Abstract
Wnt signalling cascade has developed together with multicellularity to orchestrate the development and homeostasis of complex structures. Wnt pathway components – such as β-catenin, Dishevelled, Lrp6, and Axin- are often dedicated proteins that emerged in evolution together with the Wnt signalling cascade and are believed to function primarily in the Wnt cascade. It is interesting to see that in recent literature many of these proteins are connected with cellular functions that are more ancient and not limited to multicellular organisms – such as cell cycle regulation, centrosome biology, or cell division. In this review, we summarize the recent literature describing this crosstalk. Specifically, we attempt to find the answers to the following questions: Is the response to Wnt ligands regulated by the cell cycle? Is the centrosome and/or cilium required to activate the Wnt pathway? How do Wnt pathway components regulate the centrosomal cycle and cilia formation and function? We critically review the evidence that describes how these connections are regulated and how they help to integrate cell-to-cell communication with the cell and the centrosomal cycle in order to achieve a fine-tuned, physiological response.
1. Wnt signaling pathways
Wnt signaling pathway is one of the key signaling cascades, essential for both correct embryo development and tissue homeostasis in adulthood. Research in Wnt signaling pathways started around 1980, when two groups independently reported new morphogenetic determinants in Drosophila and mouse, (Nusse and Varmus, 1982; Nusslein-Volhard and Wieschaus, 1980) respectively. Since then, Wnt signaling has been found to affect a myriad of aspects of cell behavior.
The Wnt signaling pathway is activated by Wnt ligands - secreted morphogens and drivers of embryogenesis that exert their influence over medium to long range distances. Nineteen homologs are present in the human genome and they are well conserved throughout the animal kingdom. Wnt proteins can activate several distinct pathways that are shortly introduced below.
1.1. Wnt/β-catenin pathway
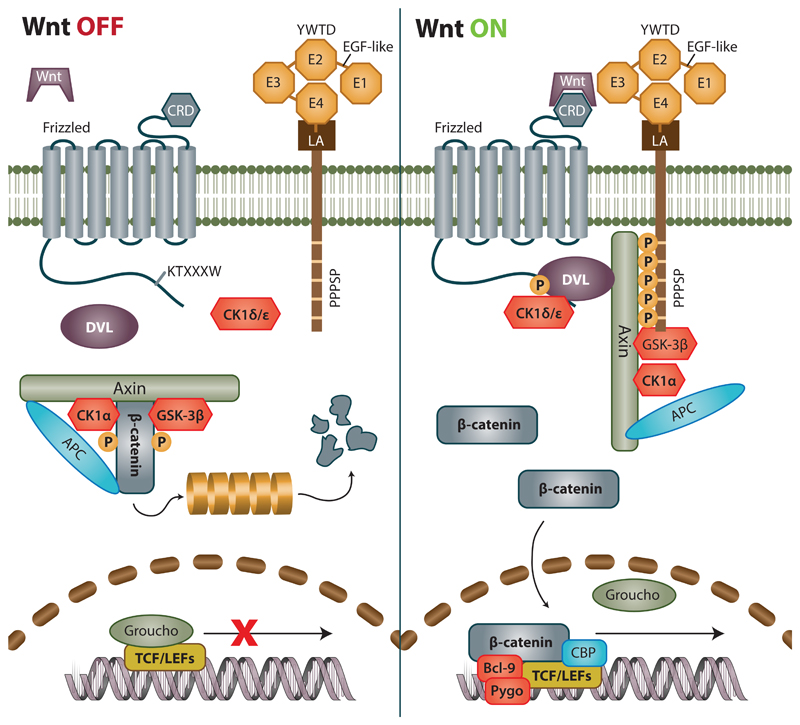
First discovered and best described, the Wnt/β-catenin pathway, also referred to as the canonical pathway, influences cell fate, proliferation and self-renewal of stem and progenitor cells throughout the lifespan of metazoa (Korinek et al., 1998; ten Berge et al., 2011). It revolves around the transcriptional co-activator β-catenin, which is present in the cell in two distinct pools. It maintains the connection to actin cytoskeleton as a component of cadherin junctions and its soluble cytoplasmic pool serves as a signaling mediator. Cytoplasmic concentration of β-catenin in the cell is kept low by multiprotein complex consisting of Axin, Adenomatous polyposis coli (APC) and Glycogen synthase kinase-3β (GSK-3β). Without a Wnt signal, this destruction complex continually phosphorylates β-catenin and targets it for degradation using the ubiquitin proteasome pathway. For a scheme of the Wnt/β-catenin pathway see Figure 1.
Figure 1. Current view of Wnt/β-catenin signaling in OFF and ON state.
During OFF state destruction complex consisting of Axin, APC and GSK-3β phosphorylates β-catenin and marks it for subsequent degradation via ubiquitin proteasome pathway. At the same time, transcription factors from the TCF/LEF family remain bound to repressors such as Groucho, blocking the transcription of Wnt target genes. Cascade is activated after binding of Wnt ligand to Frizzled (Fzd) receptor. Subsequently, both DVL and Lrp6 associate to Fzd. Intracellular residues of Lrp6 are phosphorylated and become a site of attachment for scaffold protein Axin, which can no longer serve as assembly site for destruction complex, which is thus desintegrated. It should be noted that phosphorylated Lrp6, DVL Axin together with other proteins form a structures dubbed signalosomes that ‘attract’ each other and amplify the Wnt signal. As a result, β-catenin is no longer degraded, and accumulates in the cytoplasm. After reaching a certain threshold, it is translocated to nucleus where it binds to TCF/LEF family of transcription factors, replaces resident repressors thereby co-activating transcription of its target genes.
The pathway activation’s beginning conforms to our view of standard signal transduction. Wnt protein binds the Frizzled receptor (Fz or Fzd) and Low-density-lipoprotein receptor-related proteins 5 and 6 (Lrp5/6) co-receptor forming a ternary complex. Cytoplasmic portion of this complex is phosphorylated, which prompts recruitment of Wnt cascade mediators. Dishevelled (DVL) protein binds to Fzd and initiates the phosphorylation of cytoplasmic tail of Lrp5/6 receptor, which then binds Axin. This renders the destruction complex inactive and stops the constant downregulation of β-catenin, which starts to accumulate in the cytoplasm. Upon reaching a certain threshold, β-catenin is translocated into the nucleus, where it couples with transcription factors from the T-cell-specific transcription factor/Lymphoid enhancer-binding factor (TCF/LEF) family. The final outcome of the signal cascade is the upregulation of genes connected to cell fate and cell proliferation, such as c-Myc, or Cyclin D1 and many others (Tetsu and McCormick, 1999; He et al., 1998).
1.1.1. Receptor complex – Frizzled and LRP5/6
Canonical Wnts require two receptor sets to propel the signal downstream. Frizzled are seven-pass transmembrane domain receptors belonging to class F of G-protein coupled receptors (GPCR) (Schulte and Bryja, 2007). Due to the fact that humans encode 10 Fzd receptors and 19 Wnts, their interaction, affinity, specificity and involvement in distinct cascades has been problematic to elucidate.
Lrp5/6 and Drosophila homologue Arrow are single-span transmembrane proteins that play a vital role as co-receptors in Wnt/β-catenin signaling. The intracellular part of Lrp5/6 contains highly conserved PPPS/TPxS/T motif reiterated five times (Tamai et al., 2004) whose phosphorylation is required to activate downstream signaling as described in part 1.1.3.
1.1.2. Dishevelled
Dishevelled (DVL) is a key regulator of Wnt signaling connecting the receptor complex and downstream effectors. It also stands at the branching point between Wnt/β-catenin and alternative pathways. Three DVL isoforms (DVL1, DVL2 and DVL3) are present in mammals and they have partially specific, partially overlapping functions. Even though many functions of DVL and its binding partners have been discovered and we have gained considerable insights into its regulation, the key question concerning its switching properties still remains unanswered.
DVL proteins possess a well-defined three-domain structure with interspersed unordered regions. DVL DIX domain (Dishevelled, aXin), which shares homology with a similar one present in Axin confers DVL the ability to assemble into homo- or hetero-oligomers. The ability to polymerize in a head-to-tail manner is required for Wnt/β-catenin signaling (Schwarz-Romond et al., 2007). DVL DIX domain binding to Axin inhibits Axin function (Fagotto et al., 1999; Li et al., 1999; Smalley et al., 1999) in the destruction complex. PDZ domain (Post synaptic density, Disc large, Zonula occludens-1) is the most versatile when it comes to its array of binding partners. It interacts with both canonical and non-canonical activators alike, making it an ideal candidate for a switch (Wallingford and Habas, 2005). DEP domain was thought to function predominantly in alternative Wnt pathways (Axelrod et al., 1998) but recent evidence confirmed its critical importance also in the Wnt/β-catenin pathway (Gammons et al., 2016; Paclikova et al., 2017). DEP domain helps with binding to Fzd and interacts with lipid moieties on the plasma membrane in order to stabilize this interaction (Pan et al., 2004; Tauriello et al., 2012).
DVL is a subject to a large array of post-translational modifications, as reviewed elsewhere (Bryja and Bernatik, 2014). The most important modification, with respect to its role in the Wnt pathway, is phosphorylation by casein kinase (CK1) δ/ε, required for Wnt pathway activation. CK1δ/ε phosphorylates DVL in a two-step mechanism. Initial “switch on” phosphorylation is induced by Wnt signaling, followed by a second round of phosphorylation, which act as a shutoff mechanism (Bernatik et al., 2011; Bernatik et al., 2014). Other DVL kinases have been also reported and are discussed below.
1.1.3. Cytoplasmic events and activation of transcription
The clear sequence of events happening directly below the membrane after the Wnt initiation signal arrives has not yet been characterized; nevertheless, many facts are known. Lrp5/6 and Fzd are brought into close proximity, their association alone is sufficient for Wnt signal initiation. Fzd function is linked to DVL and DVL is required for Lrp6 phosphorylation. DVL and Axin contain homologous DIX domain which confers ability to form weak homo- or heterotypic interactions leading to aggregation (Bienz, 2014). DVL homo-oligomerization promotes Fzd-Lrp6 cluster creation and also recruits Axin to the membrane, facilitating Lrp6 phosphorylation by GSK-3β and CK1γ. It creates a positive feedback loop and amplifies the signal by phosphorylating all PPPSP motifs. This model for signal transduction has been dubbed 'initiation-amplification' (Zeng et al., 2008) and signaling component aggregation creates structures named 'signalosomes' (Bilic et al., 2007).
The key regulator of cytoplasmic β-catenin levels, destruction complex, consists of Axin, APC, GSK-3β and several other proteins. Axin directly interacts with all other core components of the destruction complex (β-catenin, APC, CK1α, and GSK-3β), thus being the central scaffold (Ikeda et al., 1998; Kishida et al., 1998; Sakanaka, Weiss, and Williams, 1998).
In the absence of a Wnt signal, the role of the destruction complex is to continually phosphorylate β-catenin and target it for ubiquitination and subsequent degradation, to prevent the expression of target genes. The phosphorylation is performed by GSK-3β. In addition, CK1α binds Axin and introduces priming phosphorylation on β-catenin’s S45, which leads to it being recognized by GSK-3β. GSK-3β subsequently binds Axin as well and efficiently phosphorylates β-catenin on S33/S37/T41, which can be subsequently targeted for degradation by E3 ubiquitin ligase β-TrCP.
After deactivating the destruction complex, β-catenin is accumulated in the cytoplasm and shuttled to the nucleus by a mechanism that is not entirely understood (Henderson and Fagotto, 2002; Stadeli, Hoffmans, and Basler, 2006). When in the nucleus, β-catenin interacts with the TCF/LEF family of transcription factors, it replaces the transcriptional repressor Groucho (Daniels and Weis, 2005) and recruits co-activators such as BCL-9, Pygopus and other proteins, turning the whole complex into an activator.
1.2. β-catenin independent pathways
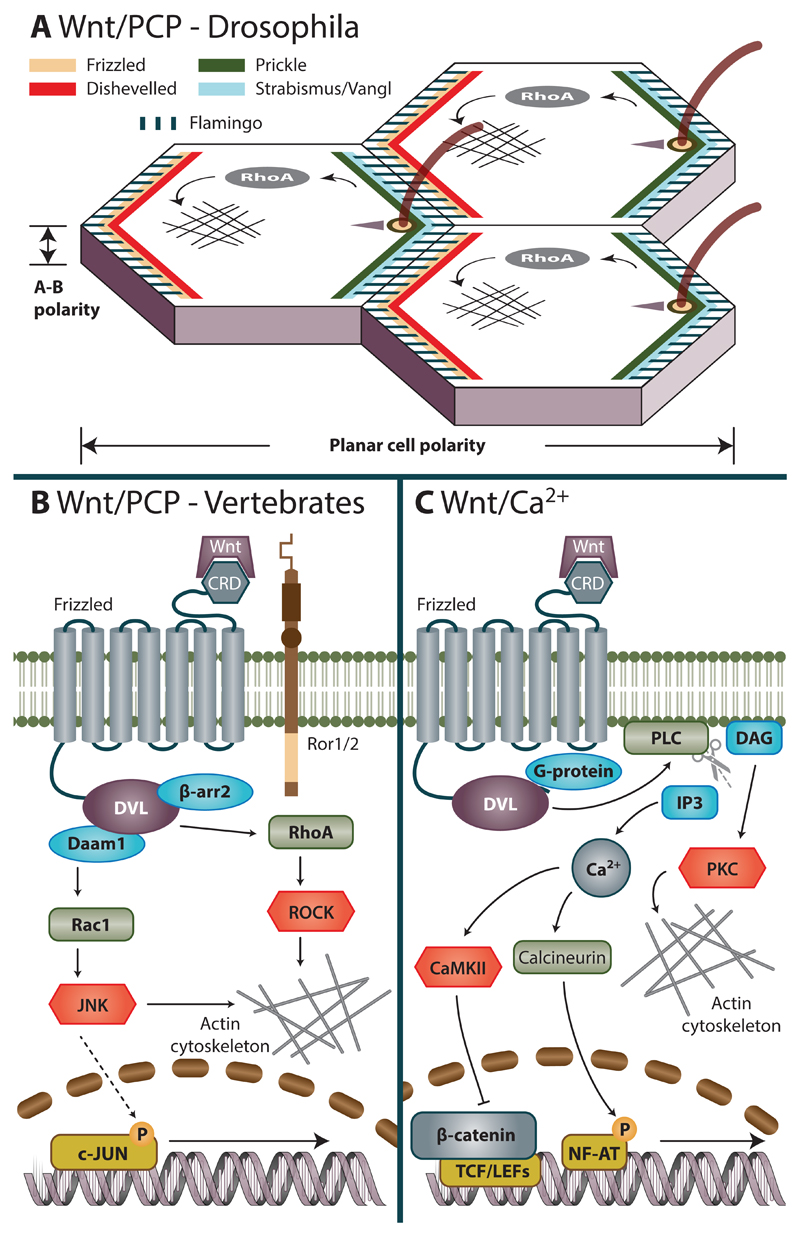
In addition to the Wnt/β-catenin pathway, Wnts can also participate in the 'alternative' or also 'non-canonical' signaling branches. They do not employ β-catenin, but rather modify the cytoskeleton. The best known non-canonical Wnt pathway is Wnt/planar cell polarity (PCP) pathway, initially described in Drosophila (Fig. 2A). Planar polarity is determined by the asymmetric localization of so-called core PCP components. Two protein subsets are located on the opposite sides of cell-cell adherent junctions (Axelrod, 2009; Zallen, 2007). The Proximal subset consists of atypical cadherin Flamingo (Fmi), LIM domain protein Prickle (Pk) and four-pass Van Gogh transmembrane protein (Vang, also known as Strabismus; mammalian homologues: Van Gogh-like proteins, Vangl1 and Vangl2). Distal subset contains Fmi, serpentine receptor Frizzled (Fz), cytoplasmic protein Dishevelled (Dsh) and ankyrin repeat protein Diego (Dgo). The asymmetric localization of core PCP proteins stems from intracellular interactions between these subsets. They display self-organizing properties and the alignment between cells constructs itself and is propagated due to asymmetric cell-to-cell contacts (Fig. 2A).
Figure 2. β-catenin-independent Wnt pathways.
A. Wnt/Planar cell polarity (PCP) in Drosophila is responsible for coordinated alignment of cells across a tissue plane. Figure shows configuration of asymmetric complexes of core PCP pathway components at the cell boundary after polarity has been established. Proximal site contains Frizzled-Dishevelled-Flamingo protein complexes and distal site contains Vang-()Prickle-Flamingo complexes. This assymetric segregation arises from both intracellular cascades that perpetrate their mutual exclusion at either proximal or distal site and from their preferrential heterotypic association extracellularly.
B. Wnt/PCP pathway in vertebrates. Activation of vertebrate PCP pathway is triggered by Wnt ligand (typically Wnt5a or Wnt11) that interact with Fzd and coreceptors (Ror1, Ror2, PTK7 or Ryk) and via DVL and β-arrestin activate members of Rho family of small GTPases. Coordinated activation of downstream effectors – JNK and ROCK – induces cytoskeletal rearrangements that in turn influence processes ranging from convergent extension movements to positioning of basal bodies or cilia.
C. Wnt/Ca2+ pathway in vertebrates. Wnts were shown to induce release of intracellular Ca2+ stores that can activate a multitude of Ca2+ dependent effectors to modulate both transcription as well as actin cytoskeleton.
Components of the PCP pathway – Fzd, DVL, Prickle, Vangl1/2 and Fmi homologues Celsr1-3 have fully conserved function also in vertebrates. However, additional proteins, namely atypical receptor kinases Ror1, Ror2 and PTK7 participate as co-receptors (Fig. 2B). Since PCP pathway activation usually results in cytoskeletal changes, its effectors in vertebrates mostly belong to the Rho family of GTPases and include RhoA and Rac1. RhoA interacts with DVL through protein Daam1 (Habas, Kato, and He, 2001) activating kinase ROCK, in turn mediating cytoskeletal rearrangements. The parallel pathway activates Rac1, leading to increased JNK activity (Fig. 2B). For a recent review on Wnt/PCP pathway see (Butler and Wallingford, 2017). In vertebrates, Wnts can in some cases activate release of intracellular calcium that subsequently triggers multitude of Ca2+ dependent events mediated via activation of CaMKII, PKC or calcineurin. This signaling cascade referred to as Wnt/Ca2+ then triggers depending on the context NFAT-mediated transcription or cytoskeletal remodeling (Fig. 2C). For further reading we refer to the reviews on this topic (Kohn and Moon, 2005; Slusarski and Pelegri, 2007).
2. Cell cycle progression and cell division from a centrosome perspective - friends with benefits
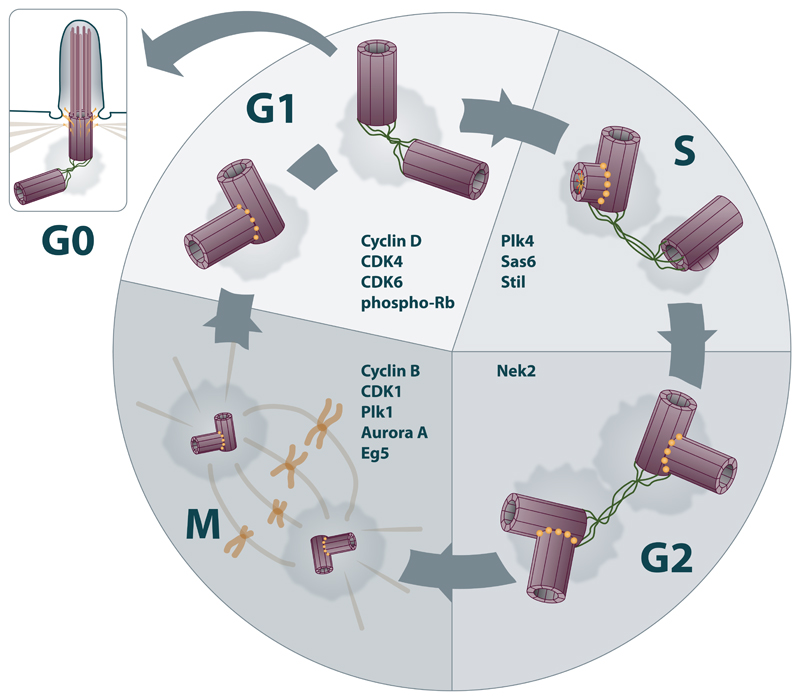
Dividing a cell to giving rise to daughter cells is one of the most fundamental cellular processes for both unicellular and multicellular organisms. In fact, “to divide” is the true purpose of proliferating cells, such as stem cells or progenitors, in order to create a sufficient pool of cells from which more specialized cells differentiate. Such a task requires the coordination of cellular metabolism, duplication of DNA, and cytoskeletal rearrangements. To ensure timely action of the aforementioned processes, the cell deploys a sophisticated cell cycle regulatory machinery, centered on cyclin dependent kinases (CDKs) and additional specialized mitotic kinases, to control its progression through the cycle via a system of checkpoints (Nurse, 1997; Khodjakov and Rieder, 2009). That said, it is not surprising that organelle, thought to play a central role in many aspects of cell cycle and division, was given the fitting name “centrosome”. In fact, it was Theodor Boveri who coined that name more than hundred years ago when observing these organelles at the poles of the bipolar mitotic spindle. At that time, he also postulated its fundamental role in cell division (Boveri, 2008). The cell cycle and centrosomal cycle are tightly connected, as depicted in Figure 3.
Figure 3. Coordination of cell cycle and centrosomal cycle.
The cell first needs to commit itself to enter new round of cell cycle (G1/S transition), then it replicates its DNA content (S phase), which is, after second gap (G2 phase) subsequently packed into chromosomes and divided into two daughter cells (M phase). Following the mitotic exit, cell typically forms primary cilium that is again disassambled before the new mitotic entry. First, cell needs to pass a 'restriction point' (G1/S) checkpoint at the end of the G1 phase. Key player regulating the G/S checkpoint is the Retinoblastoma tumor suppressor protein (Rb). Rb sequesters transcription factors that are essential for the cell cycle to progress to the S phase. Complexes of cyclin D-CDK4/6 phosphorylate Rb during early G1 phase. A cell in G1 phase typically contains one centrosome with two centrioles. After centrioles are disengaged, loose protein linker is established in-between. They are now in permissive state to duplicate. But they need to enter S phase to initiate centriole duplication. Biogenesis of new centrioles (procentrioles) is a semi-conservative process, which starts next to the proximal end of each of the two pre-existing centrioles. Key steps in the initiation of centriole biogenesis are coordinated by proteins STIL, SAS-6, and kinase PLK4, leading to formation of assembly platform called ‘cartwheel’, which recruits microtubule dimers and dictates the typical 9 fold symmetry of centrioles.. Centrioles fully mature during subsequent cell cycle, by acquisition of protein assemblies termed distal and subdistal appendages, respectivelly. Only the mature centriole is capable to transform into basal body to serve as base of cilium or flagellum. Flexible linker, formed after centriole disengagement in anaphase, allows cohesion of the two centrosomes until the onset of next mitoses. Master regulator here is a kinase NEK2, which coordinates displacement of linker proteins at G2/M and subsequent centrosome separation via phosphorylation of several linker proteins. After linker elimination, centrosomes are physically separated by action of motors. The prominent role here has kinesin motor KIF11/Eg5, action of which is fine tuned by kinases Cdk1, PLK1, and NEK family. As the cell approaches mitosis, the centriolar pairs separate from each other and migrate to the opposite poles to help organizing the mitotic spindle. Cell that is about to divide usually uses an organized array of microtubules (spindle and astral microtubules) together with microtubule motors to generate pulling force to physically segregate chromosomes into two daughters. Entry into mitosis is triggered by activity of cyclin B-CDK1 complex. Cell division leaves each daughter cell with one centrosome containing two centrioles. These are originally kept in engaged mode which restrains their re-duplication. Subsequent centriole disengagement during mitoses is controlled by PLK1 and separase, and represents critical step for licensing of centriole to duplicate in the upcoming round of cell cycle. Enrolment of these mitotic regulators in the control of centriole disengagement hence elegantly interconnects the centrosome cycle with mitotic machinery and separation of chromatids, respectively, and ensures correct timing of these events.
The first experimental evidence for the centrosome’s directing role in cell division came from classical experiments with oocytes, demonstrating that injecting purified centrosomes was sufficient to trigger parthenogenetic development in frog or fish eggs (Klotz et al., 1990; Picard et al., 1987). It has become clear that centrosomes participate in mitotic spindle formation. Further, astral microtubules connect the cell cortex to the centrosome and specify in which position the mitotic spindle will form (Bornens and Gonczy, 2014). When the centrosome is absent, a bipolar spindle is formed through the action of small GTPases Ran (Kalab and Heald, 2008). Having no anchoring point due to the lack of astral microtubules, such spindles float freely in the cytoplasm and seem to adopt a random orientation (Louvet-Vallee, Vinot, and Maro, 2005; Khodjakov and Rieder, 2001). Centrosomes also affect the position of the cleave furrow during cytokinesis by affecting spindle orientation or perhaps also by acting directly on the cytokinetic apparatus (Khodjakov and Rieder, 2001; Piel et al., 2001; Oliferenko, Chew, and Balasubramanian, 2009). Some cell types can also divide asymmetrically, meaning that the daughter cells differ in size, fate, or eventually both (Doe, 2008; Knoblich, 2008). This is especially important during embryogenesis, when a stem cell gives rise to another stem cell to replenish the niche, and one progenitor which rapidly divides further. There is growing experimental evidence from both Drosophila and mice suggesting that spindle positioning and/or asymmetry in centrosome inheritance can directly affect the fate of daughter cells (Doe, 2008; Lancaster and Knoblich, 2012).
In addition, centrosomes might also fine tune additional aspects of cell cycle progression. Signaling cascade components implicated in regulating mitotic progression, namely PLK1, Aurora A, CDK1/Cyclin B, and CDC25 have been detected in centrosomes during G2/M transition (Arquint, Gabryjonczyk, and Nigg, 2014). Further, CDK1/Cyclin B complex, essential for mitotic entry, requires a centrosome for its proper activation (Jackman et al., 2003). In addition, the actual destruction of CDK1/Cyclin B complex, marking the end of mitosis, also seems to be initiated at centrosome (Wakefield, Huang, and Raff, 2000).
2.1. Centrosome –basics
A centrosome is a non-membranous cytoplasmic organelle, formed around a core consisting of two microtubule-based cylindrical structures, the centrioles, which is surrounded by a layered protein matrix termed pericentriolar material (PCM). PCM contains proteins such as gamma-tubulin that help to organize and nucleate microtubules, explaining why centrosomes function as the main microtubule-organizing center (MTOC) in many cell types (Luders and Stearns, 2007). As the cell approaches mitoses, PCM undergoes marked changes. It significantly expands in size, but retains its inner organizations, with a pericentrin occupying layer most proximal to the centriole wall, while proteins such as CEP192 and CEP215/CDK5RAP2 are distributed in PCM’s more distal elements (Mennella et al., 2014). Through the microtubules that it organizes and the proteins it recruits, the centrosome plays an important role not only during cell division, but also participates in regulation of cell polarity, migration, and cell trafficking (Conduit, Wainman, and Raff, 2015). In addition, the centrosome serves as a hub/scaffold for proteins involved in cell cycle regulation and cell signaling (Arquint, Gabryjonczyk, and Nigg, 2014). Protein assemblies found in the centrosome’s vicinity are termed satellites, hypothesized to participate in centrosome biogenesis via transport or sequestration of key centrosome components (Tollenaere, Mailand, and Bekker-Jensen, 2014).
Centrosomes are typical for animal cells; they probably evolved from flagellar apparatus and possibly played an important role in transitioning to multicellular organisms (Bornens and Azimzadeh, 2007; Bornens, 2012). However centrosomes are not present in all multicellular organisms’ cell types. Actually, plants do not have a centrosome at all, but have developed alternative structures and mechanisms to compensate for its absence. Interestingly, the centrosome is also absent in some vertebrate cells. A typical example is an oocyte, which loses centrosomes during its meiotic maturation. Centrosome loss is also a hallmark of final stage of myoblast or cardiomyocyte differentiation, characterized by relocating centrosomal proteins to the nuclear envelope (Bornens, 2012).
Even though a mammalian cell typically contains one or two centrosomes, some specialized cell types show a surprising variability in terms of centrosome numbers (Cunha-Ferreira, Bento, and Bettencourt-Dias, 2009). However, it would be very premature to conclude that the absolute centrosome number does not matter. Besides the rather extreme examples of cell specialization, the correct number of centrosomes in a proliferating cell is under strict surveillance (Holland et al., 2012; Fava et al., 2017; Wong et al., 2015; Bazzi and Anderson, 2014), since alterations in the number of centrosomes (called numerical centrosome aberrations) have profound effects on both development and homeostasis (Gonczy, 2015; Nigg, Cajanek, and Arquint, 2014; Godinho and Pellman, 2014; Bettencourt-Dias et al., 2011). Indeed, the presence of extra centrosomes is not only sufficient to trigger genome instability (Ganem, Godinho, and Pellman, 2009) but also to initiate tumor development (Levine et al., 2017; Sercin et al., 2016; Basto et al., 2008). In addition, there is a growing body of evidence that alterations in centrosome numbers are linked to diseases such as primary microcephaly and primordial dwarfism (Nigg, Cajanek, and Arquint, 2014; Marthiens et al., 2013; Bettencourt-Dias et al., 2011).
2.2. Cilia
Cilia are microtubule-based organelles typically found protruding from the surface of non-dividing cells. They can be either motile or immotile, with immotile ones being called primary cilia. Primary cilia, given their non-motile character, were originally considered vestigial organelles. Importantly, however, an increasing number of studies appearing over the last 10-15 years have very much changed that view, and put the primary cilia into a position of important cellular antennas, with essential functions both during embrygenesis and tissue homeostasis, thanks to their employment in the Hh pathway or Calcium signaling (Goetz and Anderson, 2010; Singla and Reiter, 2006; Yoshiba and Hamada, 2014). The importance of cilia has become perhaps even more appreciated when a group of diverse human diseases was linked to ciliary structural and/or functional defects, and hence collectively termed ciliopathies (Bettencourt-Dias et al., 2011; Mitchison and Valente, 2017; Braun and Hildebrandt, 2017).
Cells usually contain one mother centriole at a time, and therefore can form only one cilium. Multiciliated cells are an exception to this rule, typically found in airway epithelium or kidneys, for example. During the course of their differentiation, these cells are able to produce up to hundreds of extra centrioles, to serve as basal bodies (Brooks and Wallingford, 2014; Meunier and Azimzadeh, 2016).
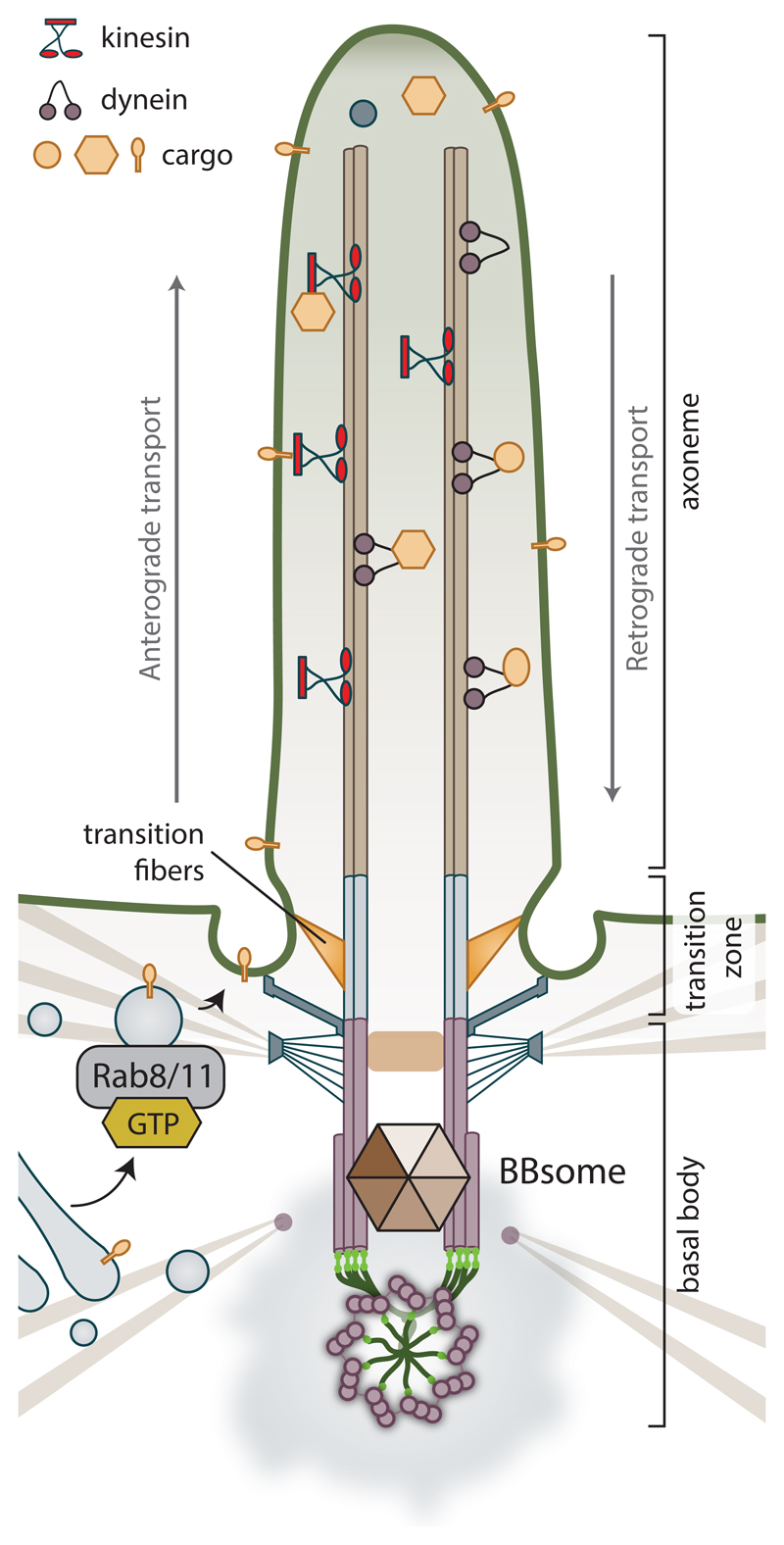
The most prominent part of the cilium is the membrane-enclosed axoneme, see Fig. 4. It comprises of nine microtubule doublets surrounding a central pair – the so-called 9+2 arrangement is typically found in the motile cilia. The central pair is usually missing in primary cilia.
Figure 4. Cilium.
The axoneme stems from the mother centriole-derived basal body, anchored to the cell membrane via its distal appendages (subsequently called transition fibers). Transition fibers, connected both to microtubules and surrounding plasma membrane, contribute to formation of transition zone, which serves as a gate controlling sorting of molecules transported in the cilium. Depositioning of membrane to the mother centriole and formation of ciliary vesicle and subsequently ciliary shaft is coordinated by interactions of Rab11/Rabin8/Rab8 pathway, BBSome (Bardet-Biedl syndrome) protein complex, and components of distal appendages. The ciliary cargo is transported along the axoneme loaded on IFT particles, which are moved by action of molecular motors. Specifically, kinesin-2 family motors mediate anterograde transport along the microtubules towards the tip of the cilium, while cytoplasmic dynein-2 mediates retrograde movement of intraflagellar transport complexes from the tip to the ciliary base.
The first insight into ciliogenesis mechanisms was revealed in pioneering work by Sorokin (Sorokin, 1962), who postulated the docking of Golgi-derived vesicles to the mother centriole as one of the key steps in cilia formation. The current model expands on that notion, see again Fig. 4. Another crucial step towards the assembled cilium is the loss of protein CP110/CEP97 protein complex from the distal end of the mother centriole, which is predicted to allow outgrowth of axonemal microtubules (Schmidt et al., 2009; Spektor et al., 2007). The precise mechanism regulating this event is not known, but it requires activity from Tau tubulin kinase 2 (TTBK2) (Goetz, Liem, and Anderson, 2012; Cajanek and Nigg, 2014) and possibly also ciliary vesicle delivery (Lu et al., 2015). TTBK2 is also implicated in the recruitment of intraflagellar transport complexes (IFTs) to the basal body (Goetz, Liem, and Anderson, 2012; Cajanek and Nigg, 2014). IFT particles are essential for cargo delivery both in and out of the cilium (Rosenbaum and Witman, 2002; Bhogaraju et al., 2013), their actual movement with their cargo is performed by motor proteins, see Fig. 4. Intraflagellar transport mediates both the assembly and resorption of the cilium, and the trafficking of key signaling molecules; in its absence cells are not able to form cilia (Rosenbaum and Witman, 2002). The assembly of cilium as well as disasembly are tigthly interconnected with the cell cycle progression. The cells form primary cilia after exiting from mitoses, in G0/G1. Conversely, resorption of cilia typically starts upon the entry to the next cell cycle.Cilia need to be fully disassembled before mitosis. In fact, proteins implicated in controlling cilium disassembly involve many well established mitotic progression regulators, such as Aurora A, PLK1, or NEK kinase family (Sanchez and Dynlacht, 2016; Nachury and Seeley, 2010).
3. Crosstalk between centrosome-controlled events and Wnt signaling
Multiple reports have observed key Wnt pathway components localized in the centrosome/basal body and/or mitotic spindle. Centrosome can be a “sticky” organelle in the immunostainings and we thus reviewed only studies that use endogenous proteins and/or overexpression of proteins tagged with fluorescent proteins. These studies and their key observations, including the methodological weaknesses and strengths, are summarized in Table 1.
Table 1. Summary of Localization of Key Wnt Pathway Components at the Centrozome and/or Mitotic Spindle.
| Protein | Localization | Method1 | Validation2 | Species/Cell Type | Note | References |
|---|---|---|---|---|---|---|
| DVL2 | spindle poles, midbody | FP tagged protein | no | Hela | also weak kinetochore staining | Kikuchi et al. (2010) |
| DVL2 | centrosome/basal body | ICC | no | Xenopus embryo, multiciliated cells | C-term sufficient | (Park et al., 2008) |
| DVL2/3 | centrosome/basal body | ICC | yes CRISPR DVL1/2/3 TKO | Hek293T | (Cervenka et al., 2016) | |
| β-catenin | spindle and spindle poles, midbody | ICC | no | L cells | (Kaplan et al., 2004) | |
| β -catenin (phospho) | centrosome/basal body | ICC | no | MEFs | phospho Ser33/37/Thr41 | (Hadjihannas, Bruckner, and Behrens, 2010) |
| β-catenin (phospho) | centrosome/basal body, midbody | ICC | no | rat and human fibroblasts | phospho Ser33/37/Thr41, total β-catenin not seen at centrosome | (Huang and Schier, 2009) |
| β-catenin | centrosome/basal body | ICC, EM | No, but multiple antibodies used | RPE-1, U2OS | localized to centriolar linker, armadillo repeat required | (Bahmanyar et al., 2008) |
| β-catenin (Sys1) | spindle poles | FP tagged protein | no | C.elegans | localization to PCM | (Vora and Phillips, 2015) |
| APC | centrosome/basal body | ICC | no | HaCa4 cells | only during mitoses | (Olmeda et al., 2003) |
| axin 1 | centrosome/basal body | ICC | Yes, siRNA | HeLa, NIH3T3 and L cells | DIX domain dependent | (Butler and Wallingford, 2017) |
| axin 1 | spindle and spindle poles | ICC | no | HaCaT cells | (Kim et al., 2009) | |
| axin2/conductin | centrosome/basal body | ICC | no | MEFs, SW480, U2OS | localized to centriolar linker | (Hadjihannas, Bruckner, and Behrens, 2010) |
| GSK3 | spindle poles and mitotic spindle | ICC | no | Hela | phospho Ser21/9 | (Wakefield, Stephens, and M., 2003) (Itoh et al., 2009) |
| Diversin | centrosome/basal body | FP tagged protein | no | Xenopus animal cap cells | ||
| Xenopus Axin-related protein | centrosome/basal body | ICC (overexp.) | no | Xenopus | DIX domain dependent | (Alexandrova and Sokol, 2010) |
| CK1δ | centrosome/basal body | ICC | no | TC-32 cells | C-term sufficient | (Greer and Rubin, 2011) |
| CK1δ | centrosome/basal body | ICC | no | SAOS-2, COS7 | (Sillibourne et al., 2002) | |
| CK1δ | centrosome/basal body | FP tagged protein | no | Jurkat cells | C-term required | (Zyss, Ebrahimi, and Gergely, 2011) |
ICC (immunocytochemical staining of endogenous protein), FP tagged protein (protein of interest visualized by addition of fluorescent protein), EM – detection by immunogold electron microscopy
Only validation based on the removal of the target protein via siRNA or Crispr/Cas9 was considered
The commonly observed localization of Wnt pathway proteins in the centrosome/basal body raised a number of questions about the relationship between the Wnt pathway and centrosome-organized events. In the chapters below we attempt to critically summarize our current understanding of this crosstalk, and how it affects key events in Wnt signaling regulation and the centrosomal cycle.
3.1. To what extent is centrosome/cilium required for the Wnt/β-catenin pathway?
Interestingly, over the years there have been an increasing number of reports arguing either for or against a possible role for centrosome, basal body or primary cilium in Wnt signaling. If one simply examines the reported localizations (Table 1), the argument of “guilty by being present” definitely applies here. This view is further supported by the reported protein-protein interactions between various Wnt pathway components (including β-catenin, DVL, Axin or APC) and bona fide centrosomal proteins that we summarize in Table 2. Recent proteomic studies that profile the protein composition of centrosomes and cilia provide additional evidence regarding the presence of Wnt pathway proteins (such as β-catenin or CK1α and δ) in the centrosome (Jakobsen et al., 2011) or their interactions with core centrosomal proteins. The work of Pelletier lab, that used the BioID approach pinpointed the widespread nature of these physical contacts and identified more than a hundred of interactions between core centrosomal and Wnt pathway proteins, is especially informative. We list these interactions in Table 2 and refer the reader to the original publication (Gupta et al., 2015) for more details.
Table 2. Protein Interactions of Wnt Pathway Components with Centrosomal/Ciliary Proteins - High Throughput Studies.
| Protein | Bait/Partner | Method of Detection | Cell Type/Species | Reference |
|---|---|---|---|---|
| DVL2 | KIAA0753 | BioID | U2OS | (Firat-Karalar et al., 2014) |
| DVL3, CK1ε | NPHP3 (1-203)/cilia APEX | APEX | IMCD3 | (Mick et al., 2015) |
| DVL1, DVL2, DVL3, β-catenin, Axin1, Lrp5, Lrp6, APC, β-arrestin, CK1α, CK1δ, CK1ε | Cep290, Cep162, LCA5, MKS1, Nek8, NIN, NINL, NPHP1, Cep162, KIAA0753, Cep44, Cep63, Cep89, Cep97, CENT2, Centrobin, DCTN1, EVC2, NPHP1, NPHP4, Sas6, SCTL1, SPICE1, SSXIP, STIL, DYNLT1, TMEM17, TMEM216, RPGF, ODF2, OFD1, PCM1, POC1a, RPGRIP1L, C3orf14, TCTN3, B9D1, CNTRL, Cep120, Cep128, Cep135, Cep152, Cep164, Cep170, Cep19, Cep104, AHI1, FBF1, B9D2, CC2D2A, Cep83, CP110, CENPJ, Cep290, TCTN1 | BioID | Hek293T, RPE-1 | (Gupta et al., 2015) |
| DVL3 | Cep164 | IP | Hek293T | (Chaki et al., 2012) |
| β-catenin, β-arrestin, CK1α, CK1δ | ? | purified centrosomes | KE37 cells | (Alexandrova and Sokol, 2010; Andersen et al., 2003) |
Given that possible connections between centrosomes/cilia and Wnt/PCP pathway have been identified earlier and discussed in several reviews (May-Simera and Kelley, 2012; Wallingford and Mitchell, 2011) primarily focus on the potential involvement of centrosome/basal body/primary cilium in the Wnt/β-catenin signaling pathway in this chapter. Probably the first reported connection to Wnt/β-catenin signaling was found in relation to Inversin/Nephrocystin-2, a mutation of which causes kidney defects (formation of cysts) typical for ciliopathies (Otto et al., 2003). Inversin was postulated to act as a molecular switch between Wnt signaling pathways, negatively regulating Wnt/β-catenin signaling (Simons et al., 2005). In addition, a later study showed that depleting Inversin increases expression of DVL1, but reduces levels of DVL2 and DVL3 at the ciliary base (Veland et al., 2013). However, the position of Inversin as a key negative regulator of the Wnt/β-catenin pathway has been challenged by the work of Sugiyama and colleagues (Sugiyama et al., 2011). The authors failed to see any upregulation of the pathway in inv mutant mice, even though the ciliopathy phenotype was present. This example illustrates what will be discussed further - there is often compelling evidence providing an argument for both scenarios (does it or does it not influence Wnt signaling?), and it is not easy to reach a definitive conclusion. However, when the reported data allow alternative interpretations, we aim to provide them.
Knock-down of BBSome components (Bbs1, Bbs4, Mkks) was reported to hyperactivate Wnt/β-catenin signaling (Gerdes et al., 2007). Furthermore, mutation/downregulation of additional regulators of ciliogenesis (Kif3a, Ift88, Ofd1) were also shown to increase Wnt/β-catenin signaling (Gerdes et al., 2007; Corbit et al., 2008; McDermott et al., 2010; Lin et al., 2003; Liu et al., 2014). However, in vivo analyses of mutants with ciliogenesis defects (IFT88, IFT172, Dync2h1, Kif3a) by other groups did not find any pronounced defects in Wnt/β-catenin signaling, even though ciliogenesis and/or Hh pathway were severely hampered (Ocbina, Tuson, and Anderson, 2009; Huang and Schier, 2009). Possible explanation for such discrepancies is the different penetrance of some phenotypes (if there is indeed a defect in Wnt signaling seen, it is often modest), or cell type specificity. One also has to consider simple explanations such as off-target effects (especially if RNAi was employed and rescue experiments were not done) or different sensitivities of used readouts.
Alternatively, one could attempt to explain at least some of the phenotypes by distinguishing strictly between those caused by the absence of protein and those by the absence of organelle (centrosome or cilia, in this case), which is a consequence of the absent protein. This view is supported by the fact that some cilia regulators also give Wnt signaling phenotype while others do not. This implies that the signaling defects may not actually be related to defective ciliogenesis but rather arise due to some additional, cilia-unrelated functions of individual ciliary proteins, that may directly or indirectly translate into cross talk with Wnt/β-catenin signaling. For illustration, several groups reported that knockdown/knockout of Kif3a, a subunit of kinesin2 molecular motor, lead to concomitant defects in ciliogenesis and Wnt/β-catenin signaling (Gerdes et al., 2007; Corbit et al., 2008; Liu et al., 2014). Interestingly, Kif3a has been also shown to affect the orientation of basal bodies by interacting with kinase PAK, independently of its role in ciliary transport (Sipe and Lu, 2011). Moreover, work reporting Kif3a and β-arrestin interaction offers an even more attractive interpretation that altered Wnt signaling observed is due to the direct effects on the β-catenin destruction coplex (Kim et al., 2016). Indeed, recent work from the Mlodzik lab (Balmer et al., 2015) has further supported this model in elegant experiments reporting the effects of several ciliary transport machinery components directly on the β-catenin level, independent of cilia formation.
Another plausible explanation of some of the conflicting results is based on well documented observation that β-catenin abundance is under the strict surveillance of proteasome machinery. Interestingly, active proteasomes have been reported to associate with centrosomes/basal bodies (Gerdes et al., 2007; Wigley et al., 1999; Fabunmi et al., 2000; Gerhardt et al., 2015; Fuentealba et al., 2007; Vora and Phillips, 2015). In addition, E3 ubiquitin ligase Jade-1, which localizes to the centrosome/basal body, is able to ubiquitinate β-catenin and hence target it for destruction by proteasome. Jade-1 activity is controlled negatively by CK1α and can be modulated by the transition zone protein Nephrocystin-4 (Chitalia et al., 2008; Borgal et al., 2014; Mollet et al., 2005). Thus, centrosome/basal body disruption could in principle lead to dysregulated proteasome/E3 ligase activity, and hence, altered Wnt/β-catenin signaling (Gerdes et al., 2007; Vora and Phillips, 2015). Importantly, such an interpretation predicts that Wnt/beta catenin signaling is fine-tuned by the membrane-anchored basal body or intact centrosome, not the presence or absence of cilium. However, there are several questions related to this model. It is not clear how relatively locally altered protein turnover (centrosome or its proximity) may lead to systemic effects on Wnt signaling.
In summary, despite some conflicting results, it seems that centrosomes/cilia are not required to activate the core Wnt/β-catenin pathway – as indicated by grossly normal Wnt signaling in multiple mouse strains lacking cilia (Ocbina, Tuson, and Anderson, 2009; Huang and Schier, 2009). In support of this conclusion, experiments where centrosomes/basal bodies were genetically or pharmacologically ablated did not reveal striking effects on either the β-catenin protein turnover or Wnt/β-catenin signaling (Wong et al., 2015; Basto et al., 2006; Bazzi and Anderson, 2014; Insolera et al., 2014). However, the possibility that centrosome/cilium fine tunes the Wnt pathway (i.e. via protein sequestration – see next chapters) and acts in a cell type-specific manner seem plausible but full understanding will require more experiments in the future.
3.2. Do Wnt pathway proteins control cell cycle progression and centrosomal cycle or vice versa?
As we have previously mentioned (see Table 1 and 2), almost all major Wnt/β-catenin pathway components have been found in the centrosome. This arrangement may to some extent facilitate the proximity of the pathway regulators and in turn may influence Wnt pathway modulation. However, when seen from the opposite perspective, one can ask how centrosomal localization of the Wnt pathway components influences the progression of either the centrosome or cell cycle.
Historically, the Wnt signaling pathway and cell cycle have been connected by Wnt target gene expression: Cyclin D1 and c-myc that are required for progression via G1/S checkpoint were one of the first discovered Wnt target genes and are surely among the most intensively studied. Recently, however, we are witnessing that Wnt signaling, the centrosome and cell cycle are probably more closely intertwined than we previously thought. In this chapter, we would like to summarize the emerging interdependencies of Wnt-centrosome-cell cycle axis. The studies discussed below have two common denominators. The fact that Wnt pathway components localize to the centrosome/basal bodies and physically interact with the core centrosomal proteins and the fact that Wnt signaling components tend to change during the cell cycle and often peak during the G2/M cell cycle phase. Cell cycle associated protein dynamics have been documented for almost all key Wnt pathway proteins (summarized in Table 3).
Table 3. Evidence for Cell Cycle Dependent Changes of Wnt Pathway Components.
| Protein | Cell Cycle/phase1 | Note | Method2 | References |
|---|---|---|---|---|
| β-catenin | G2/M | phosphorylated by NEK2, regulates centrosome cohesion | IF, WB, synchronization | (Olmeda et al., 2003; Bahmanyar et al., 2008; Kaplan et al., 2004; Mbom et al., 2014) |
| β-catenin | G2/M | rapid centrosomal turnover by proteasome, centrosomal localization negatively regulates Wnt-dependent cell fate | IF | (Vora and Phillips, 2015) |
| DVL2/DVL3 | G2/M | scaffold for centrosomal linker proteins | IF, WB, Fucci | (Cervenka et al., 2016) |
| DVL2 | G2/M | regulates spindle orientation | IF | (Kikuchi et al., 2010) |
| DVL2 | G0 | controls planar polarization and apical docking of basal bodies | IF | (Park et al., 2008) |
| Axin2 | G2/M, degraded after mitosis | alters β-catenin phosphorylation and centrosome cohesion | IF | (Hadjihannas, Bruckner, and Behrens, 2010) |
| Axin | G2/M, meiosis | regulated by Aurora A kinase, influence PLK1 and GSK3 activity | IF, FACS | (Kim et al., 2009) |
| Axin | meiosis | knockdown leads to abnormal meiotic spindles and misaligned chromosomes | IF | (He et al., 2016) |
| APC | G2/M | microtubule growth and elongation, stabilization of mitotic spindles, mitosis - stronger binding of APC to centrosome, slow and fast kinetics | IF | (Dikovskaya, Newton, and Nathke, 2004; Lui et al., 2016), |
| APC | G2/M | influences spindle orientation and asymmetric cell division | IF | (Yamashita, Jones, and Fuller, 2003) |
| APC2 | G2/M | ensures mitotic fidelity, binding to Axin is important for cytoskeletal regulation, induces ectopic furrows | IF | (Poulton et al., 2013; McCartney et al., 2001) |
| Fz2 | cytokinesis | together with DVL2, Wnt5a dependent | IF | (Fumoto et al., 2012) |
| LRP6 | G2/M | phosphorylation by Cyclin Y/Pftk1 | WB, IF, FACS | (Hadjihannas, Bruckner, and Behrens, 2010) |
| CYLD | G2/M | astral microtubule stabilization by dishevelled-NuMA-dynein/dynactin complex | synchronization, WB | (Yang et al., 2014) |
| CK1α | G2/M | IF | (Brockman et al., 1992) | |
| CK1δ | G0 | delta, blocks primary ciliogenesis, disrupts cis-Golgi organization | IF | (Greer et al., 2014) |
| GSK-3β | interphase, mitosis | phosphorylated in mitosis, inhibition affects astral microtubule length and chromosome alignment | IF, WB | (Mbom et al., 2014) |
Indicates the cell cycle phase where the highest increase was observed
Methods used for the detection of cell cycle dependent changes: IF (immunofluorescence), WB (Western blotting), FACS – flow cytometry, synchronization – chemical synchronization, Fucci - Fluorescent ubiquitination-based cell cycle indicator
3.2.1. Cell cycle modulated Wnt signaling
The obvious question raised by the cycling behaviour of key Wnt pathway proteins is: Do cells respond to Wnt ligands differently while passing through the cell cycle? There is existing evidence that this is indeed the case and the issue was recently reviewed (Niehrs and Acebron, 2012; Acebron and Niehrs, 2016). We will thus sum up this topic only briefly and focus on the open questions.
The first observation suggesting that the response to Wnt ligands can be cell cycle dependent was the identification of Cyclin-Y/CDK14 complex as the kinase phosphorylating PPPSP motifs of Lrp6. This is the key event required for signal transduction and dissolution of the destruction complex. Cyclin-Y is a membrane bound protein and its expression is regulated based on the cell cycle, peaking in G2/M (Davidson et al., 2009). Another fact hinting at the same thing shows that the cells arrested during G2/M by knock-down of CDC25 increased Wnt/β-catenin signaling (Lee et al., 2009).
Initially it was thought that this particular interaction and phosphorylation results in restricting β-catenin stabilization and thus signaling to the G2/M phase (Davidson et al., 2009). Further research suggested that this phosphorylation event is part of a signaling cascade dubbed Wnt/STOP (stabilization of proteins) signaling (Acebron et al., 2014). Wnt/STOP pathway, discovered and proposed as a signaling cascade in C. Niehrs´s laboratory, is a Wnt-induced signaling cascade that results in the inhibition of GSK-3β activity. GSK-3β was proposed to phosphorylate multiple proteins in a conserved motif, degron (found also in β-catenin), and target them for degradation. As a consequence, activating Wnt/STOP increases the cell protein content in preparation for cell division. It was proposed that this pathway protects a variety of proteins from proteasome destruction and it is claimed that this is possible solely through the inhibition of GSK-3β activity. As such, this has very interesting implications when we look at the Wnt-cell cycle connection from other angle. Although many studies reported increases in Wnt pathway molecules during the G2/M phase, little effort was invested in trying to decipher where G2/M-dependent stabilization comes from. Activating Wnt/STOP signaling offers itself as a very elegant solution, however not all Wnt pathway components are known to be targets for GSK-3β mediated proteasome degradation.
Although the studies mentioned above provide an interesting explanation to some of the experimental findings, many questions still remain open. For example – in the meantime, many other kinases, in addition to CyclinY/CDK14, were shown to phosphorylate PPPSP motif of Lrp6. These include G-protein coupled receptor kinase 5 or 6 (GRK5, GRK6 (Chen et al., 2009) and multiple mitogen activated protein kinases such as p38, JNK or ERK(Cervenka et al., 2011). They were shown to influence Wnt/β-catenin signaling amplitude (Cervenka et al., 2011; Krejci et al., 2012) which suggests that phosphorylating PPPSP motif in order to control the intensity of Wnt pathway activation can be affected by multiple factors – not only cell cycle phase but also mitogenic pathways or cellular stress activation. Similar questions accompany Wnt/STOP signaling and it has yet to be found if GSK-3 is “the” kinase or other kinases can act in a redundant fashion. Analyzing GSK-3-null cells and mice can help to fully reconcile this question.
3.2.2. Wnt pathway proteins as regulators of centrosomal cycle
The next obvious question raised by the observations summarized in Tables 1-3 is the following: Are Wnt pathway proteins involved in the regulation of the cell or centrosomal cycle? And if so, which particular events during the centrosome cycle are under such control?
β-catenin, the molecule central to Wnt/β-catenin signaling has probably been the most extensively documented when it comes to its centrosomal regulation. For the sake of simplicity, we can inspect it as a prototypic Wnt signaling component involved in the centrosome cycle, since many of the observations have been recapitulated with other proteins as well. β-catenin levels oscillate during cell cycle and peak in the G2/M phase (Olmeda et al., 2003). Reducing β-catenin levels lead to a prometaphase delay and increase in the proportion of monoastral spindles originating from unseparated centrosomes. During the interphase, β-catenin localizes to proximal centriole ends together with proteins comprising the flexible centrosomal linker – Rootletin and CNAP-1. As the cell nears mitosis and NEK2 activity peaks, both Rootletin and β-catenin are phosphorylated. Initial Rootletin-dependent localization of β-catenin to centrosome is switched to independent binding in mitosis, leading to centrosome separation (Bahmanyar et al., 2008). Interestingly, some of the β-catenin residues phosphorylated by NEK2 during these events seem to coincide with the S33/S37/T41 cluster phosphorylated by GSK-3β. The results of these phosphorylations diverge, stabilizing β-catenin in the case of NEK2 and marking it for degradation in the case of GSK-3β (Mbom 2014). Additionally, overactive β-catenin stabilization increases the formation of gamma-tubulin structures that resemble immature centrosomes, but are unable to nucleate microtubules (Bahmanyar et al., 2008).
Unlike the role of β-catenin, which seems to be confined to centrosome splitting, other key players in the Wnt pathway- namely DVL, Axin and APC - have more compound phenotypes. DVL was identified as localizing the centrosome, spindle poles and kinetochores during mitosis (Kikuchi et al., 2010; Cervenka et al., 2016). It was proposed that phosphorylation by PLK1 influences the orientation of the mitotic spindle and microtubule-kinetochore attachment. Depleting DVL reduces Mps1 autophosphorylation and localization of Bub1 and Bub1R to MT, possibly interfacing with a spindle assembly checkpoint (SAC) to mitigate the errors arising from improper chromosome segregation. Additionally, DVL acts as part of the machinery responsible for regulating centrosome cohesion. DVL localizes to the centrosome by its DIX domain, where it acts as a scaffold bridging together constituents of the intercentrosomal linker (Cervenka et al., 2016).
Maybe unsurprisingly, close DVL binding partner Axin, was found localized in the centrosome as well, both during interphase and mitosis (Fumoto et al., 2009; Kim et al., 2009). Axin2/Conductin, a negative regulator, increases during cell cycle and culminates at the G2/M boundary (Hadjihannas, Bruckner, and Behrens, 2010). Axin2/Conductin localization to the centrosome is also mediated by CNAP-1 and its loss leads to centrosome splitting due to interference with β-catenin stability and/or phosphorylation (Hadjihannas, Bruckner, and Behrens, 2010). The fact that both Axin and DVL bind to the centrosome using their respective DIX domains indicates that the DIX domain is one of the general domain signatures that may target proteins to the centrosome. Unfortunately, involvement in centrosome-related processes is plagued by purely observational evidence and poor discrimination between its effects on the centrosome itself in contrast to microtubule kinetics. Overexpressing Axin seems to influence GSK-3β and PLK1 localization, both of which have been implicated in phosphorylation cascades of β-catenin in relation to centrosome splitting (Kim et al., 2009). Although it seems to fit the bigger picture, the evidence relies heavily on antibody staining and seems slightly dubious. Moreover, Axin was itself found to be phosphorylated by PLK1 during mitosis. However, Axin phosphorylation also determines its affinity towards y-tubulin and failure to do so leads to the formation of multi-centrosomes (Ruan et al., 2012). Other centrosome related functions of Axin, such as promotion of mitotic fidelity are rather coupled to its role in cytoskeletal dynamics as well (Poulton et al., 2013). Since changes in microtubule nucleation and astral microtubule positioning can cause defects in centrosome separation, stricter deconvolution of their individual contributions towards centrosome-related defects is needed.
Two important DVL kinases, CK1δ and CK1ε, have been described to localize to the centrosome, with C-terminal part as their localization signal (Greer and Rubin, 2011). As is often the case, their participation in centrosome related events is only observational with little mechanistic insight. So far, inhibiting CK1δ in trophoblast cells was shown to result in a variety of centrosomal defects, such as multipolar spindles, centrosome amplifications and impairments in bipolar attachments as well as death by apoptosis after 24h of treatment due to mitotic failure (Greer et al., 2014).
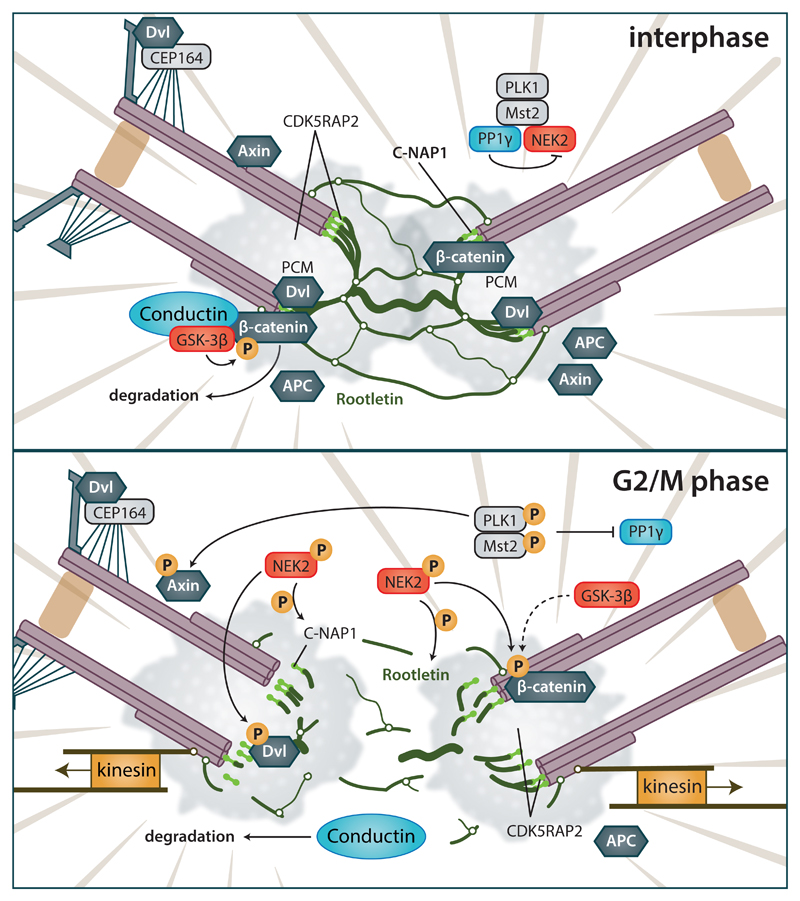
In summary, the evidence, in line with the increased Wnt pathway protein levels in the G2/M phase, suggests that many proteins – such as β-catenin, Dishevelled and Axin - participate in the process of centrosome splitting that takes place at the end of the G2 phase. The described functions of Wnt pathway components and their regulation is schematized in Figure 5. The literature suggests that during the interphase, centrosome cohesion is maintained by the action of Axin2/Conductin and GSK-3β that phosphorylates β-catenin (Bahmanyar et al., 2008; Hadjihannas, Bruckner, and Behrens, 2010). At the same time DVL interacts with distal appendage protein CEP164, perhaps to facilitate basal body docking in ciliated cells (Chaki et al., 2012). As the cell cycle progresses, DVL further accumulates in the centrosome, where it interacts with C-NAP1 (Cervenka et al., 2016). Localization of Axin and APC to the centrosome is probably mediated by microtubules. During the G2/M phase, phosphorylation cascade activates NEK2, which subsequently acts on several targets. Conductin is degraded by proteasome (Hadjihannas et al., 2012), β-catenin is protected from degradation by NEK2 phosphorylation and facilitates centrosome splitting (Bahmanyar et al., 2008). Concurrently, phosphorylation of DVL and C-NAP1 by NEK2 leads to an increase in their overall negative charge and subsequent release from the centrioles, severing the connection with Rootletin (Cervenka et al., 2016). Afterwards, the centrosomes can be pulled apart by the motor proteins’ action. Axin is probably phosphorylated by PLK1, which affects microtubule dynamics during spindle formation (Ruan et al., 2012; Poulton et al., 2013).
Figure 5. Effects of Wnt pathway components on the centrosome separation.
(Upper) Interhase cells. Centrosomes are connected by a linker that consists of fibrous Rootletin and is anchored to the proximal sides of centrioles by C-NAP1. During interphase, centrosome cohesion is maintained by action of Axin2/Conductin and GSK-3β by phosphorylation of β-catenin. DVL in ciliated cells promote basal body docking, via interaction with CEP164 and other proteins. Axin and APC can be also found at centrosome, probably due to their microtubule interactions. NEK2 is kept inactive in complex with PLK1/MST2/PP1γ. (Lower) G2/M phase. Phosphorylation cascade activates NEK2, which subsequently acts on several targets. β-catenin is protected from degradation by NEK2 phosphorylation, and promotes centrosome separation; Axin2/Conductin is degraded by proteasome. Concurrently, phosphorylation of DVL and C-NAP1 by NEK2 leads to the increase in their overall negative charge and subsequent release from centrioles. Centrosomes can be subsequently pulled apart. Axin is phosphorylated by PLK1, which affects microtubule dynamics during spindle formation. For details and references see text.
3.2.3. Control of the cell-cycle dependent functions of Wnt pathway components
The information provided above clearly indicates that individual Wnt pathway components perform multiple functions in distinct cell cycle phases. This implies that an individual Wnt pathway protein can exist in various “pools”. For the purpose of this review, we will define pool as a functional state of the protein that integrates a phosphorylation status, binding partner(s) in the complex, and subcellular localization. It is very likely that cells regulate such protein pools in a delicate manner, which subsequently allows precise control of time and space restricted activities in distinct protein pools. The characteristics of an individual protein in several cellular pools, e.g. three pools in the case of β-catenin (cytoplasmic, membrane and centrosomal) are just being discovered. Not surprisingly, pools are controlled by post-translation modifications and we summarize the available information regarding control of Wnt pathway protein pools by PTM in Table 4.
Table 4. Wnt Pathway Components as Targets of Kinases Involved in the Regulation of cell cycle.
| Substrate | Kinase | Modified Residue | Function | Method of Detection1 | Species2 | Ref. |
|---|---|---|---|---|---|---|
| β-catenin | NEK2 | S33/S37/T41/T102/T556/S675 | β-catenin stabilization, presence at mitotic centrosomes, promotion of centrosome disjunction, regulated upstream by Plk1 | MS, WB | h | (Mbom et al., 2014) |
| APC | Bub1–Bub3, BubR1–Bub3 complexes | middle and C-terminal fragments | chromosome segragation, regulation of kinetochore-microtubule attachment (speculation) | IVP | h | (Kaplan et al., 2001) |
| Dsh | NEK2 | Dsh 1–340 and Dsh 166–623 fragments | activation of Wnt/β-catenin signalling, regulation of Dsh half-life | IVP | d | (Schertel et al., 2013) |
| DVL | NEK2 | T15/S280/S643/S679 | separation of centrosomes, disengagement of centrosomal linker | MS,IVP, WB, IF | h | (Cervenka et al., 2016) |
| DVL2 | PLK1 | T206 | spindle orientation, MT-KT attachment, SAC activation | IVP, WB | h | (Kikuchi et al., 2010) |
| LRP6 | Cyclin Y/Pftk1 | S1490 | cell cycle dependent Wnt signalling activity | WB, IF | d, x, h | (Hadjihannas, Bruckner, and Behrens, 2010) |
| Axin | PLK1 | S157 | Axin-y-tubulin interaction, centrosome formation and segregation | MS, WB | h | (Mick et al., 2015) |
Methods of detection: MS (mass spectrometry), WB (Western Blotting), IVP (in vitro phosphorylation), IF (immunofluorescence with phospho-specific antibodies)
Species: h – human, d – Drosophila, x - Xenopus
The barcoding of individual protein pools is only starting to emerge and currently most information about cell cycle-dependent regulation is available for β-catenin and DVL, which will be described in further detail. Not surprisingly, NEK2 kinase – a master regulator of centrosomal separation - has a key role in regulating the centrosomal pool and the function of both these proteins.
NEK2 phosphorylates and stabilizes β-catenin, and targets it to the spindle poles by independently phosphorylating the same residues as GSK-3β (Bahmanyar et al., 2008). In direct contrast, Axin2/Conductin mediates β-catenin phosphorylation by GSK-3β, but not by NEK2. However, although engaging both the same residues and same kinase, these phosphorylations do not lead to β-catenin destruction, but increase centrosome cohesion. It is the inhibition of phosphorylation that induces centrosome splitting (Hadjihannas et al., 2010). As recent studies point out, different pools of β-catenin or APC have varying degrees of mobility and stability, which subsequently restrict their biological activity (Kafri et al., 2016; Lui, Mok, and Henderson, 2016). Route for β-catenin transport from the membrane to the centrosome has recently been discovered (Kafri et al., 2016). It takes up to 90 minutes for β-catenin to reach the_centrosome and the authors postulate that it is probably unphosphorylated β-catenin driving cell division that is transported this way. The β-catenin in the centrosome is especially short-lived when compared to other cellular compartments. If its rapid clearance (1.9 seconds) is necessary to both induce centrosome splitting and at the same time ensure its proper destruction without influencing Wnt signalling output remains to be seen. The same is true for the question as to how phosphorylation by NEK2 prevents β-catenin associating with β-TrCP and whether such stabilized β-catenin can also have signaling properties.
DVL was shown recently to be a substrate of NEK2 kinase (Cervenka et al., 2016; Schertel et al., 2013; Weber and Mlodzik, 2017). NEK2 can phosphorylate both DVL and centrosome linker proteins such as C-NAP1 or CDK5RAP2 on multiple sites – an almost unbelievable 82 (C-NAP1), 81 (CDK5RAP2) or 41 (DVL3) unique Ser/Thr sites were detected by mass spectrometry (Cervenka et al., 2016) which suggests that electrostatic repulsion or sterical exclusion proposed earlier for NEK2-driven removal of C-NAP1 from the centrosome (Hardy et al., 2014; Faragher and Fry, 2003), can represent a mechanism explaining centrosomal DVL release a complex with C-NAP1. Interestingly, NEK2-mediated DVL release from the centrosome increases the availability of cytoplasmic DVL for the Wnt/β-catenin pathway, where it has a crucial function as a component of signalosomes (Bilic et al., 2007). This effect of NEK2 on DVL pools explains the previously observed positive effects of NEK2 on Wnt/β-catenin (Cervenka et al., 2016; Schertel et al., 2013) despite the fact that NEK2 is not sufficient to trigger the Wnt/β-catenin pathway on its own. NEK2 is a substrate of anaphase-promoting complex/cyclosome (APC/C) that triggers it for degradation. It was shown that in fly retinal cells APC/C restricts retinal differentiation to the G1 phase by degradation of NEK2 and consequent time-restricted Wnt/β-catenin supression (Martins et al., 2017). The interference with NEK2 function can thus serve as a physiological mechanism that fine-tunes Wnt signaling.
Another mode how NEK2 can control Wnt pathway activation in a cell cycle-dependent manner was published recently (Weber and Mlodzik, 2017) and also builds on the fact that NEK2 is a substrate of APC/C. Weber and Mlodzik have found that NEK2 can reduce DVL stability and thus control the DVL levels required to establish planar cell polarity in the epithelium. All these examples suggest that NEK2 can act as a key integrator of DVL’s multiple roles in Wnt/β-catenin pathway, Wnt/planar cell polarity pathway, basal body docking and centrosome separation. Further tools – mainly phospho-specific antibodies and time-lapse imaging of DVL and other Wnt pathway proteins – holds the key towards the full understanding of these processes.
3.3. Role of Wnt pathway components in the regulation of (asymmetric) cell division
Another intriguing aspect we would like to discuss here is the possibility of Wnt pathway or its components to participate directly in the regulation of mitotic cell division – by the possible effects on either the centrosome or spindle positioning. These phenomena are best studied in C. elegans and Drosophila, thanks to powerful genetic tools combined with high resolution live imaging of intact, developing embryos, allowing high throughput phenotypic screening. Interestingly, such screens did indeed identify Wnt signaling components (e.g.mom-1/porc, mom-3/fz, mom-5/?, GSK3, APC, armadillo/β-catenin, kin-19/CK1, mig-5/DVL3) as important regulators of mitotic spindle positioning (Schlesinger et al., 1999; McCartney et al., 2001; Zipperlen et al., 2001; Walston et al., 2004). While the genetic evidence is convincing, the molecular understanding is somewhat lagging behind, especially in comparison with what is known about the role of the Wnt/PCP pathway in the control of oriented cell division (here we again point our readers to recent reviews - (Sawa, 2012; Wallingford, 2012; Morin and Bellaiche, 2011).
This chapter must be initiated by the short description of the function of APC (adenomatosis polyposis coli) in the cell cycle. APC is a large protein with multiple domains – some of its domains show homology with yeast proteins where they participate on cell division (eg. Kar9b in Saccharomyces cerevisiae) (Miller and Rose, 1998; Bloom, 2000). In contrast to other central Wnt pathway regulators (DVL, Axin, Fzd, Lrp6), it is thus likely that APC, or its ancient domains, functioned originally as regulators of microtubule function during cell division and only later attributed a role in the Wnt signaling. APC can interact with microtubules and multiple microtubule-associated proteins such as EB1, kinesin-associated protein 3 (KAP3) or Mitotic Centromere Associated Protein (MCAK) and is a crucial regulator of mitosis. The key findings that described the capacity and the mode of interaction of APC with microtubules and its importance for mitosis and spindle assembly checkpoint are well reviewed (Bahmanyar, Nelson, and Barth, 2009; Caldwell and Kaplan, 2009; Zhang and Shay, 2017) and we do not further focus on the APC-mediated events in this review.
There is evidence for the role of Wnt/β-catenin signaling components in oriented cell division also coming from vertebrate cells. Depleting β-catenin or DVL causes the formation of monopolar spindles (Kikuchi et al., 2010; Kaplan et al., 2004), which can be explained as a consequence of the role these proteins play in centrosome cohesion (Bahmanyar et al., 2008; Cervenka et al., 2016). Further, many other Wnt pathway components were reported to associate with spindle poles or the spindle itself (see Table 1). However, there is often only limited experimental evidence sufficiently explaining the possible functional consequences of such reported localizations for vertebrate mitosis. Moreover, one has to bear in mind that several of these observations are based purely on antibody staining, without appropriate controls (siRNA/ knock-out).
An obvious question here is if observations of Wnt components localizing to and/or affecting spindle/centrosome functions during mitoses means the direct involvement of the Wnt signaling pathway, or if these components act independently of the typical role in the Wnt/pathway. In principle, both models seem plausible and, on the level of individual components, not mutually exclusive. Nonetheless, there is evidence supporting the possibility that some of the reported observations are direct consequences to events in the receptor-ligand complexes close to the cell membrane. First, spindle positioning during mitoses is affected if Wnt secretion is perturbed, either in mom-1/porcupine mutant or Wntless mutant, in C.elegans, (Schlesinger et al., 1999; Thorpe et al., 1997; Banziger et al., 2006). Moreover, a set of experiments in mouse embryonic stem cells (mESCs) demonstrated that locally distributed Wnt/beta catenin pathway ligand, Wnt3a, induced asymmetrical LRP6 and β-catenin distribution (of note, β-catenin was enriched mainly in the cell membrane, not in the nucleus), leading to effects on both the angle of mitotic division plane and the centrosome inheritance in asymmetrically dividing cells (Habib et al., 2013). In addition, recent work by (Stolz et al., 2015) showed that inhibiting Wnt/β-catenin signaling by treatment with Dkk or SFRP effects the rate of microtubule polymerization in mitotic, but not interphase cells.
In summary, it is clear that we are still far from seeing the complete picture. However, based on both the genetic and biochemical evidence accumulated from experiments across different species and cell types, we argue that a hypothesis/model of Wnt/β-catenin signaling directly regulating mitotic cell division is plausible.
3.4. Role of Wnt proteins for basal body docking and proper function/position of cilia
There is one more aspect we feel deserves a mention in regards to Wnt signaling and centrosome/primary cilium biology. Both monocilited and multiciliated cells need to dock their mother centrioles to the plasma membrane in order to initiate ciliogenesis. There is solid evidence from multiple model systems that this step relies directly or indirectly mainly on the action of the Wnt/PCP pathway and its components. Conversely, defects in the Wnt/PCP pathway lead to either complete failure to dock basal bodies, or cause defects in cilia orientation. We have summarized current models of action for the Wnt/PCP pathway in different aspects of cilia biology in Figure 6. For a more thorough insight, we would like to point our readers to several excellent reviews comprehensively addressing this topic (May-Simera and Kelley, 2012; Wallingford and Mitchell, 2011; Carvajal-Gonzalez, Mulero-Navarro, and Mlodzik, 2016).
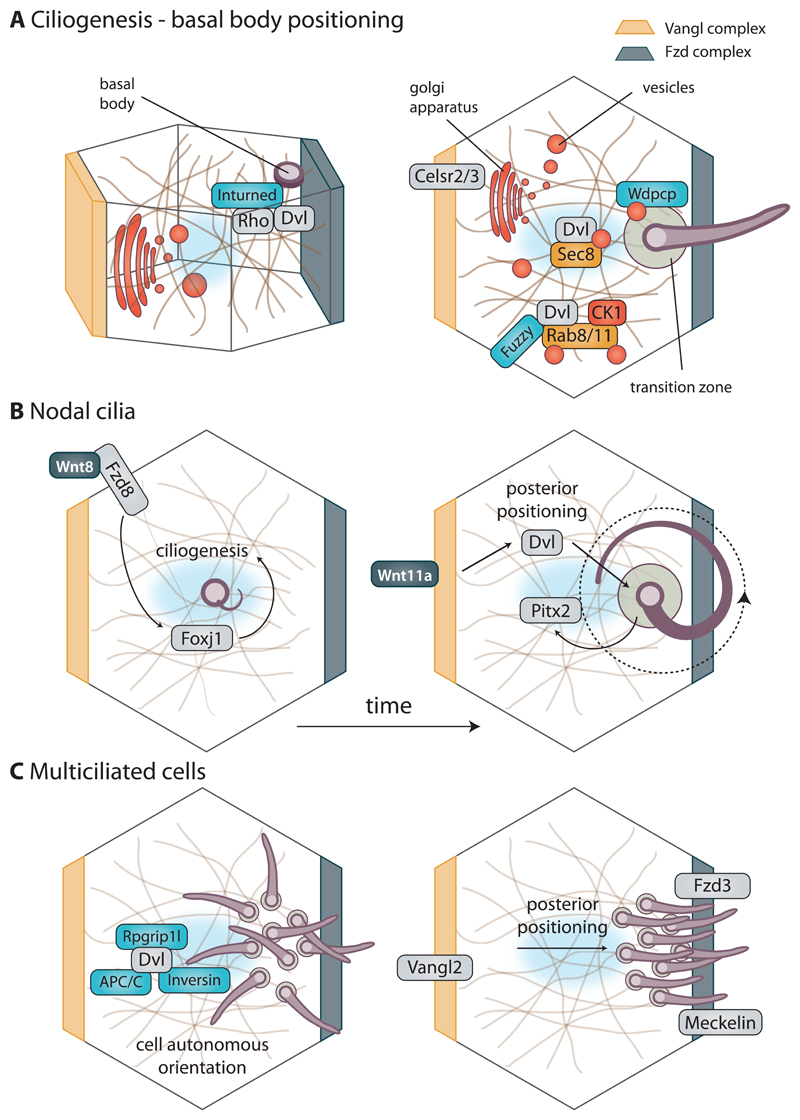
Figure 6. Modes of action of Wnt pathway components on cilia.
(A) Removal of PCP pathway components perturbs ciliogenesis and in most the cases it happens through interference with basal body docking and vesicular transport that is necessary during cilliary assembly. Side view of the cell (left) shows how PCP effector Inturned interacts with DVL in Rho-mediated actin assembly in apical positioning of basal body. At the same time in the top-down view (right), Celsr2/3, CK1, DVL and PCP effector Fuzzy influence vesicular transport towards basal body via interactions with Rab GTPases. PCP effector protein Wdpcp, located at the transition zone, influences ciliogenesis by contributing to cargo sorting and restricting diffusion into ciliary compartment.
(B) Formation of nodal cilia is initiated by Wnt8 ligand binding to Fzd8 receptor, which in turn leads to upregulation of Foxj1, a transcriptional regulator of ciliogenesis. Over time, cilia are positioned towards posterior of the cell by action of cascade involving Wnt11a, DVL, and Pitx2. Only correctly formed and positioned nodal cilia can create leftward fluid flow that determines left-right assymetry.
(C) Multiciliated cells in epithelia display cell-autonomous type of polarity that makes sure that all cilia are oriented in the same fashion. This event is influenced by stability of DVL, controlled by its interactors: Inversin, APC/C and Rpgrip1l. On the other hand, tissue-wide posterior positioning is largely controlled by PCP pathway dependently on Vangl2 and Fzd3 with help of ciliary proteins, such as Meckelin.
Nonetheless, there are still a few points, perhaps more speculative, which we would like to touch upon. One such case is whether the requirement of functional Wnt/PCP pathway for ciliogeneis lies in direct action of asymmetrically distributed PCP components, or if it rather reflects the consequences of the cytoskeletal rearrangement, downstream of the core Wnt/PCP toolkit. To this end, there is some evidence arguing that disrupting PCP does not always translate to defects in ciliogenesis (Antic et al., 2010; Borovina et al., 2010). Furthermore, many PCP proteins are involved in Rho-mediated apical actin assembly or the regulation of correct Rho localization, which may explain the reported effects on cilia basal body docking or vesicular traffic impairment in many PCP mutants (Oishi et al., 2006; Park et al., 2008; Park, Haigo, and Wallingford, 2006; Gray et al., 2009). Interestingly, recent work from the Mlodzik lab has demonstrated that actin polymerization, mediated via PCP effectors such as Inturned or Fuzzy regulate basal body docking to apical membranes via action of Rho GTPases. Conversely, centriole positioning is then one of the evolutionally conserved downstream effects of Wnt/PCP signaling (Carvajal-Gonzalez, Roman, and Mlodzik, 2016), see also Figure 6A.
In nodal cilia, two Wnt pathways interact to bring about proper cilia functioning and subsequently left-right asymmetry determination (Figure 6B). At the beginning, it is not PCP, but Wnt/β-catenin signaling, acting probably through the Wnt8-Fzd8 complex that upregulates Foxj1 expression and initiates ciliogenesis (Caron, Xu, and Lin, 2012; Stubbs et al., 2008; Walentek et al., 2012; Walentek et al., 2013). Subsequently, in order to generate a proper directed laminar flow, Wnt/PCP signaling is activated by Wnt11b to control the posterior positioning of nodal cilium (Hashimoto et al., 2010;Walentek et al., 2013). This is dependent on DVL presence, demonstrated by the phenotype in DVL1/2/3 null mice (Ohata et al., 2014). However, it is yet unclear whether in this case the absence of DVL interferes again with vesicle trafficking, rather than PCP signaling. Another study confirmed that the planar polarity established by Vangl1 and Prickle influences proper cilia positioning, which in turn generates leftward flow, leading to the induction of left-right asymmetry by expressing homeobox gene Pitx2 (Antic et al., 2010).
In agreement with the model of Wnt/PCP signaling acting upstream of ciliogenesis, DVL was also demonstrated as crucial for basal bodies docking in multiciliated cells (Park et al., 2008). DVL has been further shown to be responsible for cell-autonomous cilia orientation (Mitchell et al., 2009). The role of DVL in this type of polarity has been partially attributed to its stability, which is controlled among others by APC/C, Inversin or Rpgrip1l (Simons et al., 2005; Ganner et al., 2009; Mahuzier et al., 2012) (Figure 6C). Intriguingly, DVL2 was also proposed to play a role in resorption of primary cilia (Lee et al., 2012). In the work by Lee and collegues, DVL2 RNAi in Retinal pigment epithelial (RPE-1) cells did not cause defects in primary cilia formation, but prevented its resorption following cell cycle re-entry. The authors further identified CK1ε and PLK1 as regulators of a cilia resorption event, acting on the level of DVL2. As the formation of multiple cilia is linked to permanent cell cycle exit, it seems plausible that this function of DVL2 and its associated kinases is specific for monociliated cells. Another open question related to that is whether the link to control cilia resorption is specific for DVL2, or other DVL isoforms are involved as well. It certainly will not be a trivial task to track down all molecular processed invoving DVL in the centrosome/basal body, given the numerous interactions with various centriolar and ciliary proteins (Gupta et al., 2015; Chaki et al., 2012; Gao and Chen, 2010).
Another player in the cilia formation is GSK3β. GSK3β has been identified as promoting the assembly of the ciliary membrane and hence the initiation of ciliogenesis after the mitotic exit (Zhang et al., 2015). The suggested mechanisms involve kinase activity-dependent control of PCM component Dzip and small GTPase Rab8 interactions. However, given the multiple functions assigned to GSK3β, it is plausible it participates in ciliogenesis also by other means, as already proposed (Thoma et al., 2007).
Overall, the available evidence argues that removing PCP pathway components does typically disturb cilia formation. In most cases it seems to be linked to problems with basal body docking and vesicular transport. That being said, it should be noted that although evidence for PCP proteins participation is strong, not all studies provide sufficient mechanistic insights. For some proteins, such as DVL, CK1, or Inturned that are able to directly affect the organization of cytoskeletal apparatus, the molecular explanation seems straightforward. On the other hand, clarifying the exact role of some of the core membrane PCP proteins will require additional follow-ups to complete the picture.
4. Conclusions and open questions
The evidence for a dual role of Wnt pathway components is gradually increasing, but still remains too sporadic to formulate unifying conclusions. A lot of studies rely on antibody staining without using complementary knockdown/knockout controls for independent verification, so the results have to be taken with a grain of salt. Nevertheless, there are both interesting observations and instances of conflicting evidence available. Proteins such as DVL, Axin or APC are known scaffolds with multitude of binding partners and interact especially tightly with other Wnt pathway components. Explicit direct binding has not yet been clearly demonstrated and it is possible that the presence of one will recruit a plethora of other proteins.
In order to reach unifying conclusions one needs to carefully distinguish between the role of the organelle (cilium/centrosome) vs. protein required to form the organelle, or similarly between the pathway vs. the protein component of the pathway. Ignoring these differences often lead to oversimplification and seemingly conflicting results. However, recent advances in gene editing techniques combined with the revolution in imaging and proteomic techniques have created a platform that will help to speed up progress. Better control of experiments using Crispr/Cas9 generated knockout and analysis of cell phenotypes and protein-protein interactions in real time and in living cells gives hope that the connection between the cell and centrosomal cycle and major morphogenetic cascades will be fully elucidated in the near future.
Acknowledgements
We thank Lucie Smyčková from the Bryja lab for a help with the preparation of tables used in this review.
Footnotes
Declaration of Interest
The work in V. Bryja´s lab is supported by the Czech Science Foundation (15-21789S, 17-16680S, 17-09525S), Masaryk University (MUNI/G/1100/2016) and Neuron – Fund for Support of Science. The Work in the Cajanek lab is supported by the Federation of European Biochemical Societies (Follow up research fund), funds from the Faculty of Medicine MU (ROZV/24/LF/2016), Czech Science Foundation (16-03269Y), and Swiss National Science Foundation (IZ11Z0_166533).
References
- 1.Nusse R, Varmus HE. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 2.Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 3.Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–83. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 4.ten Berge D, Kurek D, Blauwkamp T, Koole W, Maas A, Eroglu E, Siu RK, Nusse R. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nature cell biology. 2011;13:1070–5. doi: 10.1038/ncb2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–6. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 6.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–12. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 7.Schulte G, Bryja V. The Frizzled family of unconventional G-protein-coupled receptors. Trends Pharmacol Sci. 2007;28:518–25. doi: 10.1016/j.tips.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Tamai K, Zeng X, Liu C, Zhang X, Harada Y, Chang Z, He X. A mechanism for Wnt coreceptor activation. Mol Cell. 2004;13:149–56. doi: 10.1016/s1097-2765(03)00484-2. [DOI] [PubMed] [Google Scholar]
- 9.Schwarz-Romond T, Fiedler M, Shibata N, Butler PJ, Kikuchi A, Higuchi Y, Bienz M. The DIX domain of Dishevelled confers Wnt signaling by dynamic polymerization. Nat Struct Mol Biol. 2007;14:484–92. doi: 10.1038/nsmb1247. [DOI] [PubMed] [Google Scholar]
- 10.Fagotto F, Jho E, Zeng L, Kurth T, Joos T, Kaufmann C, Costantini F. Domains of axin involved in protein-protein interactions, Wnt pathway inhibition, and intracellular localization. The Journal of cell biology. 1999;145:741–56. doi: 10.1083/jcb.145.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L, Yuan H, Weaver CD, Mao J, Farr GH, 3rd, Sussman DJ, Jonkers J, Kimelman D, Wu D. Axin and Frat1 interact with dvl and GSK, bridging Dvl to GSK in Wnt-mediated regulation of LEF-1. Embo J. 1999;18:4233–40. doi: 10.1093/emboj/18.15.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smalley MJ, Sara E, Paterson H, Naylor S, Cook D, Jayatilake H, Fryer LG, Hutchinson L, Fry MJ, Dale TC. Interaction of axin and Dvl-2 proteins regulates Dvl-2-stimulated TCF-dependent transcription. Embo J. 1999;18:2823–35. doi: 10.1093/emboj/18.10.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallingford JB, Habas R. The developmental biology of Dishevelled: an enigmatic protein governing cell fate and cell polarity. Development. 2005;132:4421–36. doi: 10.1242/dev.02068. [DOI] [PubMed] [Google Scholar]
- 14.Axelrod JD, Miller JR, Shulman JM, Moon RT, Perrimon N. Differential recruitment of Dishevelled provides signaling specificity in the planar cell polarity and Wingless signaling pathways. Genes & development. 1998;12:2610–22. doi: 10.1101/gad.12.16.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gammons MV, Rutherford TJ, Steinhart Z, Angers S, Bienz M. Essential role of the Dishevelled DEP domain in a Wnt-dependent human-cell-based complementation assay. Journal of cell science. 2016;129:3892–902. doi: 10.1242/jcs.195685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paclikova P, Bernatik O, Radaszkiewicz TW, Bryja V. N-terminal part of Dishevelled DEP domain is required for Wnt/beta-catenin signaling in mammalian cells. Molecular and Cellular Biology. 2017 doi: 10.1128/MCB.00145-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan WJ, Pang SZ, Huang T, Guo HY, Wu D, Li L. Characterization of function of three domains in dishevelled-1: DEP domain is responsible for membrane translocation of dishevelled-1. Cell Res. 2004;14:324–30. doi: 10.1038/sj.cr.7290232. [DOI] [PubMed] [Google Scholar]
- 18.Tauriello DV, Jordens I, Kirchner K, Slootstra JW, Kruitwagen T, Bouwman BA, Noutsou M, Rudiger SG, Schwamborn K, Schambony A, Maurice MM. Wnt/beta-catenin signaling requires interaction of the Dishevelled DEP domain and C terminus with a discontinuous motif in Frizzled. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E812–20. doi: 10.1073/pnas.1114802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bryja V, Bernatik O. Dishevelled at the crossroad of pathways. In: Hoppler S, Moon RT, editors. Wnt signaling in Development and Disease: Molecular Mechanisms and Biological Functions. Willey Publishers; 2014. [Google Scholar]
- 20.Bernatik O, Ganji RS, Dijksterhuis JP, Konik P, Cervenka I, Polonio T, Krejci P, Schulte G, Bryja V. Sequential activation and inactivation of Dishevelled in the Wnt/beta-catenin pathway by casein kinases. The Journal of biological chemistry. 2011;286:10396–410. doi: 10.1074/jbc.M110.169870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernatik O, Sedova K, Schille C, Ganji RS, Cervenka I, Trantirek L, Schambony A, Zdrahal Z, Bryja V. Functional analysis of dishevelled-3 phosphorylation identifies distinct mechanisms driven by casein kinase 1 and frizzled5. The Journal of biological chemistry. 2014;289:23520–33. doi: 10.1074/jbc.M114.590638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bienz M. Signalosome assembly by domains undergoing dynamic head-to-tail polymerization. Trends Biochem Sci. 2014;39:487–95. doi: 10.1016/j.tibs.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Zeng X, Huang H, Tamai K, Zhang X, Harada Y, Yokota C, Almeida K, Wang J, Doble B, Woodgett J, Wynshaw-Boris A, et al. Initiation of Wnt signaling: control of Wnt coreceptor Lrp6 phosphorylation/activation via frizzled, dishevelled and axin functions. Development. 2008;135:367–75. doi: 10.1242/dev.013540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bilic J, Huang YL, Davidson G, Zimmermann T, Cruciat CM, Bienz M, Niehrs C. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science. 2007;316:1619–22. doi: 10.1126/science.1137065. [DOI] [PubMed] [Google Scholar]
- 25.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. EMBO J. 1998;17:1371–84. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kishida S, Yamamoto H, Ikeda S, Kishida M, Sakamoto I, Koyama S, Kikuchi A. Axin, a negative regulator of the wnt signaling pathway, directly interacts with adenomatous polyposis coli and regulates the stabilization of beta-catenin. The Journal of biological chemistry. 1998;273:10823–6. doi: 10.1074/jbc.273.18.10823. [DOI] [PubMed] [Google Scholar]
- 27.Sakanaka C, Weiss JB, Williams LT. Bridging of beta-catenin and glycogen synthase kinase-3beta by axin and inhibition of beta-catenin-mediated transcription. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:3020–3. doi: 10.1073/pnas.95.6.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henderson BR, Fagotto F. The ins and outs of APC and beta-catenin nuclear transport. EMBO Rep. 2002;3:834–9. doi: 10.1093/embo-reports/kvf181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stadeli R, Hoffmans R, Basler K. Transcription under the control of nuclear Arm/beta-catenin. Curr Biol. 2006;16:R378–85. doi: 10.1016/j.cub.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 30.Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol. 2005;12:364–71. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- 31.Axelrod JD. Progress and challenges in understanding planar cell polarity signaling. Semin Cell Dev Biol. 2009;20:964–71. doi: 10.1016/j.semcdb.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Zallen JA. Planar polarity and tissue morphogenesis. Cell. 2007;129:1051–63. doi: 10.1016/j.cell.2007.05.050. [DOI] [PubMed] [Google Scholar]
- 33.Habas R, Kato Y, He X. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daaml. Cell. 2001;107:843–54. doi: 10.1016/s0092-8674(01)00614-6. [DOI] [PubMed] [Google Scholar]
- 34.Butler MT, Wallingford JB. Planar cell polarity in development and disease. Nature reviews Molecular cell biology. 2017 doi: 10.1038/nrm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohn AD, Moon RT. Wnt and calcium signaling: beta-catenin-independent pathways. Cell calcium. 2005;38:439–46. doi: 10.1016/j.ceca.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 36.Slusarski DC, Pelegri F. Calcium signaling in vertebrate embryonic patterning and morphogenesis. Developmental biology. 2007;307:1–13. doi: 10.1016/j.ydbio.2007.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nurse P. Regulation of the eukaryotic cell cycle. European journal of cancer. 1997;33:1002–4. doi: 10.1016/s0959-8049(97)00091-9. [DOI] [PubMed] [Google Scholar]
- 38.Khodjakov A, Rieder CL. The nature of cell-cycle checkpoints: facts and fallacies. Journal of biology. 2009;8:88. doi: 10.1186/jbiol195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boveri T. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. Journal of cell science. 2008;121(Suppl 1):1–84. doi: 10.1242/jcs.025742. [DOI] [PubMed] [Google Scholar]
- 40.Klotz C, Dabauvalle MC, Paintrand M, Weber T, Bornens M, Karsenti E. Parthenogenesis in Xenopus eggs requires centrosomal integrity. The Journal of cell biology. 1990;110:405–15. doi: 10.1083/jcb.110.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Picard A, Karsenti E, Dabauvalle MC, Doree M. Release of mature starfish oocytes from interphase arrest by microinjection of human centrosomes. Nature. 1987;327:170–2. doi: 10.1038/327170a0. [DOI] [PubMed] [Google Scholar]
- 42.Bornens M, Gonczy P. Centrosomes back in the limelight. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2014;369 doi: 10.1098/rstb.2013.0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalab P, Heald R. The RanGTP gradient - a GPS for the mitotic spindle. Journal of cell science. 2008;121:1577–86. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Louvet-Vallee S, Vinot S, Maro B. Mitotic spindles and cleavage planes are oriented randomly in the two-cell mouse embryo. Curr Biol. 2005;15:464–9. doi: 10.1016/j.cub.2004.12.078. [DOI] [PubMed] [Google Scholar]
- 45.Khodjakov A, Rieder CL. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. The Journal of cell biology. 2001;153:237–42. doi: 10.1083/jcb.153.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piel M, Nordberg J, Euteneuer U, Bornens M. Centrosome-dependent exit of cytokinesis in animal cells. Science. 2001;291:1550–3. doi: 10.1126/science.1057330. [DOI] [PubMed] [Google Scholar]
- 47.Oliferenko S, Chew TG, Balasubramanian MK. Positioning cytokinesis. Genes & development. 2009;23:660–74. doi: 10.1101/gad.1772009. [DOI] [PubMed] [Google Scholar]
- 48.Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–87. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- 49.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–97. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 50.Lancaster MA, Knoblich JA. Spindle orientation in mammalian cerebral cortical development. Current opinion in neurobiology. 2012;22:737–46. doi: 10.1016/j.conb.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arquint C, Gabryjonczyk AM, Nigg EA. Centrosomes as signalling centres. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2014;369 doi: 10.1098/rstb.2013.0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jackman M, Lindon C, Nigg EA, Pines J. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nature cell biology. 2003;5:143–8. doi: 10.1038/ncb918. [DOI] [PubMed] [Google Scholar]
- 53.Wakefield JG, Huang JY, Raff JW. Centrosomes have a role in regulating the destruction of cyclin B in early Drosophila embryos. Curr Biol. 2000;10:1367–70. doi: 10.1016/s0960-9822(00)00776-4. [DOI] [PubMed] [Google Scholar]
- 54.Luders J, Stearns T. Microtubule-organizing centres: a re-evaluation. Nature reviews Molecular cell biology. 2007;8:161–7. doi: 10.1038/nrm2100. [DOI] [PubMed] [Google Scholar]
- 55.Mennella V, Agard DA, Huang B, Pelletier L. Amorphous no more: subdiffraction view of the pericentriolar material architecture. Trends in cell biology. 2014;24:188–97. doi: 10.1016/j.tcb.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conduit PT, Wainman A, Raff JW. Centrosome function and assembly in animal cells. Nature reviews Molecular cell biology. 2015;16:611–24. doi: 10.1038/nrm4062. [DOI] [PubMed] [Google Scholar]
- 57.Tollenaere MA, Mailand N, Bekker-Jensen S. Centriolar satellites: key mediators of centrosome functions. Cellular and molecular life sciences : CMLS. 2014 doi: 10.1007/s00018-014-1711-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bornens M, Azimzadeh J. Origin and evolution of the centrosome. Advances in experimental medicine and biology. 2007;607:119–29. doi: 10.1007/978-0-387-74021-8_10. [DOI] [PubMed] [Google Scholar]
- 59.Bornens M. The centrosome in cells and organisms. Science. 2012;335:422–6. doi: 10.1126/science.1209037. [DOI] [PubMed] [Google Scholar]
- 60.Cunha-Ferreira I, Bento I, Bettencourt-Dias M. From zero to many: control of centriole number in development and disease. Traffic. 2009;10:482–98. doi: 10.1111/j.1600-0854.2009.00905.x. [DOI] [PubMed] [Google Scholar]
- 61.Holland AJ, Fachinetti D, Zhu Q, Bauer M, Verma IM, Nigg EA, Cleveland DW. The autoregulated instability of Polo-like kinase 4 limits centrosome duplication to once per cell cycle. Genes & development. 2012;26:2684–9. doi: 10.1101/gad.207027.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fava LL, Schuler F, Sladky V, Haschka MD, Soratroi C, Eiterer L, Demetz E, Weiss G, Geley S, Nigg EA, Villunger A. The PIDDosome activates p53 in response to supernumerary centrosomes. Genes & development. 2017;31:34–45. doi: 10.1101/gad.289728.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong YL, Anzola JV, Davis RL, Yoon M, Motamedi A, Kroll A, Seo CP, Hsia JE, Kim SK, Mitchell JW, Mitchell BJ, et al. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 2015;348:1155–60. doi: 10.1126/science.aaa5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bazzi H, Anderson KV. Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E1491–500. doi: 10.1073/pnas.1400568111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gonczy P. Centrosomes and cancer: revisiting a long-standing relationship. Nature reviews Cancer. 2015;15:639–52. doi: 10.1038/nrc3995. [DOI] [PubMed] [Google Scholar]
- 66.Nigg EA, Cajanek L, Arquint C. The centrosome duplication cycle in health and disease. FEBS letters. 2014;588:2366–72. doi: 10.1016/j.febslet.2014.06.030. [DOI] [PubMed] [Google Scholar]
- 67.Godinho SA, Pellman D. Causes and consequences of centrosome abnormalities in cancer. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2014;369 doi: 10.1098/rstb.2013.0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. Centrosomes and cilia in human disease. Trends Genet. 2011;27:307–15. doi: 10.1016/j.tig.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–82. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Levine MS, Bakker B, Boeckx B, Moyett J, Lu J, Vitre B, Spierings DC, Lansdorp PM, Cleveland DW, Lambrechts D, Foijer F, et al. Centrosome Amplification Is Sufficient to Promote Spontaneous Tumorigenesis in Mammals. Developmental cell. 2017 doi: 10.1016/j.devcel.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Serein O, Larsimont JC, Karambelas AE, Marthiens V, Moers V, Boeckx B, Le Mercier M, Lambrechts D, Basto R, Blanpain C. Transient PLK4 overexpression accelerates tumorigenesis in p53-deficient epidermis. Nature cell biology. 2016;18:100–10. doi: 10.1038/ncb3270. [DOI] [PubMed] [Google Scholar]
- 72.Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, Raff JW. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–42. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marthiens V, Rujano MA, Pennetier C, Tessier S, Paul-Gilloteaux P, Basto R. Centrosome amplification causes microcephaly. Nature cell biology. 2013;15:731–40. doi: 10.1038/ncb2746. [DOI] [PubMed] [Google Scholar]
- 74.Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. Centrosomes and cilia in human disease. Trends in genetics : TIG. 2011;27:307–15. doi: 10.1016/j.tig.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–44. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Singla V, Reiter JF. The primary cilium as the cell's antenna: signaling at a sensory organelle. Science. 2006;313:629–33. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- 77.Yoshiba S, Hamada H. Roles of cilia, fluid flow, and Ca2+ signaling in breaking of left-right symmetry. Trends in genetics : TIG. 2014;30:10–7. doi: 10.1016/j.tig.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 78.Mitchison HM, Valente EM. Motile and non-motile cilia in human pathology: from function to phenotypes. The Journal of pathology. 2017;241:294–309. doi: 10.1002/path.4843. [DOI] [PubMed] [Google Scholar]
- 79.Braun DA, Hildebrandt F. Ciliopathies. Cold Spring Harbor perspectives in biology. 2017;9 doi: 10.1101/cshperspect.a028191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brooks ER, Wallingford JB. Multiciliated cells. Current biology : CB. 2014;24:R973–82. doi: 10.1016/j.cub.2014.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meunier A, Azimzadeh J. Multiciliated Cells in Animals. Cold Spring Harbor perspectives in biology. 2016;8 doi: 10.1101/cshperspect.a028233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sorokin S. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. The Journal of cell biology. 1962;15:363–77. doi: 10.1083/jcb.15.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, Stierhof YD, Nigg EA. Control of centriole length by CPAP and CP110. Current biology : CB. 2009;19:1005–11. doi: 10.1016/j.cub.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 84.Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and CP110 suppress a cilia assembly program. Cell. 2007;130:678–90. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 85.Goetz SC, Liem KF, Jr, Anderson KV. The spinocerebellar ataxia-associated gene Tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell. 2012;151:847–58. doi: 10.1016/j.cell.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cajanek L, Nigg EA. Cepl64 triggers ciliogenesis by recruiting Tau tubulin kinase 2 to the mother centriole. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E2841–50. doi: 10.1073/pnas.1401777111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu Q, Insinna C, Ott C, Stauffer J, Pintado PA, Rahajeng J, Baxa U, Walia V, Cuenca A, Hwang YS, Daar IO, et al. Early steps in primary cilium assembly require EHDl/EHD3-dependent ciliary vesicle formation. Nature cell biology. 2015 doi: 10.1038/ncb3155. [DOI] [PubMed] [Google Scholar]
- 88.Rosenbaum JL, Witman GB. Intraflagellar transport. Nature reviews Molecular cell biology. 2002;3:813–25. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 89.Bhogaraju S, Cajanek L, Fort C, Blisnick T, Weber K, Taschner M, Mizuno N, Lamla S, Bastin P, Nigg EA, Lorentzen E. Molecular Basis of Tubulin Transport Within the Cilium by IFT74 and IFT81. Science. 2013;341:1009–12. doi: 10.1126/science.1240985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sanchez I, Dynlacht BD. Cilium assembly and disassembly. Nature cell biology. 2016;18:711–7. doi: 10.1038/ncb3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nachury MV, Seeley ES. The perennial organelle: assembly and disassembly of the primary cilium. Journal of cell science. 2010;123:511–18. doi: 10.1242/jcs.061093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jakobsen L, Vanselow K, Skogs M, Toyoda Y, Lundberg E, Poser I, Falkenby LG, Bennetzen M, Westendorf J, Nigg EA, Uhlen M, et al. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 2011;30:1520–35. doi: 10.1038/emboj.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gupta GD, Coyaud E, Goncalves J, Mojarad BA, Liu Y, Wu Q, Gheiratmand L, Comartin D, Tkach JM, Cheung SW, Bashkurov M, et al. A Dynamic Protein Interaction Landscape of the Human Centrosome-Cilium Interface. Cell. 2015;163:1484–99. doi: 10.1016/j.cell.2015.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.May-Simera HL, Kelley MW. Cilia, Wnt signaling, and the cytoskeleton. Cilia. 2012;1:7. doi: 10.1186/2046-2530-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wallingford JB, Mitchell B. Strange as it may seem: the many links between Wnt signaling, planar cell polarity, and cilia. Genes & development. 2011;25:201–13. doi: 10.1101/gad.2008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Otto EA, Schermer B, Obara T, O'Toole JF, Hiller KS, Mueller AM, Ruf RG, Hoefele J, Beekmann F, Landau D, Foreman JW, et al. Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nat Genet. 2003;34:413–20. doi: 10.1038/ng1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Simons M, Gloy J, Ganner A, Bullerkotte A, Bashkurov M, Kronig C, Schermer B, Benzing T, Cabello OA, Jenny A, Mlodzik M, et al. Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nat Genet. 2005;37:537–43. doi: 10.1038/ng1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Veland IR, Montjean R, Eley L, Pedersen LB, Schwab A, Goodship J, Kristiansen K, Pedersen SF, Saunier S, Christensen ST. Inversin/Nephrocystin-2 is required for fibroblast polarity and directional cell migration. PLoS One. 2013;8:e60193. doi: 10.1371/journal.pone.0060193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sugiyama N, Tsukiyama T, Yamaguchi TP, Yokoyama T. The canonical Wnt signaling pathway is not involved in renal cyst development in the kidneys of inv mutant mice. Kidney international. 2011;79:957–65. doi: 10.1038/ki.2010.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gerdes JM, Liu Y, Zaghloul NA, Leitch CC, Lawson SS, Kato M, Beachy PA, Beales PL, DeMartino GN, Fisher S, Badano JL, et al. Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat Genet. 2007;39:1350–60. doi: 10.1038/ng.2007.12. [DOI] [PubMed] [Google Scholar]
- 101.Corbit KC, Shyer AE, Dowdle WE, Gaulden J, Singla V, Chen MH, Chuang PT, Reiter JF. Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nature cell biology. 2008;10:70–6. doi: 10.1038/ncb1670. [DOI] [PubMed] [Google Scholar]
- 102.McDermott KM, Liu BY, Tlsty TD, Pazour GJ. Primary cilia regulate branching morphogenesis during mammary gland development. Curr Biol. 2010;20:731–7. doi: 10.1016/j.cub.2010.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lin F, Hiesberger T, Cordes K, Sinclair AM, Goldstein LS, Somlo S, Igarashi P. Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5286–91. doi: 10.1073/pnas.0836980100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu B, Chen S, Cheng D, Jing W, Helms JA. Primary cilia integrate hedgehog and Wnt signaling during tooth development. Journal of dental research. 2014;93:475–82. doi: 10.1177/0022034514528211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ocbina PJ, Tuson M, Anderson KV. Primary cilia are not required for normal canonical Wnt signaling in the mouse embryo. PLoS One. 2009;4:e6839. doi: 10.1371/journal.pone.0006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huang P, Schier AF. Dampened Hedgehog signaling but normal Wnt signaling in zebrafish without cilia. Development. 2009;136:3089–98. doi: 10.1242/dev.041343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sipe CW, Lu X. Kif3a regulates planar polarization of auditory hair cells through both ciliary and non-ciliary mechanisms. Development. 2011;138:3441–9. doi: 10.1242/dev.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim M, Suh YA, Oh JH, Lee BR, Kim J, Jang SJ. KIF3A binds to beta-arrestin for suppressing Wnt/beta-catenin signalling independently of primary cilia in lung cancer. Scientific reports. 2016;6:32770. doi: 10.1038/srep32770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Balmer S, Dussert A, Collu GM, Benitez E, Iomini C, Mlodzik M. Components of Intraflagellar Transport Complex A Function Independently of the Cilium to Regulate Canonical Wnt Signaling in Drosophila. Developmental cell. 2015;34:705–18. doi: 10.1016/j.devcel.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wigley WC, Fabunmi RP, Lee MG, Marino CR, Muallem S, DeMartino GN, Thomas PJ. Dynamic association of proteasomal machinery with the centrosome. The Journal of cell biology. 1999;145:481–90. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fabunmi RP, Wigley WC, Thomas PJ, DeMartino GN. Activity and Regulation of the Centrosome-associated Proteasome. Journal of Biological Chemistry. 2000;275:409–13. doi: 10.1074/jbc.275.1.409. [DOI] [PubMed] [Google Scholar]
- 112.Gerhardt C, Lier JM, Burmuhl S, Struchtrup A, Deutschmann K, Vetter M, Leu T, Reeg S, Grune T, Ruther U. The transition zone protein Rpgrip1l regulates proteasomal activity at the primary cilium. The Journal of cell biology. 2015;210:115–33. doi: 10.1083/jcb.201408060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fuentealba LC, Eivers E, Ikeda A, Hurtado C, Kuroda H, Pera EM, De Robertis EM. Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell. 2007;131:980–93. doi: 10.1016/j.cell.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vora S, Phillips BT. Centrosome-Associated Degradation Limits beta-Catenin Inheritance by Daughter Cells after Asymmetric Division. Current biology : CB. 2015;25:1005–16. doi: 10.1016/j.cub.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 115.Chitalia VC, Foy RL, Bachschmid MM, Zeng L, Panchenko MV, Zhou MI, Bharti A, Seldin DC, Lecker SH, Dominguez I, Cohen HT. Jade-1 inhibits Wnt signalling by ubiquitylating beta-catenin and mediates Wnt pathway inhibition by pVHL. Nature cell biology. 2008;10:1208–16. doi: 10.1038/ncb1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Borgal L, Rinschen MM, Dafinger C, Hoff S, Reinert MJ, Lamkemeyer T, Lienkamp SS, Benzing T, Schermer B. Casein Kinase 1 Alpha Phosphorylates the Wnt-Regulator Jade-1 and Modulates its Activity. The Journal of biological chemistry. 2014 doi: 10.1074/jbc.M114.562165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mollet G, Silbermann F, Delous M, Salomon R, Antignac C, Saunier S. Characterization of the nephrocystin/nephrocystin-4 complex and subcellular localization of nephrocystin-4 to primary cilia and centrosomes. Hum Mol Genet. 2005;14:645–56. doi: 10.1093/hmg/ddi061. [DOI] [PubMed] [Google Scholar]
- 118.Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. Flies without centrioles. Cell. 2006;125:1375–86. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 119.Bazzi H, Anderson KV. Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E1491–500. doi: 10.1073/pnas.1400568111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Insolera R, Bazzi H, Shao W, Anderson KV, Shi SH. Cortical neurogenesis in the absence of centrioles. Nature neuroscience. 2014;17:1528–35. doi: 10.1038/nn.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Niehrs C, Acebron SP. Mitotic and mitogenic Wnt signalling. EMBO J. 2012;31:2705–13. doi: 10.1038/emboj.2012.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Acebron SP, Niehrs C. beta-Catenin-Independent Roles of Wnt/LRP6 Signaling. Trends in cell biology. 2016;26:956–67. doi: 10.1016/j.tcb.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 123.Davidson G, Shen J, Huang Y-L, Su Y, Karaulanov E, Bartscherer K, Hassler C, Stannek P, Boutros M, Niehrs C. Cell Cycle Control of Wnt Receptor Activation. Developmental cell. 2009;17:788–99. doi: 10.1016/j.devcel.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 124.Lee G, White LS, Hurov KE, Stappenbeck TS, Piwnica-Worms H. Response of small intestinal epithelial cells to acute disruption of cell division through CDC25 deletion. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4701–6. doi: 10.1073/pnas.0900751106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Acebron SP, Karaulanov E, Berger BS, Huang YL, Niehrs C. Mitotic wnt signaling promotes protein stabilization and regulates cell size. Mol Cell. 2014;54:663–74. doi: 10.1016/j.molcel.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 126.Chen M, Philipp M, Wang J, Premont RT, Garrison TR, Caron MG, Lefkowitz RJ, Chen W. G Protein-coupled receptor kinases phosphorylate LRP6 in the Wnt pathway. The Journal of biological chemistry. 2009;284:35040–8. doi: 10.1074/jbc.M109.047456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cervenka I, Wolf J, Masek J, Krejci P, Wilcox WR, Kozubik A, Schulte G, Gutkind JS, Bryja V. Mitogen-activated protein kinases promote WNT/beta-catenin signaling via phosphorylation of LRP6. Mol Cell Biol. 2011;31:179–89. doi: 10.1128/MCB.00550-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Krejci P, Aklian A, Kaucka M, Sevcikova E, Prochazkova J, Masek JK, Mikolka P, Pospisilova T, Spoustova T, Weis M, Paznekas WA, et al. Receptor tyrosine kinases activate canonical WNT/beta-catenin signaling via MAP kinase/LRP6 pathway and direct beta-catenin phosphorylation. PLoS One. 2012;7:e35826. doi: 10.1371/journal.pone.0035826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Olmeda D, Castel S, Vilaro S, Cano A. Beta-catenin regulation during the cell cycle: implications in G2/M and apoptosis. Mol Biol Cell. 2003;14:2844–60. doi: 10.1091/mbc.E03-01-0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bahmanyar S, Kaplan DD, Deluca JG, Giddings TH, Jr, O'Toole ET, Winey M, Salmon ED, Casey PJ, Nelson WJ, Barth AI. beta-Catenin is a Nek2 substrate involved in centrosome separation. Genes & development. 2008;22:91–105. doi: 10.1101/gad.1596308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kikuchi K, Niikura Y, Kitagawa K, Kikuchi A. Dishevelled, a Wnt signalling component, is involved in mitotic progression in cooperation with Plk1. EMBO J. 2010;29:3470–83. doi: 10.1038/emboj.2010.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cervenka I, Valnohova J, Bernatik O, Harnos J, Radsetoulal M, Sedova K, Hanakova K, Potesil D, Sedlackova M, Salasova A, Steinhart Z, et al. Dishevelled is a NEK2 kinase substrate controlling dynamics of centrosomal linker proteins. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:9304–9. doi: 10.1073/pnas.1608783113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fumoto K, Kadono M, Izumi N, Kikuchi A. Axin localizes to the centrosome and is involved in microtubule nucleation. EMBO reports. 2009;10:606–13. doi: 10.1038/embor.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kim SM, Choi EJ, Song KJ, Kim S, Seo E, Jho EH, Kee SH. Axin localizes to mitotic spindles and centrosomes in mitotic cells. Exp Cell Res. 2009;315:943–54. doi: 10.1016/j.yexcr.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 135.Hadjihannas MV, Bruckner M, Behrens J. Conductin/axin2 and Wnt signalling regulates centrosome cohesion. EMBO Rep. 2010;11:317–24. doi: 10.1038/embor.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ruan K, Ye F, Li C, Liou YC, Lin SC, Lin SY. PLK1 interacts and phosphorylates Axin that is essential for proper centrosome formation. PLoS One. 2012;7:e49184. doi: 10.1371/journal.pone.0049184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Poulton JS, Mu FW, Roberts DM, Peifer M. APC2 and Axin promote mitotic fidelity by facilitating centrosome separation and cytoskeletal regulation. Development. 2013;140:4226–36. doi: 10.1242/dev.094425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Greer YE, Rubin JS. Casein kinase 1 delta functions at the centrosome to mediate Wnt-3a-dependent neurite outgrowth. The Journal of cell biology. 2011;192:993–1004. doi: 10.1083/jcb.201011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Greer YE, Westlake CJ, Gao B, Bharti K, Shiba Y, Xavier CP, Pazour GJ, Yang Y, Rubin JS. Casein kinase 1 delta functions at the centrosome and Golgi to promote ciliogenesis. Molecular biology of the cell. 2014;25:1629–40. doi: 10.1091/mbc.E13-10-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chaki M, Airik R, Ghosh AK, Giles RH, Chen R, Slaats GG, Wang H, Hurd TW, Zhou W, Cluckey A, Gee HY, et al. Exome Capture Reveals ZNF423 and CEP164 Mutations, Linking Renal Ciliopathies to DNA Damage Response Signaling. Cell. 2012;150:533–48. doi: 10.1016/j.cell.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hadjihannas MV, Bernkopf DB, Bruckner M, Behrens J. Cell cycle control of Wnt/beta-catenin signalling by conductin/axin2 through CDC20. EMBO Rep. 2012;13:347–54. doi: 10.1038/embor.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kafri P, Hasenson SE, Kanter I, Sheinberger J, Kinor N, Yunger S, Shav-Tal Y. Quantifying beta-catenin subcellular dynamics and cyclin D1 mRNA transcription during Wnt signaling in single living cells. eLife. 2016;5 doi: 10.7554/eLife.16748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lui C, Mok MT, Henderson BR. Characterization of Adenomatous Polyposis Coli Protein Dynamics and Localization at the Centrosome. Cancers. 2016;8 doi: 10.3390/cancers8050047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schertel C, Huang D, Bjorklund M, Bischof J, Yin D, Li R, Wu Y, Zeng R, Wu J, Taipale J, Song H, et al. Systematic screening of a Drosophila ORF library in vivo uncovers Wnt/Wg pathway components. Dev Cell. 2013;25:207–19. doi: 10.1016/j.devcel.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 145.Weber U, Mlodzik M. APC/CFzr/Cdh1-Dependent Regulation of Planar Cell Polarity Establishment via Nek2 Kinase Acting on Dishevelled. Developmental cell. 2017;40:53–66. doi: 10.1016/j.devcel.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hardy T, Lee M, Hames RS, Prosser SL, Cheary DM, Samant MD, Schultz F, Baxter JE, Rhee K, Fry AM. Multisite phosphorylation of C-Nap1 releases it from Cep135 to trigger centrosome disjunction. Journal of cell science. 2014;127:2493–506. doi: 10.1242/jcs.142331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Faragher AJ, Fry AM. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol Biol Cell. 2003;14:2876–89. doi: 10.1091/mbc.E03-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Martins T, Meghini F, Florio F, Kimata Y. The APC/C Coordinates Retinal Differentiation with G1 Arrest through the Nek2-Dependent Modulation of Wingless Signaling. Developmental cell. 2017;40:67–80. doi: 10.1016/j.devcel.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Schlesinger A, Shelton CA, Maloof JN, Meneghini M, Bowerman B. Wnt pathway components orient a mitotic spindle in the early Caenorhabditis elegans embryo without requiring gene transcription in the responding cell. Genes & development. 1999;13:2028–38. doi: 10.1101/gad.13.15.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.McCartney BM, McEwen DG, Grevengoed E, Maddox P, Bejsovec A, Peifer M. Drosophila APC2 and Armadillo participate in tethering mitotic spindles to cortical actin. Nature cell biology. 2001;3:933–8. doi: 10.1038/ncb1001-933. [DOI] [PubMed] [Google Scholar]
- 151.Zipperlen P, Fraser AG, Kamath RS, Martinez-Campos M, Ahringer J. Roles for 147 embryonic lethal genes on C.elegans chromosome I identified by RNA interference and video microscopy. The EMBO journal. 2001;20:3984–92. doi: 10.1093/emboj/20.15.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Walston T, Tuskey C, Edgar L, Hawkins N, Ellis G, Bowerman B, Wood W, Hardin J. Multiple Wnt signaling pathways converge to orient the mitotic spindle in early C. elegans embryos. Dev Cell. 2004;7:831–41. doi: 10.1016/j.devcel.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 153.Sawa H. Control of cell polarity and asymmetric division in C. elegans. Current topics in developmental biology. 2012;101:55–76. doi: 10.1016/B978-0-12-394592-1.00003-X. [DOI] [PubMed] [Google Scholar]
- 154.Wallingford JB. Planar cell polarity and the developmental control of cell behavior in vertebrate embryos. Annual review of cell and developmental biology. 2012;28:627–53. doi: 10.1146/annurev-cellbio-092910-154208. [DOI] [PubMed] [Google Scholar]
- 155.Morin X, Bellaiche Y. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Developmental cell. 2011;21:102–19. doi: 10.1016/j.devcel.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 156.Miller RK, Rose MD. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. The Journal of cell biology. 1998;140:377–90. doi: 10.1083/jcb.140.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bloom K. It's a kar9ochore to capture microtubules. Nature cell biology. 2000;2:E96–8. doi: 10.1038/35014089. [DOI] [PubMed] [Google Scholar]
- 158.Bahmanyar S, Nelson WJ, Barth AI. Role of APC and its binding partners in regulating microtubules in mitosis. Advances in experimental medicine and biology. 2009;656:65–74. doi: 10.1007/978-1-4419-1145-2_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Caldwell CM, Kaplan KB. The role of APC in mitosis and in chromosome instability. Advances in experimental medicine and biology. 2009;656:51–64. doi: 10.1007/978-1-4419-1145-2_5. [DOI] [PubMed] [Google Scholar]
- 160.Zhang L, Shay JW. Multiple Roles of APC and its Therapeutic Implications in Colorectal Cancer. Journal of the National Cancer Institute. 2017;109 doi: 10.1093/jnci/djw332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kaplan DD, Meigs TE, Kelly P, Casey PJ. Identification of a role for beta-catenin in the establishment of a bipolar mitotic spindle. The Journal of biological chemistry. 2004;279:10829–32. doi: 10.1074/jbc.C400035200. [DOI] [PubMed] [Google Scholar]
- 162.Thorpe CJ, Schlesinger A, Carter JC, Bowerman B. Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell. 1997;90:695–705. doi: 10.1016/s0092-8674(00)80530-9. [DOI] [PubMed] [Google Scholar]
- 163.Banziger C, Soldini D, Schutt C, Zipperlen P, Hausmann G, Basler K. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell. 2006;125:509–22. doi: 10.1016/j.cell.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 164.Habib SJ, Chen BC, Tsai FC, Anastassiadis K, Meyer T, Betzig E, Nusse R. A localized Wnt signal orients asymmetric stem cell division in vitro. Science. 2013;339:1445–8. doi: 10.1126/science.1231077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Stolz A, Neufeld K, Ertych N, Bastians H. Wnt-mediated protein stabilization ensures proper mitotic microtubule assembly and chromosome segregation. EMBO reports. 2015;16:490–9. doi: 10.15252/embr.201439410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Carvajal-Gonzalez JM, Mulero-Navarro S, Mlodzik M. Centriole positioning in epithelial cells and its intimate relationship with planar cell polarity. BioEssays : news and reviews in molecular, cellular and developmental biology. 2016;38:1234–45. doi: 10.1002/bies.201600154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Antic D, Stubbs JL, Suyama K, Kintner C, Scott MP, Axelrod JD. Planar cell polarity enables posterior localization of nodal cilia and left-right axis determination during mouse and Xenopus embryogenesis. PloS one. 2010;5:e8999. doi: 10.1371/journal.pone.0008999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Borovina A, Superina S, Voskas D, Ciruna B. Vangl2 directs the posterior tilting and asymmetric localization of motile primary cilia. Nature cell biology. 2010;12:407–12. doi: 10.1038/ncb2042. [DOI] [PubMed] [Google Scholar]
- 169.Oishi I, Kawakami Y, Raya A, Callol-Massot C, Izpisua Belmonte JC. Regulation of primary cilia formation and left-right patterning in zebrafish by a noncanonical Wnt signaling mediator, duboraya. Nature genetics. 2006;38:1316–22. doi: 10.1038/ng1892. [DOI] [PubMed] [Google Scholar]
- 170.Park TJ, Mitchell BJ, Abitua PB, Kintner C, Wallingford JB. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nature genetics. 2008;40:871–9. doi: 10.1038/ng.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Park TJ, Haigo SL, Wallingford JB. Ciliogenesis defects in embryos lacking inturned or fuzzy function are associated with failure of planar cell polarity and Hedgehog signaling. Nature genetics. 2006;38:303–11. doi: 10.1038/ng1753. [DOI] [PubMed] [Google Scholar]
- 172.Gray RS, Abitua PB, Wlodarczyk BJ, Szabo-Rogers HL, Blanchard O, Lee I, Weiss GS, Liu KJ, Marcotte EM, Wallingford JB, Finnell RH. The planar cell polarity effector Fuz is essential for targeted membrane trafficking, ciliogenesis and mouse embryonic development. Nature cell biology. 2009;11:1225–32. doi: 10.1038/ncb1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Carvajal-Gonzalez JM, Roman AC, Mlodzik M. Positioning of centrioles is a conserved readout of Frizzled planar cell polarity signalling. Nature communications. 2016;7 doi: 10.1038/ncomms11135. 11135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Caron A, Xu X, Lin X. Wnt/beta-catenin signaling directly regulates Foxj1 expression and ciliogenesis in zebrafish Kupffer's vesicle. Development. 2012;139:514–24. doi: 10.1242/dev.071746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Stubbs JL, Oishi I, Izpisua Belmonte JC, Kintner C. The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat Genet. 2008;40:1454–60. doi: 10.1038/ng.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Walentek P, Beyer T, Thumberger T, Schweickert A, Blum M. ATP4a is required for Wnt-dependent Foxj1 expression and leftward flow in Xenopus left-right development. Cell Rep. 2012;1:516–27. doi: 10.1016/j.celrep.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 177.Walentek P, Schneider I, Schweickert A, Blum M. Wnt11b is involved in cilia-mediated symmetry breakage during Xenoplus left-right development. PLoS One. 2013;8:e73646. doi: 10.1371/journal.pone.0073646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hashimoto M, Shinohara K, Wang J, Ikeuchi S, Yoshiba S, Meno C, Nonaka S, Takada S, Hatta K, Wynshaw-Boris A, Hamada H. Planar polarization of node cells determines the rotational axis of node cilia. Nature cell biology. 2010;12:170–6. doi: 10.1038/ncb2020. [DOI] [PubMed] [Google Scholar]
- 179.Ohata S, Nakatani J, Herranz-Perez V, Cheng J, Belinson H, Inubushi T, Snider WD, Garcia-Verdugo JM, Wynshaw-Boris A, Alvarez-Buylla A. Loss of Dishevelleds disrupts planar polarity in ependymal motile cilia and results in hydrocephalus. Neuron. 2014;83:558–71. doi: 10.1016/j.neuron.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mitchell B, Stubbs JL, Huisman F, Taborek P, Yu C, Kintner C. The PCP pathway instructs the planar orientation of ciliated cells in the Xenopus larval skin. Curr Biol. 2009;19:924–9. doi: 10.1016/j.cub.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ganner A, Lienkamp S, Schafer T, Romaker D, Wegierski T, Park TJ, Spreitzer S, Simons M, Gloy J, Kim E, Wallingford JB, et al. Regulation of ciliary polarity by the APC/C. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17799–804. doi: 10.1073/pnas.0909465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mahuzier A, Gaude HM, Grampa V, Anselme I, Silbermann F, Leroux-Berger M, Delacour D, Ezan J, Montcouquiol M, Saunier S, Schneider-Maunoury S, et al. Dishevelled stabilization by the ciliopathy protein Rpgrip1l is essential for planar cell polarity. The Journal of cell biology. 2012;198:927–40. doi: 10.1083/jcb.201111009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lee KH, Johmura Y, Yu LR, Park JE, Gao Y, Bang JK, Zhou M, Veenstra TD, Yeon Kim B, Lee KS. Identification of a novel Wnt5a-CK1 epsilon-Dvl2-Plk1-mediated primary cilia disassembly pathway. The EMBO journal. 2012 doi: 10.1038/emboj.2012.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gao C, Chen YG. Dishevelled: The hub of Wnt signaling. Cellular signalling. 2010;22:717–27. doi: 10.1016/j.cellsig.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 185.Zhang B, Zhang T, Wang G, Wang G, Chi W, Jiang Q, Zhang C. GSK3beta-Dzip1-Rab8 Cascade Regulates Ciliogenesis after Mitosis. PLoS biology. 2015;13:e1002129. doi: 10.1371/journal.pbio.1002129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Thoma CR, Frew IJ, Hoerner CR, Montani M, Moch H, Krek W. pVHL and GSK3beta are components of a primary cilium-maintenance signalling network. Nature cell biology. 2007;9:588–95. doi: 10.1038/ncb1579. [DOI] [PubMed] [Google Scholar]
- 187.Wakefield JG, Stephens DJ, M TJ. A role for glycogen synthase kinase-3 in mitotic spindle dynamics and chromosome alignment. J Cell Sci. 2003;116(Pt 4):637–46. doi: 10.1242/jcs.00273. [DOI] [PubMed] [Google Scholar]
- 188.Itoh K, Jenny A, Mlodzik M, Sokol SY. Centrosomal localization of Diversin and its relevance to Wnt signaling. J Cell Sci. 2009;122:3791–98. doi: 10.1242/jcs.057067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Alexandrova EM, Sokol SY. Xenopus axin-related protein: a link between its centrosomal localization and function in the Wnt/beta-catenin pathway. Dev Dyn. 2010;239:261–70. doi: 10.1002/dvdy.22125. [DOI] [PubMed] [Google Scholar]
- 190.Sillibourne JE, Milne DM, Takahashi M, Ono Y, Meek DW. Centrosomal Anchoring of the Protein Kinase CK1δ Mediated by Attachment to the Large, Coiled-coil Scaffolding Protein CG-NAP/AKAP450. Journal of Molecular Biology. 2002;322:785–97. doi: 10.1016/s0022-2836(02)00857-4. [DOI] [PubMed] [Google Scholar]
- 191.Zyss D, Ebrahimi H, Gergely F. Casein kinase I delta controls centrosome positioning during T cell activation. The Journal of cell biology. 2011;195:781–97. doi: 10.1083/jcb.201106025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Firat-Karalar EN, Rauniyar N, Yates JR, 3rd, Stearns T. Proximity interactions among centrosome components identify regulators of centriole duplication. Current biology : CB. 2014;24:664–70. doi: 10.1016/j.cub.2014.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mick DU, Rodrigues RB, Leib RD, Adams CM, Chien AS, Gygi SP, Nachury MV. Proteomics of Primary Cilia by Proximity Labeling. Developmental cell. 2015;35:497–512. doi: 10.1016/j.devcel.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–74. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 195.Mbom BC, Siemers KA, Ostrowski MA, Nelson WJ, Barth AI. Nek2 phosphorylates and stabilizes beta-catenin at mitotic centrosomes downstream of Plk1. Mol Biol Cell. 2014;25:977–91. doi: 10.1091/mbc.E13-06-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.He XQ, Song YQ, Liu R, Liu Y, Zhang F, Zhang Z, Shen YT, Xu L, Chen MH, Wang YL, Xu BH, et al. Axin-1 Regulates Meiotic Spindle Organization in Mouse Oocytes. PloS one. 2016;11:e0157197. doi: 10.1371/journal.pone.0157197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Dikovskaya D, Newton IP, Nathke IS. The adenomatous polyposis coli protein is required for the formation of robust spindles formed in CSF Xenopus extracts. Molecular biology of the cell. 2004;15:2978–91. doi: 10.1091/mbc.E03-08-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lui C, Ashton C, Sharma M, Brocardo MG, Henderson BR. APC functions at the centrosome to stimulate microtubule growth. The international journal of biochemistry & cell biology. 2016;70:39–47. doi: 10.1016/j.biocel.2015.10.028. [DOI] [PubMed] [Google Scholar]
- 199.Yamashita YM, Jones DL, Fuller MT. Orientation of Asymmetric Stem Cell Division by the APC Tumor Suppressor and Centrosome. Science. 2003;301:1547–50. doi: 10.1126/science.1087795. [DOI] [PubMed] [Google Scholar]
- 200.Fumoto K, Kikuchi K, Gon H, Kikuchi A. Wnt5a signaling controls cytokinesis by correctly positioning ESCRT-III at the midbody. J Cell Sci. 2012;125:4822–32. doi: 10.1242/jcs.108142. [DOI] [PubMed] [Google Scholar]
- 201.Yang Y, Liu M, Li D, Ran J, Gao J, Suo S, Sun SC, Zhou J. CYLD regulates spindle orientation by stabilizing astral microtubules and promoting dishevelled-NuMA-dynein/dynactin complex formation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(6):2158–63. doi: 10.1073/pnas.1319341111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Brockman JL, Gross SG, Sussman MR, Anderson RA. Cell cycle-dependent localization of casein kinase I to mitotic spindles. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:9454–58. doi: 10.1073/pnas.89.20.9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kaplan KB, Burds AA, Swedlow JR, Bekir SS, Sorger PK, Nathke IS. A role for the Adenomatous Polyposis Coli protein in chromosome segregation. Nature cell biology. 2001;3:426–32. doi: 10.1038/35070123. [DOI] [PubMed] [Google Scholar]