Abstract
Long restricted to the field of developmental biology, the use of the zebrafish (Danio rerio) has extended to the study of human pathogenesis. Fostered by the rapid adaptation of new technologies, the design and analysis of fish models of human diseases have contributed important findings that are now making their way from aquariums to clinics. Here we outline the clinical relevance of the zebrafish as a model organism.
Disease modeling: is the zebrafish the new mouse?
Understanding and fighting diseases require the right tools. Some aspects of disease biology and treatment, such as tissue homeostasis, interactions between cells and their microenvironment, and response to drugs, cannot be fully recapitulated in vitro. There is thus a clear need for reliable animal models. Over the past 20 years, the mouse (Mus musculus) has become the inescapable preclinical model. Despite its strengths as a mammalian system, the mouse has experimental limitations that ultimately restrict its utility for disease modeling. In particular, it is technically challenging to perform large-scale studies such as chemical and genetic screens. The zebrafish, by contrast, presents several logistical advantages that make it an attractive alternative for high-throughput-type analyses. Because of its small size, zebrafish require minimal facility space. Moreover, a pair of fish can produce hundreds of embryos each week that develop rapidly outside of the mother and can be easily visualized and experimentally manipulated.
Recent reports from the zebrafish genome-sequencing project have indicated that at least 70% of human protein-coding genes, including disease-associated genes, have an ortholog in fish [1], suggesting that most human physiology and pathologies can be modeled in zebrafish. The technologies available for the genetic manipulation and molecular and cellular analysis of zebrafish models have recently experienced an accelerated broadening, for example with the development of new tools for genome editing, the progress of high-throughput DNA and RNA sequencing and improvements in in vivo live imaging. Furthermore, most mouse techniques now have a fish version.
Even in domains where the mouse stands as a reference organism, such as human xenografts, zebrafish could constitute a new experimental option. Indeed, several groups have taken advantage of the optical clarity of zebrafish embryos and larvae to study the behavior of transplanted human cancer cells [2]. Finally, like mice, zebrafish can be used for drug-testing purposes. Toxicology studies have already been conducted in the fish [3]. Hence, the zebrafish is now emerging as another powerful organism for the modeling and study of human diseases and it is conceivable that zebrafish models will complement murine ones in the future.
Insights from zebrafish models
Numerous fish models of human diseases have been generated and studied, ranging from blood disorders and muscular dystrophies to neurodegenerative syndromes and cancer [4] (Figure 1). To illustrate the use of zebrafish for disease modeling, we first focus on selected blood disorders, which constitute the first human pathologies modeled in fish, in part due to the high degree of conservation of hematopoiesis between fish and human. Because cancer modeling in zebrafish has also become a field of intense investigation, we then present two fish tumor models that have proved particularly valuable in providing new mechanistic insights into cancer pathogenesis.
Figure 1.
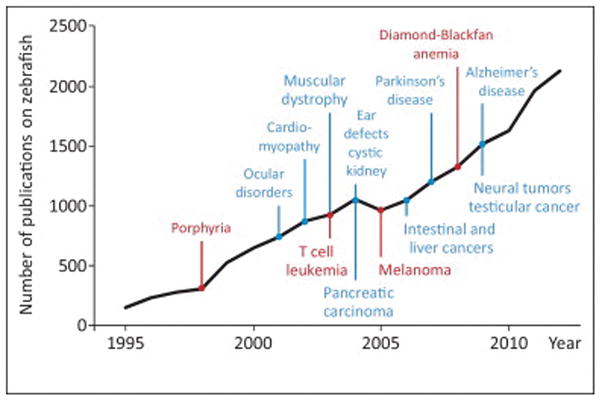
Timeline of zebrafish studies and disease models. The black line represents the evolution of the number of publications on zebrafish per year between 1990 and 2012 (PubMed search). The selected zebrafish disease models discussed in the text are displayed in red. Other important models are mentioned in blue.
Porphyria results from defects in heme biosynthesis and is characterized by skin photosensitivity and aberrant excretion of porphyrin metabolic precursors. A similar phenotype was observed in a mutant zebrafish strain obtained from an initial chemical mutagenesis screen [5]. The corresponding mutation was identified in a gene encoding uroporphyrinogen decarboxylase (UROD). Although this enzyme was previously associated with the human forms of the disease, this study in zebrafish established the enzymatic defect as the causative pathogenic event. Several similar studies of a zebrafish mutant presenting a specific developmental defect have since greatly contributed to our understanding of heme biosynthesis, iron metabolism, and associated disorders.
Recently, Diamond–Blackfan anemia (DBA) has been modeled in the zebrafish through morpholino-mediated knock down of RPS19, a ribosomal protein found recurrently mutated in the human disease [6]. RPS19 mutations are heterozygous in patients and the complete knockout of RPS19 is embryonic lethal in the mouse. Morpholinos, which allow controlled reduction in mRNA levels, more closely mimicked the human situation. Moreover, because DBA is a congenital disease, it was important to study the effect of RPS19 downregulation during embryogenesis. Zebrafish, whose embryonic stages can be tightly monitored, provided a unique tool to model DBA. Like humans, fish with reduced RPS19 display defects in erythropoiesis, indicating that zebrafish models can faithfully recapitulate human diseases, and offer new opportunities to investigate pathological mechanisms and design novel therapeutic approaches. Indeed, the zebrafish DBA model was subsequently utilized to test in vivo the therapeutic potential of L-leucine, an amino acid that enhances mRNA translation, following the idea that DBA may result from diminished translation due to incomplete ribosome biogenesis [7]. The use of L-leucine to treat DBA patients is currently in clinical trial, suggesting that zebrafish models can also serve as preclinical tools.
Several tumor types have been successfully modeled in the fish. T cell leukemias arise fromthe overexpression of the Myc oncogene driven by the lymphoid-specific rag2 promoter [8]. A conditional version of the same transgenic model allowed the investigation of the molecular determinants of the progression from a preleukemic stage to acute leukemia [9]. Elevated expression of cell-adhesion molecules such as S1P1 and ICAM1 inhibited the intravasation and dissemination of malignant cells. Release of this block was proposed to underlie the transition to a more aggressive disease.
Solid tumors also have faithful zebrafish equivalents (Figure 1). A model of melanoma has been generated by expressing human BrafV600E, a mutant allele found in more than 50% of melanomas, under the control of the melanocyte-specific promoter of the mitfa gene. Combined with p53 deficiency, the overexpression of BrafV600E drives aggressive melanomas in zebrafish [10]. This model has served as a basis to screen for new oncogenes frequently amplified in human melanomas. Tol2 transposon technology was used to insert an engineered DNA construct into the zebrafish genomeon coinjection of an appropriate vector and mRNA encoding the Tol2 transposase into one-cell stage embryos. Each human gene present in a region on chromosome 1, which is frequently gained in human melanomas, was thus overexpressed in BrafV600E, p53−/−fish in a melanocyte-specific manner. This approach revealed that the amplification of SETDB1, a gene encoding a chromatin modulator, accelerates tumor onset, thus uncovering its role as a cooperating oncogene in melanoma and identifying the SETDB1 protein as a new potential therapeutic target [11].
These models greatly augment our knowledge of the pathogenic mechanisms of human diseases, but also provide invaluable experimental platforms to accelerate the development of new therapies.
From tank to bedside
The possibility of collecting thousands of synchronized zebrafish embryos as well as their easy handling make them particularly amenable to chemical treatment. Accordingly, numerous chemical screens have been successfully performed in the fish [12]. One such screen looking for chemicals that would block formation of the neural crest, from which melanocytes arise, led to the identification of leflunomide, an inhibitor of pyrimidine synthesis [13]. Leflunomide was shown to trigger a pause in the transcriptional elongation of neural crest genes. Importantly, melanoma cells, which present neural crest features, also appear exquisitely sensitive to leflunomide treatment in vitro and in vivo. The combination of leflunomide and Braf inhibitors in the treatment of advanced metastatic melanoma is now in Phase II clinical trial.
Another chemical screen aimed at increasing the pool of hematopoietic stem cells (HSC) in the aorta–gonad–mesonephros, an early hematopoietic site in zebrafish and mammals, uncovered the crucial role of prostaglandin E2 (PGE2) signaling in stem cell regulation [14]. While PGE2 synthesis was revealed to be necessary for HSC homeostasis, this study also demonstrated that treatment with a stable form of PGE2 could enhance HSC formation in vivo. Interestingly, concordant observations were made in mice, where PGE2 also increased stem cell numbers in transplantation settings, opening vistas for clinical use. Indeed, the efficacy of treating cord blood with PGE2 to improve engraftment was tested in a Phase I trial and a Phase II trial is under way.
Concluding remarks: the translational future of the zebrafish
The generation of faithful zebrafish models is fueled by our knowledge of the genetics of human diseases. The recent development of deep-sequencing strategies on large sets of human tumor samples may yield novel insights into the genetic determinants of malignant transformation. Accordingly, the number of zebrafish models of cancer will continue to rise. The range of available techniques for the manipulation of zebrafish is increasing, thus offering new options to build models. Similar to zinc-finger nucleases and transcription activator-like effector nucleases (TALENs), CRISPR/Cas technology has been shown to efficiently cause mutations at targeted genomic sites in zebrafish embryos [15] and is emerging as another powerful tool for genome editing. Because they facilitate the rapid generation of large numbers of knockout strains, it is foreseeable that these new technologies will change the shape of loss-of-function approaches in the fish.
Other tools, such as in situ hybridization and microarrays, enable analysis of the genomic landscape of zebra-fish cells while the list of antibodies against zebrafish proteins is expanding, allowing in depth investigation of the molecular mechanisms during disease development or in response to treatment. Consequently, additional mechanistic studies on zebrafish disease models can be performed, leading to new therapeutic targets as well as to the development of novel therapies. The zebrafish is on its way to earning the trust of the scientific community as a reliable preclinical model.
Acknowledgments
The authors thank Elliott J. Hagedorn for critical reading of the manuscript. This work was supported by NIH grant 5 R01 CA103846-10. In addition, L.I.Z. is a Howard Hughes Medical Institute Investigator and J.A. received support from the Foundation Bettencourt–Schueller.
Footnotes
Disclaimer statement
L.I.Z. is a founder and stockholder of Fate, Inc. and Scholar Rock and a scientific advisor for Stemgent.
References
- 1.Howe K, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Konantz M, et al. Zebrafish xenografts as a tool for in vivo studies on human cancer. Ann N Y Acad Sci. 2012;1266:124–137. doi: 10.1111/j.1749-6632.2012.06575.x. [DOI] [PubMed] [Google Scholar]
- 3.Peterson RT, Macrae CA. Systematic approaches to toxicology in the zebrafish. Annu Rev Pharmacol Toxicol. 2012;52:433–453. doi: 10.1146/annurev-pharmtox-010611-134751. [DOI] [PubMed] [Google Scholar]
- 4.Santoriello C, Zon LI. Hooked! Modeling human disease in zebrafish. J Clin Invest. 2012;122:2337–2343. doi: 10.1172/JCI60434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, et al. A zebrafish model for hepatoerythropoietic porphyria. Nat Genet. 1998;20:239–243. doi: 10.1038/3041. [DOI] [PubMed] [Google Scholar]
- 6.Danilova N, et al. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112:5228–5237. doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
- 7.Payne EM, et al. L-Leucine improves the anemia and developmental defects associated with Diamond–Blackfan anemia and del(5q) MDS by activating the mTOR pathway. Blood. 2012;120:2214–2224. doi: 10.1182/blood-2011-10-382986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langenau DM, et al. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299:887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- 9.Feng H, et al. T-lymphoblastic lymphoma cells express high levels of BCL2, S1P1, and ICAM1, leading to a blockade of tumor cell intravasation. Cancer Cell. 2010;18:353–366. doi: 10.1016/j.ccr.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patton EE, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol. 2005;15:249–254. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 11.Ceol CJ, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan JL, Zon LI. Chemical screening in zebrafish for novel biological and therapeutic discovery. Methods Cell Biol. 2011;105:493–516. doi: 10.1016/B978-0-12-381320-6.00021-7. [DOI] [PubMed] [Google Scholar]
- 13.White RM, et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471:518–522. doi: 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.North TE, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR–Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]


