Abstract
Depression is a prevalent and complex psychiatric syndrome. Epigenetic mechanisms bridge the genetic and environmental factors that contribute to the pathophysiology of depression. A surge of research over the last decade has identified changes in DNA methylation, histone modifications, histone organization, and noncoding RNAs associated with depression and stress-induced depression-like behavior in animal models. We focus here on associations of epigenetic factors concurrent with depression and depression-like behavior, although risk for depression and some of the associated epigenetic changes are known to have developmental origins. Finally, emerging technology may enable breakthroughs in the ability to rescue depression-associated epigenetic modifications at specific genes, greatly enhancing specificity of future potential therapeutic treatments.
1. INTRODUCTION
Major depressive disorder is characterized by persistence of sadness, loss of interest or pleasure, feelings of guilt or low self-worth, and other symptoms, including sleep and circadian changes, changes in weight or appetite, and diminished concentration. More than 300 million people worldwide are living with depression, making it the leading global cause of disability.1 Depression can affect people at any age, and can persist or reoccur throughout the lifespan. Previous experience of depression increases the likelihood of future episodes. Women are approximately 50% more likely than men to experience depression, with increased risk associated with puberty, peripartum periods, and menopause.
As with other psychiatric disorders, depression is a complex, heterogeneous syndrome. The causes of depression cannot be pinned down to any one biological or external factor, and instead result from the interplay of genetic, neurobiological, environmental, and cultural factors. The genetic heritability of depression is estimated to be from 35%–40%.2,3 While heritable genetic factors clearly play a role, a significant proportion of depression risk is attributed to life experience and environment, particularly previous and/or recent stress experience. In the last decade, there has been a considerable focus on the role of epigenetics as a possible bridge between genes and experience in the pathophysiology of depression.
The epigenetic landscape, originally envisioned by Waddington more than 60 years ago, sits beautifully at the intersection of genetics, development, environmental influence, and inheritance.4 The word “epigenetic” means “above the genome” and epigenetic mechanisms refer to structural and molecular factors that alter expression of genes without altering the sequence of DNA. These include many types of chemical modifications to histone proteins, DNA, and RNA, as well as alterations in other regulatory proteins and noncoding RNAs.5–7 While epigenetic mechanisms were previously defined as stable through cell division, the dynamic nature of epigenetic modifications is now understood. As primary regulators of gene expression, epigenetic mechanisms are ideal molecular candidates to flexibly encode and translate environmental factors that increase risk for depression and other highly complex psychiatric syndromes.
This chapter discusses major advances in the study of epigenetics and depression, with a focus on recent findings. While early life events are known to increase lifelong risk for depression, the epigenetic consequences of early life stress in mediating depression and other psychiatric disease is discussed in other chapters of this text.8 We focus here on associations of epigenetic factors concurrent with depression and depression-like behavior. Work to date implicating epigenetic regulation in the context of depression and depression-like behavior has come from studies of both postmortem human brain and animal models.
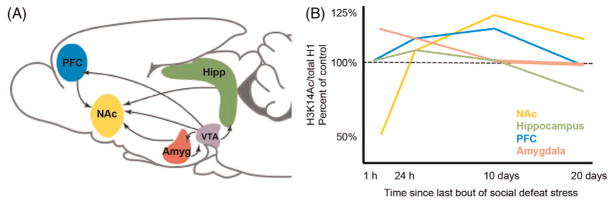
Research discussed in this chapter focuses on a few key regions of the brain (Fig. 1). Impaired motivation and anhedonia are core symptoms of depression, which implicate the brain’s limbic circuitry, including ventral tegmental area (VTA) and nucleus accumbens (NAc).9 Cognitive dysfunction and impaired attention, also common symptoms of depression, implicate the prefrontal cortex (PFC), hippocampus, and amygdala, among other regions. The hippocampus and amygdala are also known to be highly sensitive to the effects of stress and to control the hypothalamic-pituitary-adrenal axis and the secretion of glucocorticoids. The NAc is of particular interest as it is a site of integration for glutamatergic signals from the PFC, hipocampus, and amygdala, as well as dopaminergic projections from VTA.9
Fig. 1.
(A) Brain regions implicated in major depressive disorder and response to stress, shown on sagittal view of mouse brain. (B) H3K14Ac levels in mouse brain after chronic social defeat stress. Chronic fluoxetine treatment normalizes hippocampal H3K14Ac at 20 days (not shown). Amyg, Amygdala; Hipp, hippocampus; NAc, nucleus accumbens; PFC, prefrontal cortex; VTA, ventral tegmental area. Adapted from Covington HE, Maze I, LaPlant QC, Vialou VF, Ohnishi YN, Berton O, et al. Antidepressant actions of histone deacetylase inhibitors. J Neurosci. 2009;29(37):11451–60.53 Covington HE, Vialou VF, Laplant Q, Ohnishi YN, Nestler EJ. Hippocampal-dependent antidepressant-like activity of histone deacetylase inhibition. Neurosci Lett. 2011;493(3):122–126.54 Covington HE III, Maze I, Vialou V, Nestler EJ. Antidepressant action of HDAC inhibition in the prefrontal cortex. Neuroscience. 2015;298:329–335.56
2. OVERVIEW OF EPIGENETIC MECHANISMS
The human genome contains approximately 6 billion nucleotides of DNA neatly packaged into 23 pairs of chromosomes.10 The fundamental unit of chromatin is the nucleosome, which allows the extraordinary organization and compaction of DNA into a microscopic cell nucleus. The nucleosome consists of ~146 bp of DNA wrapped around a core histone octamer (~1.65 turns). Each octamer contains two copies each of the histones H2A, H2B, H3, and H4. Epigenetic mechanisms control the spacing of nucleosomes and the degree to which they are tightly condensed or loosely wound. In simplified terms, chromatin exists across a continuum between an inactivated, condensed state (heterochromatin), which represses gene transcription, and an activated, open state (euchromatin), which is permissive for gene transcription. The three-dimensional (3D) structure and packaging of DNA thus regulates gene expression, and is controlled by complex biochemical processes.
2.1 DNA Methylation
DNA methylation is one way in which the structure of DNA can be altered by covalent attachment of a methyl group to the C5 position of cytosine (5mC) predominantly at cytosine-guanine dyads (CpG sites) to affect transcription without altering the overall sequence.11 CpG dinucleotides cluster within or around gene promoters, referred to as “CpG islands,” and tend to be hypomethylated in normal somatic cells.11 Gene promoter hypermethylation generally represses gene expression. It can either prevent access of transcriptional activators and RNA polymerase II to the transcription start site, or bind to methyl-CpG-binding proteins to recruit transcriptional corepressors to modify the surrounding chromatin into a silenced state.11–13 The process of methylation is dependent on the presence of methyl donors (provided by nutrients, such as folic acid, methionine, and choline) and DNA methyltransferases, which catalyze either de novo DNA methylation (i.e., DNMT3) or maintenance of methylation across cell division and DNA repair (i.e., DNMT1). More recent findings have indicated that a significant portion of DNA methylation occurs at non-CpG sites and that DNA methylation can either induce or suppress gene expression depending on other factors.14
DNA methylation is considered to be a relatively stable epigenetic modification, making it an attractive target in the study of long-lasting epigenetic signatures of psychiatric disease. Until recently, DNA methylation was thought to be a permanent modification that could only be reduced in tissue by passive mechanisms, such as failure to maintain methylation in daughter cells. This view was supported by evidence of an overall accumulation of DNA methylation across the lifespan of an organism. However, DNA methylation is now known to be a reversible process with the discovery of active DNA demethylase enzymes, namely the ten–eleven translocation (TET) family proteins. TET proteins oxidize 5mC into 5-hydroxymethylcytosine (5hmC), and subsequently into 5-formylcytosine and 5-carboxylcytosine.15 Through deamination, glycosylation, or base excision repair, these newly discovered forms of cytosine modification can then be converted back into an unmethylated state. In contrast to the generally repressive effect of 5mC on gene expression, 5hmC is more correlated with transactivation.16 Interestingly, 5mC oxidation derivatives are expressed at highest levels in neurons. Most studies of DNA methylation in psychiatric disorders to date have not distinguished between 5mC and 5hmC, which is clearly a major need in the field.
2.2 Chromatin Modifications
Posttranslational modifications to histone proteins, most commonly on amino acid residues within the N-terminal tails that protrude from the nucleosome core, represent a more dynamic process of gene regulation. Such modifications include acetylation, ubiquitination, or SUMOylation at lysine (K) residues, methylation at lysine or arginine residues, and phosphorylation at serine or threonine residues, among many others. Histone acetylation is generally associated with chromatin decondensation and increased transcriptional activity, by negating the positive charge of K residues in histone tails and increases spacing between nucleosomes. The acetylation state of these histones is controlled by histone acetyltransferases (HATs) and histone deacetylases (HDACs). There is a considerable interest in the therapeutic potential of HDAC inhibitors, which effectively increase gene expression by shifting histones to an acetylated state.17 Histone tails can be methylated by histone methyltransferases (HMTs), and demethylated by histone demethylases (HDMs). The exact location and valence of histone tail methylation (mono-, di- or trimethylation) either promotes euchromatin or heterochromatin, and are thus associated with either activation or repression of gene transcription, respectively.18 The specificity of numerous HATs and HDACs for specific K residues remains incompletely understood. In contrast, distinct HMTs and HDMs control the methylation of specific K and arginine residues and even the valence of methylation. The complexity of the “histone code” is illustrated by the number of possible posttranslational modifications to H3 lysines alone. Human H3 has 13 lysine residues that can potentially be methylated with one, two, or three methyl groups. This results in huge potential combinatorial power over the level of gene transcription, with more than 67 million (4^13) different H3K-methylation patterns, in one nucleosome.
The functional consequences of histone modifications are mediated in part through “readers”—proteins that bind to specific modified residues and effect transcriptional change.5,6 For example, different families of chromatin remodeling proteins, which use ATP-derived energy to alter nucleosome spacing and condensation, recognize specific forms of modified histones and enhance or repress the activity of nearby genes. The involvement of this diverse family of proteins is just now being studied in the nervous system.19 Ultimately, hundreds of proteins are thought to be recruited to a gene in concert with its activation or repression, again emphasizing the extraordinary complexity of epigenetic mechanisms. Despite this complexity, strong evidence links chromatin modifications with stress and depression.
2.3 Noncoding RNAs
Only 20% of the human genome is associated with protein-coding genes, with less than 1% transcribed into mRNA, while the rest is thought to contribute to regulation of those genes. Interestingly, higher order species have increasingly greater proportions of noncoding DNA, which includes a range of noncoding RNAs. The diversity of noncoding RNAs and their role in transcriptional regulation is just beginning to be understood. Small non-coding RNAs, including microRNAs (miRNAs), are found in most eukaryotic organisms. They are produced either from their own gene, or commonly from introns of their target or other genes.7 MicroRNAs are first transcribed from a longer pre-miRNA, including a 5′-cap and poly-A tail, after which a hairpin loop is formed, trimmed, and exported from the cell nucleus to cytoplasm, and processed to a mature single-stranded ~22 nucleotide by a series of enzymes, including DROSHA and DICER. Within an RNA-induced silencing complex (RISC), miRNAs base-pair with complementary mRNA sequences which typically results in silencing by sequestration, degradation, or translational suppression of target mRNAs. Other small noncoding RNAs include short interfering RNAs (siRNAs) Piwi-interacting RNAs (piRNAs), and small nucleolar RNAs (snoRNAs). Long noncoding RNAs (lncRNAs) are longer than 200 nucleotides and also associated with silencing of target genes. LncRNAs are largely species- and tissue-specific, with increasing proportions of the genome occupied by lncRNAs in higher order species.20 LncRNAs are also highly expressed in the brain, making them attractive as potential mechanisms of gene regulation in complex neuropsychiatric syndromes, such as depression. Given our relatively recent understanding of noncoding RNAs, a major frontier in the neurobiology of psychiatric disease is to understand the role of specific small and lncRNAs as biomarkers or in the onset and maintenance of depression.21
2.4 3D Chromatin Structure
Beyond modifications of DNA, histones, and chromatin remodeling, an important new effort in neuroepigenetics research concerns the 3D organization of the genome in neurons and glia. For example, chromosomal loop formations—which often require CCCTC-binding factor (CTCF), cohesins, and other proteins assembled into scaffolds and anchors—potentially bypass many kilobases, even megabases, of linear genome, thereby repositioning promoter-distal regulatory elements next to their target promoters. Exploration of regulatory DNA elements in the context of chromosomal loopings and higher order chromatin is beginning to assign regulation of genes and complex behavior for some of the noncoding sequences in the human genome.22
3. METHODS FOR STUDYING EPIGENETICS AND DEPRESSION
As stated earlier, depression is a heterogeneous disorder, and the difficulty in defining specific subsets of depression has made clear identification of its multiple etiologies very difficult. On top of this, to study the molecular correlates of human depression requires access to relevant biological tissue from human patients. Many studies utilize easily obtainable peripheral tissue, such as blood or saliva. However, the main substrate of depression is the brain; and few studies have provided robust evidence for consistent epigenetic modification of genes between peripheral and central tissues, the main exception being for the gene encoding the glucocorticoid receptor (NR3C1). Studies utilizing human brain tissue samples pose different challenges as tissue is necessarily collected postmortem and studies must control for age, cause of death, variables related to sample handling and quality, and genetic background which must be accounted for to understand the additional contribution of epigenetic modifications. Often, information on life history is absent and at best it is difficult to use such information to infer causality.
Stress paradigms in animals have enabled considerable progress in understanding the neurobiology of depression. The wide use of several standardized rodent chronic stress paradigms, and the objective measurement of depression-like behavior, has also enabled comparison of epigenetic factors in depression across studies. While acute stress paradigms are designed to evaluate an animal’s initial coping response, chronic stress paradigms involve prolonged exposure to either physical or psychological stressors, such as social subordination or variable, daily environmental stress. In the chronic social defeat stress paradigm, an experimental mouse is subjected to “bullying” by a new larger more aggressive mouse each day for 10 days.23 A variation of this paradigm removes the physical component by testing whether depression-like behavior develops in animals that “witness” another mouse being physically aggressed.24 Such chronic stress paradigms successfully recapitulate certain behavioral features of human depression. For example, chronic social stress in adult male mice results in anhedonia (reduced preference for a sucrose solution), social avoidance, circadian changes, weight changes, and anxiety-like behaviors, such as decreased exploration of a novel environment.25 Recently, social defeat has been adapted to female mice, which display a similar range of behavioral and neural abnormalities.26 These depression-like behaviors are effectively reversed by chronic but not acute treatment with existing antidepressant medications, a treatment course comparable to that required in humans.27 Not all mice that experience chronic social defeat stress succumb to depression-like behavior, as not all humans who experience stress become depressed. Approximately, half to two-thirds of socially stressed mice are “susceptible” and exhibit depression-like behavior, whereas remaining mice are “resilient.”25 This enables researchers to explore molecular and environmental factors that increase or decrease susceptibility and resilience.
4. DNA METHYLATION IN DEPRESSION
Human and animal studies converge to support a role for DNA methylation in mediating part of the impact of stress on the brain. At a broad level, the denovo methyltransferase DNMT3a is increased in NAc among humans suffering from major depression.28 These findings are replicated in mouse models of chronic social defeat stress and subchronic variable stress.28,29 Dnmt3a mRNA is higher in female mouse NAc than male, although there are no sex differences in human DNMT3a expression in NAc, and depression—or stress exposure—increases DNMT3a in both male and female humans and mice, respectively. Artificially, increasing Dnmt3a mRNA expression in mouse NAc increases depression-like behavior in male and female mice.28,29 Knockout of Dnmt3a mRNA in NAc selectively reduces depression-like behavior in female mice after subchronic variable stress,28 although depression-like behavior was rescued in male mice exposed to chronic defeat stress by infusion of the general DNA methylation inhibitor, RG108, into Nac.29 Interestingly, NAc DNMT3a levels are partially rescued (reduced) with antidepressant treatment in both men and women,28 although it remains to be seen whether peripheral DNMT3a may be used as a predictive biomarker.
DNMT3b, which exhibits higher expression during embryonic development, and DNMT1, which has a higher affinity for hemimethylated DNA and is necessary for maintenance methylation, are not different in depressed NAc. Among male and female suicide completers, however, DNMT1 mRNA is decreased in frontopolar cortex and amygdala, and DNMT3b is increased in frontopolar cortex and decreased in brain stem regions.30
Expression of the DNA demethylaseTet1, but not Tet2 orTet3, is down-regulated in NAc of mice susceptible to chronic social defeat stress.31 Paradoxically, knocking out Tet1 from mouse NAc relieved depression-like behaviors, suggesting a complicated relationship between target gene methylation and behavior. It remains to be seen whether TET enzymes may be involved in broad DNA methylation changes associated with human depression, and whether similar changes are observed in other brain regions.
Increases in DNMTs and decreases in demethylases would suggest an overall hypermethylation state in the brains of depressed individuals and mice. However, genome-wide 5mC and 5hmC analyses instead find regions of both hyper- and hypomethylation in postmortem PFC among depressed compared to control subjects, indicating specificity in DNA methylation changes.32 5mC and 5hmC genome-wide profiling indicate enrichment of differentially methylated regions in genes related to nervous system development, mitochondria function, and immune system regulation.32–34 Genome-wide methylation analyses have yet to be published for other brain regions directly testing association with adult depression, although several important studies have examined the association with history of child abuse, which is positively associated with depression.
DNA methylation at several candidate genes has been examined directly. Among these candidate genes altered in stress and depression are two key neurotrophic factors, brain-derived neurotrophic factor (BDNF) and glial cell-derived neurotrophic factor (GDNF). At least 20 studies have now examined the association between DNA methylation of BDNF in human blood or saliva samples and depression in humans. A majority of these studies have found increased methylation within BDNF promoter I or VI, and three studies found reversal by antidepressant treatment (reviewed by Chen et al.35). Adult chronic social defeat stress in C57BL/6 mice does not alter Bdnf promoter P3 methylation in NAc, although other promoters and brain regions have not been examined.36 Chronic unpredictable stress also increases Gdnf mRNA levels in NAc of C57BL/6 mice, although the effects of Gdnf methylation may be mediated by methyl-binding proteins in a complex, strain-specific manner.37 BDNF is also highly regulated in depression by histone modifications (Box 1), and is exemplar of how individual genes are regulated at multiples levels.
BOX 1. A closer look at epigenetic regulation of BDNF-TRKB signaling in depression.
Many of the histone modifications in stress and depression reviewed in this chapter are at a global rather than gene-specific level. One prominent focus of targeted studies has been BDNF. BDNF is repeatedly found to be decreased in serum of depressed patients and in PFC and hippocampus of depressed subjects and suicide victims examined postmortem.38 Chronic stress in rodents similarly decreases BDNF in hippocampus.36 In contrast, chronic social defeat stress in mice increases BDNF in NAc, and removing NAc BDNF prevents development of depression-like behavior in response to this form of stress.27 Indeed, the effects of manipulating BDNF levels on depression-like behavior are sex-, brain region-, and developmental timing-specific.39 BDNF is also an attractive target as its levels are responsive to antidepressant treatment by conventional SSRI’s and by ketamine.39 Within the hippocampus, decreased BDNF mRNA is accompanied by increased H3K27me2 at two promoters (associated with transcriptional repression).36 However, H3K27me2 was unaffected by chronic imipramine treatment and therefore unlikely to mediate the antidepressant’s effect to normalize BDNF and behavior. Instead, chronic stress and chronic imipramine treatment interact to increase H3 acetylation and H3K4me2 at two BDNF promoters (an effect not seen after either stress or imipramine alone).36
Consistent with evidence from animal models, postmortem studies in human brains suggest that antidepressants promote open chromatin structures by decreasing H3K27me3 levels at certain BDNF promoters in PFC of a depression sample.40 Follow up studies in peripheral blood reveal higher peripheral BDNF expression in treatment responders compared to nonresponders, with H3K27me3 levels being inversely correlated with both BDNF IV expression levels and with symptom severity.41 Decreased expression of BDNF’s receptor, tyrosine receptor kinase B (TRKB), in the PFC of depressed cases is also associated with an enrichment of H3K27me3 levels in the promoter of both TRKB and its astrocytic variant, TRKB.T1.42 The elevated H3K27me3 levels associate with changes in DNA methylation in the promoter suggesting the presence of dual epigenetic control over TRKB.T1 expression, as reported for many genes in simpler systems. In addition, mice overexpressing TRKB.T1 are more susceptible to chronic social stress than wild type mice suggesting that epigenetic changes at the TRKB.T1 promoter could define the vulnerability to chronic social stress and the development of depression.43
Serotonin signaling in the brain is undeniably linked to antidepressant mechanisms and is regulated in part by the serotonin transporter encoded by SLC6A4. Methylation of a CpG “island” in the SLC6A4 promoter in vitro reduces gene expression, after accounting for genetic variation in the serotonin transporter-linked polymorphic region.44 Nearly 10 studies to date have found a positive association between SLC6A4 methylation and depression in humans.35,45 Antidepressant treatment has also been found to reverse SLC6A4 methylation changes.35 SLC6A4 methylation is also positively linked with frontal gray matter brain volume and resting-state connectivity between cortical brain regions.46 These studies have used peripheral tissue samples, suggesting that SLC6A4 methylation may be a useful biomarker.46,47
Finally, DNA methylation changes are found in several genes critical along the HPA axis response to stress in human depression and animal models. Corticotropin releasing factor (CRF) in the paraventricular nucleus (PVN) of the hypothalamus is increased in mice susceptible to chronic social defeat stress, accompanied by decreased DNA methylation of the Crf promoter.48 Both effects are reversed by chronic imipramine treatment.48 DNA methylation is also increased at the Crf promoter in the PVN of female rats subjected to chronic unpredictable stress, suggesting that DNA methylation may play a role in determining sex-specific regulation of HPA-axis function.49 Increased DNA methylation of hippocampal glucocorticoid receptor (NR3C1) is also associated with depression and depression-like behaviors in human and animal models, particularly with a history of early life adversity.50,51
The examples summarized here are certainly not exhaustive of the work done to uncover how aberrant DNA methylation may either contribute to the pathophysiology of depression, or serve as a biomarker. There is a complementary body of work on epigenetic mechanisms that mediate developmental origins of depression and other psychiatric syndromes (see other Chapters 1, 7, 10 and 13, in this volume). As in genome-wide association studies searching for genetic alterations that increase risk for depression, it is clear that stress effects on DNA methylation are not restricted to a small number of candidate genes. Additional genome-wide DNA methylation studies in humans and animals will help to uncover how changes at dozens or more genes act in concert to promote susceptibility or resilience to environmental stress.
5. HISTONE MODIFICATIONS IN DEPRESSION
5.1 Histone Acetylation
The potential importance of chromatin modifications in depression was initially suggested by observations that HDAC inhibition—alone or in combination with antidepressant treatment—ameliorated depression-like behavior in rodents.36,52–54 Histone 3 lysine 14 (H3K14) acetylation is decreased in NAc of depressed humans and transiently decreased in NAc of mice that experience chronic social defeat stress.53 Blocking class I HDACs with the inhibitor MS275, infused directly into mouse NAc, reduces depression-like behavior after chronic stress, suggesting that histone acetylation in NAc mediates the effects of chronic stress on behavior.53 In support of this, overexpression of a dominant-negative form of the class I Hdac2 in NAc prevented depression-like behavior in a chronic unpredictable mouse stress paradigm, while expression of a form of Hdac2 with increased chromatin binding affinity was prodepressant.37 In contrast, expression of Hdac5, a class II HDAC, is decreased in mice susceptible to social defeat stress and increased by chronic imipramine treatment suggesting a proresiliency effect of this HDAC.55 Furthermore, Hdac5 knock-out mice display exaggerated depression-like behavior after chronic social defeat stress in adulthood.55 The opposing effects of HDAC2 and HDAC5 are difficult to reconcile with a simplistic role of histone acetylation in depression and stress adaptation. However, it is conceivable that HDAC2 and HDAC5 may regulate distinct populations of genes. Additionally, it is important to note that HDAC5 could also regulate nonhistone targets due to its cytoplasmic, as well as nuclear localization. Genome-wide studies of NAc gene expression in defeated mice treated systemically with fluoxetine or intra-NAc with MS275 demonstrated that both treatments reverse a large proportion of defeat-induced differential gene expression. Although each treatment also regulate subsets of unique genes, there was also significant overlap, suggesting that antidepressant effects of fluoxetine may in part be mediated by affecting histone acetylation.53
Stress does not regulate histone modifications uniformly across brain regions. In contrast to the NAc, chronic social defeat stress transiently increases H3K14Ac in the PFC, amygdala, and hippocampus, 24 h after stress.54,56 Within the amygdala, H3K14Ac levels normalize within 10 days after stress ceases, while hippocampal levels are persistently decreased thereafter, and PFC levels are persistently increased.54,57 The long-lasting decrease in hippocampal H3K14Ac likely contributes to depressive-like behaviors, as chronic fluoxetine treatment rescues decreased hippocampal H3K14Ac but has no effect on amygdala levels.54 Infusion of the HDAC inhibitor MS275 into the hippocampus rescues sucrose preference behavior (anhedonia) but not social avoidance, while infusion into the amygdala rescues social avoidance behavior but not sucrose preference.54 These results reflect how PFC, amygdala, and hippocampus regulate different aspects of depression-like behavior. Interestingly, intrahippocampal HDAC inhibition by MS275 restores social avoidance if paired with social housing conditions, suggesting that H3K14Ac in the hippocampus facilitates adaptation.54
If H3K14Ac levels reflect increased permissiveness for transcriptional changes in response to stimuli, it is possible that the increase in H3K14Ac within NAc, PFC, hippocampus, and amygdala 24 h after stress represents a natural window of behavioral therapeutic intervention. Fluoxetine appears to restore this epigenetic-mediated ability to adapt to environmental experience and update behavioral response, but only within hippocampus. Supplementing traditional serotonin reuptake inhibitor (SSRI)-based anti-depressant treatment with HDAC inhibitors increases efficacy in mice.58 Indeed, the possibility of using HDAC inhibitors in patients with treatment-resistant depression has recently been suggested.59
The regional specificity in histone acetylation response to stress may be explained in context of work elucidating the circuit-level relationships of these regions in mice susceptible or resilient to depression-like behavior after chronic social defeat stress. Chronic stress increases measures of regional activity in NAc and ventral hippocampus, but decreases activity measures in mPFC among susceptible mice.60–62 In support of a causal role for pathway-specific activity in depression-like behavior, optogenetic activation of ventral hippocampus-NAc projections is prodepressive, while activation of mPFC-NAc or basolateral amygdala-NAc projections are antidepressant.61 Together with the regional acetylation data, a pattern emerges in which the basolateral amygdala works in opposition to NAc during and immediately following stress or artificial activation, and at long-term time points engagement of the hippocampus positively regulates NAc and can either produce prodepressive or proresilient effects if paired with stress or enrichment, respectively.54,61
One illustration of how genetics and epigenetic processes work in concert to influence response to stress and depression-like behavior comes from rats that have been selectively bred to be high responders (HR) or low responders (LR) based on their locomotor response in a novel environment. Cross-fostering of pups from HR to LR litters demonstrates that stress-response is genetic. In unstressed conditions, HR rats have high preference for a sucrose solution (low anhedonia-like behavior) compared to LR rats, while after repeated social defeat stress anhedonia increases among HR rats and decreases among LR rats.63 These behavioral changes are mirrored in global H3 and H2B acetylation levels in hippocampus, which in turn may be mediated by similar changes in the histone acetyl transferase, CREB-binding protein.63 Thus, epigenetic processes are at the intersection of genetic factors and environmental stress experiences.
5.2 Histone Methylation
Histone methylation is implicated in genome-wide association studies of human depression.64 Chronic social defeat stress downregulates the histone methyltransferases G9a and G9a-like protein, which catalyze H3K9me2, a major repressive mark, in NAc.65 Overexpression of G9a in NAc is antidepressant-like, and increased H3K9me2 at specific gene promoters is implicated in the antidepressant effect of fluoxetine.65,66 Indeed, chronic exposure to fluoxetine reduces Camkii expression in NAc in association with reduced H3Ac and increased H3K9me2 levels at the Camkiia promoter in NAc. Interestingly, these effects are found in the NAc of depressed humans exposed to antidepressants suggesting that the stress-induced loss of repressive methylation is maladaptive and that the reinstatement of these marks at specific gene loci might contribute to the therapeutic effects of antidepressant drugs. Another gene, which illustrates this mode of regulation, is Ras. Reduced H3K9me2 at this gene in the NAc of susceptible mice results in increased Ras expression, induction of ERK signaling, and ultimately CREB activation which in this brain region acts to induce depression-like behavior.65
The observed decreases in H3K9me2 in NAc would be expected to mediate a more permissive transcriptional state similar to the effect of the global increases in H3 acetylation described earlier. Curiously, however, manipulations that decrease repressive methylation induce susceptibility, whereas manipulations that increase acetylation induce resilience. To reconcile and integrate these findings, genome-wide approaches are required to examine regulation of histone modifications at specific gene loci to understand the precise coordinated regulation of depression-related target genes.
Additional methylation at the H3K9 locus, catalyzed by SETDB1 to H3K9me3, is also associated with transcriptional repression. Within the hippocampus, genome-wide profiling of H3K9me3-assocaited genes by ChIP-seq found that restraint stress dramatically enriched this mark at repetitive elements, nontranscriped regions of the genome.67,68 Such an effect may influence genomic instability. Finally, whole forebrain overexpression of Setdb1 reduced depression-like behavior, suggesting that the increase in H3K9me3 after acute stress may represent an adaptive response.69 These modifications are associated with changes in the 3D structure of chromatin as described in a later section.
H3K27 also can be mono-, di-, or trimethylated and these states are associated with activation, bivalency, or repression. ChIP-chip analysis (chromatin immunoprecipitation followed by genome-wide promoter microarrays) examined stress-induced redistribution of H3K27me2 in NAc of mice subjected to chronic social defeat or protracted social isolation. Significant and dynamic changes in repressive histone methylation were observed in upstream regulatory regions in both stress models with more genes implicating increased histone methylation. Interestingly, approximately 20% of genes are similarly regulated in both models H3K27me3 alterations have not been described in rodent stress models at a genome-wide level to date, although this mark is associated with repression of specific genes, including Rac1.70 Rac1 influences characteristic dendritic spine morphology changes in depressed human brain and socially defeated mice.71
Aside from the few examples cited earlier, human postmortem studies examining histone modifications in depression are sparse. Elevated levels of H3K4me3—a mark of gene activation—were reported at the synapsin gene family in PFC of depressed humans.72 There are also reports of altered H3K4me3 or H3K27me3 in promoter regions of several candidate genes, including BDNF (Box 1) and others (e.g., OAZ, SYN2,TRKB) in postmortem PFC.40 In general, however, there is a major need for genome-wide profiles for prominent histone modifications in depression and in response to antidepressant treatment, across brain regions, in order to define the genes and molecular pathways that mediate these responses. Thus far, there are no genome-wide analyses of histone modifications in depressed human brain. This is a high priority for future research.
6. CHROMATIN REMODELING IN DEPRESSION
Chromatin remodeling, not to be confused with chromatin modifications, refers to the actual movement of nucleosomes along DNA.73 Chromatin remodeling complexes use the energy of ATP hydrolysis to alter the packing state of chromatin and work in concert with chromatin modifying enzymes to direct nucleosomal dynamics. Very little is known concerning the role of chromatin remodeling complexes in depression or any other psychiatric syndrome. However, our group demonstrated that chronic social defeat stress or other forms of chronic stress induces a repressive chromatin remodeling complex in NAc, which by ChIP-seq was shown to mediate stress-repression of a set of genes important for mediating stress susceptibility.74 Induction of the same complex was found in NAc of depressed humans, providing translational validation. Induction of this repressive complex at suppressed genes correlates with lower levels of activating histone marks (e.g., H3K4me3 and H4K16ac) and increased levels of certain repressive histone marks (e.g., H3K9me2), thus emphasizing the coordinated nature of epigenetic regulation.74 These findings underscore the importance of mapping numerous epigenetic mechanisms at a genome-wide scale in order to define the combinatorial code of epigenetic changes associated with depression or antidepressant responses.
Histone variant dynamics may also reflect stress experience across the lifespan and play a role in susceptibility or resilience to depression. The four canonical histone subunits within the nucleosome (H2A, H2B, H3, and H4) also come in several variants. Histone variants, such as H2A.Z and H3.1–3 are incorporated into chromatin independently of DNA replication; this process therefore requires removal of existing nucleosomes. In particular, H3.3-containing nucleosomes at genomic enhancer and promoter regions can undergo rapid turnover, and such turnover is further associated with histone modifications marking active gene expression.75 It was recently found that H3.3 dynamics are increased in depressed human NAc and in response to chronic social defeat stress in mice, and reversed by antidepressant treatment.76 Moreover, H3.3 was found to accumulate across development to near-saturation by midadolescence, and chronic stress in a postnatal sensitive period of mouse development accelerates H3.3 accumulation in NAc and increases susceptibility to later stress.76–78 Preventing H3.3 dynamics promotes resilience to chronic social defeat stress in mice.76 These findings represent a novel frontier in understanding epigenetic mechanisms of depression.
7. REGULATION OF THE 3D GENOME IN DEPRESSION
There is now an increasing appreciation of the degree to which the 3D structure of chromatin is under dynamic regulation in the developing and adult brain, and this mode of regulation is beginning to be considered in neuropsychiatric disorders.22 One recent study found that knockout of Setdb1 (an H3K9me3 HMT) in forebrain neurons induces a dramatic reorganization of ~1.2 Mb of DNA around the protocadherin gene cluster.79 This reorganization is associated with altered levels of CTCF binding, DNA methylation, and several histone modifications (e.g., H3K27ac) and with altered expression levels of specific protocadherin isoforms. As noted earlier, Setdb1 knockout mice exhibit increased depression-like behavioral abnormalities, raising the important goal for future research to directly link changes in neuronal function and behavior to the disordered 3D genome at the protocadherin locus. Of note, the 3D structure of this locus as well as patterns of CTCF binding and H3K9me3 enrichment are highly homologous between mouse forebrain neurons and human glutamate-like neurons induced from pluripotent stem cells, supporting the validity of using the mouse for epigenetic explorations of depression mechanisms.79
8. NONCODING RNAS IN DEPRESSION
Increasing evidence implicates a role for microRNAs (miRNAs) in depression or as potential biomarkers for diagnosing and targeting treatment for this complex mood disorder.80 microRNAs are both spatially and temporally regulated, similar to other transcripts. miRNAs are relatively conserved evolutionarily, suggesting that they play an essential role in transcriptional regulation.7 miRNAs have been estimated to target up to hundreds of genes. As depression and other complex psychiatric syndromes are now accepted to involve many subtle transcriptional changes, miRNAs represent an attractive potential therapeutic tool to achieve broad regulatory ability over entire gene networks.81
Although many miRNAs are conserved across species, miR-1202 appears to be primate-specific and enriched within the brain. Depression decreases miR-1202 in postmortem human PFC, and history of antidepressant treatment partially restores its expression.39 miR-1202 can be measured in human blood and similarly reflects depression status and response to chronic but not acute antidepressant treatment.79,80 Moreover, peripheral baseline miR-1202 levels predict resting state connectivity between frontal regions, and correlate with activity changes during a response inhibition task in other relevant brain regions.82 One identified target of miR-1202 is metabotropic glutamate receptor-4 (GRM4), which is inversely correlated with miR-1202, increased in depressed postmortem PFC, and rescued with a history of antidepressant treatment.39 GRM4 pre- and postsynaptically modulates glutamatergic, dopaminergic, GABAergic, and serotonergic signaling throughout the brain. miR-1201 may be a useful biomarker to detect depression and treatment response and altered neurotransmission. Other possible biomarkers of major depression and antidepressant response that were recently identified include miR-146b-5p, miR-425-3p, miR-24-3p, miR-185, and miR-491-3p.83 Another study found 21 miRNAs down-regulated in PFC of depressed suicide cases.84 These miRNAs are predicted to bind several targets, including DNMT3B and TRKB.T1. Additionally, miR-218 is reduced in PFC of depressed humans and mice susceptible to chronic social defeat stress.85 One of its targets is the developmental netrin-1 guidance cue receptor DCC, and both are expressed in pyramidal neurons in humans and mice. Dcc overexpression increases vulnerability to chronic stress, thus demonstrating functional relevance for miR-218.85 Human studies of miRNAs in depression have predominately focused on changes within PFC, and more research is needed to understand how miRNAs in other brain regions contribute to depression.
Let-7a is one of the first two identified miRNAs and is found in a wide range of species. It is thought to regulate genes involved in developmental patterning and cell-cycle regulation. Mice exposed to acute restraint stress have increased expression of Let-7a in PFC and amygdala, but not hippocampus.86,87 Let-7a increases were maintained in amygdala after chronic stress, but not in PFC. Additional miRNAs, including miR-9 and miR 26-a/b, were increased acutely in PFC, and miR-376b and miR-208 were increased in hippocampus following acute or chronic stress, while miR-9-1 was decreased in hippocampus.87
Other reports implicate miRNAs in the posttranscriptional regulation of Nr3c1 (glucocorticoid receptor). Compared to Sprague-Dawley rats, F344 stress-sensitive rats have lower Nr3c1 protein but not mRNA levels in the hypothalamic PVN and corresponding increases in miR-18a levels. Overexpression of miR-18a downregulates NR3C1 protein levels in cultured neurons suggesting that the exaggerated HPA stress response in F344 rats may be mediated by miR-18a regulation of Nr3c1 translation.88 Interestingly, in amygdala, acute stress induced expression of several miRNAs is implicated in regulation of anxiety through effects on the mineralocorticoid receptor and Crfr1 expression.89,90
A significant downregulation of several miRNAs has been reported in the hippocampus of rats resistant to “learned helplessness”—a subchronic stress model—after inescapable foot shock compared to those rats that develop learned helplessness.91 These miRNA-mediated effects coincide with robust changes in hippocampus gene expression and are believed to represent a potential coping mechanism that allows the development of physiological responses to overcome the effects of stress.92
miRNAs may also be essential components of antidepressant efficacy and of resilience. Chronic fluoxetine treatment in mice increases miR-16 levels in serotonergic raphe nuclei, which downregulates serotonin transporter (Slc6a4) levels, thus altering serotonergic activity in the raphe.93 Intraraphe administration of fluoxetine induces the release of S100β, a known inhibitor of miR-16. Via the reciprocal connections between dorsal raphe and locus coeruleus (LC), S100β is believed to migrate to noradrenergic neurons in the LC and decrease expression of miR-16. By decreasing miR-16 in the LC, S100β turned on the expression of serotonergic functions in noradrenergic neurons. Moreover, intraraphe and -intra-LC miR-16 administration alleviated the behavioral deficits induced by 6 weeks of chronic unpredictable stress suggesting that the therapeutic effects of fluoxetine may be mediated via its actions on miRNA expression in the brain.93 A different study identified dicer, an miRNA-processing protein, as a major target of the transcription factor, β-catenin, in the NAc, where both proteins contribute to a state of resilience.94 Small RNA sequencing identified numerous miRNAs whose induction in this region associates with resilience to chronic social defeat stress.
Finally, miRNAs may help explain sex differences in the pathophysiology of depression. Male and female mice exposed to subchronic variable stress each had broad changes in miRNA expression in NAc.95 In this stress paradigm, only female mice developed depression-like behavior after sub-chronic variable stress, while male mice developed depression-like behavior after chronic stress. miRNA alterations were also nearly completely non-overlapping between the sexes.95 Predicted functions of stress-altered female miRNA-mRNA networks include neurotransmitter receptor activity and binding, gated channel activity, and cytokine activity.95 More research is needed to understand sex differences in miRNAs in depression in human brain and peripheral tissue.
The earlier findings suggest the involvement of miRNAs in the pathophysiology of depressive disorders. A key challenge moving forward is to directly test the functional relevance of miRNAs in depression by altering specific miRNAs in animal brain and testing effects on susceptibility to stress. In addition, while in silico tools to predict miRNA regulation of mRNA targets have improved in recent years, more work is needed to empirically test predictions in vitro and in vivo.
9. CONCLUSIONS
Looking toward the future, it will be important for studies to integrate peripheral epigenetic modifications—DNA methylation and noncoding RNAs in particular—with changes within the brain, across relevant brain regions. An essential challenge in neuropsychiatry moving forward is to increase the cell-type specificity of studies of epigenetic modifications within the brain. Epigenetic changes are known to be specific to developmental stage, tissue or region, and cell type, and studies are just now beginning to measure and manipulate epigenetic mechanisms with this resolution. Furthermore, in order to push the field forward constant dialogue is needed between human and animal researchers. Model systems enable researchers to test the functional relevance of findings from human studies to determine whether identified epigenetic alterations are correlative or causative. Finally, there is an urgent demand for increased specificity in animal studies testing the causative nature of epigenetic changes. Manipulation of DNMTs or HATs, for example, will have genome-wide impact, yet sequencing studies are increasingly revealing specificity in epigenetic regulation. Recent developments in molecular tools, including zinc finger proteins, transcription activator-like effectors (TALES), and CRISPR now enable researchers to target individualgenes for a single type of epigenetic modification.96–98 These technologies enable brain-region-specific, gene-specific, and potentially, temporal targeting of epigenetic modifiers to determine whether epigenetic regulation at a specific locus is responsible for life-long perturbations in psychiatric disorders. This technology also offers therapeutic promise for the future.
Acknowledgments
This work is supported in part by K99 MH115096 to C.J.P and P50 MH096890 and Hope for Depression Research Foundation funding to E.J.N.
References
- 1.World Heath Organization (WHO) Depression and Other Common Mental Disorders. Available from: http://apps.who.int/iris/bitstream/10665/254610/1/WHO-MSD-MER-2017.2-eng.pdf.
- 2.Kendler KS, Prescott CA. A population-based twin study of lifetime major depression in men and women. Arch Gen Psychiatry. 1999;56(1):39–44. doi: 10.1001/archpsyc.56.1.39. [DOI] [PubMed] [Google Scholar]
- 3.Geschwind DH, Flint J. Genetics and genomics of psychiatric disease. Science. 2015;349(6255):1489–1494. doi: 10.1126/science.aaa8954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waddington CH. Canalization of development and genetic assimilation of acquired characters. Nature. 1959;183(4676):1654–1655. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 5.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 6.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(3s):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 7.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 8.Jonson-Reid M, Kohl PL, Drake B. Child and adult outcomes of chronic child maltreatment. Pediatrics. 2012;129(5):839–845. doi: 10.1542/peds.2011-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russo SJ, Nestler EJ. The brain reward circuitry in mood disorders. Nat Rev Neurosci. 2013;14(9):609–625. doi: 10.1038/nrn3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Annunziato L, Di Renzo GF, Lombardi G, Preziosi P, Scapagnini U. Proceedings: catecholaminergic control of thyroid stimulating hormone (TSH) and adrenocroticotrophic hormone (ACTH) secretion. Br J Pharmacol. 1974;52(3):442P–443P. [PMC free article] [PubMed] [Google Scholar]
- 11.Bird AP, Wolffe AP. Methylation-induced repression: belts, braces, and chromatin. Cell. 1999;99(5):451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 12.Razin A. CpG methylation, chromatin structure and gene silencing—a three-way connection. EMBOJ. 1998;17(17):4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. 2005;15(5):490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341(6146):1237905–1237915. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo JU, Ma DK, Mo H, Ball MP, Jang M-H, Bonaguidi MA, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14(10):1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szulwach KE, Li X, Li Y, Song C-X, Wu H, Dai Q, et al. 5-hmC–mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14(12):1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116(2):259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 18.Ha M, Ng DW, Li W-H, Chen ZJ. Coordinated histone modifications are associated with gene expression variation within and between species. GenomeRes. 2011;21(4):590–598. doi: 10.1101/gr.116467.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun HH, Kennedy PJP, Nestler EJE. Epigenetics of the depressed brain: role of histone acetylation and methylation. Neuropsychopharmacology. 2013;38(1):124–137. doi: 10.1038/npp.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10(6):925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang X, Luo Y-L, Mao Y-S, Ji J-L. The link between long noncoding RNAs and depression. Prog Neuropsychopharmacol Biol Psychiatry. 2017;73:73–78. doi: 10.1016/j.pnpbp.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell AC, Javidfar B, Bicks LK, Neve R, Garbett K, Lander SS, et al. Longitudinal assessment of neuronal 3D genomes in mouse prefrontal cortex. Nat Commun. 2016;7:12743. doi: 10.1038/ncomms12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golden SA, Covington HE, Berton O, Russo SJ. A standardized protocol for repeated social defeat stress in mice. Nat Protoc. 2011;6(8):1183–1191. doi: 10.1038/nprot.2011.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warren BL, Vialou VF, Iñiguez SD, Alcantara LF, Wright KN, Feng J, et al. Neurobiological sequelae of witnessing stressful events in adult mice. Biol Psychiatry. 2013;73(1):7–14. doi: 10.1016/j.biopsych.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnan V, Han M-H, Graham DL, Berton O, Renthal W, Russo SJ, et al. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell. 2007;131(2):391–404. doi: 10.1016/j.cell.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi A, Chung J-R, Zhang S, Zhang H, Grossman Y, Aleysin H, Flanigan ME, Pfau ML, Menard C, Dumitriu D, Hodes GE, McEwen BS, Nestler EJ, Han M-H, Russo SJ. Establishment of a repeated social defeat stress model in female mice. Sci Rep. 2017;7:12838. doi: 10.1038/s41598-017-12811-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berton O, Mcclung CA, Dileone RJ, Krishnan V, Renthal W, Russo SJ, et al. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science. 2006;311(5762):864–868. doi: 10.1126/science.1120972. [DOI] [PubMed] [Google Scholar]
- 28.Hodes GE, Pfau ML, Purushothaman I, Ahn HF, Golden SA, Christoffel DJ, et al. Sex differences in nucleus accumbens transcriptome profiles associated with susceptibility versus resilience to subchronic variable stress. J Neurosci. 2015;35(50):16362–16376. doi: 10.1523/JNEUROSCI.1392-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaPlant QQ, Vialou VV, Covington HEH, Dumitriu DD, Feng JJ, Warren BLB, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13(9):1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poulter MO, Du L, Weaver ICG, Palkovits M, Faludi G, Merali Z, et al. GABAA receptor promoter hypermethylation in suicide brain: implications for the involvement of epigenetic processes. Biol Psychiatry. 2008;64(8):645–652. doi: 10.1016/j.biopsych.2008.05.028. [DOI] [PubMed] [Google Scholar]
- 31.Feng J, Peña CJ, Purushothaman I, Engmann O, Walker D, Brown AN, et al. Tet1 in nucleus accumbens opposes depression- and anxiety-like behaviors. Neuropsychopharmacology. 2017;42(8):1657–1669. doi: 10.1038/npp.2017.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gross JA, Pacis A, Chen GG, Drupals M, Lutz P-E, Barreiro LB, et al. Gene-body 5-hydroxymethylation is associated with gene expression changes in the prefrontal cortex of depressed individuals. Transl Psychiatry. 2017;7(5):e1119. doi: 10.1038/tp.2017.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Córdova-Palomera A, Fatjó-Vilas M, Gastó C, Navarro V, Krebs M-O, Fañanás L. Genome-wide methylation study on depression: differential methylation and variable methylation in monozygotic twins. Transl Psychiatry. 2015;5(4):e557. doi: 10.1038/tp.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy TM, Crawford B, Dempster EL, Hannon E, Burrage J, Turecki G, et al. Methylomic profiling of cortex samples from completed suicide cases implicates a role for PSORS1C3 in major depression and suicide. Transl Psychiatry. 2017;7(1):e989. doi: 10.1038/tp.2016.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen D, Meng L, Pei F, Zheng Y, Leng J. A review of DNA methylation in depression. J Clin Neurosci. 2017;43:39–46. doi: 10.1016/j.jocn.2017.05.022. [DOI] [PubMed] [Google Scholar]
- 36.Tsankova NMN, Berton OO, Renthal WW, Kumar AA, Neve RLR, Nestler EJE. Sustained hippocampal chromatin regulation in a mouse model of depression and anti-depressant action. Nat Neurosci. 2006;9(4):519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 37.Uchida S, Hara K, Kobayashi A, Otsuki K, Yamagata H, Hobara T, et al. Epigenetic status of Gdnf in the ventral striatum determines susceptibility and adaptation to daily stressful events. Neuron. 2011;69(2):14–24. doi: 10.1016/j.neuron.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 38.Castrén E, Kojima M. Brain-derived neurotrophic factor in mood disorders and antidepressant treatments. Neurobiol Dis. 2017;97(Pt. B):119–126. doi: 10.1016/j.nbd.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 39.Lopez JP, Lim R, Cruceanu C, Crapper L, Fasano C, Labonté B, et al. miR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and anti-depressant treatment. Nat Med. 2014;20(7):764–768. doi: 10.1038/nm.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen ESE, Ernst CC, Turecki GG. The epigenetic effects of antidepressant treatment on human prefrontal cortex BDNF expression. Int J Neuropsychopharmacol. 2011;14(3):427–429. doi: 10.1017/S1461145710001422. [DOI] [PubMed] [Google Scholar]
- 41.Lopez JP, Mamdani F, Labonte B, Beaulieu M-M, Yang JP, Berlim MT, et al. Epigenetic regulation of BDNF expression according to antidepressant response. Mol Psychiatry. 2012;18(4):398–399. doi: 10.1038/mp.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ernst C, Chen ES, Turecki G. Histone methylation and decreased expression of TrkB.T1 in orbital frontal cortex of suicide completers. Mol Psychiatry. 2009;14(9):830–832. doi: 10.1038/mp.2009.35. [DOI] [PubMed] [Google Scholar]
- 43.Ernst C, Deleva V, Deng X, Sequeira A, Pomarenski A, Klempan T, et al. Alternative splicing, methylation state, and expression profile of tropomyosin-related kinase B in the frontal cortex of suicide completers. Arch Gen Psychiatry. 2009;66(1):22–32. doi: 10.1001/archpsyc.66.1.22. [DOI] [PubMed] [Google Scholar]
- 44.Philibert R, Madan A, Andersen A, Cadoret R, Packer H, Sandhu H. Serotonin transporter mRNA levels are associated with the methylation of an upstream CpG island. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(1):101–105. doi: 10.1002/ajmg.b.30414. [DOI] [PubMed] [Google Scholar]
- 45.Philibert RA, Sandhu H, Hollenbeck N, Gunter T, Adams W, Madan A. The relationship of 5HTT (SLC6A4) methylation and genotype on mRNA expression and liability to major depression and alcohol dependence in subjects from the Iowa Adoption Studies. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(5):543–549. doi: 10.1002/ajmg.b.30657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ismaylova E, Di Sante J, Szyf M, Nemoda Z, Yu W-J, Pomares FB, et al. Serotonin transporter gene promoter methylation in peripheral cells in healthy adults: neural correlates and tissue specificity. Eur Neuropsychopharm. 2017;27(10):1032–1041. doi: 10.1016/j.euroneuro.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 47.Olsson CA, Foley DL, Parkinson-Bates M, Byrnes G, McKenzie M, Patton GC, et al. Prospects for epigenetic research within cohort studies of psychological disorder: a pilot investigation of a peripheral cell marker of epigenetic risk for depression. Biol Psychol. 2010;83(2):159–165. doi: 10.1016/j.biopsycho.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 48.Elliott EE, Ezra-Nevo GG, Regev LL, Neufeld-Cohen AA, Chen AA. Resilience to social stress coincides with functional DNA methylation of the Crf gene in adult mice. Nat Neurosci. 2010;13(11):1351–1353. doi: 10.1038/nn.2642. [DOI] [PubMed] [Google Scholar]
- 49.Sterrenburg L, Gaszner B, Boerrigter J, Santbergen L, Bramini M, Elliott E, et al. Chronic stress induces sex-specific alterations in methylation and expression of corticotropin-releasing factor gene in the rat. PLoS One. 2011;6(11):e28128. doi: 10.1371/journal.pone.0028128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mcgowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonté B, Szyf M, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12(3):342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weaver ICG, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 52.Schroeder FA, Lin CL, Crusio WE, Akbarian S. Antidepressant-like effects of the histone deacetylase inhibitor, sodium butyrate, in the mouse. BPS. 2007;62(1):55–64. doi: 10.1016/j.biopsych.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 53.Covington HE, Maze I, LaPlant QC, Vialou VF, Ohnishi YN, Berton O, et al. Antidepressant actions of histone deacetylase inhibitors. J Neurosci. 2009;29(37):11451–11460. doi: 10.1523/JNEUROSCI.1758-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Covington HE, Vialou VF, Laplant Q, Ohnishi YN, Nestler EJ. Hippocampal-dependent antidepressant-like activity of histone deacetylase inhibition. Neurosci Lett. 2011;493(3):122–126. doi: 10.1016/j.neulet.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Renthal W, Maze I, Krishnan V, Covington HE, Xiao G, Kumar A, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56(3):13–23. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 56.Covington HE, III, Maze I, Vialou V, Nestler EJ. Antidepressant action of HDAC inhibition in the prefrontal cortex. Neuroscience. 2015;298:329–335. doi: 10.1016/j.neuroscience.2015.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hinwood M, Tynan RJ, Day TA, Walker FR. Repeated social defeat selectively increases δFosB expression and histone H3 acetylation in the infralimbic medial prefrontal cortex. Cerebral Cortex. 2011;21(2):262–271. doi: 10.1093/cercor/bhq080. [DOI] [PubMed] [Google Scholar]
- 58.Schmauss C. An HDAC-dependent epigenetic mechanism that enhances the efficacy of the antidepressant drug fluoxetine. Sci Rep. 2015;5(1):8. doi: 10.1038/srep08171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fuchikami M, Yamamoto S, Morinobu S, Okada S, Yamawaki Y, Yamawaki S. The potential use of histone deacetylase inhibitors in the treatment of depression. Prog Neuropsychopharmacol Biol Psychiatry. 2016;64:320–324. doi: 10.1016/j.pnpbp.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 60.Christoffel DJ, Golden SA, Dumitriu D, Robison AJ, Janssen WG, Ahn HF, et al. IκB kinase regulates social defeat stress-induced synaptic and behavioral plasticity. J Neurosci. 2011;31(1):314–321. doi: 10.1523/JNEUROSCI.4763-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bagot RC, Parise EM, Peña CJ, Zhang H-X, Maze I, Chaudhury D, et al. Ventral hippocampal afferents to the nucleus accumbens regulate susceptibility to depression. Nat Commun. 2015;6:7062–7072. doi: 10.1038/ncomms8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Covington HE, Lobo MK, Maze I, Vialou V, Hyman JM, Zaman S, et al. Antidepressant effect of optogenetic stimulation of the medial prefrontal cortex. J Neurosci. 2010;30(48):16082–16090. doi: 10.1523/JNEUROSCI.1731-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hollis F, Duclot F, Gunjan A, Kabbaj M. Individual differences in the effect of social defeat on anhedonia and histone acetylation in the rat hippocampus. Hor Behav. 2011;59(3):331–337. doi: 10.1016/j.yhbeh.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O’Dushlaine C, Rossin L, Lee PH, Duncan L, Parikshak NN, Newhouse S, et al. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci. 2015;18(2):199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Covington HE, Maze I, Sun H, Bomze HM, DeMaio KD, Wu EY, et al. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron. 2011;71(4):656–670. doi: 10.1016/j.neuron.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robison AJ, Vialou V, Sun H-S, Labonté B, Golden SA, Dias C, et al. Fluoxetine epigenetically alters the CaMKIIα promoter in nucleus accumbens to regulate ΔFosB binding and antidepressant effects. Neuropsychopharmacology. 2014;39(5):1178–1186. doi: 10.1038/npp.2013.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, Mcewen BS. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc Natl Acad Sci USA. 2009;106(49):20912–20917. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hunter RG, Murakami G, Dewell S, Seligsohn M, Baker MER, Datson NA, et al. Acute stress and hippocampal histone H3 lysine 9 trimethylation, a retrotransposon silencing response. Proc Natl Acad Sci USA. 2012;109(43):17657–17662. doi: 10.1073/pnas.1215810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jiang Y, Matevossian A, Huang H-S, Straubhaar J, Akbarian S. Isolation of neuronal chromatin from brain tissue. BMC Neurosci. 2008;9:42–52. doi: 10.1186/1471-2202-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilkinson MB, Xiao G, Kumar A, Laplant Q, Renthal W, Sikder D, et al. Imipramine treatment and resiliency exhibit similar chromatin regulation in the mouse nucleus accumbens in depression models. J Neurosci. 2009;29(24):7820–7832. doi: 10.1523/JNEUROSCI.0932-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Golden SA, Christoffel DJ, Heshmati M, Hodes GE, Magida J, Davis K, et al. Epigenetic regulation of RAC1 induces synaptic remodeling in stress disorders and depression. Nat Med. 2013;19(3):337–344. doi: 10.1038/nm.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cruceanu C, Alda M, Nagy C, Freemantle E, Rouleau GA, Turecki G. H3K4 tri-methylation in synapsin genes leads to different expression patterns in bipolar disorder and major depression. Int J Neuropsychopharmacol. 2013;16(2):289–299. doi: 10.1017/S1461145712000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vogel-Ciernia A, Wood MA. Neuron-specific chromatin remodeling: a missing link in epigenetic mechanisms underlying synaptic plasticity, memory, and intellectual disability disorders. Neuropharmacology. 2014;80:18–27. doi: 10.1016/j.neuropharm.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun H, Damez-Werno DM, Scobie KN, Shao NY, Dias C, Rabkin J, et al. ACF chromatin-remodeling complex mediates stress-induced depressive-like behavior. Nat Med. 2015;21(10):1146–1153. doi: 10.1038/nm.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kraushaar DC, Jin W, Maunakea A, Abraham B, Ha M, Zhao K. Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3. Genome Biol. 2013;14(10):R121. doi: 10.1186/gb-2013-14-10-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lepack AE, Bagot RC, Peña CJ, Loh Y-HE, Farrelly LA, Lu Y, et al. Aberrant H3.3 dynamics in NAc promote vulnerability to depressive-like behavior. Proc Natl Acad Sci USA. 2016;113(44):12562–12567. doi: 10.1073/pnas.1608270113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maze I, Wenderski W, Noh K-M, Bagot RC, Tzavaras N, Purushothaman I, et al. Critical role of histone turnover in neuronal transcription and plasticity. Neuron. 2015;87(1):77–94. doi: 10.1016/j.neuron.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peña CJ, Kronman HG, Walker DM, Cates HM, Bagot RC, Purushothaman I, et al. Early life stress confers lifelong stress susceptibility in mice via ventral tegmental area OTX2. Science. 2017;356(6343):1185–1188. doi: 10.1126/science.aan4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jiang Y, Loh Y-HE, Rajarajan P, Hirayama T, Liao W, Kassim BS, et al. The methyltransferase SETDB1 regulates a large neuron-specific topological chromatin domain. Nat Genet. 2017;49(8):1239–1250. doi: 10.1038/ng.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Issler O, Chen A. Determining the role of microRNAs in psychiatric disorders. Nat Rev Neurosci. 2015;16(4):201–212. doi: 10.1038/nrn3879. [DOI] [PubMed] [Google Scholar]
- 81.O’Connor RM, Dinan TG, Cryan JF. Little things on which happiness depends: microRNAs as novel therapeutic targets for the treatment of anxiety and depression. Mol Psychiatry. 2012;17(4):359–376. doi: 10.1038/mp.2011.162. [DOI] [PubMed] [Google Scholar]
- 82.Lopez JP, Pereira F, Richard-Devantoy S, Berlim M, Chachamovich E, Fiori LM, et al. Co-variation of peripheral levels of miR-1202 and brain activity and connectivity during antidepressant treatment. Neuropsychopharmacology. 2017;42(10):2043–2051. doi: 10.1038/npp.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maussion G, Yang J, Yerko V, Barker P, Mechawar N, Ernst C, et al. Regulation of a truncated form of tropomyosin-related kinase B (TrkB) by Hsa-miR-185* in frontal cortex of suicide completers. PLoS One. 2012;7(6):e39301–e39311. doi: 10.1371/journal.pone.0039301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smalheiser NR, Lugli G, Rizavi HS, Torvik VI, Turecki G, Dwivedi Y. MicroRNA expression is down-regulated and reorganized in prefrontal cortex of depressed suicide subjects. PLoS One. 2012;7(3):e33201. doi: 10.1371/journal.pone.0033201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Torres-Berrío A, Lopez JP, Bagot RC, Nouel D, Dal Bo G, Cuesta S, et al. DCC confers susceptibility to depression-like behaviors in humans and mice and is regulated by miR-218. Biol Psychiatry. 2017;81(4):306–315. doi: 10.1016/j.biopsych.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rinaldi A, Vincenti S, De Vito F, Bozzoni I, Oliverio A, Presutti C, et al. Stress induces region specific alterations in microRNAs expression in mice. Behav Brain Res. 2010;208(1):265–269. doi: 10.1016/j.bbr.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 87.Meerson A, Cacheaux L, Goosens KA, Sapolsky RM, Soreq H, Kaufer D. Changes in brain MicroRNAs contribute to cholinergic stress reactions. J Mol Neurosci. 2010;40(1–2):47–55. doi: 10.1007/s12031-009-9252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Uchida S, Nishida A, Hara K, Kamemoto T, Suetsugi M, Fujimoto M, et al. Characterization of the vulnerability to repeated stress in Fischer 344 rats: possible involvement of microRNA-mediated down-regulation of the glucocorticoid receptor. Eur J Neurosci. 2008;27(9):2250–2261. doi: 10.1111/j.1460-9568.2008.06218.x. [DOI] [PubMed] [Google Scholar]
- 89.Haramati S, Navon I, Issler O, Ezra-Nevo G, Gil S, Zwang R, et al. MicroRNA as repressors of stress-induced anxiety: the case of amygdalar miR-34. J Neurosci. 2011;31(40):14191–14203. doi: 10.1523/JNEUROSCI.1673-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mannironi C, Camon J, De Vito F, Biundo A, De Stefano ME, Persiconi I, et al. Acute stress alters amygdala microRNA miR-135a and miR-124 expression: inferences for corticosteroid dependent stress response. PLoS One. 2013;8(9):e73385. doi: 10.1371/journal.pone.0073385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Smalheiser NR, Lugli G, Rizavi HS, Zhang H, Torvik VI, Pandey GN, et al. MicroRNA expression in rat brain exposed to repeated inescapable shock: differential alterations in learned helplessness vs. non-learned helplessness. Int J Neuropsychopharmacol. 2011;14(10):1315–1325. doi: 10.1017/S1461145710001628. [DOI] [PubMed] [Google Scholar]
- 92.Kohen R, Kirov S, Navaja GP, Happe HK, Hamblin MW, Snoddy JR, et al. Gene expression profiling in the hippocampus of learned helpless and nonhelpless rats. Pharmacogenomics J. 2005;5(5):278–291. doi: 10.1038/sj.tpj.6500322. [DOI] [PubMed] [Google Scholar]
- 93.Baudry A, Mouillet-Richard S, Schneider B, Launay J-M, Kellermann O. miR-16 targets the serotonin transporter: a new facet for adaptive responses to antidepressants. Science. 2010;329(5998):1537–1541. doi: 10.1126/science.1193692. [DOI] [PubMed] [Google Scholar]
- 94.Dias C, Feng J, Sun H, Shao NY, Mazei-Robison MS, Damez-Werno D, et al. β-Catenin mediates stress resilience through Dicer1/microRNA regulation. Nature. 2014;516(7529):51–55. doi: 10.1038/nature13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pfau ML, Purushothaman I, Feng J, Golden SA, Aleyasin H, Lorsch ZS, et al. Integrative analysis of sex-specific microRNA networks following stress in mouse nucleus accumbens. Front Mol Neurosci. 2016;9:144. doi: 10.3389/fnmol.2016.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Heller EA, Cates HM, Peña CJ, Sun H, Shao N, Feng J, et al. Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors. Nat Neurosci. 2014;17(12):1720–1727. doi: 10.1038/nn.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Anton T, Bultmann S. Site-specific recruitment of epigenetic factors with a modular CRISPR/Cas system. Nucleus. 2017;8(3):1–8. doi: 10.1080/19491034.2017.1292194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brocken DJW, Tark-Dame M, Dame RT. dCas9: a versatile tool for epigenome editing. Curr Issues Mol Biol. 2018;26:15–32. doi: 10.21775/cimb.026.015. [DOI] [PubMed] [Google Scholar]