Abstract
There is a current interest in the development of biodosimetric methods for rapidly assessing radiation exposure in the wake of a large-scale radiological event. This work was initially focused on determining the exposure dose to an individual using biological indicators. Gene expression signatures show promise for biodosimetric application, but little is known about how these signatures might translate for the assessment of radiological injury in radiosensitive individuals, who comprise a significant proportion of the general population, and who would likely require treatment after exposure to lower doses. Using Parp1−/− mice as a model radiation-sensitive genotype, we have investigated the effect of this DNA repair deficiency on the gene expression response to radiation. Although Parp1 is known to play general roles in regulating transcription, the pattern of gene expression changes observed in Parp1−/− mice 24 h postirradiation to a LD50/30 was remarkably similar to that in wild-type mice after exposure to LD50/30. Similar levels of activation of both the p53 and NFκB radiation response pathways were indicated in both strains. In contrast, exposure of wild-type mice to a sublethal dose that was equal to the Parp1−/− LD50/30 resulted in a lower magnitude gene expression response. Thus, Parp1−/− mice displayed a heightened gene expression response to radiation, which was more similar to the wild-type response to an equitoxic dose than to an equal absorbed dose. Gene expression classifiers trained on the wild-type data correctly identified all wild-type samples as unexposed, exposed to a sublethal dose or exposed to an LD50/30. All unexposed samples from Parp1−/− mice were also correctly classified with the same gene set, and 80% of irradiated Parp1−/− samples were identified as exposed to an LD50/30. The results of this study suggest that, at least for some pathways that may influence radiosensitivity in humans, specific gene expression signatures have the potential to accurately detect the extent of radiological injury, rather than serving only as a surrogate of physical radiation dose.
INTRODUCTION
In this time of heightened concerns over the possibility of accidental releases of radioactive material or deliberate radiological or nuclear attacks, the development of biodosimetry approaches has been recognized as an important component within a broader national preparedness campaign (1–3). Radiological triage currently relies on clinical signs, such as daily blood counts, which would be impractical in a large-scale event, and time to emesis, which is imprecise and prone to a high-false-positive rate (4). Having a biologically based method for rapid triage of individuals with radiological injuries would be crucial in a resource-limited environment. In addition, the ability to reassure the “worried well” who received no appreciable radiation exposure could help to reduce panic in a chaotic situation. Areas of focus for biodosimetry include the automation of traditional cytogenetic assays, such as the scoring of micronuclei (5, 6), dicentrics (7, 8) or breaks in prematurely condensed chromosomes (8, 9), and the development of molecular signatures based on transcriptomic (10, 11), proteomic (12, 13) or metabolomic (14–16) profiling.
It has become clear from clinical studies that the same dose of radiation does not translate to an identical level of radiological injury in all individuals, with 2–4% of the population estimated to have some form of enhanced radiation sensitivity (17). The results of cell-based studies have further suggested broad variations in radiation sensitivity exist among the seemingly normal population (18, 19), with defects in DNA repair potentially affecting approximately 1 in 3 individuals and resulting in up to a 50% reduction in efficiency (20). In other studies, an interindividual variation in base excision repair (BER) up to 3.4-fold has been reported, depending on the assay used (21).
While the factors underlying general radiosensitivity have not been fully elucidated, the extreme sensitivity of patients with DNA repair deficiency syndromes, such as ataxia telangiectasia (22), suggests that DNA repair capacity may be an important factor. Although members of the general population with previously undetected radiosensitivity are unlikely to have the complete loss of function, as represented in mutant mouse models, such models nevertheless help to define the type and upper limits of the effects that DNA repair-related sensitivity might be expected to have on gene expression in response to radiation exposure.
In a recent study, we used genetically engineered radiosensitive mouse models with deficiency in DNA double-strand break (DSB) pathways [ataxia telangiectasia mutated (Atm−/−) or DNA-PKcs (PrkdcScid)] to show that disruption of these pathways dramatically changed the radiation-induced gene expression response (23). Despite differences from the human radiation response, including DNA repair pathway preference and specific gene expression changes, genetically engineered mouse models representing deficiencies in DNA repair can be useful both for characterizing radiation response at the molecular level and for guiding the development of biodosimetry models. It should also be noted that while mouse models are often used to address radiation biodosimetry questions that cannot be studied directly in humans, mouse signatures are not developed to be directly informative in humans. Indeed, even results from non-human primate studies appear not to be directly applicable to human biodosimetry, although careful selection of signature genes and application of interspecies conversion algorithms show promise for the translation of the results of such experiments (24).
In addition to DSB repair, BER is an important DNA repair pathway relevant to radiation exposure. BER repairs single-strand breaks and helps protect cells from oxidative damage caused by radiation (25). Poly (ADP-ribose) polymerases play a crucial role in the BER pathway, with poly(ADP-ribose) polymerase-1 (Parp1) accounting for the majority of Parp activity in mouse cells (26). The Parp polymerases catalyze synthesis of poly (ADP-ribose) from NAD+ on a wide array of target proteins in a process known as poly ADP-ribosylation (PARylation). PARylation of chromatin proteins helps to signal DNA breaks and makes the chromatin accessible to repair factors. Although Parp1 was initially associated with the BER pathway, more recent evidence suggests it also plays a role in other repair pathways, including the DSB repair pathways (27).
In addition to its roles in DNA repair, Parp1 is involved in many other cellular processes, including the regulation of chromatin structure, cell death pathways and transcription (28). Parp1 may help to drive decisions of cell fate towards cell cycle arrest and DNA repair after low levels of DNA damage, or towards apoptosis and other modes of cell death after high levels of DNA damage (29). Human mutations in Parp1, or alterations in its expression, have also been associated with an increased cancer risk (21, 30, 31) and improper DNA repair (32). Similarly, BER pathway mutations in the DNA polymerase beta (POLB) and X-ray repairing cross-complementing 1 (XRCC1) genes have also been studied extensively and have been associated with variations in DNA repair levels from 37–60%, and with increased cancer risks (33–35).
Parp1−/− mice are sensitive to radiation exposure, with an LD50/30 of approximately 6 Gy (36), compared to an LD50/30 of approximately 8.8 Gy in their wild-type (WT) parental strain (37). The enhanced radiosensitivity of the Parp1−/− mice is associated with an extended cellular G2 arrest and accumulation of unrepaired DNA damage, followed by mitotic catastrophe. Increased Parp activity has been linked to longevity across species (38, 39), however, the mutant Parp1 mouse does not have a shortened life span (40). In a recently reported study, these mice were found to have differences from WT mice in their urinary metabolome after radiation exposure at times relevant to radiological triage (37). Although we would not expect humans undergoing triage to include individuals with a complete lack of Parp1, this model serves as an extreme case to test the potential for defects in BER and the Parp pathway to influence determination of radiation dose by gene expression.
In previous studies, we have used a transcriptomic approach to identify gene signatures for radiation biodosimetry (41–47). We have examined the effect of dose, dose rate, radiation quality and internally deposited radionuclides on the expression of genes for potential use in biodosimetry (41, 45–51)
More recently, we have begun to investigate the effect of deficiencies in major DNA repair pathways relevant to radiation survival on the gene expression response to radiation, with initial studies focused on DNA DSB repair using Atm−/− and PrkdcScid mouse models (23). In these studies, we have compared responses at LD50/30, since this represents a defined level of physiological response to allow direct comparison between strains with different radiation sensitivities. Here, we report on the changes observed in global gene expression one day postirradiation of Parp1−/− or WT mice to an approximately LD50/30 of gamma rays (6 Gy for Parp1−/− and 8.8 Gy for WT mice). We also measured the response of WT mice to 6 Gy gamma rays for comparison of a dose equal to the LD50/30 of Parp1−/− mice. In Parp1−/− mice, we found a heightened gene expression response to radiation, which was more similar to the wild-type response to an equitoxic dose than to an equal absorbed dose.
METHODS
Mice and Irradiations
Mice with a homozygous deletion of Parp1 (129S-Parp1tm1Zqw/J, referred to henceforth as Parp1−/−) (40) and the corresponding 129S wild-type strain were purchased from the Jackson Laboratory (Bar Harbor, ME) and bred at Georgetown University (Washington, DC). All mouse work was conducted at Georgetown University under Protocol 13-003 approved by the Georgetown University Institutional Animal Care and Use Committee, and in accord with NIH guidelines. The animals had access to food and water ad libitum and were maintained with a standard 12 h light/dark cycle.
Male mice at 8–10 weeks of age were exposed to approximately LD50/30 137Cs gamma rays, corresponding to 6 Gy for the Parp1−/− animals (36) and 8.8 Gy for the WT mice (37). The dose rate was approximately 1.67 Gy/min. A second group of WT animals were also exposed to 6 Gy, and for both genotypes a group of control animals were sham irradiated.
At 24 h postirradiation animals were sacrificed, and blood was collected by cardiac puncture into PAXgene™ blood RNA stabilization solution (PreAnalytiX; QIAGEN®, Valencia, CA). After mixing thoroughly, the samples were stored at −80°C prior to shipping to Columbia University (New York, NY) for RNA analysis.
RNA Isolation and Microarray Expression Analysis
RNA isolation, microarray hybridization and data analysis were performed as described elsewhere (45). Briefly, RNA was extracted from the stabilized blood with the PAXgene Blood RNA kit (QIAGEN), and globin transcripts were reduced with the GLOBIN-clear™-Mouse/Rat Kit (Thermo Fisher Scientific™ Inc., Rockford, IL). The Agilent Bioanalyzer (Santa Clara, CA) was used to assess RNA quality, and samples with RIN ≥7 (average 8.5) were used for single-color hybridization with Agilent Whole Mouse Genome Microarrays (v2 4×44K) according to the manufacturer’s protocol. After hybridization and washing, slides were imaged on the Agilent DNA Microarray Scanner, and data were extracted using the default settings of the Agilent Feature Extraction Software version 11.5.1. The final dataset comprised five animals from each genotype-dose group (except for the 8.8 Gy irradiated WT group, which contained samples from four animals) and is available from the Gene Expression Omnibus (accession no. GSE101402).
For each genotype, genes with significantly different expression levels between the control and irradiated groups were identified using the BRB-ArrayTools class comparison tool (52). P < 0.001 was considered statistically significant, and the Benjamini-Hochberg false discovery rate (FDR) was used to correct for multiple comparisons and limit false discoveries to below 5% (53). Genes meeting both these criteria are referred to as “regulated”. BRB-ArrayTools was also used to generate clustered heat maps and multidimensional scaling (MDS) plots to illustrate gene expression patterns.
Class prediction was also performed with BRB-ArrayTools. Genes were selected using a statistical cutoff of P < 10−7, and the wild-type expression data were used as a training set with four classification methods (diagonal linear discriminant analysis, 1- and 3-nearest neighbors, nearest centroid) used to classify samples as exposed to 0, 6 or 8.8 Gy. These four classifiers were then used to predict the radiation status of the Parp1−/− samples. The algorithm was run using the stated parameters without optimizing or minimizing the number of genes included in the classifier.
Gene ontology analysis was performed on lists of differentially expressed genes using the Statistical Overrepresentation Test in PANTHER Tools with default parameters (54, 55). GO-Slim biological processes or PANTHER pathways with P < 0.05 (after applying Bonferroni correction for multiple-comparison testing) were considered significant.
Expression values of the differentially expressed genes were imported into the Ingenuity Pathway Analysis software [IPA; QIAGEN (formerly Ingenuity Systems), Redwood City, CA], for each genotype-dose condition. The IPA core analysis was run to predict putative upstream regulators of the observed gene expression responses. IPA makes use of a database of relationships between gene products curated from the published literature to predict activation or inhibition of potential regulators upstream of the observed changes in gene expression. A z-score ≥ 2 or ≤ −2 was considered statistically significant for prediction of regulatory factor activation or inhibition, respectively.
Quantitative Real-Time RT-PCR
Total RNA was reverse transcribed (RT) using the manufacturer’s protocol of the High-Capacity cDNA Reverse Transcription Kit with RNase Inhibitor (Thermo Fisher Scientific). Real-time RT-PCRs were performed using the Applied Biosystems® 7900 HT system (Carlsbad, CA) with TaqMan® assays for Cdkn1a (assay ID: Mm01303209_m1), Phlda3 (assay ID: Mm00449846_m1), Il1B (assay ID; Mm00434228_m1) and Actb (assay ID: Mm00607939_s1). Relative fold changes were calculated using the ΔΔCt method, as described elsewhere (56) and normalized to Actb. Fold changes were the mean of three independent pools formed by pooling RNA from 2–4 different biological replicates for each condition. Each reaction was performed in duplicate to provide technical replicates.
RESULTS
Differentially Expressed Genes
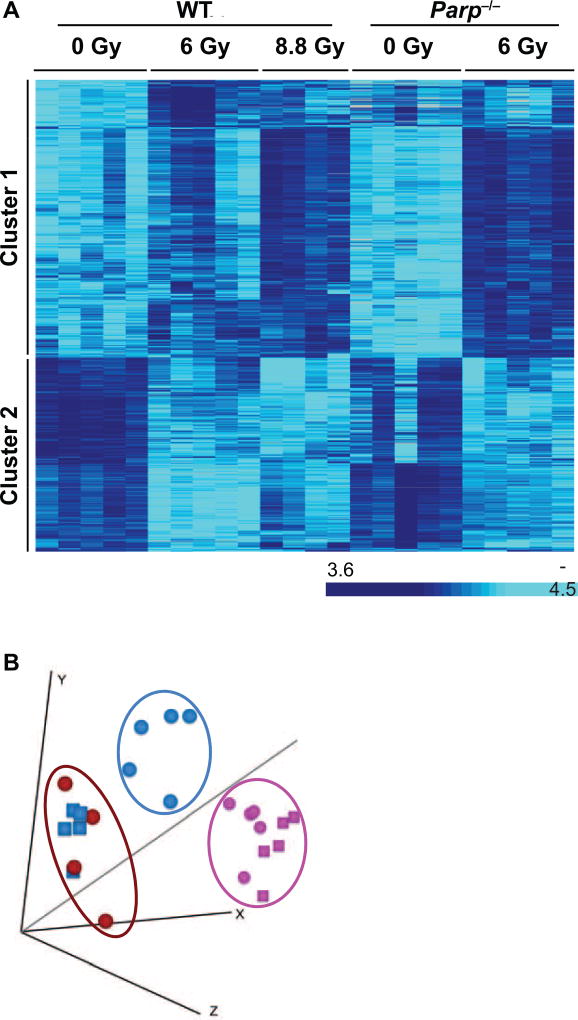
Equitoxic doses (approximately LD50/30) of gamma rays were given to Parp1−/− (6 Gy) and WT (8.8 Gy) mice, with a second group of WT mice also receiving 6 Gy. Sham-irradiated mice of each genotype were used as controls. Blood was collected 24 h postirradiation for whole-genome RNA analysis using Agilent microarrays. The expression data were analyzed using BRB-ArrayTools to identify genes significantly differentially expressed after irradiation. Compared to controls, there were 1,370 differentially expressed genes [P < 0.001, false discovery rate (FDR) <5%] in WT mice after 8.8 Gy irradiation and 1,177 after 6 Gy irradiation (Supplementary File S1; http://dx.doi.org/10.1667/RR14990.1.S1). In Parp1−/− mice, 1,677 genes were differentially expressed after 6 Gy irradiation (Supplementary File S1). Altogether, 3,009 genes were differentially expressed in at least one dose-genotype condition (Fig. 1A and Supplementary File S2; http://dx.doi.org/10.1667/RR14990.1.S2). Of these, 1,332 were regulated in WT but not Parp1−/− mice and 847 were regulated in Parp1−/− but not WT mice (Supplementary File S2). The overall response pattern of all 3,009 radiation-responsive genes in each sample is summarized visually in the MDS plot shown in Fig. 1B.
FIG. 1.
Panel A: Heatmap of the 3,009 genes (rows) differentially expressed in any of the dose-genotype combinations tested in this study. Each column represents an individual mouse. Relative expression level is indicated by color intensity according to the scale bar to the right. Clusters 1 and 2 represent up- and downregulated genes, respectively, 24 h postirradiation. No genes were identified that responded in opposite directions in WT and Parp1−/− mice. Panel B: MDS plot of the 3,009 differentially expressed genes. Each circle represents a sample from an individual WT mouse, each square a Parp1−/− mouse. The symbols are color coded by dose: pink (0 Gy controls), blue (6 Gy) and red (8.8 Gy). The distance between points on the plot represents the overall difference in expression of the 3,009 differentially expressed genes.
Gene ontology analysis was performed in PANTHER to look for common biological processes or pathways that were over-represented among the differentially expressed genes. Since 80% of the genes differentially expressed in Parp1−/− mice after irradiation were upregulated, the analysis focused on upregulated genes. The results (Table 1) are indicative of a relatively typical radiation response, with broad similarity evident between the pathways responding to LD50/30 in WT (8.8 Gy) and Parp1−/− (6 Gy) mice. The lists of differentially expressed genes were also imported into IPA for prediction of activation or inhibition of regulatory factors potentially upstream of the observed gene expression (Fig. 2 and Supplementary File S2; http://dx.doi.org/10.1667/RR14990.1.S2).
TABLE 1.
Significantly Overrepresented PANTHER Pathways and Biological Processes among Upregulated Genes
| Genotype, radiation dose | |||
|---|---|---|---|
|
|
|||
| Parp1−/−a (6 Gy) | WT (6 Gy) | WT (8.8 Gy) | |
| PANTHER pathways | |||
| Integrin signaling pathway (P00034) | 2.03 × 10−4 (26) | NS | 3.78 × 10−8 (24) |
| Blood coagulation (P00011) | 4.72 × 10−3 (11) | NS | 1.18 × 10−3 (9) |
| p53 pathway (P00059) | 1.70 × 10−3 (15) | NS | 1.04 × 10−2 (10) |
| Ras Pathway (P04393) | 3.26 × 10−4 (15) | NS | 1.84 × 10−2 (9) |
| Gonadotropin-releasing hormone receptor pathway (P06664) | 4.58 × 10−2 (24) | NS | 2.05 × 10−2 (17) |
| Inflammation mediated by chemokine and cytokine signaling pathway (P00031) | 3.13 × 10−4 (31) | NS | 2.13 × 10−2 (18) |
| Apoptosis signaling pathway (P00006) | 2.16 × 10−3 (18) | NS | 4.08 × 10−2 (11) |
| p53 pathway feedback loops 2 | NS | 4.58 × 10−2 (5) | NS |
| CCKR signaling map | 4.88 × 10−5 (25) | NS | NS |
| Heterotrimeric G-protein signaling pathway-Gq alpha and Go alpha mediated pathway | 7.44 × 10−3 (17) | NS | NS |
| Heterotrimeric G-protein signaling pathway-Gi alpha and Gs alpha mediated pathway | 8.83 × 10−3 (20) | NS | NS |
| EGF receptor signaling pathway | 1.18 × 10−2 (18) | NS | NS |
| Oxidative stress response | 4.82 × 10−2 (10) | NS | NS |
| PANTHER GO-Slim biological process | |||
| Cell communication | 4.32 × 10−2 (177) | NS | 6.48 × 10−3 (110) |
| Signal transduction | NS | NS | 1.50 × 10−2 (100) |
| MAPK cascade | 8.89 × 10−3 (31) | NS | NS |
| Intracellular signal transduction | 9.73 × 10−3 (75) | NS | NS |
| Protein transport | 1.54 × 10−2 (73) | NS | NS |
| Intracellular protein transport | 4.05 × 10−2 (70) | NS | NS |
Note. Numbers in parentheses are the number of differentially expressed genes contributing to each category.
Numbers are Bonferroni-corrected P values. NS = not significant (Bonferroni P > 0.05).
FIG. 2.
Heat map illustrating the patterns of upstream regulator activation in response to LD50/30 as predicted by IPA analysis. Each row across represents a potential regulator of the gene expression patterns observed. Each column represents a genotype exposed to LD50/30: WT (8.8 Gy) and Parp1−/− (6 Gy). The responses are shaded according to the scale bar to represent the z-score associated with the prediction of response. A z-score ≥ 2 represents a significant prediction of activation (red) and a z-score ≤ −2 represents a significant prediction of inhibition (blue).
Quantitative Real-Time RT-PCR (qRT-PCR)
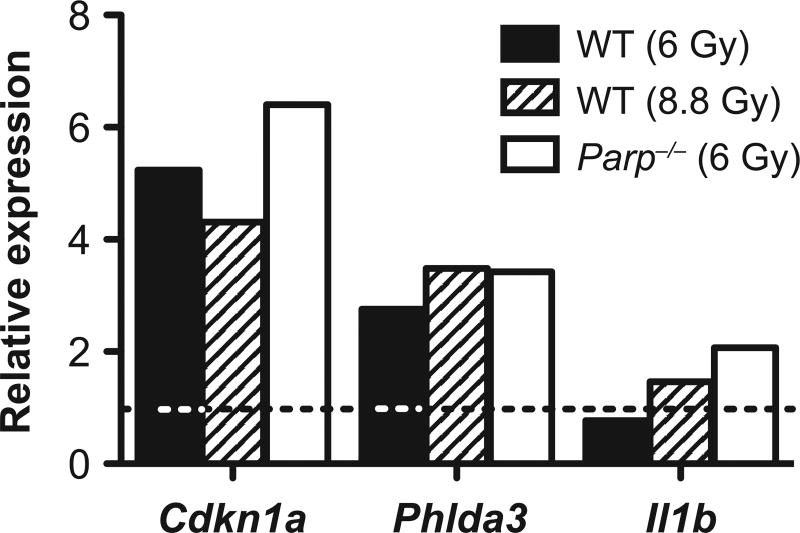
Relative expression of Cdkn1a, Phlda3 and Il1b was measured by qRT-PCR (Fig. 3). The results agreed with the pattern of expression seen in the array data, supporting the observed trend of heightened gene expression responses among both p53-regulated genes (Cdkn1a and Phlda3) and mediators of inflammatory response (Il1b) in the Parp1−/− mice responding to radiation.
FIG. 3.
Relative expression by quantitative real-time RT-PCR of Cdkn1a and Phlda3, two p53-regulated radiation response genes, and Il1b, an immune response gene regulated by NFκB. Data are the mean of three independent replicates and are normalized to Actb. Fold changes are relative to the sham-irradiated samples of the corresponding genotype (indicated by the dashed line).
Class Prediction
BRB-ArrayTools was also used to train classifier algorithms using the gene expression data from the WT mice, and then to test the ability of those algorithms to classify the radiation status of the Parp1−/− samples. The resulting classifier was composed of 36 genes (Supplementary File S3; http://dx.doi.org/10.1667/RR14990.1.S3). IPA upstream analysis revealed multiple potential upstream regulators for these 36 genes, with the largest number of genes regulated by interferon gamma (IFNG, 11 genes), interleukin-2 (IL2, 9 genes) and tumor protein p53 (TP53, 9 genes) and 7 genes each regulated by signal transducer and activator of transcription 1 (STAT1), interleukin-4 (IL4), interleukin-1B (IL1B), tumor necrosis factor (TNF) and hepatocyte nuclear factor 4 alpha (HNF4A) (Supplementary File S3). All four algorithms tested (diagonal linear discriminant analysis, 1- and 3-nearest neighbors and nearest centroid) performed with 100% accuracy during leave-one-out cross validation on the WT samples. The performance of all four algorithms on the data from the Parp1−/− samples was highly consistent and agreed with our earlier clustering results (Fig. 1). Each algorithm classified all Parp1−/− control samples correctly as controls, and 4 of the 5 irradiated Parp1−/− samples as exposed to 8.8 Gy (the biological or LD50/30), with one irradiated sample being classified as exposed to 6 Gy (the physical dose delivered to the mice).
DISCUSSION
In this study, we have compared the gene expression response to radiation in Parp1−/− and wild-type mice at 24 h postirradiation. This time was selected for initial studies as it represents the earliest time in planning scenarios when outside assistance is expected to arrive and sample collection for large-scale biodosimetry can be initiated (57, 58). Since gene expression responses to radiation exposure are known to be highly dynamic (45, 59–62), and biodosimetry methods may be needed through the first week after an event, future studies will be needed to fully elucidate the effect of DNA repair deficiencies on gene expression throughout this time.
We observed a pattern of similarity in the gene expression response between the two genotypes in all analyses (Fig. 1). Although 61% of the genes responding significantly to radiation in WT mice were not significantly responsive in Parp1−/− mice, and 51% of those responding in Parp1−/− mice were not significantly radiation responsive in WT mice, the clustered heat maps in Supplementary File S2 (Worksheet 2; http://dx.doi.org/10.1667/RR14990.1.S2) clearly illustrate that the majority of these “unique” response genes showed the same trend of response in both genotypes.
Similarly, gene ontology analysis of the genes responding in each genotype-dose group (Table 1) suggested broad similarities in the pathways activated by radiation in both genotypes. Integrin, p53, Ras, inflammation and apoptosis pathways were over-represented among differentially expressed genes responding to the LD50/30 in both genotypes, but not in the WT response to 6 Gy irradiation. Additional pathways, including EGF receptor signaling and oxidative stress response were significantly over-represented only among the genes responding in Parp1−/− mice.
Many factors in addition to DNA repair status can affect gene expression levels, leading to variation among individuals in both endogenous expression levels (63) and radiation response (64, 65). Factors of concern in addition to genetic differences include disease status, especially conditions involving chronic inflammation (66–69), and lifestyle factors, such as smoking (42, 66, 70). A few studies have been undertaken to explore the potential effect of such confounding factors on gene expression biodosimetry. For instance, using lipopolysaccharide (LPS) injection in mice as a model for inflammatory response, several groups have found that while both the baseline and postirradiation levels of some radiation-responsive genes are altered by LPS, it is still possible to select genes for biodosimetry that appear unaffected by inflammation (66, 67). We drew similar conclusions from a study using ex vivo irradiation of human peripheral blood from smokers or non-smokers. While the radiation response of a small number of genes was different in the two groups, smoking status did not affect the ability of a previously defined gene expression signature to detect radiation exposures as low as 0.1 Gy (42). While the combined effects of multiple factors, such as DNA repair deficiency and active smoking status, are currently unknown, this remains an area of active interest in research, and may be especially important when discrimination of lower doses (≤2 Gy) is needed.
In addition to its roles in DNA repair, Parp1 is involved in several aspects of the regulation of gene expression, including regulating chromatin accessibility (28), acting as a transcriptional co-activator (71, 72), RNA metabolism and interactions with RNA binding proteins (73), and modifying transcription factors to inhibit their promoter binding activity (74, 75). We therefore applied IPA upstream regulator analysis to our gene expression results, and again found a broad similarity of predicted transcription factor activation by radiation in Parp1−/− and WT mice (Fig. 2 and Supplementary Table S2; http://dx.doi.org/10.1667/RR14990.1.S2). For instance, two of the major transcription factors involved in the radiation response, p53 and NFκB, were predicted to be significantly activated in both genotypes after LD50/30 (p53: WT z-score = 5.8, Parp1−/− z-score = 6.1; and NFκB complex: WT z-score = 1.8, Parp1−/− z-score = 3.8).
Both p53 and NFκB have been shown to be transiently PARylated in response to various stresses (76), generally reducing their affinity for binding to the promoters of downstream effector genes (76). PARylation of p53 promotes stabilization and accumulation of the protein, but induction of Bax and Fas was not seen until PARylation levels declined. In the absence of functional Parp1, non-PARylated p53 retains a high affinity for its consensus sequence (77), possibly due to compensatory activity of other Parp family members. This, in conjunction with prolonged damage signaling from unrepaired DNA lesions in the Parp1−/− mice (78), may also contribute to higher levels of stress signaling and a heightened or more prolonged induction of p53 target genes in the Parp1−/− mice. This is reflected in our study by the trend of larger magnitude induction observed, for example, for the p53 targets Cdkn1a and Phlda3 (Fig. 3). Furthermore, this pattern is consistent with the changes in energy metabolism recently reported in the urinary metabolome of irradiated Parp1−/− mice (37), which were attributed in part to altered timing and modes of radiation-induced cell death, and induction of mitotic catastrophe as a result of unrepaired DNA damage.
Consistent with our observations of gene expression patterns, Beneke et al. (79) reported a larger induction of p53, as well as increased apoptosis, in thymocytes from Parp1−/− mice within the first 36 h after 2 Gy X-ray irradiation. They also demonstrated increased radiation-induced transcriptional transactivation of an Mdm2 reporter construct by p53 when Parp1 activity was ablated. A similar increase in apoptosis and p53 accumulation has also been reported after DNA damage in splenocytes from Parp1−/− mice (78). In contrast, mouse embryo fibroblasts from Parp1−/− mice were reported to show reduced accumulation and Ser-15 phosphorylation of p53 in response to gamma rays, along with reduced responses of Cdkn1a and Mdm2 mRNA (80). Inhibition of Parp1 function in two transformed human cell lines also resulted in delayed and attenuated induction of Cdkn1a and Mdm2 proteins and suppressed G1 arrest after exposure to ionizing radiation (81). Together, these results may suggest that without active Parp1, the p53-driven transcriptional response to radiation may be ablated in fibroblasts or cells of epithelial origin, with Parp1 loss having the opposite effect in cells of lymphoid/myeloid origin. This may be a result of different expression of other Parp family members in different tissues, or due to the activity of different p53 cofactors in white blood cells compared to less radiosensitive tissues.
Parp1 also contributes directly to NFκB activation and its regulation of gene transcription (82, 83). Activation of NFκB and its DNA binding/transactivation activity were reported to be dependent on wild-type Parp1 activity when assayed at 1–2 h postirradiation in several cell types derived from Parp1−/− or WT mice (82, 84). These findings contrast with our observation of an apparent enhancement of radiation-induced NFκB activation in the Parp1−/− mice (Supplementary File S2; http://dx.doi.org/10.1667/RR14990.1.S2) at 24 h postirradiation. However, the difference in the times at which measurements were made suggest that in the absence of functional Parp1 there could be an initial delay in the response of NFκB and its effector genes, such as Il1b (Fig. 3), which is overcome by 24 h postirradiation.
The similarity between the gene expression response to LD50/30 in both WT and Parp1−/− mice was evident across all 3,009 genes that responded significantly in either genotype. This broad similarity of responses contrasts with the findings of our recently published experiments in irradiated Atm−/− and Scid mice, in which we reported strong overall ablation of the gene expression response (23). The similarity of responses in WT and Parp1−/− mice is illustrated in Fig. 1B by a MDS plot. The MDS plot represents each individual sample as a point, with the distance between two points representing the relative difference in expression of all 3,009 genes in those samples. Figure 1B shows all the control samples cluster together regardless of genotype, but the samples from Parp1−/− mice exposed to 6 Gy cluster with the WT samples exposed to an equitoxic 8.8 Gy dose, rather than with the 6 Gy irradiated WT equidose samples. This suggests that gene expression in Parp1−/− mice after radiation exposure is more reflective of the level of radiological injury than the level of physical dose, which could be useful in a biodosimetry setting.
To further test the ability of gene expression to detect an LD50/30 exposure in Parp1−/− radiosensitive mice, we trained classifiers using the WT data, and used them to predict the radiation status of Parp1−/− mice. Although the classifiers were not optimized for the minimum number of genes required, unexposed Parp1−/− mice were predicted as such with 100% accuracy, while 80% of the irradiated Parp1−/− mice were classified as receiving a dose equivalent to the WT LD50/30, rather than their actual physical dose of 6 Gy. This basic classification exercise further supports the notion that at least for some underlying genotypes, radiation sensitivity with abnormalities in the BER pathway may affect the magnitude of gene expression in response to a given physical dose, such that the resulting gene expression profile may provide an accurate reflection of the extent of individual radiological injury. Taken together with the results of our recently reported study with DSB repair mutants (23), the findings suggest that genes selected and tested on a diverse population of humans should likely perform appropriately across a broad range of radiosensitivities, but this will require testing and signature development in a human model. Further studies are needed to more fully explore the effect of DNA repair-deficient phenotypes, subtle variations in repair capacity or the possible interactions between DNA repair deficiencies and other potential confounding factors on biodosimetry, using gene expression signatures at a range of doses and times.
Supplementary Material
File S1. Lists of genes identified as significantly differentially expressed after irradiation. As shown in the first, second and third tables, respectively, compared to controls, in WT mice after 8.8 Gy irradiation, there were 1,370 differentially expressed genes [P < 0.001, false discovery rate (FDR) <5%] and after 6 Gy, there were 1,177; and in Parp1−/− mice after 6 Gy, 1677 genes were differentially expressed.
File S2. A total of 3,009 genes were differentially expressed in at least one dose-genotype condition. Of these, 1,332 were regulated in WT but not Parp1−/− mice, and 847 were regulated in Parp1−/− but not WT mice. The first table shows genes in the clustered order in which they appear in the heatmap in Fig. 1A. The second table provides lists of genes only responding significantly in WT mice (list A) and Parp1−/− mice (list B).
The lists of differentially expressed genes were imported into IPA for prediction of activation or inhibition of regulatory factors potentially upstream of the observed gene expression. The third table is an annotation of Fig. 2.
File S3. BRB-ArrayTools was used to train classifier algorithms using the gene expression data from the WT mice, and then to test the ability of those algorithms to classify the radiation status of the Parp1−/− samples. The resulting classifier was composed of 36 genes (first table). Top upstream regulators of classifier genes, as determined by IPA analysis, are also listed (second table).
Acknowledgments
We thank Pelagie Ake, Steven Strawn, Bo-Hyun Moon, Maria Elena Diaz-Rubio and Yi-Wen Wang for breeding and genotyping of the mice and sample collection, Dr. Sunirmal Paul for beginning the RNA work, and Dr. Norman J. Kleiman, Mr. Jeffrey D. Knotts and Ms. Mashkura Chowdhury for further technical assistance. Analyses were performed using BRB-ArrayTools developed by Dr. Richard Simon and the BRB-ArrayTools Development Team. This work was supported by the Center for High-Throughput Minimally-Invasive Radiation Biodosimetry, National Institute of Allergy and Infectious Diseases (grant no. U19AI067773).
References
- 1.Pellmar TC, Rockwell S. Priority list of research areas for radiological nuclear threat countermeasures. Radiat Res. 2005;163:115–23. doi: 10.1667/rr3283. [DOI] [PubMed] [Google Scholar]
- 2.Koenig KL, Goans RE, Hatchett RJ, Mettler FA, Schumacher TA, Noji EK, et al. Medical treatment of radiological casualties: current concepts. Ann Emerg Med. 2005;45:643–52. doi: 10.1016/j.annemergmed.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 3.Sullivan JM, Prasanna PG, Grace MB, Wathen LK, Wallace RL, Koerner JF, et al. Assessment of biodosimetry methods for a mass-casualty radiological incident: medical response and management considerations. Health Phys. 2013;105:540–54. doi: 10.1097/HP.0b013e31829cf221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demidenko E, Williams BB, Swartz HM. Radiation dose prediction using data on time to emesis in the case of nuclear terrorism. Radiat Res. 2009;171:310–9. doi: 10.1667/RR1552.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fenech M. The lymphocyte cytokinesis-block micronucleus cytome assay and its application in radiation biodosimetry. Health Phys. 2010;98:234–43. doi: 10.1097/HP.0b013e3181b85044. [DOI] [PubMed] [Google Scholar]
- 6.Repin M, Pampou S, Karan C, Brenner DJ, Garty G. RABiT-II: Implementation of a high-throughput micronucleus biodosimetry assay on commercial biotech robotic systems. Radiat Res. 2017;187:492–8. doi: 10.1667/RR011CC.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garty G, Bigelow AW, Repin M, Turner HC, Bian D, Balajee AS, et al. An automated imaging system for radiation biodosimetry. Microsc Res Tech. 2015;78:587–98. doi: 10.1002/jemt.22512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.M’kacher R, El Maalouf E, Terzoudi G, Ricoul M, Heidingsfelder L, Karachristou I, et al. Detection and automated scoring of dicentric chromosomes in nonstimulated lymphocyte prematurely condensed chromosomes after telomere and centromere staining. Int J Radiat Oncol Biol Phys. 2015;91:640–9. doi: 10.1016/j.ijrobp.2014.10.048. [DOI] [PubMed] [Google Scholar]
- 9.Terzoudi GI, Pantelias G, Darroudi F, Barszczewska K, Buraczewska I, Depuydt J, et al. Dose assessment intercomparisons within the RENEB network using G0-lymphocyte prematurely condensed chromosomes (PCC assay) Int J Radiat Biol. 2017;93:48–57. doi: 10.1080/09553002.2016.1234725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amundson SA, Bittner M, Meltzer P, Trent J, Fornace AJ. Induction of gene expression as a monitor of exposure to ionizing radiation. Radiat Res. 2001;156:657–61. doi: 10.1667/0033-7587(2001)156[0657:iogeaa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 11.Lucas J, Dressman HK, Suchindran S, Nakamura M, Chao NJ, Himburg H, et al. A translatable predictor of human radiation exposure. PLoS One. 2014;9:e107897. doi: 10.1371/journal.pone.0107897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guipaud O. Serum and plasma proteomics and its possible use as detector and predictor of radiation diseases. Adv Exp Med Biol. 2013;990:61–86. doi: 10.1007/978-94-007-5896-4_4. [DOI] [PubMed] [Google Scholar]
- 13.Sharma M, Moulder JE. The urine proteome as a radiation biodosimeter. Adv Exp Med Biol. 2013;990:87–100. doi: 10.1007/978-94-007-5896-4_5. [DOI] [PubMed] [Google Scholar]
- 14.Johnson CH, Patterson AD, Krausz KW, Kalinich JF, Tyburski JB, Kang DW, et al. Radiation metabolomics. 5. Identification of urinary biomarkers of ionizing radiation exposure in nonhuman primates by mass spectrometry-based metabolomics. Radiat Res. 2012;178:328–40. doi: 10.1667/rr2950.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laiakis EC, Mak TD, Anizan S, Amundson SA, Barker CA, Wolden SL, et al. Development of a metabolomic radiation signature in urine from patients undergoing total body irradiation. Radiat Res. 2014;181:350–61. doi: 10.1667/RR13567.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pannkuk EL, Fornace AJ, Laiakis EC. Metabolomic applications in radiation biodosimetry: exploring radiation effects through small molecules. Int J Radiat Biol. 2017;93:1151–76. doi: 10.1080/09553002.2016.1269218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Health risks from exposure to low levels of ionizing radiation: BEIR VII phase 2. Washington, DC: National Research Council, National Academies Press; 2006. [PubMed] [Google Scholar]
- 18.Deschavanne PJ, Fertil B. A review of human cell radiosensitivity in vitro. Int J Radiat Oncol Biol Phys. 1996;34:251–66. doi: 10.1016/0360-3016(95)02029-2. [DOI] [PubMed] [Google Scholar]
- 19.Geara FB, Peters LJ, Ang KK, Wike JL, Sivon SS, Guttenberger R, et al. Intrinsic radiosensitivity of normal human fibroblasts and lymphocytes after high- and low-dose-rate irradiation. Cancer Res. 1992;52:6348–52. [PubMed] [Google Scholar]
- 20.Kato TA, Wilson PF, Nagasaw H, Peng Y, Weil MM, Little JB, et al. Variations in radiosensitivity among individuals: a potential impact on risk assessment. Health Phys. 2009;97:470–80. doi: 10.1097/HP.0b013e3181b08eee. [DOI] [PubMed] [Google Scholar]
- 21.Wilson DM, Kim D, Berquist BR, Sigurdson AJ. Variation in base excision repair capacity. Mutat Res. 2011;711:100–12. doi: 10.1016/j.mrfmmm.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavin MF. Radiosensitivity and oxidative signalling in ataxia telangiectasia: an update. Radiother Oncol. 1998;47:113–23. doi: 10.1016/s0167-8140(98)00027-9. [DOI] [PubMed] [Google Scholar]
- 23.Rudqvist N, Laiakis EC, Ghandhi SA, Kumar S, Knotts JD, Chowdhury M, et al. Global gene expression response in mouse models of DNA repair deficiency after gamma irradiation. Radiat Res. 2018;189:337–44. doi: 10.1667/RR14862.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park JG, Paul S, Briones N, Zeng J, Gillis K, Wallstrom G, et al. Developing human radiation biodosimetry models: Testing cross-species conversion approaches using an ex vivo model system. Radiat Res. 2017;187:708–21. doi: 10.1667/RR14655.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallace SS. Base excision repair: a critical player in many games. DNA Repair (Amst) 2014;19:14–26. doi: 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shieh WM, Amé JC, Wilson MV, Wang ZQ, Koh DW, Jacobson MK, et al. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J Biol Chem. 1998;273:30069–72. doi: 10.1074/jbc.273.46.30069. [DOI] [PubMed] [Google Scholar]
- 27.Martin-Hernandez K, Rodriguez-Vargas JM, Schreiber V, Dantzer F. Expanding functions of ADP-ribosylation in the maintenance of genome integrity. Semin Cell Dev Biol. 2017;63:92–101. doi: 10.1016/j.semcdb.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 28.Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–67. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- 29.Elkholi R, Chipuk JE. How do I kill thee? Let me count the ways: p53 regulates PARP-1 dependent necrosis. Bioessays. 2014;36:46–51. doi: 10.1002/bies.201300117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alanazi M, Pathan AA, Abduljaleel Z, Arifeen Z, Shaik JP, Alabdulkarim HA, et al. Association between PARP-1 V762A polymorphism and breast cancer susceptibility in Saudi population. PLoS One. 2013;8:e85541. doi: 10.1371/journal.pone.0085541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green AR, Caracappa D, Benhasouna AA, Alshareeda A, Nolan CC, Macmillan RD, et al. Biological and clinical significance of PARP1 protein expression in breast cancer. Breast Cancer Res Treat. 2015;149:353–62. doi: 10.1007/s10549-014-3230-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hua RX, Li HP, Liang YB, Zhu JH, Zhang B, Ye S, et al. Association between the PARP1 Val762Ala polymorphism and cancer risk: evidence from 43 studies. PLoS One. 2014;9:e87057. doi: 10.1371/journal.pone.0087057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rzeszowska-Wolny J, Polanska J, Pietrowska M, Palyvoda O, Jaworska J, Butkiewicz D, et al. Influence of polymorphisms in DNA repair genes XPD, XRCC1 and MGMT on DNA damage induced by gamma radiation and its repair in lymphocytes in vitro. Radiat Res. 2005;164:132–40. doi: 10.1667/rr3400. [DOI] [PubMed] [Google Scholar]
- 34.Yamtich J, Speed WC, Straka E, Kidd JR, Sweasy JB, Kidd KK. Population-specific variation in haplotype composition and heterozygosity at the POLB locus. DNA Repair (Amst) 2009;8:579–84. doi: 10.1016/j.dnarep.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sterpone S, Cozzi R. Influence of XRCC1 genetic polymorphisms on ionizing radiation-induced DNA damage and repair. J Nucleic Acids. 2010;2010 doi: 10.4061/2010/780369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masutani M, Nozaki T, Nakamoto K, Nakagama H, Suzuki H, Kusuoka O, et al. The response of Parp knockout mice against DNA damaging agents. Mutat Res. 2000;462:159–66. doi: 10.1016/s1383-5742(00)00033-8. [DOI] [PubMed] [Google Scholar]
- 37.Laiakis EC, Pannkuk EL, Diaz-Rubio ME, Wang YW, Mak TD, Simbulan-Rosenthal CM, et al. Implications of genotypic differences in the generation of a urinary metabolomics radiation signature. Mutat Res. 2016;788:41–9. doi: 10.1016/j.mrfmmm.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burkle A, Muller M, Wolf I, Kupper JH. Poly(ADP-ribose) polymerase activity in intact or permeabilized leukocytes from mammalian species of different longevity. Mol Cell Biochem. 1994;138:85–90. [PubMed] [Google Scholar]
- 39.Grube K, Burkle A. Poly(ADP-ribose) polymerase activity in mononuclear leukocytes of 13 mammalian species correlates with species-specific life span. Proc Natl Acad Sci U S A. 1992;89:11759–63. doi: 10.1073/pnas.89.24.11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, et al. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995;9:509–20. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- 41.Paul S, Amundson SA. Development of gene expression signatures for practical radiation biodosimetry. Int J Radiat Oncol Biol Phys. 2008;71:1236–44. doi: 10.1016/j.ijrobp.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paul S, Amundson SA. Gene expression signatures of radiation exposure in peripheral white blood cells of smokers and non-smokers. Int J Radiat Biol. 2011;87:791–801. doi: 10.3109/09553002.2011.568574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paul S, Barker CA, Turner HC, Mclane A, Wolden SL, Amundson SA. Prediction of in vivo radiation dose status in radiotherapy patients using ex vivo and in vivo gene expression signatures. Radiat Res. 2011;175:257–65. doi: 10.1667/RR2420.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Templin T, Paul S, Amundson SA, Young EF, Barker CA, Wolden SL, et al. Radiation-induced micro-RNA expression changes in peripheral blood cells of radiotherapy patients. Int J Radiat Oncol Biol Phys. 2011;80:549–57. doi: 10.1016/j.ijrobp.2010.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paul S, Ghandhi SA, Weber W, Doyle-Eisele M, Melo D, Guilmette R, et al. Gene expression response of mice after a single dose of 137Cs as an internal emitter. Radiat Res. 2014;182:380–9. doi: 10.1667/RR13466.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paul S, Smilenov LB, Elliston CD, Amundson SA. Radiation dose-rate effects on gene expression in a mouse biodosimetry model. Radiat Res. 2015;184:24–32. doi: 10.1667/RR14044.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Broustas CG, Xu Y, Harken AD, Garty G, Amundson SA. Comparison of gene expression response to neutron and x-ray irradiation using mouse blood. BMC Genomics. 2017;18:2. doi: 10.1186/s12864-016-3436-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paul S, Smilenov LB, Amundson SA. Widespread decreased expression of immune function genes in human peripheral blood following radiation exposure. Radiat Res. 2013;180:575–83. doi: 10.1667/RR13343.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghandhi SA, Weber W, Melo D, Doyle-Eisele M, Chowdhury M, Guilmette R, et al. Effect of 90Sr internal emitter on gene expression in mouse blood. BMC Genomics. 2015;16:586. doi: 10.1186/s12864-015-1774-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghandhi SA, Smilenov LB, Elliston CD, Chowdhury M, Amundson SA. Radiation dose-rate effects on gene expression for human biodosimetry. BMC Med Genomics. 2015;8:22. doi: 10.1186/s12920-015-0097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Broustas CG, Xu Y, Harken AD, Chowdhury M, Garty G, Amundson SA. Impact of neutron exposure on global gene expression in a human peripheral blood model. Radiat Res. 2017;187:433–40. doi: 10.1667/RR0005.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simon R, Lam A, Li M-C, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 2007;2:11–7. [PMC free article] [PubMed] [Google Scholar]
- 53.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 54.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–66. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, et al. PANTHER version 11: expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–9. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 57.Murrain-Hill P, Coleman CN, Hick JL, Redlener I, Weinstock DM, Koerner JF, et al. Medical response to a nuclear detonation: creating a playbook for state and local planners and responders. Disaster Med Public Health Prep. 2011;5:S89–97. doi: 10.1001/dmp.2011.13. [DOI] [PubMed] [Google Scholar]
- 58.Coleman CN, Koerner JF. Biodosimetry: medicine, science, and systems to support the medical decision-maker following a large scale nuclear or radiation incident. Radiat Prot Dosimetry. 2016;172:38–46. doi: 10.1093/rpd/ncw155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barenco M, Brewer D, Papouli E, Tomescu D, Callard R, Stark J, et al. Dissection of a complex transcriptional response using genome-wide transcriptional modelling. Mol Syst Biol. 2009;5:327. doi: 10.1038/msb.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghandhi SA, Sinha A, Markatou M, Amundson SA. Time-series clustering of gene expression in irradiated and bystander fibroblasts: an application of FBPA clustering. BMC Genomics. 2011;12:2. doi: 10.1186/1471-2164-12-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heinonen M, Guipaud O, Milliat F, Buard V, Micheau B, Tarlet G, et al. Detecting time periods of differential gene expression using Gaussian processes: an application to endothelial cells exposed to radiotherapy dose fraction. Bioinformatics. 2015;31:728–35. doi: 10.1093/bioinformatics/btu699. [DOI] [PubMed] [Google Scholar]
- 62.Ghandhi SA, Turner HC, Shuryak I, Dugan GO, Bourland JD, Olson JD, et al. Whole thorax irradiation of non-human primates induces persistent nuclear damage and gene expression changes in peripheral blood cells. PLoS One. 2018;13:e0191402. doi: 10.1371/journal.pone.0191402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen KY, Morley M, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33:422–5. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 64.Correa CR, Cheung VG. Genetic variation in radiation-induced expression phenotypes. Am J Hum Genet. 2004;75:885–90. doi: 10.1086/425221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smirnov DA, Morley M, Shin E, Spielman RS, Cheung VG. Genetic analysis of radiation-induced changes in human gene expression. Nature. 2009;459:587–91. doi: 10.1038/nature07940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Budworth H, Snijders AM, Marchetti F, Mannion B, Bhatnagar S, Kwoh E, et al. DNA repair and cell cycle biomarkers of radiation exposure and inflammation stress in human blood. PLoS One. 2012;7:e48619. doi: 10.1371/journal.pone.0048619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tucker JD, Grever WE, Joiner MC, Konski AA, Thomas RA, Smolinski JM, et al. Gene expression-based detection of radiation exposure in mice after treatment with granulocyte colony-stimulating factor and lipopolysaccharide. Radiat Res. 2012;177:209–19. doi: 10.1667/rr2749.1. [DOI] [PubMed] [Google Scholar]
- 68.Georgakilas AG, Pavlopoulou A, Louka M, Nikitaki Z, Vorgias CE, Bagos PG, et al. Emerging molecular networks common in ionizing radiation, immune and inflammatory responses by employing bioinformatics approaches. Cancer Lett. 2015;368:164–72. doi: 10.1016/j.canlet.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 69.Schaue D, Micewicz ED, Ratikan JA, Xie MW, Cheng G, Mcbride WH. Radiation and inflammation. Semin Radiat Oncol. 2015;25:4–10. doi: 10.1016/j.semradonc.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paul S, Amundson SA. Differential effect of active smoking on gene expression in male and female smokers. J Carcinog Mutagen. 2014;5:198. doi: 10.4172/2157-2518.1000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kannan P, Yu Y, Wankhade S, Tainsky MA. PolyADP-ribose polymerase is a coactivator for AP-2-mediated transcriptional activation. Nucleic Acids Res. 1999;27:866–74. doi: 10.1093/nar/27.3.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meisterernst M, Stelzer G, Roeder RG. Poly(ADP-ribose) polymerase enhances activator-dependent transcription in vitro. Proc Natl Acad Sci U S A. 1997;94:2261–5. doi: 10.1073/pnas.94.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Isabelle M, Moreel X, Gagné JP, Rouleau M, Ethier C, Gagne P, et al. Investigation of PARP-1, PARP-2, and PARG interactomes by affinity-purification mass spectrometry. Proteome Sci. 2010;8:22. doi: 10.1186/1477-5956-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oei SL, Griesenbeck J, Schweiger M, Ziegler M. Regulation of RNA polymerase II-dependent transcription by poly(ADP-ribosyl)ation of transcription factors. J Biol Chem. 1998;273:31644–7. doi: 10.1074/jbc.273.48.31644. [DOI] [PubMed] [Google Scholar]
- 75.Soldatenkov VA, Chasovskikh S, Potaman VN, Trofimova I, Smulson ME, Dritschilo A. Transcriptional repression by binding of poly(ADP-ribose) polymerase to promoter sequences. J Biol Chem. 2002;277:665–70. doi: 10.1074/jbc.M108551200. [DOI] [PubMed] [Google Scholar]
- 76.Smulson ME, Simbulan-Rosenthal CM, Boulares AH, Yakovlev A, Stoica B, Iyer S, et al. Roles of poly(ADP-ribosyl)ation and PARP in apoptosis, DNA repair, genomic stability and functions of p53 and E2F-1. Adv Enzyme Regul. 2000;40:183–215. doi: 10.1016/s0065-2571(99)00024-2. [DOI] [PubMed] [Google Scholar]
- 77.Simbulan-Rosenthal CM, Rosenthal DS, Luo RB, Samara R, Jung M, Dritschilo A, et al. Poly(ADP-ribosyl)ation of p53 in vitro and in vivo modulates binding to its DNA consensus sequence. Neoplasia. 2001;3:179–88. doi: 10.1038/sj.neo.7900155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci U S A. 1997;94:7303–7. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beneke R, Geisen C, Zevnik B, Bauch T, Muller WU, Küpper JH, et al. DNA excision repair and DNA damage-induced apoptosis are linked to Poly(ADP-ribosyl)ation but have different requirements for p53. Mol Cell Biol. 2000;20:6695–703. doi: 10.1128/mcb.20.18.6695-6703.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Valenzuela MT, Guerrero R, Nunez MI, Ruiz De Almodovar JM, Sarker M, De Murcia G, et al. PARP-1 modifies the effectiveness of p53-mediated DNA damage response. Oncogene. 2002;21:1108–16. doi: 10.1038/sj.onc.1205169. [DOI] [PubMed] [Google Scholar]
- 81.Wieler S, Gagne JP, Vaziri H, Poirier GG, Benchimol S. Poly(ADP-ribose) polymerase-1 is a positive regulator of the p53-mediated G1 arrest response following ionizing radiation. J Biol Chem. 2003;278:18914–21. doi: 10.1074/jbc.M211641200. [DOI] [PubMed] [Google Scholar]
- 82.Stilmann M, Hinz M, Arslan SC, Zimmer A, Schreiber V, Scheidereit C. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IkappaB kinase activation. Mol Cell. 2009;36:365–78. doi: 10.1016/j.molcel.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 83.Zerfaoui M, Errami Y, Naura AS, Suzuki Y, Kim H, Ju J, et al. Poly(ADP-ribose) polymerase-1 is a determining factor in Crm1-mediated nuclear export and retention of p65 NF-kappa B upon TLR4 stimulation. J Immunol. 2010;185:1894–902. doi: 10.4049/jimmunol.1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hunter JE, Willmore E, Irving JA, Hostomsky Z, Veuger SJ, Durkacz BW. NF-kappaB mediates radio-sensitization by the PARP-1 inhibitor, AG-014699. Oncogene. 2012;31:251–64. doi: 10.1038/onc.2011.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
File S1. Lists of genes identified as significantly differentially expressed after irradiation. As shown in the first, second and third tables, respectively, compared to controls, in WT mice after 8.8 Gy irradiation, there were 1,370 differentially expressed genes [P < 0.001, false discovery rate (FDR) <5%] and after 6 Gy, there were 1,177; and in Parp1−/− mice after 6 Gy, 1677 genes were differentially expressed.
File S2. A total of 3,009 genes were differentially expressed in at least one dose-genotype condition. Of these, 1,332 were regulated in WT but not Parp1−/− mice, and 847 were regulated in Parp1−/− but not WT mice. The first table shows genes in the clustered order in which they appear in the heatmap in Fig. 1A. The second table provides lists of genes only responding significantly in WT mice (list A) and Parp1−/− mice (list B).
The lists of differentially expressed genes were imported into IPA for prediction of activation or inhibition of regulatory factors potentially upstream of the observed gene expression. The third table is an annotation of Fig. 2.
File S3. BRB-ArrayTools was used to train classifier algorithms using the gene expression data from the WT mice, and then to test the ability of those algorithms to classify the radiation status of the Parp1−/− samples. The resulting classifier was composed of 36 genes (first table). Top upstream regulators of classifier genes, as determined by IPA analysis, are also listed (second table).