction.
RNA isolation and real-time PCR
The protocols were carried out as previously described (19,21). TaqMan probes used in this study were purchased from ThermoFisher Scientific; USP9X, #Hs01126052; CTGF, #Hs00170014; CYR61, #Hs00155479; TEAD1, #Hs00173359; TEAD2, #Hs00366217; TEAD3, #Hs00243231; TEAD4, #Hs01125032 and 18S, #Hs03003631.
siRNA
Transfections were carried out as previously described (22). The following siRNAs were purchased from Dharmacon: Control (non-target), #D-001810-10-50; USP9X, #L-006099-00-0005; YAP, #L-012200-00-0005; TAZ, #L-016083-00-0005; TEAD1, #J-012603-05; TEAD2, #J-012611-10; TEAD3, #J-012604-08; TEAD4, #J-019570-09.
Short hairpin (sh) RNA-mediated knockdown
The sequences for the sense oligonucleotides for the knockdown constructs were as follows: USP9X shRNA #1, CCTTAGAGATGGAGCAAGA; USP9X shRNA #2, CCAAAGGAATGGTGGAGAT; USP9X shRNA #3, GAGAGAAATCGCTGGTATA; USP9X shRNA #4, GGGTCGTGATTCAGAGTAA; TEAD1 shRNA GTAAGGAAGACGAGTGAAA; TEAD2 shRNA GAAGCCATTCTCACAGACA; TEAD3 shRNA TGAAGGAGCTCTATGAGAA; TEAD4 shRNA GGGCAGACCTCAACACCAA; YAP shRNA, GGTCAGAGATACTTCTTAA and TAZ shRNA (#652) was previously described (23).
Deubiquitination assay
HEK293 cells were transiently transfected with the indicated plasmids and treated with MG-132 (40μM) for 8 hours prior to cell lysis. Subsequently, cells were washed with ice cold-PBS and lysed in ice-cold lysis buffer (150 mM NaCl, 50 mM Tris-HCL at pH 7.4, 0.5% NP-40, protease inhibitor cocktail [Roche]). Next, cell lysates were cleared by a spin (13,000 rpm, 10 min, 4°C) and a fixed amount of whole-cell lysates was incubated with the appropriate agarose conjugated antibodies overnight. The agarose resin was washed twice with lysis buffer with 500 mM NaCl, followed by three times with lysis buffer. Deubiquitination was assessed by subjecting the samples to Western blot analysis with anti-HA antibodies (ubiquitin).
Immunohistochemistry
Human pancreatic cancer tissue arrays (#PAN801, #PA961b, US Biomax, Inc.) and pancreatic cancer patient tissue samples (Singapore Health Services Pte Ltd.) were used to examine protein expression of USP9X, LATS1/2 kinases and YAP/TAZ in normal and cancer tissues. Immunohistochemistry was done using Leica Bond Polymer Refine Detection kit. The slides were deparaffinised in Bond™ Dewax Solution and rehydrated through 100% ethanol to 1X Bond™ Wash Solution followed by antigen retrieval using Bond™ Epitope Retrieval Solution for 40 min at 100°C. After cooling to room temperature, the slides were rinsed with 1X Bond™ Wash Solution for 4 times. Subsequently, endogenous peroxidase was blocked for 15 min in 3-4% H2O2. After rinsing with Bond™ Wash Solution, the slides were blocked for 30 min in 10% goat serum and incubated with the primary antibody for 15 min at room temperature. Next, the slides were washed 5 times in 1X Bond™ Wash Solution followed by 5 min incubation with the polymer. After series of washes in 1X Bond™ Wash Solution, Bond™ Mixed DAB Refine was applied for 7 minutes and rinsed off in deionized water to terminate DAB reaction. The nuclei were counterstained with haematoxylin for 5 min. Subsequently, slides were dehydrated and mounted in synthetic mounting media. Tissues were digitalized using 3D Histech Pannoramic 250 Flash II slide scanner and analysed using ImageJ and CaseViewer software. Protein expression levels were scored as grades accordingly to the immunostaining intensity. Tissue arrays (#PAN801, #PA961b) were scored as three grades (negative, weak and strong expression), whereas samples derived from pancreatic cancer patients were scored as four grades (0-negative expression; 1-weak expression; 2-moderate expression; 3-strong expression). The studies were conducted in accordance with the International Ethical Guidelines for Biomedical Research Involving Human Subjects (CIOMS) and were approved by an institutional review board (SingHealth Institution). SingHealth Institution obtained consents from the patients.
Extraction of clinical and microarray gene expression data from pancreatic cancer patient datasets. Extraction of clinical and microarray gene expression data from pancreatic cancer patient datasets
One pancreatic cancer patient dataset, GSE21501 (24), available in the Gene Expression Omnibus (GEO) Database (http://www.ncbi.nlm.nih.gov/gds) was identified in this study based on searching the key words “pancreatic cancer” and “survival” with a return with the highest number of samples, available gene expression array data and patient survival data. The GEO website has standardized URLs for its individual datasets, e.g. the overall summary information about the microarray dataset GSE21501 can be accessed at http://www.ncbi.nlm.gov/geo/query/acc.cgi?acc=GSE21501. The Series Matrix File(s), which contain the expression values for each gene (probeset) can be accessed with the URL ftp://ftp.ncbi.nlm.nih.gov/pub/geo/DATA/SeriesMatrix/GSE21501. The file in gzip format was then unzipped to the tab-delimited text format, which contains detailed information for statistical analysis. The GSE21501 dataset is the largest pancreatic cancer patient dataset in the GEO database comprising 102 patients. Microarray gene expression data were retrieved from the data matrix deposited to the GEO database by the original authors. R scripting was used to extract the expression values of genes (probesets) of interest and the clinical data from the data matrix downloaded from GEO.
Correlations of gene expression levels and clinical data
All statistical analyses were performed using SPSS19.0. The associations between expression levels of genes were analyzed by Spearman’s rank test. ANOVA was used to test the differential gene expression between groups of patients. Kaplan-Meier survival analysis was performed to investigate the association between expression and survival; results of Kaplan-Meier analysis were compared by log-rank test for statistical analysis. P < 0.05 was considered significant in all statistical analyses.
Results
USP9X interacts with Hippo pathway components
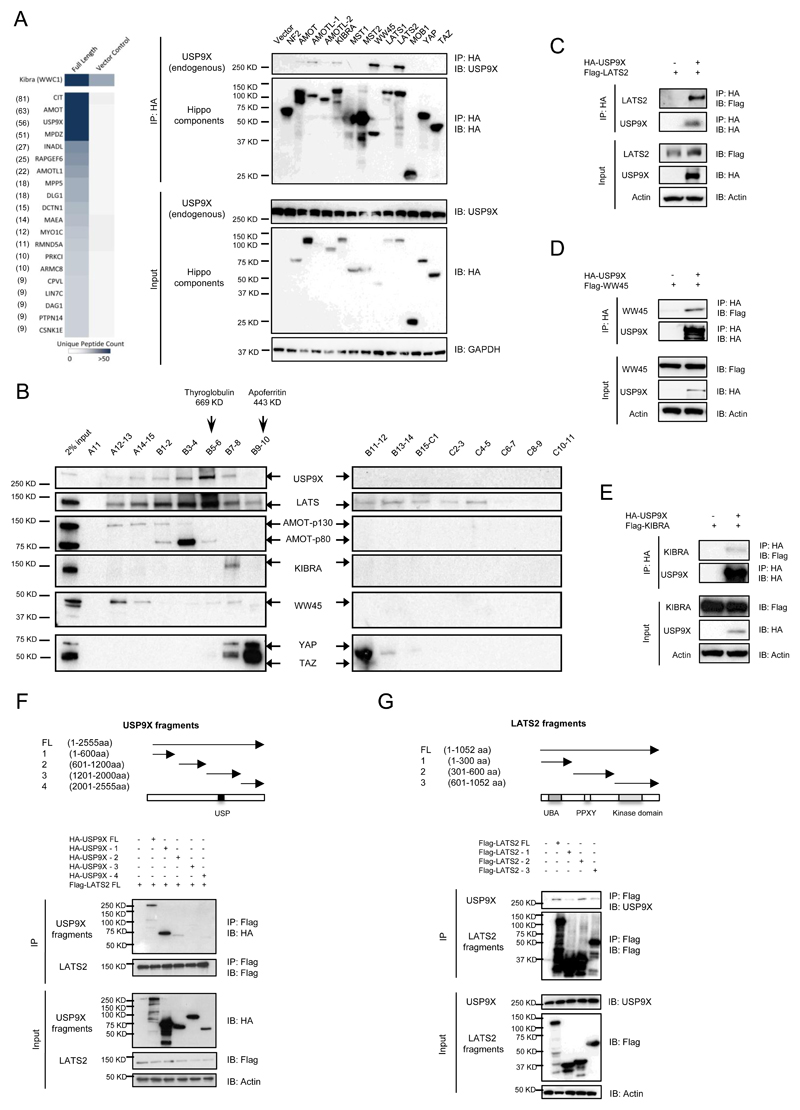
To understand how the protein stability of the Hippo pathway components is regulated, we identified the ubiquitin E3 ligases and deubiquitinating enzymes from the protein interaction network of the Hippo pathway generated in our group. Among them, the deubiquitinating enzyme, USP9X, turned out to be one of the top confidence interactions (Fig. 1A, left panel). USP9X was also identified from protein interaction network of the Hippo pathway by another group (25). In order to verify whether USP9X truly interacts with the Hippo pathway, we expressed HA-tagged version of different Hippo pathway components in HEK293 and checked if endogenous USP9X is indeed associated with each of them. Figure 1A, right panel, shows that among the 13 Hippo components tested, LATS2 and WW45 interact with USP9X strongly. AMOT family members (AMOT, AMOTL1, and AMOTL2) and KIBRA also interact with USP9X, but only weakly. To further examine if USP9X forms complexes with any of these interactors, we used gel filtration analysis through Fast Protein Liquid Chromatography (FPLC). Most of the USP9X was found to form large complexes (> 669kDa) with LATS, lesser with WW45, KIBRA, AMOT, YAP and TAZ (Fig. 1B). AMOT was shown recently to be a USP9X-interacting protein (26,27). Since LATS, KIBRA, and WW45 are found as new interactors of USP9X, we performed reciprocal immunoprecipitation experiments. Indeed, USP9X was found in the immunoprecipitates, indicating that LATS, WW45, and KIBRA are genuine interactors of USP9X, with LATS showing the strongest interaction (Fig. 1C-E). These results indicate that USP9X may regulate the Hippo pathway through its strong interactions with LATS as well as with WW45, KIBRA, and AMOT. Subsequently, N- and C-terminal-truncated USP9X and LATS2 mutants were constructed to map the binding region for LATS2 (Fig. 1F) and USP9X (Fig. 1G) respectively. The results suggest that the first 600 amino acids of USP9X are mostly responsible for interaction with LATS2, whereas the USP9X-interacting domain of LATS2 is mainly present between 300 to 600 amino acids. Although other regions of the proteins may also contribute to the bindings, the interactions were less significant. Since USP9X interacts strongly with LATS and LATS kinases are involved in the direct regulation of downstream targets YAP and TAZ, in this report, we concentrated our study on the regulation of LATS by USP9X.
Figure 1. USP9X interacts with Hippo pathway components.
A, USP9X was identified as the top interactor of KIBRA. The chart for the unique peptide counts is presented (left panel). HA-tagged components of the Hippo pathway were transiently expressed in 293 cells and cell lysates were immunoprecipitated with an anti-HA antibody.The co-precipitation of endogenous USP9X with various components was done using USP9X antibody (right panel). B, FPLC analysis of 293 cell lysate.45 fractions (A11-15, B1-15 & C1-11) were collected and proteins combined from two fractions were pooled and run on each lane. Fractions starting from A11 to C11 were shown. Only LATS was found to be strongly co-migrated in the same fractions as USP9X (fractions B5-6). C, D, E, Analytic reciprocal co-immunoprecipitation experiments showing the interaction of USP9X with LATS2, WW45, and KIBRA with LATS2 being the strongest interactor of USP9X. F, Mapping of the LATS2-interacting domain of USP9X. G, Mapping of the USP9X-interacting domain of LATS2.
USP9X stabilizes Hippo pathway upstream components
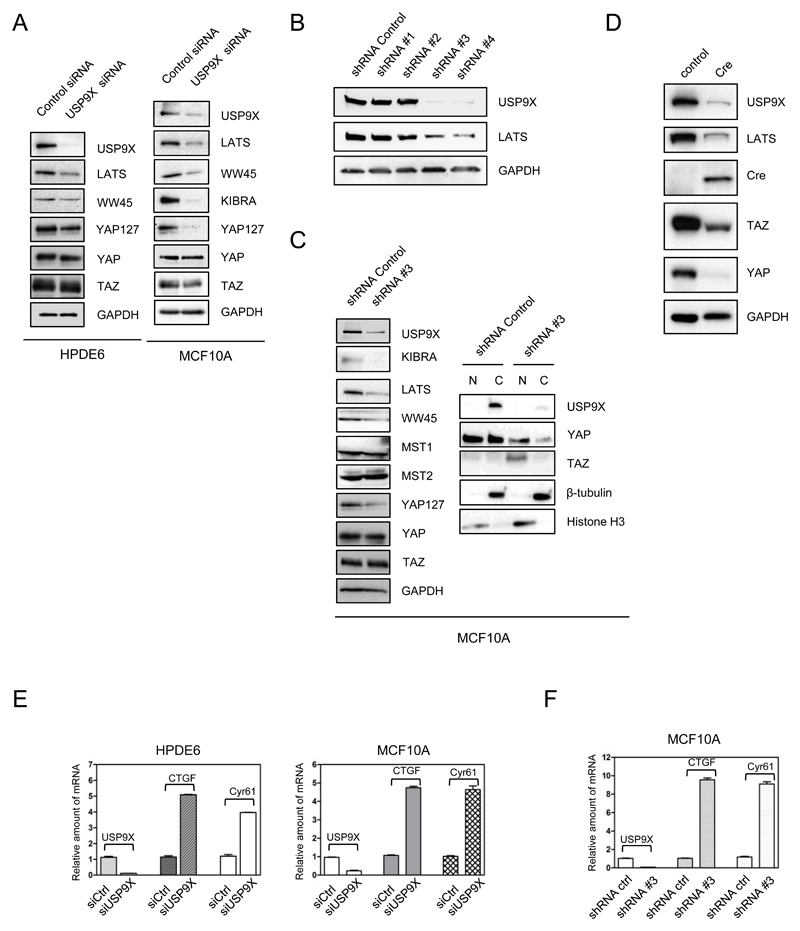
USP9X belongs to the USP family of deubiquitinating enzymes and stabilizes proteins by preventing their proteasomal degradation (13). In order to investigate the effect of USP9X on the protein stability of Hippo components, USP9X was knocked down by siRNA in immortal pancreatic duct epithelial HPDE6 cells followed by the investigation of the protein levels of Hippo members. HPDE6 were chosen because USP9X was originally identified as a tumor suppressor in pancreatic tissues (14) and interaction of USP9X with the Hippo pathway suggests a potential molecular mechanism for its tumor suppressor role. Following the knockdown of USP9X in HPDE6 cells, the protein levels of LATS kinase, WW45 and phospho-YAP were significantly down-regulated (Fig. 2A, left panel). USP9X knockdown was additionally performed in MCF10A cells (Fig. 2A, right panel) with similar results to HPDE6. In MCF10A, KIBRA was significantly downregulated as well. These results indicate that USP9X is important for the maintaining stability of Hippo components especially LATS, WW45, and KIBRA.
Figure 2. USP9X stabilizes upstream Hippo components.
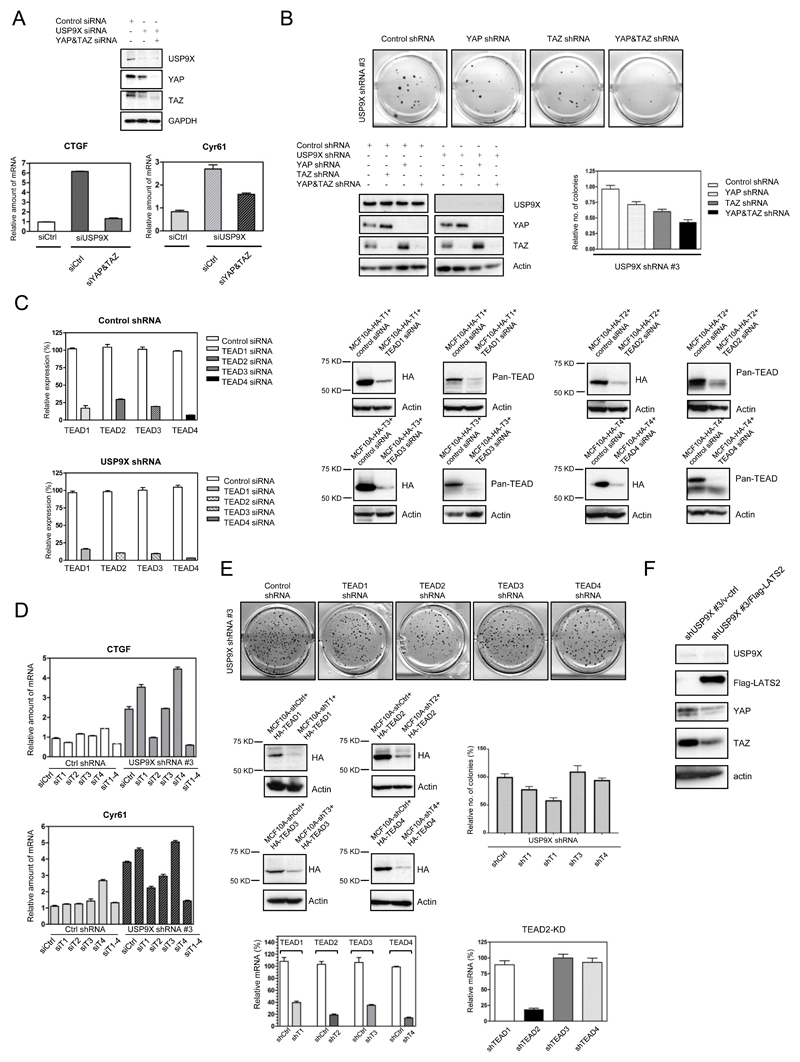
A, Knockdown of USP9X by siRNA in HPDE6 (left) and MCF10A (right) cells resulted in downregulation of LATS, WW45, and phospho-YAP. Down-regulation of KIBRA was seen in MCF10A cells. B, Screening of USP9X shRNA in MCF10A cells. Co-downregulation of LATS with USP9X was seen with shRNA #3 and #4. C, MCF10A were stably knocked down with USP9X shRNA #3. LATS, WW45, KIBRA, and phospho-YAP were downregulated (left panel) and TAZ was significantly translocated to the nucleus (right panel) in response to USP9X silencing. D, LATS was downregulated in Floxed USP9X MEF cells after Cre expression. E, F, Knockdown of USP9X by siRNA (E) and shRNA (F) in various cell lines resulted in upregulation of YAP/TAZ target genes, CTGF and CYR61 as accessed by real-time PCR.
In addition, we screened for shRNA which could be used for stable knockdown of USP9X. As shown in Fig. 2B, out of four shRNAs designed for USP9X, shRNA#3 and shRNA#4 showed significant down-regulation of LATS in MCF10A. shRNA#3 was chosen to knockdown USP9X stably in cells. As expected, sustainable knockdown of USP9X in MCF10A cells significantly downregulates LATS, WW45, and KIBRA (Fig. 2C). Subsequently, the subcellular localization of YAP and TAZ was investigated. USP9X knockdown MCF10A cells were fractionated into nuclear and cytoplasmic fractions. As shown in Fig. 2C, right panel, most of the YAP and TAZ is found in the nucleus, much less in the cytoplasm and this is consistent with the fact that there is less phosphorylated YAP in all cell types with the USP9X knockdown by either siRNA or shRNA (YAP127 panels). In our previous study, we have shown that nuclear form of TAZ (S89A) is less stable and expressed less as compared to wild type (22). Furthermore, Pan D lab has also shown that nuclear form of YAP is less stable (28). This might explain why after the USP9X knockdown, YAP/TAZ was activated and translocated to the nucleus, while the total amount of YAP/TAZ was reduced. To further corroborate the results, depletion of USP9X by Cre resulted in a decrease of LATS significantly, verifying that the expression of USP9X and LATS is positively correlated (Fig. 2D). Unexpectedly, the YAP and TAZ expression was downregulated. We think it is due to negative feedback regulation of the Hippo pathway, especially in the primary cells. The negative feedback is mediated through LATS/YAP/TAZ/TEAD as shown previously (28,29). When cells express highly activated YAP and TAZ, LATS is activated to counter the hyperactivity of YAP and TAZ. In order to sustain homeostasis of cells, YAP and TAZ expression is controlled, especially in primary cells where the feedback is intact. In cancer cells, this negative feedback might be compromised.
Taken together, these results suggest that down-regulation of USP9X leads to destabilization of LATS and other Hippo components, which in turn results in translocation of YAP and TAZ to the nucleus.
Knockdown of USP9X enhances expression of YAP/TAZ target genes CTGF and Cyr61
We also investigated if the increase in nuclear YAP and TAZ due to USP9X knockdown would result in the increment of their well-documented target genes, CTGF and Cyr61. Since the antibodies for CTGF and Cyr61 are not specific in our hands, their mRNA expression was analyzed by quantitative PCR. Fig. 2E shows that depletion of USP9X by siRNA in HPDE6 and MCF10A induced CTGF and Cyr61 mRNA expression between 3-5 fold. A similar trend was observed in MCF10A cells stably expressing USP9X shRNA#3, CTGF and Cyr61 mRNA levels were upregulated (Fig. 2F). These results suggest that USP9X indeed acts upstream of YAP and TAZ to suppress their activity.
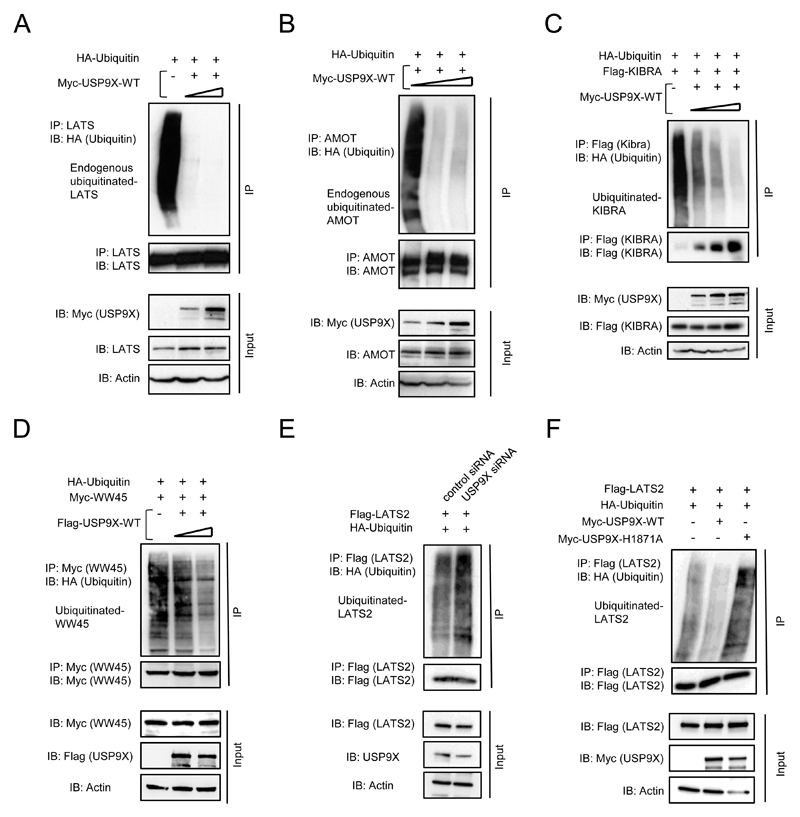
USP9X deubiquitinates Hippo pathway components
USP9X may potentially stabilize Hippo pathway components through its deubiquitinating activity to prevent their proteasomal degradation. In order to investigate whether USP9X is able to deubiquitinate the Hippo pathway components LATS, WW45, KIBRA and AMOT, ubiquitination and deubiquitination assays were performed. The increasing amount of USP9X was expressed in 293 followed by the assessment of endogenous LATS ubiquitination status. Indeed, as shown in Fig. 3A, endogenous-ubiquitinated LATS levels decreased in the presence of USP9X. Furthermore, deubiquitination assays were performed for AMOT, KIBRA, and WW45, the other USP9X substrates. Fig. 3B, C & D, shows that there are less endogenous-ubiquitinated AMOT, exogenous KIBRA, and WW45 in the presence of USP9X. Furthermore, increased LATS ubiquitination was observed in USP9X knockdown 293 cells, as compared to that of control cells (Fig. 3E). In order to verify that the deubiquitinating enzyme activity of USP9X is indeed responsible for the stability and deubiquitination of the Hippo components, we constructed enzyme defective mutant of USP9X (USP9X-H1871A) by changing the crucial amino acid 1871 histidine to alanine of USP9X. Fig. 3F shows that USP9X-H1871A did loose deubiquitinating activity towards LATS2. These results indicate that USP9X stabilizes Hippo pathway components by deubiquitination and prevention of their proteasome-mediated degradation.
Figure 3. USP9X deubiquitinates Hippo components LATS, AMOT, KIBRA, and WW45 in 293 cells.
Expression of increasing amount of USP9X resulted in decreasing amount of endogenous ubiquitinated LATS (A), AMOT (B), exogenous Kibra (C) and WW45 (D). E, Knockdown of USP9X augmented ubiquitinated LATS2. F, Enzyme defective USP9X (Myc-USP9X-H1871A) lost the ability to protect Lats2 from ubiquitination.
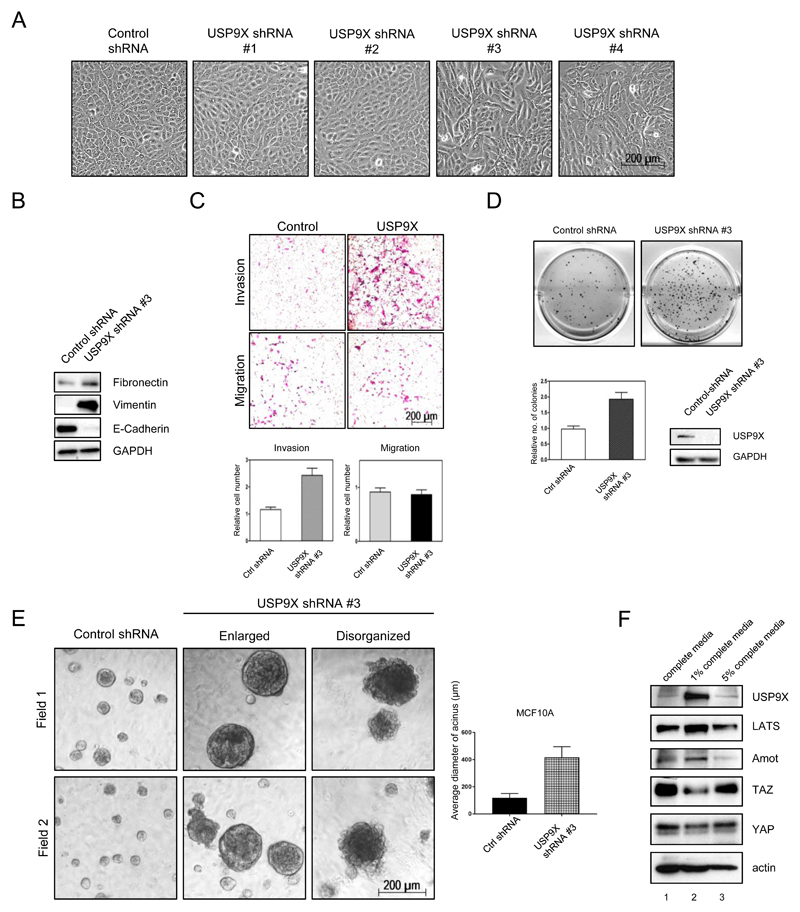
USP9X knockdown cells are oncogenic and knockdown of YAP and TAZ, TEAD2, and LATS kinase rescue the phenotypes
MCF10A cells stably expressing USP9X shRNA#3 and shRNA#4 displayed elongated, fibroblastic morphology, reminiscent of epithelial to mesenchymal transition (EMT) (Fig. 2B and 4A). Indeed, EMT markers fibronectin and vimentin were increased, whereas the epithelial marker E-cadherin was downregulated significantly in USP9X shRNA#3 MCF10A cells compared to the control cells (Fig. 4B). Moreover, transwell migration and invasion assays were used to investigate if the USP9X knockdown MCF10A cells acquired these characteristics. Fig. 4C shows that after knockdown of USP9X in MCF10A, cell invasion was increased, whereas migration was not obviously different from the control cells. Furthermore, knockdown of USP9X in MCF7 breast cancer cells significantly increased the number and the sizes of soft agar colonies in an anchorage-independent growth assay (Fig. 4D). In addition, a three-dimensional acini assay in matrigel was performed using USP9X knockdown MCF10A (16). The acini formed by MCF10A control cells in matrigel were well-organized, round and smooth (Fig. 4E), but the acini formed by USP9X knockdown MCF10A in matrigel were enlarged and disorganized, indicating that USP9X knockdown cells were transformed.
Figure 4. Knockdown of USP9X leads to oncogenic transformation.
A, Knockdown of USP9X by shRNA #3 & #4 in MCF10A cells resulted in cell morphological change reminiscent of EMT. B, Increased expression of fibronectin, vimentin and decreased expression of E-Cadherin in USP9X knockdown MCF10A cells. C, Increased invasion activity of USP9X shRNA #3 MCF10A cells in matrigel transwell assays. D, USP9X shRNA #3 MCF7 cells displayed enhanced anchorage-independent growth in soft agar. E, Three-dimensional culture of USP9X shRNA #3 MCF10A in matrigel. USP9X shRNA #3 MCF10A acini have enlarged and disorganized morphology. F, USP9X and LATS expression are highly regulated by growth signal. MCF10A grown in different percentage of serum were assessed for the expression of USP9X, LATS, AMOT, YAP, and TAZ.
Next, the correlation of USP9X and LATS in serum-starved condition was examined. As shown in Figure 4F, in 1% serum condition (lane 2), expression of USP9X was significantly increased as compared to the complete media condition (lane 1). The upregulation of USP9X was accompanied by increased LATS expression verifying their positive correlation under reduced serum conditions. Strong expression of USP9X and LATS was accompanied by the reduced YAP and TAZ expression. In cells grown in 5% serum, the expression of USP9X and LATS were, like AMOT, downregulated to the level of that of the cells grown in complete media with accompanying increase of YAP and TAZ. These results indicate that USP9X and LATS expression, like Hippo pathway, are highly regulated by growth signal.
In order to show that USP9X indeed works with Hippo pathway to inhibit YAP, TAZ and their transcription factor TEAD, we investigated whether the upregulated YAP/TAZ target genes, CTGF and Cyr61, in USP9X knockdown cells could be rescued by either knockdown of YAP and TAZ, four of the TEAD isoforms or LATS. As shown in Fig. 5A, the upregulated CTGF, and Cyr61 in USP9X shRNA#3 MCF10A cells could be rescued efficiently by knockdown of both YAP and TAZ, especially in the case of CTGF. As previously shown, the knockdown of USP9X enhanced anchorage-independent growth (Fig. 4D), therefore we investigated if knockdown of YAP and/or TAZ could rescue the anchorage-independent growth property of USP9X knockdown cells. As shown in Fig. 5B, left lower panel, knockdown of YAP resulted in upregulation of TAZ and vice versa, indicating the redundancy between YAP and TAZ. As reflected in Fig. 5B, only when both YAP and TAZ were knocked down were the number of soft agar colonies reduced (Fig. 5B, upper panel, and right lower panel).
Figure 5. YAP/TAZ and TEAD are downstream mediators of USP9X.
A, The activation of YAP/TAZ target genes, CTGF and Cyr61, due to USP9X knockdown could be rescued by knockdown of YAP/TAZ. The mRNAs of CTGF and Cyr61 were accessed by RT-PCR. B, The enhanced anchorage-independent growth by the USP9X knockdown in MCF7 cells could be rescued by knockdown of YAP and TAZ. C, D, TEAD2 was identified as one of the major TEAD isoforms downstream of USP9X. The activation of YAP/TAZ target genes, CTGF and Cyr61, due to USP9X knockdown could be rescued by knockdown of TEAD2. siRNA of the four TEAD isoforms could be effectively knocked down in cells (C) and TEAD2 knockdown could significantly suppress increased CTGF and Cyr61 mRNA in USP9X knockdown cells (D). E, TEAD2 is able to rescue the anchorage-independent growth of USP9X knockdown cells. F, LATS2 is able to rescue the YAP and TAZ expression induced by the USP9X knockdown.
Next, we investigated which isoform(s) of TEAD is involved in the activation of YAP/TAZ target genes in USP9X knockdown cells. Individual TEAD isoform or all four TEAD isoforms were knocked down in the control shRNA or USP9X shRNA#3 MCF10A cells by siRNA. All four of the TEAD isoforms could be knocked down efficiently in these cells (Fig. 5C), verified by RT-PCR and Western blotting. As shown in Fig. 5D, knockdown of TEAD2 alone could efficiently reduce the activation of CTGF and Cyr61 in response to USP9X knockdown. However, other TEADs might be involved since knockdown of all four isoforms of TEAD further downregulated activation of CTGF and, to a lesser extent, Cyr61. Furthermore, knockdown of TEAD2 by shRNA significantly reduced the number of soft agar colonies induced by USP9X knockdown (Fig. 5E). The four isoforms of TEAD could be knocked down efficiently by their respective shRNA, verified by RT-PCR as well as Western blotting (Fig. 5E, left lower panel) and knockdown of TEAD2 in the cells did not obviously affect the expression of other isoforms of TEAD (Fig. 5E, right lower panel). In addition, expression of LATS kinase rescued the phenotype of USP9X knockdown by reducing the expression of YAP and TAZ (Fig. 5F).
Taken together, these results indicate that USP9X works with the components of Hippo pathway to inhibit oncogenic growth in cells by stabilization of Hippo components especially LATS to restrict the activity of TAZ and YAP.
USP9X expression is positively correlated with LATS kinase and negatively with YAP/TAZ in cancer cells and cancer tissues
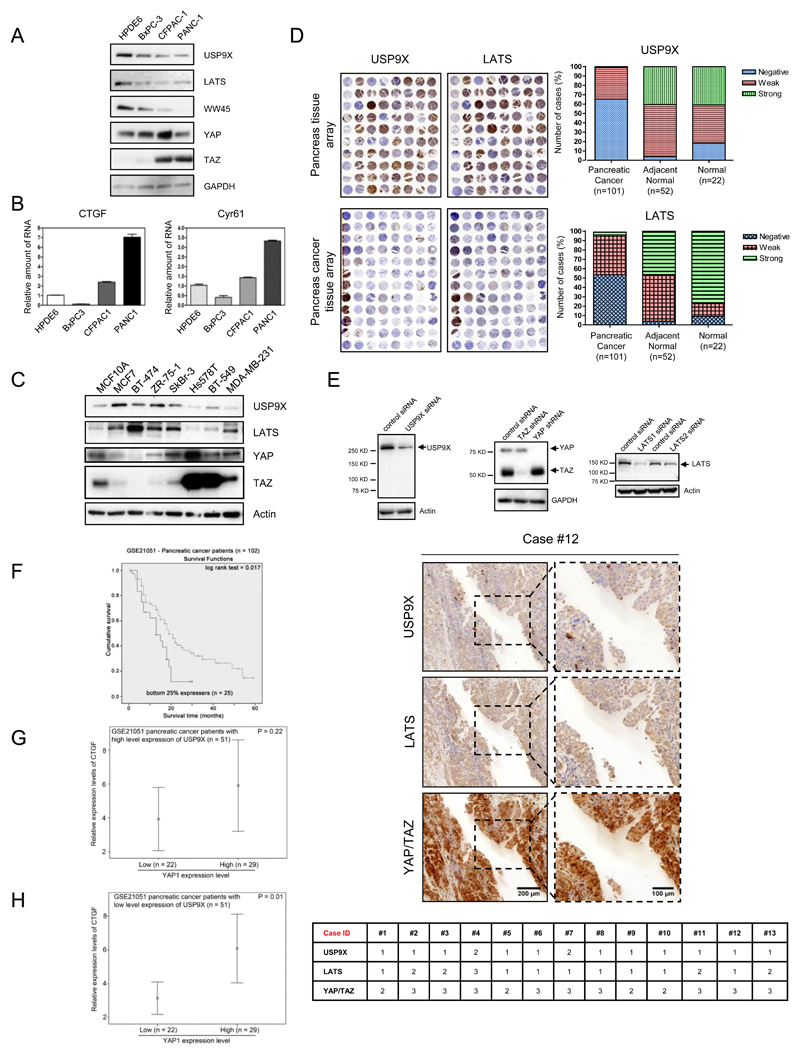
USP9X was identified as a tumor suppressor in pancreatic tissues (14) and we have shown that USP9X is able to stabilize Hippo pathway components especially LATS kinase to restrict YAP/TAZ/TEAD activity. This suggests that USP9X could act as a tumor suppressor via the Hippo pathway to inhibit cell growth and proliferation. Subsequently, a panel of pancreatic cancer cell lines was used to examine the potential correlation of USP9X expression with Hippo components. Fig. 6A shows that USP9X protein level is lower in invasive cells, BxPC3, CFPAC1, and PANC1, than in non-transformed HPDE6 cells. The expressions of LATS and WW45 show similar profiles to USP9X. YAP/TAZ protein expression is upregulated in a number of cancer types (4,6,23,30). TAZ expression is significantly higher in CFPAC1 and PANC1 cells, but barely detectable in HPDE6 and in the weakly invasive BxPC3 cells. Furthermore, CTGF and Cyr61 (Fig. 6B) are also highly expressed in CFPAC1 and PANC1. The positive correlation of USP9X with Hippo pathway component LATS, WW45 and negative correlation with YAP and TAZ and their target genes was revealed in this pancreatic cell line panel.
Figure 6. USP9X and LATS are positively correlated in cancer cell lines and pancreatic cancer tissues.
A, Decreased protein expression of USP9X, LATS, WW45, YAP, and TAZ was detected in pancreatic cancer cells. B, Increased mRNA levels of CTGF (left) and Cyr61 (right) in pancreatic cancer cells quantified by RT-PCR. C, USP9X, and LATS protein levels were downregulated in triple negative breast cancer cells. D, Immunohistochemical (IHC) staining of USP9X and LATS in normal and pancreatic cancer tissue microarrays (left panel). USP9X and LATS protein expression were predominantly negative in pancreatic cancer tissues (right panel). E, Characterization of USP9X, LATS and YAP/TAZ antibodies used for IHC tissue staining (top). USP9X and LATS were expressed weakly as opposed to strong YAP/TAZ expression in analogous tissue sections derived from pancreatic cancer patients (bottom). F-H, Analysis of USP9X, YAP and YAP/TAZ target genes using pancreatic cancer patient dataset, GSE21501. F, Tumors expressing USP9X lower than the lower quartile cut off had shorter life-span. G-H, Positive correlation of YAP and its target gene CTGF only observed in the low expression of USP9X.
USP9X is also oncogenic in some cell types (31,32), but as shown in Fig. 6C, USP9X expression is significantly lower in triple-negative breast cancer cells, Hs578T, BT549 and MBA-MD-231 and MCF10A cells but expressed more in weakly invasive cell lines MCF7, SKBR3, BT-474, ZR75.1, and SKBR3. The LATS expression profile is similar to USP9X, and both USP9X and LATS expression is inversely correlated with those of YAP and TAZ especially TAZ, which is highly expressed in invasive breast cells Hs578T, BT-549, and MDA-MB-231 (23). Taken all together, these results indicate that USP9X may cooperate with Hippo pathway component LATS to inhibit YAP and TAZ, especially TAZ, in pancreatic and breast cells.
Next, we examined their expression levels in pancreatic tissue microarrays (Fig. 6D) and consecutive tissue sections obtained from pancreatic cancer patients (Fig. 6E). USP9X, LATS, YAP/TAZ antibodies were verified to be specific after knocking down each of them in the cells by their respective siRNA (Fig. 6E, upper panel). Notably, negative expression of USP9X was observed in 65.4% (66 of 101) of pancreatic tumors, 3.9% (2 of 52) of cancer adjacent normal and 18.2% (4 of 22) of normal pancreatic tissues, demonstrating a significant correlation between USP9X downregulation and pancreatic tumorigenesis (Fig. 6D, upper right panel). Moreover, negative expression of LATS in the same tissue microarrays was detected in 53.3% (54 of 101) of pancreatic tumors, 3.9% (2 of 52) of cancer adjacent normal and 9.1% (2 of 22) of normal pancreatic tissues (Fig. 6D, bottom right panel). YAP and TAZ expression levels and activation was previously reported to be markedly higher in pancreatic cancer cells and tissues (33,34). Collectively, these findings suggest a possible link between low USP9X, LATS expression and high YAP/TAZ expression in human pancreatic tumors. Moreover, the analysis of the consecutive sections obtained from pancreatic cancer patients confirmed this relationship. Fig. 6E shows that the expression profiles of USP9X in pancreatic cancer tissues are also negatively correlated with that of YAP/TAZ, but positively correlated with LATS1/2.
In addition, in pancreatic cancer patient dataset (GSE21501), as shown in Table 1, by Spearman’s rank test, we found that expression level of YAP1 was significantly positively correlated with the expression levels of TEAD2 (r = 0.351, p < 0.001) and TEAD4 (r = 0.257, p = 0.00), while the expression level of USP9X was significantly negatively correlated with YAP1 (r -0.217, p = 0.029), TEAD2 (r = -0.328, p = 0.001) and TEAD4 (r = -0.397, p < 0.001). These results suggest that YAP1, TEAD2 and TEAD4 may be regulated in an opposite direction to that of USP9X.
Table 1. GSE21051 dataset (n = 102) Correlations between YAP1, USP9X and TEADs.
| YAP1 | USP9X | TEAD1 | TEAD2 | TEAD3 | TEAD4 | |||
|---|---|---|---|---|---|---|---|---|
| Spearman's rho | YAP1 | Correlation Coefficient | 1.000 | -.217* | .433** | .351** | .202* | .257** |
| P value | . | 0.029 | < 0.001 | < 0.001 | 0.042 | 0.001 | ||
| USP9X | Correlation Coefficient | -.217* | 1.000 | -.058 | -.328** | -.031 | -.397** | |
| P value | 0.029 | . | 0.559 | 0.001 | 0.759 | < 0.001 | ||
| TEAD1 | Correlation Coefficient | .433** | -.058 | 1.000 | .121 | .160 | -.053 | |
| P value | < 0.001 | 0.559 | . | 0.226 | 0.109 | 0.599 | ||
| TEAD2 | Correlation Coefficient | .351** | -.328** | .121 | 1.000 | -.032 | .407** | |
| P value | < 0.001 | 0.001 | 0.226 | . | 0.749 | < 0.001 | ||
| TEAD3 | Correlation Coefficient | .202* | -.031 | .160 | -.032 | 1.000 | .232* | |
| P value | 0.042 | 0.759 | 0.109 | 0.749 | . | 0.019 | ||
| TEAD4 | Correlation Coefficient | .257** | -.397** | -.053 | .407** | .232* | 1.000 | |
| P value. | 0.009 | < 0.001 | 0.599 | < 0.001 | 0.019 | . |
Correlation is significant at the 0.01 level (2-tailed).
Correlation is significant at the 0.05 level (2-tailed).
By Kaplan-Meier analysis, we found that 25% lowest USP9X expressing patients had a mean survival time of 14.2 months, which was significantly shorter than the mean survival time of 26.6 months for the rest of patients whose tumors expressed a higher level of USP9X (p = 0.017; Fig. 6F). This result suggests that a high level expression of USP9X may be a favourable prognostic marker for pancreatic cancer. Moreover, a significant positive association between YAP1 and its downstream target, CTGF, could only be observed in a background where USP9X was expressed at a low level (p = 0.01) but not in a background where USP9X was expressed at a high level (p = 0.22) (Fig. 6 G&H). These results suggest that a high level expression of USP9X may impair YAP activity.
Discussion
In this study, we identified USP9X deubiquitinating enzyme as a synergizing component of the Hippo pathway that interacted with and stabilized LATS kinase, WW45, KIBRA and AMOT to negatively regulate YAP/TAZ, transcription factor TEAD and their target genes to suppress tumor growth.
The post-translational modifications such as phosphorylation are well known to play an essential role in the regulation of this tumor suppressive pathway. Nonetheless, regulation through the covalent attachment of the ubiquitin molecule by ubiquitin ligases or its removal through deubiquitinating enzymes has not been explored in great detail so far. In recent times, increasing number of reports describing the regulation of the Hippo pathway through ubiquitination has emerged (8,10,35). However, none of the deubiquitinating peptidases were ascribed to the Hippo pathway regulation. Through proteomics approach, we identified USP9X as one of the candidate deubiquitinating enzymes regulating the Hippo pathway. During the preparation of the manuscript, two other groups reported USP9X as an interactor of Hippo components (26,27). In these two reports, USP9X was found to regulate and cooperate with Angiomotin family members, though with opposite effects on Hippo pathway. These findings further verify the importance of USP9X in the Hippo pathway.
Strikingly, we found USP9X to interact with the four fundamental components of the Hippo pathway. FPLC analysis revealed that among these interactors only LATS was found to interact strongly with USP9X in the same fractions. Through immunoprecipitation assays, LATS and WW45 were also demonstrated to be the two strongest interactors of USP9X. Even though USP9X was shown to deubiquitinate and stabilize all of the four Hippo components, LATS and WW45 were revealed to be the most responsive substrates for USP9X in our experiments. As USP9X is a large protein of ~270 kDa, it could potentially simultaneously interact with all the four Hippo components. In order to maximize the Hippo signaling effect, it is likely that USP9X associated with more than one Hippo components to stabilize them and exert their inhibition on the downstream effectors YAP/TAZ/TEAD.
One issue that is important in this study is the feedback regulation of the Hippo pathway. Prolonged USP9X knockdown will lead to downregulation of YAP/TAZ/TEAD target genes CTGF and Cyr61 instead of their upregulation. It is also true for LATS; prolonged USP9X knockdown will result in an increase of LATS or vice versa. LATS kinase has been previously shown to be upregulated after YAP or/and TAZ depletion (29,36). This feedback loop might explain why there is a conflicting outcome after the USP9X knockdown from the other two studies (26,27).
Dysregulation of the Hippo pathway signaling has been previously implicated in pancreatic cancer. YAP and TAZ have been revealed to play essential roles in the advancement of pancreatic tumors (33,34,37). Interestingly, there are not many reports suggesting a possible role for LATS in pancreatic tumorigenesis. This kinase has been previously implicated to function as a tumor suppressor in other cancer types including breast cancer, hepatocellular carcinoma and ovarian cancer (9,38,39). However, not many studies conducted a comprehensive investigation of its importance in pancreatic cancer. In this study, we demonstrated that USP9X expression was markedly downregulated in pancreatic cancer cells and in pancreatic tumors. Furthermore, the protein expression of USP9X and LATS was shown to be positively correlated in normal and cancerous tissue samples. Moreover, we hypothesized that USP9X regulation of WW45 adaptor protein may also play a crucial role in pancreatic cancer. Expression levels of these two proteins were demonstrated to be positively correlated in pancreatic cells. In conjunction with our findings, the lower expression of WW45 has been recently linked to pancreatic ductal adenocarcinoma progression further supporting our conclusions (40). In addition, we extended our studies to the analysis of mRNA dataset in which USP9X was shown to be negatively correlated with YAP, TEAD, and CTGF, but not TAZ and LATS kinase. TAZ has been previously demonstrated to function as an oncoprotein in a number of cancer types and its expression was established to be regulated at the protein level, but not mRNA level (23). This might explain why in the mRNA dataset findings, there was no negative correlation between TAZ and USP9X. This might also explain why LATS, although positively correlated with USP9X in pancreatic cells and tissues at the protein level, does not show a positive correlation with USP9X in the results obtained from mRNA dataset.
Another important finding from this study is that the protein expression level of USP9X and LATS is downregulated in highly invasive breast cancer cells as compared to weakly invasive cells and their expression is in reversed relationship with TAZ, which has been already shown to be upregulated in these highly cancerous cells (23). The panel of breast cancer cell lines used in this report has been previously established to recapitulate characteristics of the breast cancer disease (41,42). Despite the fact that USP9X has been earlier reported to be overexpressed in breast cancer tissues (43), the results revealed in this report suggest that USP9X might indeed act as tumor suppressor similarly to its function in pancreatic cells and tissues. Further investigation of clinical samples of invasive breast carcinoma will provide a better understanding of the USP9X regulation of the Hippo pathway components and its role in this deadly disease.
In conclusion, we have identified one of the first deubiquitinating enzymes to stabilize core Hippo component LATS kinase and to indirectly stimulate YAP, TAZ and transcription factor TEAD. Strikingly, USP9X not only was revealed to regulate LATS kinase but also was demonstrated to interact and stabilize WW45, KIBRA, and AMOT, the other essential members of the Hippo pathway. LATS kinase and WW45 adaptor protein are implicated as tumor suppressors in various cancers and a number of oncogenic E3 ubiquitin ligases have been found to destabilize their protein levels and alter their functions resulting in cell transformation and further tumorigenesis. USP9X might act as a deubiquitinating enzyme opposing the effect of E3 ligases in order to stabilize LATS kinase and WW45, therefore, promoting their tumor suppressive roles. There are certainly more deubiquitinating enzymes and ubiquitin ligases regulating the Hippo pathway components yet to be discovered. Further investigation of their functional interactions and implications in the progression of various tumors will hopefully provide valuable insights for the future cancer therapeutics.
Acknowledgements
We thank Magdalene Koh Hui Keng, Khin Sabai, SingHealth Tissue Repository, and Muhammad Kairos Bin Kosnan, Farazeela Binte Mohd Ibrahim, Histopathology Laboratory, Institute of Molecular and Cell Biology, A*STAR, for their generous help with patient tissues and staining.
Financial Support: This work was funded by Agency for Science, Technology and, Research (A*STAR).
References
- 1.Harvey K, Tapon N. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nature reviews Cancer. 2007;7:182–91. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- 2.Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–97. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- 3.Saucedo LJ, Edgar BA. Filling out the Hippo pathway. Nat Rev Mol Cell Biol. 2007;8:613–21. doi: 10.1038/nrm2221. [DOI] [PubMed] [Google Scholar]
- 4.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–57. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 5.Yu FX, Zhao B, Guan KL. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell. 2015;163:811–28. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the Roots of Cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho KC, Zhou Z, She YM, Chun A, Cyr TD, Yang X. Itch E3 ubiquitin ligase regulates large tumor suppressor 1 stability [corrected] Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4870–5. doi: 10.1073/pnas.1101273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeung B, Ho KC, Yang X. WWP1 E3 ligase targets LATS1 for ubiquitin-mediated degradation in breast cancer cells. PLoS One. 2013;8:e61027. doi: 10.1371/journal.pone.0061027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma B, Chen Y, Chen L, Cheng H, Mu C, Li J, et al. Hypoxia regulates Hippo signalling through the SIAH2 ubiquitin E3 ligase. Nature cell biology. 2015;17:95–103. doi: 10.1038/ncb3073. [DOI] [PubMed] [Google Scholar]
- 10.Bae SJ, Kim M, Kim SH, Kwon YE, Lee JH, Kim J, et al. NEDD4 controls intestinal stem cell homeostasis by regulating the Hippo signalling pathway. Nat Commun. 2015;6:6314. doi: 10.1038/ncomms7314. [DOI] [PubMed] [Google Scholar]
- 11.Wang C, An J, Zhang P, Xu C, Gao K, Wu D, et al. The Nedd4-like ubiquitin E3 ligases target angiomotin/p130 to ubiquitin-dependent degradation. Biochem J. 2012;444:279–89. doi: 10.1042/BJ20111983. [DOI] [PubMed] [Google Scholar]
- 12.Wood SA, Pascoe WS, Ru K, Yamada T, Hirchenhain J, Kemler R, et al. Cloning and expression analysis of a novel mouse gene with sequence similarity to the Drosophila fat facets gene. Mech Dev. 1997;63:29–38. doi: 10.1016/s0925-4773(97)00672-2. [DOI] [PubMed] [Google Scholar]
- 13.Murtaza M, Jolly LA, Gecz J, Wood SA. La FAM fatale: USP9X in development and disease. Cell Mol Life Sci. 2015;72:2075–89. doi: 10.1007/s00018-015-1851-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez-Mancera PA, Rust AG, van der Weyden L, Kristiansen G, Li A, Sarver AL, et al. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature. 2012;486:266–70. doi: 10.1038/nature11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Liu Y, Yang B, Cao H, Yang CX, Ouyang W, et al. Elevated expression of USP9X correlates with poor prognosis in human non-small cell lung cancer. J Thorac Dis. 2015;7:672–9. doi: 10.3978/j.issn.2072-1439.2015.04.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–68. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 17.Stegeman S, Jolly LA, Premarathne S, Gecz J, Richards LJ, Mackay-Sim A, et al. Loss of Usp9x disrupts cortical architecture, hippocampal development and TGFbeta-mediated axonogenesis. PLoS One. 2013;8:e68287. doi: 10.1371/journal.pone.0068287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Behringer R, Gertsenstein M, Vintersten Nagy K, Nagy A, Nagy KV. Manipulating the mouse embryo : a laboratory manual. :814. xxii. [Google Scholar]
- 19.Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C, Hong W. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J Biol Chem. 2011;286:7018–26. doi: 10.1074/jbc.C110.212621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan SW, Lim CJ, Guo F, Tan I, Leung T, Hong W. Actin-binding and cell proliferation activities of angiomotin family members are regulated by Hippo pathway-mediated phosphorylation. The Journal of biological chemistry. 2013;288:37296–307. doi: 10.1074/jbc.M113.527598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan SW, Lim CJ, Huang C, Chong YF, Gunaratne HJ, Hogue KA, et al. WW domain-mediated interaction with Wbp2 is important for the oncogenic property of TAZ. Oncogene. 2011;30:600–10. doi: 10.1038/onc.2010.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan SW, Lim CJ, Loo LS, Chong YF, Huang C, Hong W. TEADs mediate nuclear retention of TAZ to promote oncogenic transformation. J Biol Chem. 2009;284:14347–58. doi: 10.1074/jbc.M901568200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan SW, Lim CJ, Guo K, Ng CP, Lee I, Hunziker W, et al. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 2008;68:2592–98. doi: 10.1158/0008-5472.CAN-07-2696. [DOI] [PubMed] [Google Scholar]
- 24.Stratford JK, Bentrem DJ, Anderson JM, Fan C, Volmar KA, Marron JS, et al. A six-gene signature predicts survival of patients with localized pancreatic ductal adenocarcinoma. PLoS Med. 2010;7:e1000307. doi: 10.1371/journal.pmed.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Couzens AL, Knight JD, Kean MJ, Teo G, Weiss A, Dunham WH, et al. Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci Signal. 2013;6:rs15. doi: 10.1126/scisignal.2004712. [DOI] [PubMed] [Google Scholar]
- 26.Kim M, Park SJ, Lee C, Lim DS. Role of Angiomotin-like 2 mono-ubiquitination on YAP inhibition. EMBO Rep. 2016;17:64–78. doi: 10.15252/embr.201540809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thanh Nguyen H, Andrejeva D, Gupta R, Choudhary C, Hong X, Eichhorn PJ, et al. Deubiquitylating enzyme USP9x regulates hippo pathway activity by controlling angiomotin protein turnover. Cell Discov. 2016;2:16001. doi: 10.1038/celldisc.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Q, Zhang N, Xie R, Wang W, Cai J, Choi KS, et al. Homeostatic control of Hippo signaling activity revealed by an endogenous activating mutation in YAP. Genes & development. 2015;29:1285–97. doi: 10.1101/gad.264234.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moroishi T, Park HW, Qin B, Chen Q, Meng Z, Plouffe SW, et al. A YAP/TAZ-induced feedback mechanism regulates Hippo pathway homeostasis. Genes & development. 2015;29:1271–84. doi: 10.1101/gad.262816.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nature reviews Cancer. 2015;15:73–9. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–7. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 32.Boise LH. DUB-ling down on B-cell malignancies. Blood. 2015;125:3522–3. doi: 10.1182/blood-2015-04-638262. [DOI] [PubMed] [Google Scholar]
- 33.Morvaridi S, Dhall D, Greene MI, Pandol SJ, Wang Q. Role of YAP and TAZ in pancreatic ductal adenocarcinoma and in stellate cells associated with cancer and chronic pancreatitis. Sci Rep. 2015;5:16759. doi: 10.1038/srep16759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie D, Cui J, Xia T, Jia Z, Wang L, Wei W, et al. Hippo transducer TAZ promotes epithelial mesenchymal transition and supports pancreatic cancer progression. Oncotarget. 2015;6:35949–63. doi: 10.18632/oncotarget.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salah Z, Itzhaki E, Aqeilan RI. The ubiquitin E3 ligase ITCH enhances breast tumor progression by inhibiting the Hippo tumor suppressor pathway. Oncotarget. 2014;5:10886–900. doi: 10.18632/oncotarget.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jukam D, Xie B, Rister J, Terrell D, Charlton-Perkins M, Pistillo D, et al. Opposite feedbacks in the Hippo pathway for growth control and neural fate. Science. 2013;342:1238016. doi: 10.1126/science.1238016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diep CH, Zucker KM, Hostetter G, Watanabe A, Hu C, Munoz RM, et al. Down-regulation of Yes Associated Protein 1 expression reduces cell proliferation and clonogenicity of pancreatic cancer cells. PLoS One. 2012;7:e32783. doi: 10.1371/journal.pone.0032783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng S, Pan W, Jin Y, Zheng J. MiR-25 promotes ovarian cancer proliferation and motility by targeting LATS2. Tumour Biol. 2014;35:12339–44. doi: 10.1007/s13277-014-2546-0. [DOI] [PubMed] [Google Scholar]
- 39.Guo C, Wang X, Liang L. LATS2-mediated YAP1 phosphorylation is involved in HCC tumorigenesis. Int J Clin Exp Pathol. 2015;8:1690–7. [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L, Wang Y, Li PP, Wang R, Zhu Y, Zheng F, et al. Expression profile and prognostic value of SAV1 in patients with pancreatic ductal adenocarcinoma. Tumour Biol. 2016 doi: 10.1007/s13277-016-5457-4. [DOI] [PubMed] [Google Scholar]
- 41.Sommers CL, Byers SW, Thompson EW, Torri JA, Gelmann EP. Differentiation state and invasiveness of human breast cancer cell lines. Breast Cancer Res Treat. 1994;31:325–35. doi: 10.1007/BF00666165. [DOI] [PubMed] [Google Scholar]
- 42.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng S, Zhou H, Xiong R, Lu Y, Yan D, Xing T, et al. Over-expression of genes and proteins of ubiquitin specific peptidases (USPs) and proteasome subunits (PSs) in breast cancer tissue observed by the methods of RFDD-PCR and proteomics. Breast Cancer Res Treat. 2007;104:21–30. doi: 10.1007/s10549-006-9393-7. [DOI] [PubMed] [Google Scholar]