Abstract
Altered metabolism is a distinct feature of cancer cells. During transformation, the entire metabolic network is rewired to efficiently convert nutrients to biosynthetic precursors to sustain cancer cell growth and proliferation. Whilst the molecular underpinnings of this metabolic reprogramming have been described, its role in tumor progression is still under investigation. Importantly, the mitochondria is a central actor in many of the metabolic processes that are altered in tumors. Yet, we have only begun to understand the dualities of mitochondrial function during cancer metastasis and therapy resistance. Paradoxically, mitochondrial metabolism can be both advantageous and detrimental to these processes, highlighting the need for a better understanding of the molecular and micro-environmental cues that define the role of this fascinating organelle. In this review article, we present an updated view on the different mitochondrial metabolic strategies adopted by cancer cells to overcome the many hurdles faced during tumor progression.
Introduction
Cancer is a multifaceted disease whose pathogenesis remains elusive, despite the series of breakthroughs since the discovery of the first oncogene, SRC, more than 50 years ago (see glossary) [1]. Although it was initially thought that a set of mutations in specific oncogenes and tumor suppressors was sufficient to drive tumorigenesis, the spontaneous accumulation of such mutations in otherwise healthy tissue [2] indicates that cell transformation is a process exceeding this initial simplistic view. Indeed, tumorigenesis is supported by acquisition of cell-and non-cell-autonomous traits, known as the hallmarks of cancer [3]. Among these, altered energy metabolism has recently gained some attention. Bolstered by the development of a variety of techniques to assess the cellular metabolome, it has been shown that during transformation cancer cells undergo profound metabolic changes, including activation of glycolysis, altered utilization of amino acids, and dysregulation of mitochondrial function [4]. The availability of large-scale datasets from The Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov/) has allowed the identification of the underpinning genetic determinants of these metabolic changes, showing that they are orchestrated by well-known oncogenes and tumor suppressors [5]. Furthermore, it has been observed that tumors share a subset of metabolic gene signatures independent of their tissue of origin, and upregulate genes that encode for glycolysis and nucleotide biosynthesis enzymes [6, 7]. On the other hand, recent experiments using elegant mouse models showed that cancers also retain metabolic features from their tissue of origin [8].
Glossary.
Metastasis The process whereby, during tumor progression, cancer cells can leave the primary tumor mass and disseminate to other tissues and organs. Metastasis to vital organs is considered the main cause of death for cancer patients.
Mitochondria Intracellular organelles at the core of cell metabolism involved in the coupling of oxygen consumption and nutrient catabolism to produce energy and metabolic intermediates for the cell. These are the sites where OXPHOS and TCA cycle occur.
Metabolism It is the set of life-sustaining chemical reactions/transformations that occur within the cells of living organisms.
Metabolic adaptation The intrinsic ability of the network of metabolic reactions to adapt to external stimuli (e.g. nutrient availability, pharmacological treatment) or internal alterations (e.g. mutations) in order to maintain cell homeostasis. Metabolic adaptation can allow to quickly change cellular phenotype and function.
EMT Epithelial-to-Mesenchymal Transition; a process in which epithelial cells loss adhesion properties and become mesenchymal cells with invasive and migratory capacities.
FH Fumarate Hydratase; mitochondrial enzyme that catalyzes the reversible hydration/dehydration of fumarate to malate in the TCA cycle.
MtDNA Mitochondrial DNA
OXPHOS Oxidative Phosphorylation; metabolic pathway that occurs in the mitochondria in which nutrients are oxidized releasing energy that is then converted into ATP.
PC Pyruvate Carboxylase; an enzyme that catalyzes the irreversible carboxylation of pyruvate to oxaloacetate (OAA).
PDAC Pancreatic Ductal Adenocarcinoma.
PGC1 α Peroxisome proliferator-activated receptor γ co-activator 1 α; a transcriptional co-factor implicated in energy metabolism and the principal regulator of mitochondrial biogenesis.
SDHA Succinate Dehydrogenase A; Subunit of the succinate-ubiquinone oxidoreductase as part of the mitochondrial respiratory chain. Contains the FAD binding site where succinate is deprotonated and converted to fumarate.
SRC Proto-Oncogene Tyrosine-Protein Kinase.
TCA Tricarboxylic acid; Series of biochemical reactions used by all aerobic organisms to release energy (ATP) through oxidation of nutrients.
TCGA The Cancer Genome Atlas.
Mitochondria co-ordinate a large fraction of metabolic, energetic, and physiological processes, and their integrity is a central checkpoint for cancer cells [4]. In this respect, the finding that genetic alterations of mitochondrial metabolic enzymes, such as Fumarate Hydratase (FH), Succinate Dehydrogenase (SDH), and Isocitrate Dehydrogenase (IDH1/2) [9], can predispose or contribute to cancer shook the field. In fact, investigations on these models revealed that metabolites of mitochondrial origin accumulated in these tumors can activate oncogenic signaling cascades, making them bona fide oncometabolites [9]. In addition to exacerbated or inhibited metabolite production, cancer cells also exploit the reversible nature of many metabolic reactions. In fact, beyond aberrant activation of glycolysis, the use of the tricarboxylic acid cycle in reversed mode (reductive carboxylation) enables the use of glutamine for biosynthetic purposes in cells with dysfunctional mitochondria [10, 11]. Interestingly, a compilation of experimental evidence suggest that mitochondrial dysfunction can reach a threshold where it turns from advantageous to detrimental for cancer cells. In this line, depletion of mitochondrial DNA (mtDNA) upon disruption of the mitochondrial transcription factor A (TFAM) inhibits mutant Kras-driven tumorigenesis in mice [12]. Furthermore, mtDNA-depleted cancer cells transplanted in mice acquire whole mitochondria from host cells via horizontal transfer as an strategy to restore their mitochondrial function [13]. These results indicate that changes in mitochondrial metabolism are not only mere consequences of transformation and that fine regulation of mitochondrial function is required to drive tumorigenesis.
While the metabolic underpinnings of tumor initiation have been described for a wide range of tumor types, little is known on the metabolic adaptations that can occur at later phases of cancer progression. The aim of this review article is to focus on the initial evidence gathered in recent years about the role of mitochondrial metabolism during the stages following cancer initiation and accompanying cancer progression. Understanding how tumor cells rewire and adapt their mitochondrial metabolism during tumor progression could be instrumental both for the development of novel anticancer strategies and for the identification of aggressiveness signs of prognostic value
Altered mitochondrial metabolism contributes to tumor progression
The importance of metabolism in cancer is illustrated by the fact that tumors can be clustered based on their metabolic signature, with important implications for cancer diagnosis and patient stratification [7]. However, tumors are far from being a static entity. Environmental constraints, such as nutrient and oxygen availability, and the exposure to anti-tumor agents, inevitably challenge the survival of cancer cells and force their evolution and/or selection within the tumor [14]. Indeed, recent evidence indicates that the metabolic phenotype of cancer varies at different disease stages, and it is a contributing factor for tumor progression (Figure 1). For instance, an increase in glycolytic enzymes and a decrease in mitochondrial transcriptional programs has been observed at different stages of prostate cancer progression [15, 16]. Similarly, stage-specific metabolic traits were identified in breast [17], renal cancer [18], and lung cancer [19]. In this context, mitochondrial metabolism seems to play a key role. Transcriptional analysis of 21 tumor types collected by the TCGA revealed that the repression of genes related to mitochondrial metabolism is strikingly associated with poor clinical outcome and is associated with the presence of an epithelial to mesenchymal transition (EMT) gene signature, a molecular pathway linked to tumor initiation, invasion, and metastasis [24]. These results suggest that during tumor progression, mitochondrial dysfunction can be advantageous and could make cancer cells more motile and invasive, predisposing to metastasis. Complementary studies have showed that mutations of enzymes from the TCA cycle, SDH and FH, are linked to the activation of EMT and invasive phenotype in pheochromocytoma and paraganglioma [25] and renal cancer [26]. Similarly, mtDNA abundance displays high heterogeneity among human tumors and mtDNA depletion, which is generally associated with bioenergetics defects, is linked with poor patient prognosis in several human cancers [27].
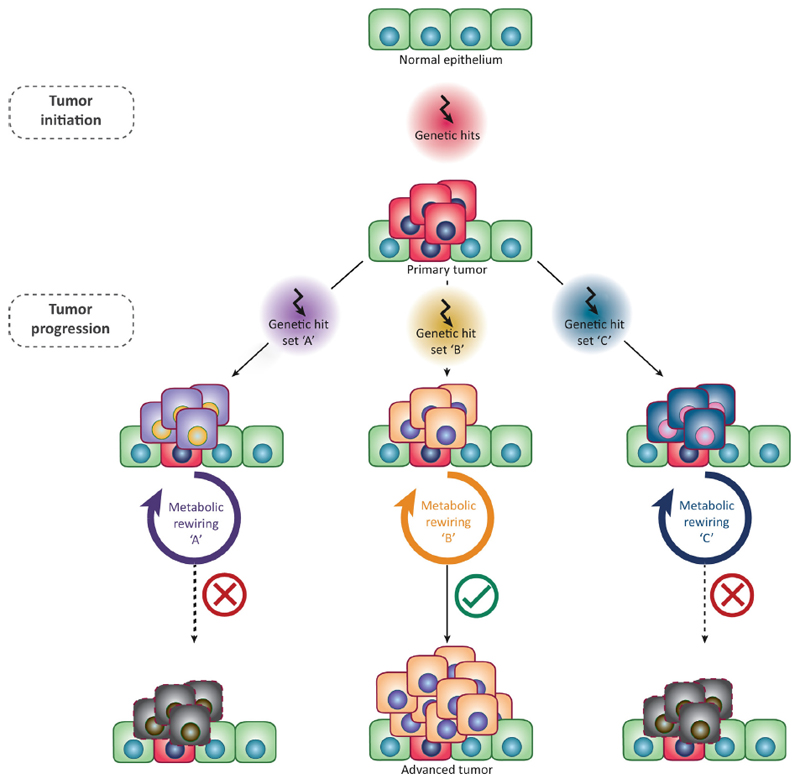
Figure 1. Metabolic adaptations during cancer progression.
Schematic representation of the evolution and progression of cancer, based on specific metabolic rewiring. Upon the establishment of an initial tumor mass, the acquisition of additional genetic mutations can lead to metabolic changes that will confer cancer cells different proliferative/survival capabilities. Survival of cancer cells and subsequent tumor progression is dependent on the acquisition of a successful metabolic state, defined here as metabolic flexibility. Please note that genetic mutations have been depicted as single consecutive events for simplicity, but various cumulative mutations can occur at each stage.
Overall, these results indicate that the suppression of TCA cycle enzymes provides a distinct advantage to the cancer cells during tumor progression, and it can contribute to the clinical outcome of patients. However, there are also reported examples of mitochondrial dysfunction being detrimental for cancer aggressiveness. Studies performed on renal oncocytoma, a benign tumor characterized by aberrant accumulation of dysfunctional mitochondria, showed that defects in mitochondrial function can inhibit the autophagic machinery, thus creating a metabolic checkpoint that inhibits tumor progression [20]. Conversely, activation of autophagy is fundamental to support mutant Ras-driven cancer cells by providing substrates for mitochondrial metabolism and clearing dysfunctional mitochondria [21–23]. Consistently, inhibition of autophagy leads to mitochondrial dysfunction and blocks malignant tumor progression by reprogramming tumor fate towards benign neoplasms [21].
With this new perspective of mitochondrial function as friend and foe for tumor progression, it will be important to define in future studies to which extent is this phenomenon the consequence of tissue or tumor-specific mitochondrial requirements. Alternatively, it could be the result of a byphasic contribution of this organelle to tumour progression, being the reduction in mitochondrial function progressively advantegous to cancer cells until it reaches a “minimal integrity point”, below which this alteration becomes detrimental.
The dual role of mitochondrial metabolism in cancer cell dissemination and metastasis
During tumor progression, epithelial cells face loss of anchorage to their native location, which triggers a stress response that leads to cell death in a process termed anoikis [28]. The bypass of this phenomenon requires a profound adaptation of cellular metabolism, which has been reviewed elsewhere [29]. In addition, once detached, cancer cells remain exposed to additional micro-environmental stresses in their journey to a foreign distant tissue to establish metastatic lesions. These challenges include the invasion beyond the basal membrane, intravasation (with the potential requirement of EMT), survival in circulation, extravasation, homing in a secondary organ and tumor regrowth [30] (Figure 2). This process is tremendously inefficient as the majority of maladapted cells die in circulation or upon reaching a hostile microenvironment. Yet, a small percentage of cells with intrinsic abilities or sufficient plasticity to adapt and survive upon intravasation can successfully colonize a secondary organ [30]. Once seeded, cancer cells re-enter a proliferative stage (a process that can take from weeks to years) and overcome the hostile environmental conditions to generate a disseminated tumor. Importantly, these cells will need to optimize their metabolic state according to the characteristics of the target tissue, as well as to their own mutational background.
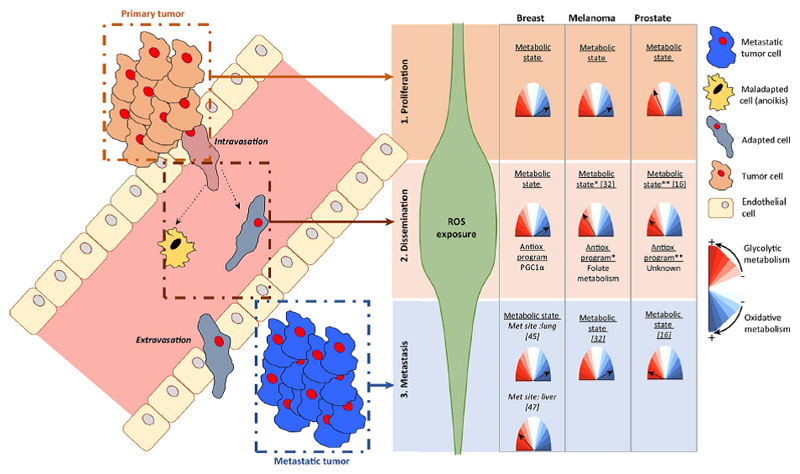
Figure 2. Metabolic modulation during metastatic dissemination.
Schematic representation of the stages of metastatic dissemination (proliferation, dissemination and metastasis). Right panel provides a summary of the metabolic adaptation exhibited by different tumor types and the estimated exposure to reactive oxygen species (ROS). *, glycolytic activity is presumed from results indicating reduced mitochondrial mass **, glycolytic activity is presumed from the progressive decrease in mitochondrial oxidative transcriptional programs from primary to metastatic cells. References: 1- Piskounova E. et al., Nature 2015; 2- Torrano V. et al., Nature Cell Biology 2016; 3- Le Bleu V.S. et al., Nature Cell Biology 2014; 4- Dupuy F. et al., Cell Metabolism 2015). Met: Metastasis.
The determinants of the metabolic adaptations during dissemination and metastasis are only partially known. In line with the increase in oxidative stress during anchorage-independent growth [28, 31], it was recently shown that metastatic cells from primary melanoma require the activation of mitochondrial antioxidant pathways to survive [32]. In support of these findings, it was recently shown that cell detachment leads to a complex rewiring of nutrient utilization, with reductive carboxylation being a key pathway to generate antioxidant molecules [33, 34]. Overall, cancer cells that detach from the primary tumor experience oxidative stress and, by activating antioxidant pathways, they can overcome this challenge and eventually metastasize. This view is supported by the many examples with isolated antioxidant genes or chronic therapies with antioxidant properties that promote metastasis [35–41].
The activation of specific transcriptional programs that regulate mitochondrial activity during cancer cell dissemination and metastasis underpins these metabolic changes. In prostate cancer, active mitochondrial oxidative metabolism represents a disadvantageous metabolic state for cancer cells and leads to tumor suppression [16]. Moreover, deletion of the master transcriptional regulator of mitochondrial oxidative metabolism, Peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC1α), in the mouse prostate induces a glycolytic switch and promotes cancer progression and metastasis, in agreement with the down-regulation of PGC1α observed in human specimens [16]. Of note, striking similarities exist between prostate cancer specimens that will eventually recur and the metastatic lesions, in terms of the PGC1α-dependent metabolic program. These results suggest that the selection process for metabolically fit metastatic clones could start in primary tumors long before prostate cancer disseminates (Figure 2). Interestingly, in a complementary study in melanoma it was shown that PGC1α suppresses metastasis through the regulation of an adhesion and invasion transcriptional program [42]. In support of a role of mitochondrial dysfunction in promoting metastasis, partial inhibition of mitochondrial respiratory chain with rotenone is sufficient to induce cell migration and clonogenicity in vitro and to support lung metastasis in vivo [43]. Finally, mtDNA mutations affecting complex I have been found to support breast cancer metastasis in vivo via de-regulation of NAD+/NADH and activation of autophagy. Importantly, rescue of mitochondrial function through enhancement of complex I activity could inhibit formation of metastasis[44].
Despite these consistent lines of evidence, the role of mitochondrial function in metastasis remains controversial. For instance, increased mitochondrial function was detected in circulating cells from orthotopically-implanted breast cancer [45], suggesting that tumor-specific reprogramming might occur during metastasis. The same study showed that establishment of metastases was accompanied by decrease expression of OXPHOS genes, thus adding complexity to the role of mitochondrial function in the survival of matrix-detached cells and distant tissue colonization [45]. Of note, differential use of pyruvate in the mitochondria has been recently shown to dictate the site of metastasis in breast cancer [46, 47]. Breast-cancer-derived lung metastases showed increase dependency on pyruvate conversion into oxaloacetate through Pyruvate Carboxylase activation [46]. Liver-metastatic cells, however, divert pyruvate into lactate production as a result of increased activity of Pyruvate Dehydrogenase Kinase 1 [47]. These results suggest that metastatic cells undergo adaptations that are dictated by the different metabolic landscapes of target tissues and call for a careful analysis of metastatic site-dependent metabolic rewiring.
Changes in mitochondrial function are inherently linked with the increased requirement of antioxidant molecules of disseminated cells that we described above. Multiple strategies can be adopted to increase antioxidant power (Figure 2). On the one hand, a decrease in mitochondrial activity would result in reduced production of mitochondrial free radicals. This is the proposed mechanism in melanoma [32], where cells activate compensatory antioxidant mechanisms (folate metabolism, the main producer of reducing power [48]). On the other hand, activation of PGC1α pathway would result in increased mitochondrial metabolism and activation of an antioxidant program, which would counteract oxidative stress. This strategy is observed in breast cancer [45], and is in line with other studies pointing at the enhanced antioxidant response activated upon induction of PGC1α [49]. Therefore, cancer cells with different genetic drivers or tissue-of-origin constrains might differ in the optimal metabolic state that counteracts oxidative stress and supports the sequential process of metastasis. Finally, a recent study highlighted the link between epigenetic changes occurring in PDAC metastases and reprogramming of the oxidative branch of the pentose phosphate pathway [50], thus indicating that epigenetic mechanisms might control the antioxidant capacity of metastatic cells. Overall, these studies increase the complexity of the oncogenic activity of metabolic programs and call for caution when defining a dogmatic and static oncogenic metabolic reprogramming.
Metabolic adaptation contributes to therapy resistance
As discussed above, the ability to adapt the metabolic phenotype at different stages of the disease is a clear advantage for cancer cells. This metabolic modulation is also key when cancer cells are subject to the toxic effects of anticancer drugs. Indeed, specific metabolic features of cancer cells can overcome the toxic effects of anticancer drugs, leading to chemoresistance [51–57] or support lipid synthesis and mutagenesis in these challenging conditions [58] (Figure 3). For instance, cisplatin resistance is driven by a metabolic reprogramming that supports the generation of antioxidant molecules, including NADPH via the pentose phosphate pathway [51] and glutathione biosynthesis from glutamine [53]. Resistance to oncogene addiction [55] has been also shown to rely on metabolic adaptation. In this model, the extinction of mutant KRas in established tumors leads to the selection of a subpopulation of resistant cells dependent on mitochondrial metabolism [55]. Similar findings have been made in melanoma, in which inhibition of BRafV600 in BRaf V600-driven melanoma induces an oxidative phosphorylation switch orchestrated by PGC1α [49, 52], enhancing the detoxification capacities of these cells. In addition, resistance to MAPK inhibitors in this type of cancer is associated with increase mitochondrial DNA content and oxidative phosphorylation [57]. These results are in support of a role of activated mitochondrial function as a key determinant of therapy resistance (Figure 3).
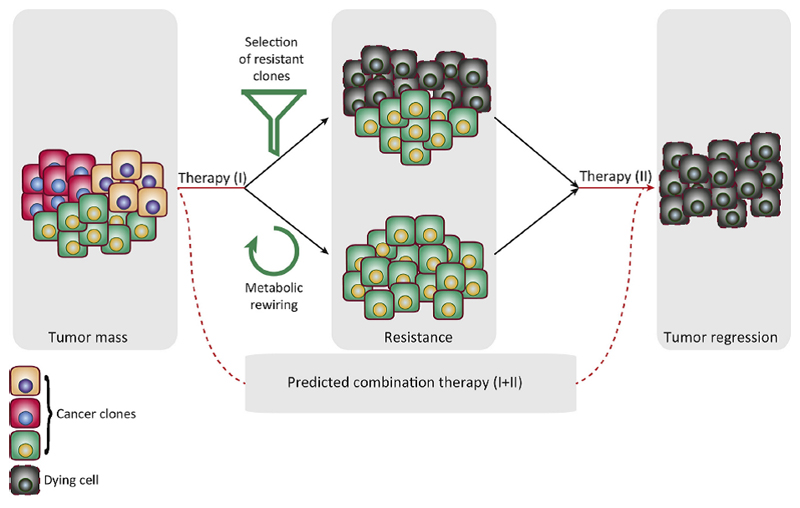
Figure 3. Metabolic modulation as a phenomenon underpinning therapy resistance.
Schematic representation of how metabolic adaptations can enable resistance to anticancer therapies. The acquisition of drug resistance induces new metabolic essentialities that can be harnessed to specifically target resistant cells.
Of note, core cellular processes such as genomic integrity pathways and cell cycle progression are tightly associated to mitochondrial function. On the one hand, mitochondrial dysfunction is accompanied by depletion of nucleotide pools and this has been linked with the induction of DNA damage [59]. Beyond the well-established role of DNA damage in elicting genome instability and support tumorigenesis, high mutational burden has been linked to increased sensitivity to the checkpoint inhibitors anti-CTLA-4 [60] and anti-PD1 [61]. On the other hand, cell cycle checkpoint regulators Cyclin D3-CDK6 negatively regulate mitochondrial metabolism by drifting glucose-derived carbons away from the TCA and into the pentose-phosphate pathway, hence supporting the production of antioxidant power [62]. This profound interplay between mitochondrial metabolism and core cellular checkpoints reveals the potential of targeting the activity of this organelle in combination therapies to harness the vulnerabilities of tumor cells.
Mitochondrial metabolism is heterogeneous within and between tumors [63], as evidenced in studies on cancer stem cell biology and antiangiogenic therapies [64–67]. These results suggest that the efficacy of anticancer therapy may depend on the intrinsic metabolic features of cancer cells. It also supports the notion that cancer cells endowed with higher adaptive metabolic capacity, or benefiting from the metabolic support of the microenvironment [68] can more easily escape drug toxicity. Furthermore, due to the association of cancer-initiating and therapy-resistant cells to a more oxidative metabolic program, it is tempting to speculate that: i) the emergence of therapy-resistant cancer clones relies on the newly acquired metabolic state, and ii) this metabolic plasticity can be therapeutically exploited through the combination of standard and anti-metabolic therapies (Figure 3).
Concluding Remarks
Metabolic rewiring is a hallmark of cancer. Well-characterized changes in mitochondrial metabolism generally provide a growth advantage in an environment that supports cell proliferation. However, this program needs to be complemented by additional metabolic strategies to support anchorage-independent growth, metastatic dissemination, or pharmacological challenges. In this scenario, mutations in oncogenes and tumor suppressors can be seen as ways to reach new metabolic landscapes. Given that this metabolic reprogramming is tissue-specific, cancer cells might need to activate different gene networks depending on their tissue of origin. This hypothesis could explain why most of the well-characterized oncogenes and tumor suppressors exhibit potent regulatory functions on the metabolic network, and why they are selected in specific tissues. The benefit of specific mitochondrial metabolic features at different stages of tumor progression and in response to therapy are yet to be clarified. Interestingly, changes in metabolism are frequently associated to epigenetic alterations through transcriptional or post-translational modifications, rather than irreversible genomic events. It is therefore possible that a specific metabolic landscape would enable epigenetic flexibility to control gene expression.
Finally, although these metabolic adaptations might provide a selective advantage to cancer cells, they will likely unveil novel therapeutic vulnerabilities. Restricting the metabolic modulation of cancer cells and cornering them into a specific metabolic state, either by pharmacological strategies or changing nutrient availability, could be a strategy to hamper tumor progression and increase therapeutic efficacy. Understanding the different metabolic states that cancer cells can acquire upon therapeutic treatment is therefore crucial for the development of successful anticancer strategies.
Acknowledgements
Apologies to those whose related publications were not cited due to space limitations. We would like to acknowledge the Carracedo and the Frezza lab for the continuous input and discussions. The work of A.C. is supported by the Ramón y Cajal award, the Basque Department of Industry, Tourism and Trade (Etortek) and the department of education (IKERTALDE IT1106-16), ISCIII (PI10/01484, PI13/00031), FERO VIII Fellowship, the BBVA foundation, the MINECO (SAF2016-79381-R) and the European Research Council Starting Grant (336343). The participation of A.C. and V.T. as part of CIBERONC was co-funded with FEDER funds. L. V-J is supported by Basque Government of Education. V.T. is founded by Fundación Vasca de Innovación e Investigación Sanitarias, BIOEF (BIO15/CA/052), the AECC J.P. Bizkaia and the Basque Department of Health (2016111109). EG and CF are supported by the Medical Research Council, core fund to the MRC Cancer Unit SKAG106.
Footnotes
The authors declare no conflict of interest.
References
- 1.Martin GS. The hunting of the Src. Nat Rev Mol Cell Biol. 2001;2(6):467–475. doi: 10.1038/35073094. [DOI] [PubMed] [Google Scholar]
- 2.Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349(6255):1483–9. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cairns RA, et al. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 6.Hu J, et al. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat Biotechnol. 2013;31(6):522–9. doi: 10.1038/nbt.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaude E, Frezza C. Tissue-specific and convergent metabolic transformation of cancer correlates with metastatic potential and patient survival. Nat Commun. 2016;7:13041. doi: 10.1038/ncomms13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayers JR, et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science. 2016;353(6304):1161–5. doi: 10.1126/science.aaf5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan LB, et al. Altered metabolite levels in cancer: implications for tumour biology and cancer therapy. Nat Rev Cancer. 2016;16(11):680–693. doi: 10.1038/nrc.2016.85. [DOI] [PubMed] [Google Scholar]
- 10.Mullen AR, et al. Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Rep. 2014;7(5):1679–90. doi: 10.1016/j.celrep.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mullen AR, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2011;481(7381):385–8. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107(19):8788–93. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong LF, et al. Horizontal transfer of whole mitochondria restores tumorigenic potential in mitochondrial DNA-deficient cancer cells. Elife. 2017;6 doi: 10.7554/eLife.22187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lipinski KA, et al. Cancer Evolution and the Limits of Predictability in Precision Cancer Medicine. Trends in Cancer. 2(1):49–63. doi: 10.1016/j.trecan.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pertega-Gomes N, et al. A glycolytic phenotype is associated with prostate cancer progression and aggressiveness: a role for monocarboxylate transporters as metabolic targets for therapy. J Pathol. 2015;236(4):517–30. doi: 10.1002/path.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torrano V, et al. The metabolic co-regulator PGC1alpha suppresses prostate cancer metastasis. Nat Cell Biol. 2016;18(6):645–56. doi: 10.1038/ncb3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denkert C, et al. Metabolomics of human breast cancer: new approaches for tumor typing and biomarker discovery. Genome Medicine. 2012;4(4):37–37. doi: 10.1186/gm336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hakimi AA, et al. An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer Cell. 2016;29(1):104–16. doi: 10.1016/j.ccell.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerr EM, et al. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature. 2016;531(7592):110–3. doi: 10.1038/nature16967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joshi S, et al. The Genomic Landscape of Renal Oncocytoma Identifies a Metabolic Barrier to Tumorigenesis. Cell Rep. 2015;13(9):1895–908. doi: 10.1016/j.celrep.2015.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo JY, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25(5):460–70. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo JY, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27(13):1447–61. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo JY, et al. Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells. Genes Dev. 2016;30(15):1704–17. doi: 10.1101/gad.283416.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nieto MA, et al. EMT: 2016. Cell. 2016;166(1):21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 25.Loriot C, et al. Epithelial to mesenchymal transition is activated in metastatic pheochromocytomas and paragangliomas caused by SDHB gene mutations. J Clin Endocrinol Metab. 2012;97(6):E954–62. doi: 10.1210/jc.2011-3437. [DOI] [PubMed] [Google Scholar]
- 26.Sciacovelli M, et al. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature. 2016;537(7621):544–547. doi: 10.1038/nature19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reznik E, et al. Mitochondrial DNA copy number variation across human cancers. Elife. 2016;5 doi: 10.7554/eLife.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schafer ZT, et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461(7260):109–13. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carracedo A, et al. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer. 2013;13(4):227–32. doi: 10.1038/nrc3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Massague J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature. 2016;529(7586):298–306. doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeon SM, et al. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485(7400):661–5. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piskounova E, et al. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527(7577):186–91. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang L, et al. Reductive carboxylation mediated oxidative stress defense supports anchorage independent cell growth. Cancer & Metabolism. 2014;2(Suppl 1):P30–P30. [Google Scholar]
- 34.Jiang L, et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532(7598):255–8. doi: 10.1038/nature17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen EI, et al. Adaptation of energy metabolism in breast cancer brain metastases. Cancer Res. 2007;67(4):1472–86. doi: 10.1158/0008-5472.CAN-06-3137. [DOI] [PubMed] [Google Scholar]
- 36.Dey S, et al. ATF4-dependent induction of heme oxygenase 1 prevents anoikis and promotes metastasis. J Clin Invest. 2015;125(7):2592–608. doi: 10.1172/JCI78031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamarajugadda S, et al. Manganese superoxide dismutase promotes anoikis resistance and tumor metastasis. Cell Death Dis. 2013;4:e504. doi: 10.1038/cddis.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu X, et al. Metabolomic changes accompanying transformation and acquisition of metastatic potential in a syngeneic mouse mammary tumor model. J Biol Chem. 2010;285(13):9317–21. doi: 10.1074/jbc.C110.104448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qu Y, et al. Thioredoxin-like 2 regulates human cancer cell growth and metastasis via redox homeostasis and NF-kappaB signaling. J Clin Invest. 2011;121(1):212–25. doi: 10.1172/JCI43144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, et al. NRF2 activation by antioxidant antidiabetic agents accelerates tumor metastasis. Sci Transl Med. 2016;8(334):334ra51. doi: 10.1126/scitranslmed.aad6095. [DOI] [PubMed] [Google Scholar]
- 41.Le Gal K, et al. Antioxidants can increase melanoma metastasis in mice. Sci Transl Med. 2015;7(308):308re8. doi: 10.1126/scitranslmed.aad3740. [DOI] [PubMed] [Google Scholar]
- 42.Luo C, et al. A PGC1α-mediated transcriptional axis suppresses melanoma metastasis. Nature. 2016;537(7620):422–426. doi: 10.1038/nature19347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Porporato PE, et al. A mitochondrial switch promotes tumor metastasis. Cell Rep. 2014;8(3):754–66. doi: 10.1016/j.celrep.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 44.Santidrian AF, et al. Mitochondrial complex I activity and NAD+/NADH balance regulate breast cancer progression. J Clin Invest. 2013;123(3):1068–81. doi: 10.1172/JCI64264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.LeBleu VS, et al. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16(10):992–1003. doi: 10.1038/ncb3039. 1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christen S, et al. Breast Cancer-Derived Lung Metastases Show Increased Pyruvate Carboxylase-Dependent Anaplerosis. Cell Rep. 2016;17(3):837–848. doi: 10.1016/j.celrep.2016.09.042. [DOI] [PubMed] [Google Scholar]
- 47.Dupuy F, et al. PDK1-Dependent Metabolic Reprogramming Dictates Metastatic Potential in Breast Cancer. Cell Metab. 2015;22(4):577–89. doi: 10.1016/j.cmet.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 48.Fan J, et al. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510(7504):298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vazquez F, et al. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23(3):287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McDonald OG, et al. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet. 2017 doi: 10.1038/ng.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Catanzaro D, et al. Inhibition of glucose-6-phosphate dehydrogenase sensitizes cisplatin-resistant cells to death. Oncotarget. 2015;6(30):30102–14. doi: 10.18632/oncotarget.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haq R, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell. 2013;23(3):302–15. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hudson CD, et al. Altered glutamine metabolism in platinum resistant ovarian cancer. Oncotarget. 2016;7(27):41637–41649. doi: 10.18632/oncotarget.9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Samatar AA, Poulikakos PI. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat Rev Drug Discov. 2014;13(12):928–942. doi: 10.1038/nrd4281. [DOI] [PubMed] [Google Scholar]
- 55.Viale A, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514(7524):628–32. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ying H, et al. Oncogenic Kras Maintains Pancreatic Tumors through Regulation of Anabolic Glucose Metabolism. Cell. 2012;149(3):656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang G, et al. Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J Clin Invest. 2016;126(5):1834–56. doi: 10.1172/JCI82661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Havas KM, et al. Metabolic shifts in residual breast cancer drive tumor recurrence. J Clin Invest. 2017 doi: 10.1172/JCI89914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mannava S, et al. Depletion of deoxyribonucleotide pools is an endogenous source of DNA damage in cells undergoing oncogene-induced senescence. Am J Pathol. 2013;182(1):142–51. doi: 10.1016/j.ajpath.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–99. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang H, et al. The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature. 2017;546(7658):426–430. doi: 10.1038/nature22797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hensley CT, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell. 2016;164(4):681–94. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sancho P, et al. MYC/PGC-1alpha Balance Determines the Metabolic Phenotype and Plasticity of Pancreatic Cancer Stem Cells. Cell Metab. 2015;22(4):590–605. doi: 10.1016/j.cmet.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 65.Allen E, et al. Metabolic Symbiosis Enables Adaptive Resistance to Anti-angiogenic Therapy that Is Dependent on mTOR Signaling. Cell Rep. 2016;15(6):1144–60. doi: 10.1016/j.celrep.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jimenez-Valerio G, et al. Resistance to Antiangiogenic Therapies by Metabolic Symbiosis in Renal Cell Carcinoma PDX Models and Patients. Cell Rep. 2016;15(6):1134–43. doi: 10.1016/j.celrep.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pisarsky L, et al. Targeting Metabolic Symbiosis to Overcome Resistance to Anti-angiogenic Therapy. Cell Rep. 2016;15(6):1161–74. doi: 10.1016/j.celrep.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gui DY, et al. Environment Dictates Dependence on Mitochondrial Complex I for NAD+ and Aspartate Production and Determines Cancer Cell Sensitivity to Metformin. Cell Metab. 2016;24(5):716–727. doi: 10.1016/j.cmet.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]