SUMMARY
As organisms age, cells accumulate genetic and epigenetic errors that eventually lead to impaired organ function or catastrophic transformation such as cancer. Because aging reflects a stochastic process of increasing disorder, cells in an organ will be individually affected in different ways, thus rendering bulk analyses of postmitotic adult cells difficult to interpret. Here, we directly measure the effects of aging in human tissue by performing single-cell transcriptome analysis of 2,544 human pancreas cells from eight donors spanning six decades of life. We find that islet endocrine cells from older donors display increased levels of transcriptional noise and potential fate drift. By determining the mutational history of individual cells, we uncover a novel mutational signature in healthy aging endocrine cells. Our results demonstrate the feasibility of using single-cell RNA sequencing (RNA-seq) data from primary cells to derive insights into genetic and transcriptional processes that operate on aging human tissue.
In Brief
Aging is associated with increased transcriptional dysregulation and loss of identity at the single-cell level
INTRODUCTION
Aging in higher-order metazoans is the result of a gradual accumulation of cellular damage, which eventually leads to a decline in tissue function and fitness (López-Otín et al., 2013). Because the fundamental processes involved in aging affect single cells in a stochastic manner, they have been difficult to study systematically in primary human tissue. Studies of selected genes in mice indicate that aging postmitotic cells of the heart display a transcriptional instability (Bahar et al., 2006) that is not observed in actively renewing cell populations such as those of the hematopoietic system (Warren et al., 2007). An accumulation of genetic aberrations has been suggested to underlie transcriptional dysregulation by affecting promoter and enhancer elements as well as exonic sequences (Vijg, 2004). However, due to technical constraints, it has previously been difficult to study these processes in human tissue or at the whole transcriptome level. In particular, little is known about the mutational load on post-mitotic cells that cannot be expanded in culture. Studies on CAG repeats in mouse brain (Gonitel et al., 2008) have shown that age-dependent somatic mutation rates in post-mitotic cells might be higher than previously anticipated. Because these mutational processes operate in chronological time rather than number of cell divisions, an analysis of human cells from a large age span rather than from short-lived model organisms is needed. However, such a systematic survey of human tissue from different ages has not been performed.
The pancreas functions both as an endocrine and an exocrine gland and is associated with illnesses such as type II diabetes, that have a considerable age-related disease risk. The exocrine function is mediated by acinar cells producing enzymes for the digestive system, while the endocrine function is mediated by islets of Langerhans, where the major cell types are α-cells, β-cells, δ-cells, and pancreatic polypeptide (PP) cells. Previously, single-cell RNA sequencing (scRNA-seq) on primary tissue has been used to study heterogeneity within cell types and to further refine them—for the pancreas, see Muraro et al. (2016), Segerstolpe et al. (2016), Li et al. (2016), and Wang et al. (2016). However, scRNA-seq also provides an ideal framework to study noisy processes that act on single cells, such as aging. Thus, to overcome the previous technical difficulties in studying cellular aging, we analyzed single human cells from donors of a wide spectrum of ages. Using this approach allows us to detect features of aging that are not coordinated across many cells but rather affect different cells randomly and to quantify them with high precision.
RESULTS
A Comprehensive Survey of Single Pancreatic Cells from Human Donors across Different Ages
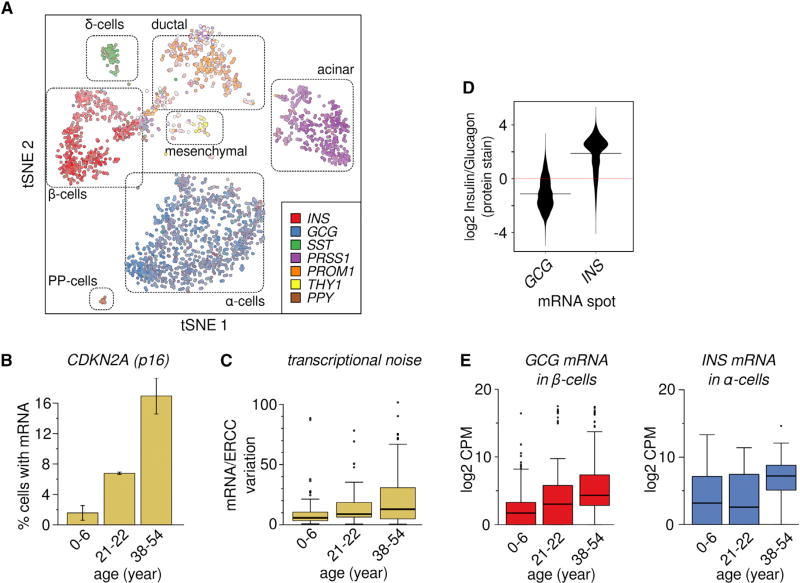
To investigate the effect of physiological aging on pancreatic epithelial cells, we obtained pancreata from eight previously healthy donors operationally defined as juvenile (ages 1 month, 5 years, and 6 years), young adult (ages 21 years and 22 years), and adult/middle aged (ages 38 years, 44 years, and 54 years). Single pancreatic cells were purified by flow cytometry and their mRNA expression analyzed using scRNA-seq (Picelli et al., 2014) with transcript abundance expressed as counts per million (CPM) and the quality of individual cells assessed using an automated quality control pipeline (see STAR Methods for details). Dimensionality reduction analysis (tSNE) of data from all donors led to consistent clustering of different cell types into distinct regions (Figure 1A), indicating an absence of donor- or sequencing-related batch effects.
Figure 1. A Comprehensive Survey of Single Cells Sampled from Human Pancreas across Different Ages.
(A) tSNE plot of 2,544 successful scRNA-seq libraries from eight donors. Each point represents one cell and points are positioned to retain pairwise distances as determined by Pearson correlation of the 500 most highly expressed genes. Cell identity is indicated by marker gene expression.
(B) Fraction of cells that express the aging associated gene CDKN2A (p16) in juvenile (0–6 years), young adult (21–22 years), and middle-aged (38–54 years) donors increases with age (p = 3.1E-3, n = 8, linear regression.) Bars are mean ± SEM (n = 2–3).
(C) Boxplot of transcriptional noise in β-cells, plotted by age group. Higher age is associated with increased whole-transcriptome cell-to-cell variability within cell type (p = 6.67E-9, n = 384). Boxes indicate the middle quartiles, separated by median line. Whiskers indicate last values within 1.5 × the interquartile range for the box.
(D) Violin plots show the ratio of Insulin–Glucagon protein staining at the sites of Insulin (INS, n = 5,801) and Glucagon (GCG, n = 3,254) RNA hybridization spots.
(E) Boxplot of Log2 counts per million (CPM) of cell-atypical glucagon transcript in β cells (left), and insulin transcripts in α-cells (right), in cells from juvenile (0–6 years), young adult (21–22 years) and middle-aged (38–54) donors. Boxes indicate the middle quartiles, separated by median line. Whiskers indicate last values within 1.5 × the interquartile range for the box.
See also Figure S1 and Tables S1, S2, and S3.
Transcriptional Instability and Fate Drift in Cells from Older Donors
The large span of donor ages (≈6 decades), allowed us to assess the effect of organismal aging at the single-cell level. The fraction of cells expressing known markers of organismal aging, such as CDKN2A (p16INK4A), were associated with age (Figure 1B) consistent with prior studies using bulk RNA-seq on larger donor cohorts (Arda et al., 2016; Chen et al., 2011); however, overall we observed only modest systematic age-dependent transcriptional changes for many age-specific genes (Figures S1A and S1B; Tables S2 and S3). From investigations on a small panel of genes in the mouse heart (Bahar et al., 2006), it has previously been suggested that aging is the result of an increase in transcriptional instability rather than a coordinated transcriptional program. To test whether this observation can be generalized to a full transcriptional profile in human tissue, we measured the transcriptional noise within cell types and donors using estimates based on Euclidean distance (Figure S1C) and Pearson correlation as a fraction of technical error (Figure 1C). Both methods indicated increased transcriptional noise in samples from older donors compared to samples from young adults and children, demonstrating age-dependent transcriptional noise (p = 3.01E−11, n = 2,544, for Pearson and p < 1E-16, n = 80,000 for Euclidean, linear regression) without changes in cellular composition (Figure S1D).
A subset of α-cells and β-cells simultaneously expressed both Insulin (INS) and Glucagon (GCG) mRNA—a result that is consistent with prior studies (Blodgett et al., 2015; Xin et al., 2016; Katsuta et al., 2010) and that we verified using in situ RNA staining (Figures 1D and S2). scRNA-seq revealed that the fraction of α- or β-cells co-expressing both Insulin and Glucagon mRNA increased significantly with advancing age (Figure 1E, GCG in β-cells: p = 1.74E-27, n = 348; INS in α-cells: p = 5.38E-10, n = 998, linear regression). As expected, cells with high levels of transcriptional noise also express more cell-atypic hormone (Figure 2A). Thus, increasing numbers of cells with “atypical” hormone mRNA expression is emblematic of age-dependent transcriptional instability, and such “fate drift” suggests a physiological basis for declining endocrine function, in spite of increased hormone secretion, in the aging pancreas (Chang and Halter, 2003; De Tata, 2014).
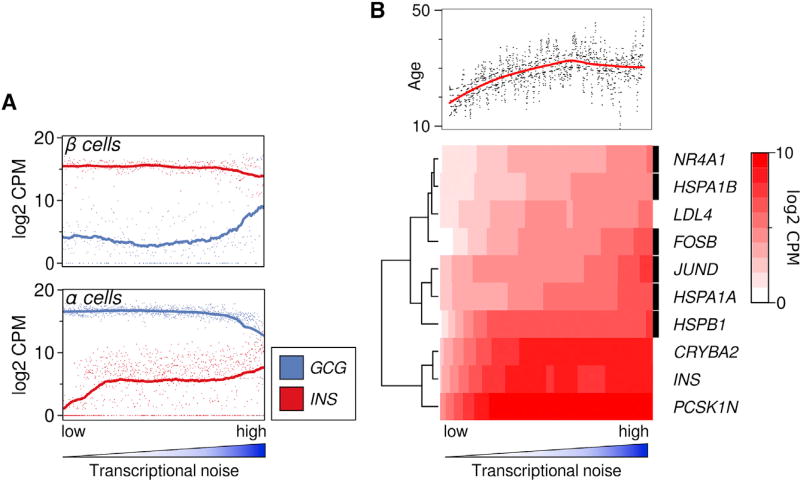
Figure 2. Gene Expression Changes Associated with Transcriptional Noise.
(A) Expression of cell-typical (INS for β-cells, GCG for α-cells) and non-typical hormone in cells, ranked by transcriptional noise. Dots represent individual cells, line is running mean, with k = n/5 (k = 69 for β cells and 199 for α cells).
(B) Organismal age and expression of stress-related genes are strongly associated with transcriptional noise. All genes were tested for association with transcriptional noise (linear rank regression), shown are the top genes by coefficient, with FDR <1E-3. Heatmap shows loess fit. Rows marked with a black box indicate genes that are associated with response to stress (Yu et al., 2015; Daugaard et al., 2007; Paneni et al., 2013; Toone et al., 2001).
We performed linear regression on gene expression levels as a function of noise rank (batch corrected and within cell type) to investigate whether any systematic gene expression differences accompany an increase in transcriptional noise. As shown in Figure 2B, stress response genes such as FOSB, HSPA1A, and JUND were most highly associated with increasing transcriptional noise, supporting an aging paradigm that implicates cellular stress in age-related pathology (Harman, 1965).
Analysis of Single Nucleotide Variants in scRNA-Seq Data Reveals Cell-Type-Specific Somatic Substitutions and Neuronal mRNA Editing
Aging is accompanied by the accumulation of somatic DNA substitutions, and the pattern of somatic substitutions in a cell depends on the mutational processes that cause them. A growing body of data from tumor genomes has uncovered a multitude of such mutational signatures (Alexandrov et al., 2013b; Nik-Zainal et al., 2014, 2016; Kasar and Brown, 2016), many of which can be linked to specific mutational processes. However, these signatures are dominated by processes associated with tumor growth and only 3 out of 21 such signatures have been linked to aging in tumors or organoid cultures of stem cells (Alexandrov et al., 2015; Blokzijl et al., 2016). Post-mitotic cells are especially difficult to study, because they cannot be clonally expanded. Thus, very little is known about the mutational processes that operate on the terminally differentiated cells that make up most of our body. To directly study mutational signatures that are active in healthy tissue, we developed a computational method for determining genetic variation within single cells using scRNA-seq data and validated the method using deep whole-genome sequencing (see STAR Methods). Using this method, we compiled a catalog of putative somatic and constitutional (donor-specific germline) mutations from the 2,544 pancreas cells together with 398 previously published single cells from adult human brain (Darmanis et al., 2015). We also compiled a similar catalog of clonal variation within 73 cells from GP5d colon cancer cells cultured in vitro (Figure 3A). We used synthetic spike-in RNA (ERCC control) as an internal control, which allowed us to sift out technical artifacts, removing 92.6% of these false positive calls (Figure S3C). Further, we used whole genome sequencing data to benchmark our method of separating somatic substitutions from germline variation, with the majority (67.4%) of putative somatic mutations being absent from genomic calls. Somatic substitutions were enriched in untranslated regions of transcripts such as the 3′UTR (p = 1.40E-32, paired t test, n = 73) and also enriched for mutations resulting in codons that do not alter the amino acid sequence (Figures 3B and S5H). As expected, the vast majority of putative somatic substitutions were observed in only one cell each (Figure S3A), indicating that the method is specific to somatic variation. Substitution calls were very rare in low copy-number transcripts and greatly enriched in high copy-number transcripts, while ERCC calls were not (Figures S3C–S3E), precluding the possibility of library preparation artifacts being a major source of substitution calls. Whereas low expressed transcripts often showed allelic imbalance at heterozygous alleles, highly expressed genes did not (Figure S3G), suggesting that the main driver of allelic imbalance was bursty gene expression rather than early cycle PCR errors. Somatic mutation rates exceed the technical error rates due to amplification and sequencing error, as measured by internal spike-in controls of synthetic RNA included in each single-cell experiment (Figure 3A).
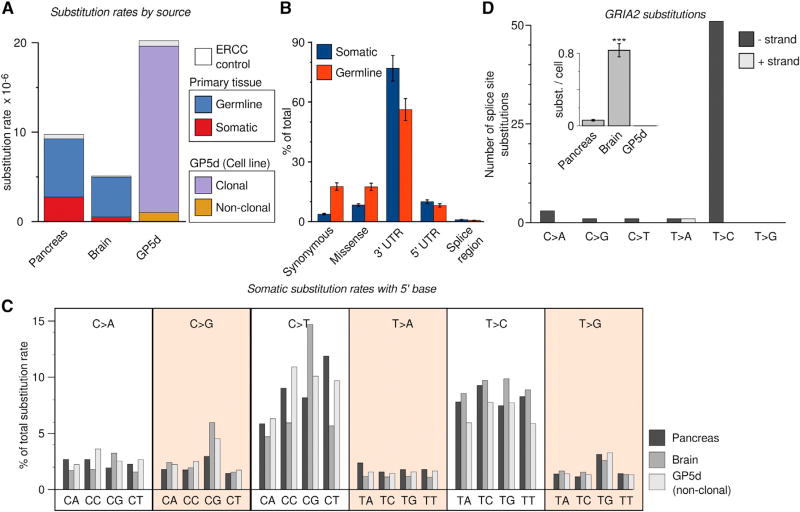
Figure 3. Somatic Mutation Profiles Derived from Single Primary Human Cells.
(A) Substitution rates for each type of substitution in the three datasets. Somatic substitution rates were more than five times as high in pancreas as in brain (2.74 × 10−6 versus 0.52 × 10−6), whereas germline substitution rates were similar between the two. As expected, the rate of clonal substitutions in the tumor cell-line (GP5d) is several fold higher than germline rates in primary tissue.
(B) Somatic substitutions are strongly enriched on untranslated regions compared to germline substitutions. Bars are mean ± SEM, n = 73.
(C) Comparison of relative mutation rates of single-nucleotide substitutions in the context of the nucleotide immediately 5′ of the altered base. Different substitution types are separated by boxes with the substitution type indicated (e.g., C > A: C to A transversion). The relative substitution rate for C > T substitutions within a CpG context, and T > C substitutions is higher in brain than in the other tissues tested (p = 6.38E-61 and p = 1.89E-17, respectively; Wilcoxon test, n = 2,544 for pancreas, n = 73 for gp5d, and n = 332 for brain).
(D) Detecting mRNA editing in brain samples. Shown is the number of splice site substitutions in the GRIA2 gene. T > C substitutions mapping to the transcribed (−) strand, corresponding to adenine substituted for guanine in the transcribed RNA, are highly enriched whereas other substitution types remain at baseline levels. Inlay shows mean number of GRIA2 substitutions per cell for the three datasets, brain is highly enriched in such substitutions (p = 5.40E-19. Bars are mean ± SEM, n = 2,544 for pancreas, n = 3323 for brain, and n = 73 for GP5d).
See also Figure S3.
To investigate patterns of somatic mutations, we determined the rates (substitutions per base pair) of the six possible single nucleotide substitutions in each cell. Single cells from pancreas had a markedly higher overall rate (> 5-fold) of somatic variation compared to brain tissue (Figure 3A), and there were considerable differences also between cell types in the pancreas (Figure S3B), whereas we only observed small fluctuations in the number of substitutions on ERCC control RNA from the same cells (Figure S3C, red bars). However, rates of C > T substitutions in a CpG dinucleotide context, known to deaminate spontaneously when methylated, and T > C substitutions were relatively higher in brain compared to pancreas (Figure 3C), in line with what was previously found for postmitotic brain cells (Lodato et al., 2015). Synthetic control RNA substitution rates were similar between cell types of the pancreas and represent a lower level of technical noise in the measurement. Thus, analyzing the raw sequence reads from scRNA-seq data allows us to determine the mutational history of primary tissues as well as the clonal variation in a tumor cell line.
Because we are analyzing processed mRNA rather than DNA, our method can potentially be used to uncover systematic mRNA editing events in addition to DNA substitutions. mRNA editing is a controlled cellular process found in neuronal lineage cells, where adenosine residues are converted to inosine, resulting in T > C substitutions on the transcribed strand. To determine whether mRNA editing can be detected using our method, we analyzed substitutions in the glutamate receptor GRIA2 gene, which is a well-known target for mRNA editing at splice junctions (Higuchi et al., 1993). This gene is expressed in both endocrine cells and brain cells, making a direct comparison possible. Consistent with mRNA editing being specific to neurons, T > C substitutions in GRIA2 occurred almost exclusively in brain cells. A more precise analysis of the GRIA2 splice sites confirmed this because these sites were highly enriched in T > C substitutions on the transcribed strand (Figure 3D).
Endocrine Cells Display a Specific Mutational Signature Related to Oxidative Stress
To identify the mutational signatures (S1–S3, SC4–SC7) that underlie the observed substitution rates, we used non-negative matrix factorization (NMF) followed by hierarchical clustering (similar to Alexandrov et al. [2013a], see STAR Methods for details) on the substitution rates of single cells (Figures 4A and S4). The NMF analysis also acts as a second filter for false-positive substitution calls by ordering substitutions due to technical artifacts such as PCR errors into their own signatures. Thus, we excluded signatures with a high degree of similarity to the substitution rates of the negative control RNA, lacking cell-type specificity or positive age association, or with a very low signal (excluded signatures SC4–SC7 in Figure 3A, see STAR Methods for details).
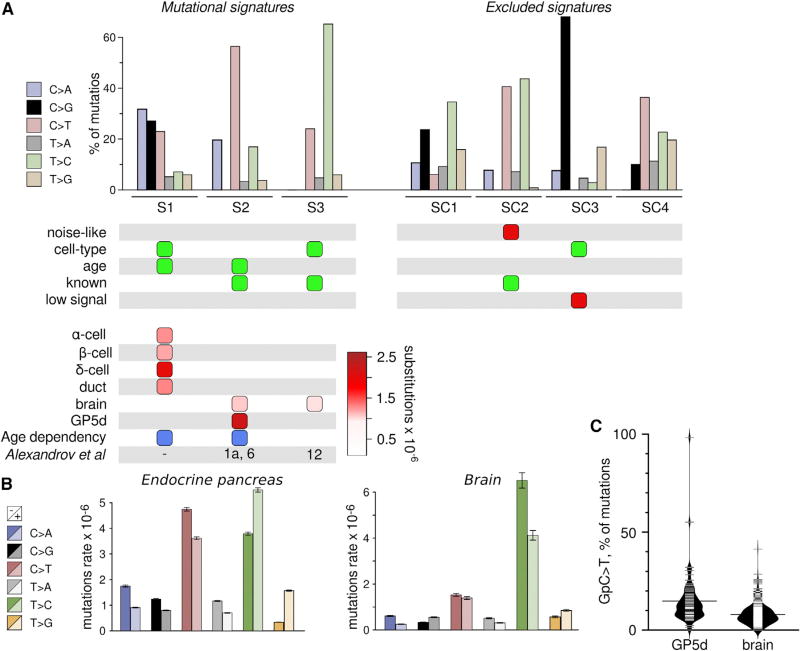
Figure 4. Mutational Signatures Derived from scRNA-Seq Data.
(A) Single-nucleotide substitutions in 3,003 cells from pancreas, brain, and the colon cancer cell line GP5d were organized into mutational signatures using non-negative matrix factorization followed by agglomerative hierarchical clustering. Bar plot illustrates the percent of mutations attributed to each substitution type in each of the three signatures (S1–S3, left) and the four excluded signatures (SC1–SC4, right). Colors as in (A). Panel below the bar plot indicates selection items for determining whether to exclude the signature. Green, cause for inclusion; red, cause for exclusion. Bottom panel denotes the presence of a signature (columns) in a cell type (rows), with color scale indicating strength of signature as median substitution rate for cells of the indicated type. Blue boxes denote significant association between signature load and donor age. Bottom row indicates equivalent signatures from Alexandrov et al. (2013b).
(B) Strand specificity differs between cell types. Mutations were annotated based on whether the mutated pyrimidine occurred on the transcribed (−) or untranscribed (+) strand. Bars represent mean ± SEM of raw substitution counts in endocrine cells (left) and brain cells (right). Note that endocrine cells have a strong strand bias for the transcribed strand for C > A, C > G, and C > T substitutions (p = 1.00E-79, 1.37e-28, and 6.40E-34, respectively; Wilcoxon test, n = 1,429) previously observed in oxidative stress-related tumor signatures, while brain has a bias for T > C substitutions on the transcribed strand (p = 3.41E-11; Wilcoxon test, n = 466) similar to tumor signature 12 (Alexandrov et al., 2013b).
(C) Signature S2 is composed of two sub-signatures corresponding to cancer signatures 1 and 6. Violin plot show C > T substitutions with a preceding G as a fraction of all substitutions in a cell, which is a hallmark of cancer signature 6 and that separates GP5d and brain cells (p = 7.156E-11; Wilcoxon test, n = 73 for GP5d and n = 332 for brain cells).
The S1 signature (high rate of C > A, followed by C > G and C > T substitutions), and S3 signature (highly elevated rate of T > C substitutions), were cell-type-specific signatures, with S1 found in the endocrine pancreas and S3 in the brain. The S2 signature was highly enriched in clonal variation within the mismatch repair-deficient GP5d cell line, with weaker signal in brain. The pancreas-specific signature S1 was characterized by C > A substitutions, with C > G and C > T substitutions at progressively lower rates. C > A and C > G substitutions are attributed to oxidation of the guanine base, creating 8-Oxo-2′-deoxyguanosine (8-Oxo) that mispairs with adenine and can be further oxidized to mispair with guanine (Moriya et al., 1991; Kino and Sugiyama, 2005), whereas C > T substitutions are attributed to oxidation of the cytosine base (Kreutzer and Essigmann, 1998).
Consistent with oxidation of guanosine driving the mutational signature of β cells, 8-hydroxyguanosine levels were markedly elevated in the DNA of β cells compared to non-islet cells, while only modestly elevated in RNA (Figure 5). 8-Oxo substitutions preferentially occur when the guanine is on the non-transcribed strand (Park et al., 2012; Alexandrov et al., 2013b), possibly due to transcription-coupled nuclear excision repair of adducts on the transcribed strand (Banerjee et al., 2011). In order to determine if transcriptional strand bias occurred in our data, we annotated the single-base substitutions with whether the mutated pyrimidine was on the transcribed (−) or untranscribed (+) strand. As expected, C > A and C > G substitutions had a strong preference to occur on the transcribed strand in endocrine cells, but not in brain cells, consistent with guanine oxidation driving signature S1 (Figure 4B). Taken together, signature S1 appears to be a novel, strand-specific mutational signature that is enriched in transcribed genes and that bears the hallmarks of oxidative damage.
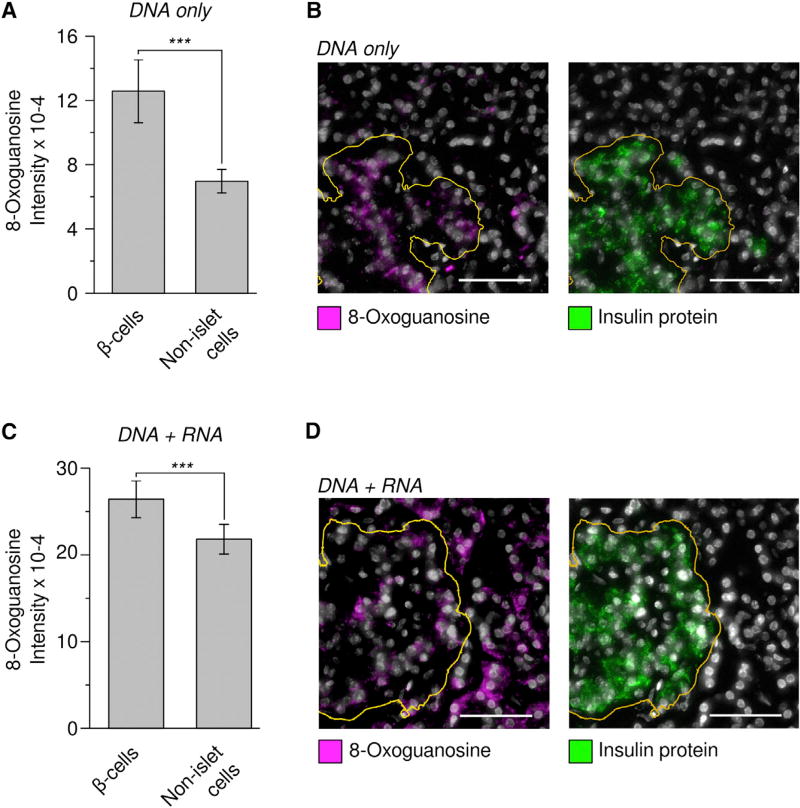
Figure 5. The Genomic DNA in Pancreatic Islets Are Highly Enriched in Oxidized Guanine.
(A) Pancreatic β-cell DNA is enriched in oxidized guanosine. Nuclear staining intensity of anti 8-Oxoguanosine antibody was quantified for INS-positive or INS-negative cells, from the same images. Slides were treated with RNase so as to only measure oxidized bases on DNA. Bar plot indicates mean ± SEM (p = 7.30E-57; Wilcoxon test, n = 769 β-cells, 10,713 non-islet cells.).
(B) Left: representative micrograph with 8-Oxoguanosine in magenta and nuclear stain (DAPI) in gray (scale bar, 50 µm). Right: insulin protein staining of the same region. Insulin-positive islet cell mass is at bottom left, boundary indicated with orange line.
(C) Pancreatic β-cell RNA is marginally enriched in oxidized guanosine. Cytoplasmic staining intensity of anti 8-Oxoguanosine antibody was quantified for INS-positive β cells and INS-negative cells from the same slides. Bar plot indicates mean ± SEM (p = 9.5E-22, 1,239 β-cells, 21,048 surrounding cells).
(D) Left: representative micrograph with 8-Oxoguanosine in magenta and nuclear stain (DAPI) in gray. Right: insulin protein staining of the same region. INS-positive islet cell mass boundary indicated with orange line. Scale bar, 50 µm.
See also Figure S5.
Previous large-scale efforts to decipher cancer-specific mutational signatures in bulk tumor genomes (Alexandrov et al., 2013b) discovered 21 unique signatures based on the substitution type and the surrounding two bases. We reasoned that our signatures might have been also detected in the tumor data and compared the signatures by collapsing their probabilities into single-base substitution probabilities. Signature S3 found in this study was very similar to tumor signature 12 from Alexandrov et al. (2013b) (Figure S5D, Pearson correlation 0.971), and the characteristic T > C substitutions in brain display a similar degree of strand specificity to tumor signature 12 (Figure 4B). Signature S2 was almost identical to both the age-dependent tumor signature 1 and the mismatch repair-associated tumor signature 6 (Figure S5D, Pearson correlation 0.975 and 0.987, respectively). The major distinguishing feature between the two tumor signatures is the rate of C > T substitutions within a GpC context. As shown in Figure 4B, this distinguishing feature clearly separates the two tissues in our data, suggesting that non-clonal substitutions in GP5d mainly stem from faulty mismatch repair, whereas somatic substitutions in brain are caused by the same age-dependent process as tumor signature 1.
Interestingly, tumor signature 5, which is of unknown etiology and is found at low levels in all tumor types, is highly reminiscent of our false positive signature (Figure S5D, Pearson correlation 0.990)—suggesting that it is either a product of false-positive calls in the tumor datasets or caused by a mechanism shared between human replication and enzymes used for nucleic acid amplification. None of the 21 tumor signatures found to date is directly related to endogenous oxidative stress, and the endocrine signature S1 has no direct counterpart among the tumor signatures.
The strongest correlation was to tumor signature 3 (Pearson correlation 0.769), which has been found in pancreatic, breast, and ovarian cancers, followed by signature 24 (Pearson correlation 0.756), which is found in cancers resulting from aflatoxin exposure via oxidative stress-induced DNA damage. However, signature S1 only bears a passing resemblance to these two, and further investigation into mutational signatures of healthy tissues will be needed to elucidate whether signature S1 is emblematic of mainly post mitotic cells with high rate of metabolism, which rarely form tumors, or if it is specific to endocrine pancreatic cells.
Mutational Load of Signature S1 Is Higher in Endocrine Cells from Older Donors and Correlate with Induction of Protein Synthesis-Related Genes
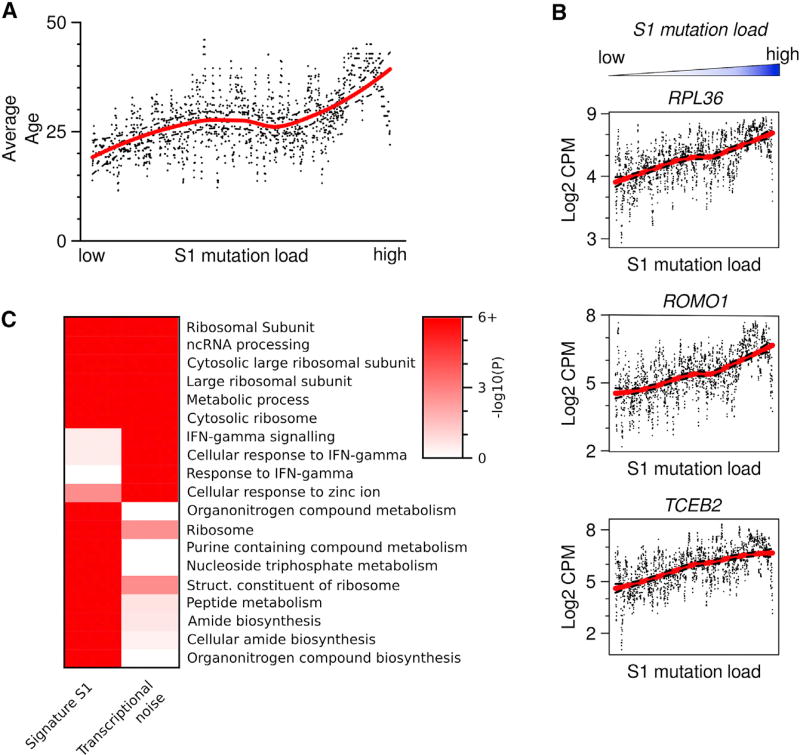
Ranking of cells by signature-specific mutational load indicated that signatures S1 and S2 were highly correlated with age, with S1 showing the highest significance (p = 5.95E-23, Figures 6A and S5). Signature S2 showed none or little effect on gene expression—only 45 genes were significantly affected with false discovery rate (FDR) <1E-3, none of which were upregulated. PON2 (a membrane protein with a putative antioxidant activity) and EGR1 displayed the highest upregulation associated with mutational load of the age-dependent S2 (at FDR <0.05) (Figure S5; Table S6). Signature S1, on the other hand, was associated with a considerable transcriptional effect (1,595 genes at FDR <1E-3). The genes most highly associated with high S1 load were involved in transcription (TCEB2), protein synthesis (RPL36), and modulation of ROS (ROMO1) (Figure 6B, see also Table S5 for an expanded list).
Figure 6. Transcriptional Correlates of Mutational Signatures.
Endocrine pancreas cells were ordered according to the fraction of mutations attributed to Signature S1.
(A) Average age is higher in cells with high S1 load (p = 5.95E-23, linear rank regression). Points are running mean, k = 10, and line is Loess fit, dotted lines indicate ± 0.999 confidence interval.
(B) Each gene was tested for association with signature S1 (linear rank regression), shown are the top genes by coefficient, with p < 1E-15 (FDR corrected). Points are individual mRNA measurements, line loess fit as in (A).
(C) Comparison of the top ten gene ontology (GO) categories positively correlated with signature S1 and transcriptional noise. Categories related to protein production, such as ribosomal proteins, recur in both. Color scale indicates FDR-adjusted p value, winsorized at 10−6.
See also Table S5.
Gene set enrichment analysis (Subramanian et al., 2005) indicated that pathways involved in protein synthesis were altered in both cells with high S1 load and cells with high transcriptional noise (Figure 6C). Further, signature S1 correlated with higher abundance of the tumor suppressor CDKN2A (p16) (Figure S4D, p = 0.024, n = 1,425, linear regression), a correlation that was not observed between transcriptional noise and CDKN2A expression (Figure S4C, p = 0.17, n = 1,425, linear regression) and that suggests that even low levels of mutational load might activate the cell’s tumor suppressive response.
DISCUSSION
Cellular aging in long-lived organisms appears to be a complex stochastic process of gradual accumulation of errors (López-Otín et al., 2013). Using single-cell data, we find that aging is accompanied by both increased transcriptional noise and an accumulation of genetic errors. It has been previously suggested that DNA substitutions have a direct causative role in transcriptional instability (Vijg, 2004). However, as shown in this work and by others (Lodato et al., 2015), the mutational burden in single cells is on the order of one to a few thousand substitutions genome-wide and is unlikely to affect the expression of a large enough number of genes or regulatory elements to have an impact on overall transcriptional noise. If there were a causal link between mutational load and transcriptional noise, we would expect the correlation between these two features to be considerably stronger than a correlation of either feature with organismal age. By contrast, we would expect similar correlations between all three of these features if mutational load and transcriptional noise were independently acquired with age. Our data support the absence of a causal link between mutational load and transcriptional noise. In fact, the correlation of either transcriptional noise or signature S1 with age was slightly stronger than the correlation between mutational load and transcriptional noise (age–noise: p = 2.94E-11, age–S1: p = 5.29E-16, noise–S1: p = 4.83E-11. Two-sided Pearson correlation test, n = 1,429). Thus, our single-cell approach seems to suggest that aging is characterized by a gradual accumulation of both epigenetic and genetic errors in a stochastic and independent fashion.
Importantly, the accrual of epigenetic errors is likely to cause a drift in cell fate, as suggested by an increase in non-cell-type-specific hormone expression in endocrine cells. Such “fate drift” could help explain the decrease in fitness and organ function associated with aging. In addition to identifying age-dependent mutational signatures and transcriptional noise, our findings refined previous results on age-dependent increase in CDKN2A gene expression. We identified CDKN2A expression in a higher fraction of cells in pancreata from older donors, rather than an increase of transcript abundance in every cell. Such cellular heterogeneity suggests that the previously observed age-dependent changes in CDKN2A expression (Arda et al., 2016) are due to events affecting a subset of cells rather than an intrinsic program dictating cellular aging.
Age-dependent decline in function and regenerative potential has been attributed partially to the activity of reactive oxygen species produced by cellular metabolism (Harman 1965). The age-dependent mutational signature in the endocrine pancreas is characterized by a high rate of C > A and C > G substitutions, which are selectively induced by reactive oxygen species (Figure S5E) (Kino and Sugiyama, 2001, 2005; Kamiya et al., 2009). Pancreatic islet cells are sensitive to reactive oxygen species due to low expression of antioxidant enzymes such as SOD1 (Tiedge et al., 1997), a relatively high rate of ATP-dependent processes such as protein production and secretion, and the requirements for reducing power to keep insulin disulfide bonded. Our results thus suggest that the age-specific mutational signature observed in the endocrine pancreas is due to ROS-dependent lesions on DNA. Interestingly, oxidative damage is part of the pathology of type II diabetes, and plasma 8-hydroxyguanosine is a good correlate to endocrine dysfunction (Shin et al., 2001).
Current methods used to study somatic mutations rely either on single-cell genomic sequencing or on sequencing DNA from many cells that stem from a clone that has been expanded in vitro (Blokzijl et al., 2016; Lodato et al., 2015; Gawad et al., 2016). Both families of methods are very costly, precluding large-scale experiments on thousands of cells, and analysis of a specific cell type requires pre-selection of the cells because the information on cell identity provided by mRNA-sequencing is lost. Our methods for determining transcriptional noise and for identifying mutational signatures from scRNA-seq data provide a means to study these features in arbitrarily specific cell populations from primary tissue, irrespective of the replicative potential of the cells. Such methods applied to much larger donor cohorts, and different tissue types could be a crucial tool for understanding aging and other stochastic processes that act on single cells.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
CONTACT FOR REAGENT AND RESOURCE SHARING
EXPERIMENTAL MODELS AND SUBJECT DETAILS
- METHOD DETAILS
-
◦Flow Cytometry
-
◦Single-Cell RNA-Seq
-
◦Genomic sequencing
-
◦In situ RNA and protein staining
-
◦
- QUANTIFICATION AND STATISTICAL ANALYSIS
-
◦Number of replicates used
-
◦Single-cell RNA-seq Data Analysis
-
◦Somatic mutational signatures in single-cell RNA-seq data
-
◦Estimation of transcriptional noise
-
◦
DATA AND SOFTWARE AVAILABILITY
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Stephen R. Quake (quake@stanford.edu).
EXPERIMENTAL MODELS AND SUBJECT DETAILS
All studies involving human pancreas or islets were conducted in accordance with Stanford University Institutional Review Board guidelines, including informed consent for tissue donation from all subjects. De-identified human pancreata or islets were obtained from previously healthy, non-diabetic organ donors with BMI < 30, less than 15 hr of cold ischemia time, and deceased due to acute trauma or anoxia. Organs and islets were procured through Integrated Islet Distribution Network (IIDP), National Diabetes Research Institute (NDRI), UCSF Islet Isolation Core (San Francisco, CA USA) and International Institute for the Advancement of Medicine (IIAM). For FACS, scRNA-seq studies islets from three juvenile (ages 1 month-old, 5, 6), and five adult donors (ages 21, 22, 38, 44, 54 years) were used. For immunostaining studies pancreatic tissue sections from a 31-year-old donor were used.
Tissue from both male and female donors were used, an analysis of systematic influence of sex on the results is included in Figure S1B. Subjects were not involved in previous studies. Further donor details are provided in Table S1.
Verified GP5d cells (colon adenocarcinoma from human female Caucasian) were obtained from Sigma-Aldrich (95090715), and only first-passage cells were used in this study.
METHOD DETAILS
Flow Cytometry
Isolated human islets were dissociated into single cells by enzymatic digestion using Accumax (Invitrogen). Prior to antibody staining, cells were incubated with blocking solution containing FACS buffer (2% v/v fetal bovine serum in PBS and goat IgG [Jackson Labs], 11.2 µg per million cells). LIVE/DEAD Fixable Aqua Dead Cell Dye (Life Technologies) was used as a viability marker. Cells were then stained with appropriate antibodies at 1:100 (v/v) final concentration. The following antibodies were used for FACS experiments: HPx1-Dylight 488 (Novus, NBP1-18951G), HPi2-Dylight 650 (Novus, NBP1-18946C), CD133/1 - Biotin (Miltenyi Biotec 130-090-664), CD133/2 - Biotin (Miltenyi Biotec 130-090-852), streptavidin-eFluor780 (eBioscience, 47-4317-82), streptavidin-APC (eBioscience, 17-4317-82), anti human EpCAM-APC (Biolegend, 324208). Cells were sorted on a special order 5-laser FACS Aria II (BD Biosciences) using a 100 m nozzle following doublet removal. Sorted single cells were collected directly into 96-well plates (Bio-Rad cat #: HSP9601) containing 4 µL of lysis buffer with dNTPs (Picelli et al., 2014) for downstream single-cell RNA-seq assays.
Single-Cell RNA-Seq
Single-cell RNA-seq libraries were generated as described (Picelli et al., 2014). Single-cells collected in 96-well plates were lysed, followed by reverse transcription with template-switch using an LNA-modified template switch oligo to generate cDNA. After 21 cycles of pre-amplification, DNA was purified and analyzed on an automated Fragment Analyzer (Advanced Analytical). Each cell’s cDNA fragment profile was individually inspected and only wells with successful amplification products (concentration higher than 0.06 ng/ul) and with no detectable RNA degradation were selected for final library preparation. Tagmentation assays and barcoded sequencing libraries were prepared using Nextera XT kit (Illumina) according to the manufacturer’s instructions. Barcoded libraries were pooled and subjected to 75 bp paired-end sequencing on the Illumina NextSeq instrument.
Genomic sequencing
Genomic variants were determined from whole genome sequencing data following GATK Best Practices (Van der Auwera et al., 2013). Adapters and low quality bases were trimmed using cutadapt v1.9 (Van der Auwera et al., 2013; Martin, 2011). Reads were aligned to hg19 using BWA-MEM 0.7.12 (Li and Durbin, 2010). Duplicates were removed using Picard tools v1.119 followed by indel realignment and base recalibration using GATK v3.5 (McKenna et al., 2010). Variants were called using haplotype caller and recalibrated using VQSR. Default software parameters were used and reference files downloaded from the GATK Resource Bundle 2.8/hg19.
In situ RNA and protein staining
Multiplex RNA staining was performed on 10 µm thick, formalin-fixed, tissue sections using barcoded transcript-specific padlock probes and rolling circle amplification (RCA) as described before (Ke et al., 2013). The primer sequences were
GCG: G+TC+TC+TC+AA+AT+TC+ATCGTGACGTTT
INS: G+CA+CC+AG+GGC+CCC+CGCCCAGCTCCA
Padlock probes
-
GCG: Phosp-GAATAACATTGCCAAACGTGTGTCTATTTAGTGGATCCCGTGCG
CCTGGTAGCAATTAGCTCCACTGTTACTAGATTGGAATACCAAGAGGAACAG
-
INS: Phosp-AGGTGGGGCAGGTGGAGCCTCAATGCTGCTGCTGTACTCTACG
ATTTTACCAGTTGCCCTAGATGTTCCGCTATTGTCCGGGAGGCAGAGGACCTGC
Detection probes
DO_1_FITC: AGUCGGAAGUACTACTCUCT_FITC
DO_1_Cy3: CCUCAATGCUGCTGCTGUAC_Cy3
DO_1_Cy5: TGUGTCTATUTAGTGGAUCC_Cy5
DO_2_FITC: CGUGCGCCUGGTAGCAAUTA_FITC
DO_2_Cy3: AGUAGCCGUGACTATCGUCT_Cy3
DO_2_Cy5: TCUACGATUTTACCAGTUGC_Cy5
DO_3_FITC: CCUAGATGTUCCGCTATUGT_FITC
DO_3_Cy3: GCUCCACTGUTACTAGAUTG_Cy3
DO_3_Cy5: CTUGTGCTGUATGATCGUCC_Cy5
The RCA products were stained by sequential hybridization of three uracil-containing fluorescent oligonucleotides following a modified protocol from Ke 2013 (Ke et al., 2013). The three reported probes were mixed 0.1 mM each with hybridization buffer (20% formamide in 2× SSC) and incubated with the tissue at 37°C for 30’. After incubation, tissue section was washed in PBS 5′ and nuclei were counterstained with DAPI 300nM in PBS at room temperature for 15’. The tissue was washed in ethanol 70, 85 and 100% 5′ each, air-dried and mounted in Antifade gold (Invitrogen) before imaging. After imaging, the fluorescent probes were removed by digestion with 0.02 U/µl UNG (Thermo) in UNG buffer and 0.2µg/µl BSA at 37°C for 30’ followed by two washes in 65% formamide pre-warmed at 55°C. Consecutive staining of the RCA products were performed, in the same way, with different set of fluorescent probes.
After RNA, immunofluorescent staining was done on the same tissue section. The tissue was washed twice in PBS with 0.025% Triton X-100 at room temperature and blocked with 1%BSA in PBS for 2 hr at room temperature. Antibodies against human Insulin (DAKO, A0564, guinea pig) and glucagon (Sigma, G 2654, mouse) were diluted 1% in PBS containing 1% BSA and applied to the tissue and incubated at 4°C overnight. The tissue was washed twice in PBS with 0.025% Triton X-100 before incubation with 1% anti-guinea pig GFP labeled and anti-mouse Cy5 secondary antibody, 1% BSA in hybridization buffer for 1 hr at room temperature. Cy3-labeled RCA reporter probes were also added at 0.1 µM concentration to stain all the RCA products and used to align immunofluorescence images to previous RNA staining. After incubation in secondary antibody the section was washed 3 times in 1×PBS at room temperature before mounting in Antifade gold and imaging. For 8-hydroxyguanosine staining, 8-oxo-dG Ab (MyBioSource, MBS606843, mouse) was used, which binds to the oxidized based both in DNA and RNA. To measure the levels of oxidized genomic guanine, cells were treated with RNaseA before staining according to the protocol provided by the manufacturer. Briefly, sections were incubated in PBS buffer containing 500 µg/ml RNaseA (ThermoFisher), 150 mM NaCl and 15 mM sodium citrate for 1 hr at 37°C. After washing the sample twice in PBS the DNA was denatured by incubating with HCl 2N for 5′ at room temperature and then neutralized by incubation with Tris-base 5′ at room temperature followed by two washes in PBS. Blocking and antibody staining against human insulin and 8-Hydroxy-2′-deoxyguanosine was performed as described before (anti 8-oxo-dG was used at 1:250 dilution).
Multidimensional imaging was done with a Zeiss Axioplan epifluorescence microscope equipped with filter-cubes for DAPI, FITC, Cy3 and Cy5, a Axiocam 506 mono camera (Zeiss), automated filter-cube wheel and a motorized stage. Z stacks of 15 images were acquired with a Plan-Apochromat 63× objective and check objective) several field of view of each region of interest were projected (maximum intensity projection) and automatically stitched using the Axiovision software (Zeiss).
Images were exported as single-channel 16-bit grayscale and analyzed as described before (Ke et al., 2013). Briefly, single channels images from staining cycle one were combined and used as mask to align images from subsequent cycles based on nuclei and RCA staining. Image alignment was done using MultiStackReg module of ImageJ (version 1.50e). Pre-aligned RNA images were analyzed with CellProfiler 2.1.1 (rev 6c2d896) and intensity and position of RCA products were measured using the same pipeline as in Mignardi et al. (2015). The barcode decoding was obtained using the same MATLAB script as described before (Ke et al., 2013). Lowering the quality threshold to zero (Qt = 0) allowed us to increase sensitivity of detection while the fraction of insulin and glucagone signals detected outside the islets (false positives) was still negligible (less than 0.3% of all GCG and INS signals). Object-based measurement of immunostaining intensity was done with CellProfiler on the corresponding images using the identified RCA products as mask.
QUANTIFICATION AND STATISTICAL ANALYSIS
Number of replicates used
The number of biological and/or technical replicates for each experiment is stated in the “Method Details” section and the figure legends.
Single-cell RNA-seq Data Analysis
Sequencing reads were trimmed, adaptor sequences removed and the reads aligned to the hg19 reference assembly using STAR (Dobin et al., 2013) with default parameters. Duplicate reads were removed using picard (McKenna et al., 2010). Raw transcript counts were obtained using HT-Seq (Anders et al., 2014) and hg19 UCSC exon/transcript annotations. Transcript counts were normalized into log transformed counts per million (CPM), by applying the formula log2(cij * 1 000 000 / tcj + 1, where cij is the transcript counts for gene i in cell j, and tcj is the total number of transcript counts for cell j. Single cell profiles with the following features were deemed to be of poor quality and removed: 1) cells with less than 100.000 total number of valid counts on exonic regions. 2) cells with very low actin CPM. To determine a cutoff for actin CPM, we used the normal distribution with empirical mean and standard deviation from actin. The cutoff was set to the 0.01 quantile (e.g., the lower 0.01% of the bell curve).
Pairwise distances between cells were estimated using pearson correlation on the 500 most highly expressed genes (by CPM) in any one cell. Dimensionality reduction of the pairwise correlation matrix was performed using the t-SNE method (van der Maaten and Hinton, 2008).
To determine Gene Ontology categories that were associated with transcriptional noise or signature specific mutational load, we used Gene Set Enrichment Analysis (GSEA), using the coefficients of association to noise/rank of significantly altered genes (p < 1E-5, linear model, FDR corrected). Coefficients were used as a preranked list in the GSEA software using default parameters with the gene set database “c5.all.v5.2.symbols.gmt,” which includes all GO categories. Statistical overrepresentation of gene sets was performed using the PANTHER overrepresentation test (pantherdb.org) using the full GO biological process categorization.
Somatic mutational signatures in single-cell RNA-seq data
To explore mutational signatures in single postmitotic cells, we analyzed the raw sequence reads from mRNA-seq. Previously, mutational signatures have been successfully extracted from exome sequencing; however, using single-cell data poses a number of additional challenges. First, we need to deal with the higher error rate associated with reverse transcription and a higher number of PCR cycles. We do this in two ways - by including positive and negative internal controls for each cell, that are used to derive a meaningful cutoff when calling substitutions, and by performing an additional post-selection of signatures, discarding potential false-positives. Second, the sequence space in a single-cell RNA-seq experiment is typically fairly limited, even compared to exome sequencing. We mitigate this issue by sequencing long reads (75 bp paired-end), and by sequencing deeper than typically needed for scRNA-seq (approx. 1M mapped reads per cell). Further, we calculate substitution rates based on the actual number of sequenced kmers in each cell, to account for differences in base distribution. Finally, the limited number of substitutions in each cell means that the sequence context cannot be reliably included in all cases, which is why we generally restricted ourselves to analyzing single-base substitutions.
Raw variation calls were made using the Haplotype Caller (GATK pipeline) (McKenna et al., 2010; Van der Auwera et al., 2013) on the BAM files after applying SplitNCigarReads to remove overhangs into intronic regions. Variants were filtered to remove clusters (> 3 SNPs within 35 bases), as well as variants with QD < 2.0 and FS > 30.0. Germline mutations were called using a merged set of all single-cell profiles from each patient. Subsequently, we filtered the raw variation calls by applying variant quality score recalibration using the GATK pipeline. To reliably call substitutions we need internal controls for each cell, corresponding to a true-positive and true-negative set. We used known variants (dbSNP release 138) from our germline calls that mapped to transcribed regions of the genome as a true positive set (phred-scaled prior: 15.0) and variants that map to ERCC control reads as a false positive set (ERCC controls are synthetic RNA sequences and therefore devoid of systematic variation). To filter somatic substitutions, a strict cutoff, allowing 10% false negative rate was used. Variants also found in the germline were flagged as germline mutations and not used for somatic signatures. In all subsequent analysis, only single-nucleotide substitutions were considered.
For each cell, we extracted the genomic context of each mutation and created a catalog of the frequency of mutation types. We then divided these frequencies with the kmer counts derived from fastq sequences for the cell to obtain the final substitution rates. Negative control ERCC sequences were processed in parallel, to give accurate substitution rates that reflect the different sequence background. Substitution rates in these ERCC samples were 4.8E-7. Assuming that false-positive substitutions stem exclusively from somatic calls (e.g., that the germline calls are completely devoid of false positives), this result indicates a false discovery rate of 15.05% for somatic substitutions (excluding transcriptional errors, which are not accounted for by the ERCC controls). Thus, we estimate that the upper bound of our false discovery rate is 15%. To further validate our method we performed 25× whole genome sequencing (WGS) of GP5d and compared the overlapping substitution calls from single-cell mRNA seq and bulk genomic sequencing. A total of 151,030 genomic positions were determined to have single-base substitutions from the reference genome based on mRNA-seq. Out of these 151,030 substitution calls, 105,673 were also found in WGS and 105,543 were identical (concordant). 45 357 substitutions, or 30.0% of total, were not found in WGS calls; these calls include somatic substitutions, false negative calls from WGS and technical errors. These numbers are in line with the previously determined false-positive rate (≤ 15%), and somatic substitution rates on highly transcribed DNA (~15%, see below for discussion).
It would be of interest to estimate the absolute number of somatic substitutions in the different tissues. On average, we find that 73.5% of our raw substitutions calls are called as germ-line with the rest consisting mainly of somatic substitutions, false-positive calls and germline substitutions that were erroneously called somatic. Based on the ERCC error rate and NMF filterning, we estimate the non-germline error rate to be 7%–15%, and based on WGS sequencing the rate of germline substitutions erroneously called somatic is 32.6%. Thus, the final number of somatic substitutions in our mRNA data is approximately 15%, which, if extrapolated linearly, would still indicate a total number of somatic substitutions significantly higher than even the mutational burden of many tumors. However, we have to take into account that we can only call substitutions in highly expressed genes. Coding regions are depleted in germ-line mutations because of negative selection against non-silent mutations. In our GP5d WGS data, for example, we observe one substitution from the hg19 reference genome per 510 bp genome-wide, but only one per 886 bp in exonic sequences. However, the transcribed genome generally has a considerably higher substitution rate than the non-transcribed genome with increases of between ~2-fold and 50-fold reported depending on the cell types/species and the level of transcriptional activity (Subramanian and Kumar, 2003; Alexander et al., 2013). This bias is so strong that it is detectable using mRNA-seq data alone – the sensitivity to detect somatic substitutions is significantly more dependent on gene expression levels than the sensitivity to detect germline substitutions is (p < 1E-16, linear model n = 316234), even though the sensitivity to call both types is highly dependent on expression levels. Because of this intrinsic limitation of the method, we avoid absolute quantification of substitution rates and limit ourselves to relative quantification between samples. DNA-sequencing of brain single brain cells indicated that neurons contain between 1458 and 1580 somatic single nucleotide variants, which were mostly acquired during active transcription in post-mitotic cells (Lodato et al., 2015), similarly to what we find for endocrine pancreas cells. The somatic substitution rate in our endocrine pancreas cells was 5.2-fold higher than the rate in our brain data (2.74E-6 and 0.52E-6 substitutions per base, respectively), which would indicate a somatic mutational load of between 7582 and 8216 substitutions per genome in endocrine pancreatic cells, given that the association with active transcription is similar between the two mutational processes.
As described above, classification of substitutions as either germline or somatic is done based on scRNA-seq data merged over all cells from a donor. Because of the sparsity of the data, some germline substitutions will appear to be somatic (e.g., be called in a single cell, but not in the merged data). To determine how well our method identifies somatic substitutions, we used germline substitutions called from bulk WGS of GP5d colon cancer cells as a gold standard. This analysis indicated that 32.6% of the putative somatic substitutions were actually germline SNPs.
Thus, we estimate the overall false discovery rate for somatic substitutions in our data (before applying nonnegative matrix factorization and signature selection) to be approximately 40%, which includes ~30% that represent real variation stemming from germline rather than somatic events and ~10% substitution calls that were erroneously called due to technical errors such as PCR or sequencing artifacts. This should be compared to previous single-cell DNA-sequencing approaches, where the error rate is around 20%–30% (Lodato et al., 2015).
To further explore structure within the somatic substitution calls, we examined the effect of substitutions on protein sequence. Because of the degeneracy of the exon code, a fraction of exonic substitutions will give rise to a DNA sequence which codes for the same amino acid sequence. Such synonymous (or silent) substitutions are enriched in germline SNPs, and given that a subset of amino acid substitutions will negatively affect fitness of the cells, we would expect some enrichment of synonymous substitutions also among somatic substitutions. Also, we would expect this enrichment to be similar in different cell types, irrespective of the mutational load. Substitution calls due to technical errors, however, will not be enriched in silent substitutions. We annotated the substitution calls based on genomic notation (hg19), and calculated the fraction of calls that result in a codon for the same amino acid. As a comparison, we calculated the fraction of synonymous substitutions based on random DNA mutation. The average fraction of synonymous substitutions was 40% higher than expected by random chance (0.32 in pancreas compared to 0.23 expected by random, p = 3.34E-125, Wilcoxon test. Figure S5H). Importantly, this number did not correlate with mutational load; cells with higher number of mutations in fact had a somewhat increased fraction of synonymous substitutions (Slope = 3.25E-5, p = 0.08, linear regression), and pancreas cells had almost identical fraction of silent mutations compared to brain even though the substitution rate was 5-fold higher in pancreas (Figure S5I). Thus, the differences in substitution rates likely reflect genetic alterations in the cells, rather than technical error.
To decipher the underlying mutational signatures, we applied non-negative matrix factorization using the NMF R package (Gaujoux and Seoighe, 2010) to the substitution rates of single-nucleotide substitutions (e.g., the mean of the rates for a substitution type over all contexts) for each cell type separately. The highest scoring solution out of 10000 independent runs of the algorithm was used for the final result. The number of possible signatures (5) was chosen to be higher than the number of unique signatures actually found by the algorithm, and duplicate signatures were merged together. We applied hierarchical clustering on the full set of mutational signatures (“basis matrices”) to identify distinct mutational signatures (Figure S4A). Finally, we selected signatures based on five criteria (summarized below and in Figure 4A). To find the signatures that likely represent cell type specific processes that were active in the healthy cell during the donor’s lifetime, we determined cell type specificity and age dependence of each signature. Also, because of the relatively high level of noise in the data, a signature might represent errors that arose systematically during reverse transcription. Thus, to arrive at the final three signatures (S1–S3), removed mutational signatures with a high degree of similarity to the substitution rates of the negative control RNA, with no cell-type specificity, positive age dependence, or with a very low signal. We also determined the similarity of the signatures to the COSMIC tumor signatures (Alexandrov et al., 2013b). Figure 4A, bottom panel, summarizes the association of signatures with these traits. It should be noted that we cannot formally rule out the possibility that the excluded signatures were due to a cell-type specific process active during the lifetime of the donor. Further investigation on much larger panels of tissues will be needed to determine the origin of these signatures.
Figure 4A show the geometric median signature of each cluster. Mutational load of a signature on a cell was determined as the fraction of somatic substitutions of that cell attributed to the signature in question. To obtain a signature load ranking, cells were ordered according to the fraction of mutations that are attributed to a specific signature. Statistical significant association was determined using linear regression.
Estimation of transcriptional noise
In order to ascertain the robustness of age dependent transcriptional noise, we computed three measurements of transcriptional instability each of which displayed a strong statistical significance and positive coefficient to age. As a main measure, we used a correlation based method where noise is expressed as biological variation over technical variation. First, we calculated the biological variation bijk = 1−cor(xijk, uij), where ui is the mean expression vector in cell type i, patient j and xijk is the expression vector of cell k in that cell type i, patient j. Next, we calculated the corresponding technical variation tijk = 1−cor(xcontrijk, ucontr) where xcontrijk and ucontr are the expression vector and mean expression vector of the ERCC spike-in controls. The final measurement is bijk/tijk - the biological noise as a fraction of technical noise. The cells were ordered by this distance within cell type, and their normalized ranking used for linear regression.
For per-donor measurements we also first divided the cells into cell types and computed the mean expression vector for each cell type. We then calculated the Euclidean distance between each cell and its corresponding celltype mean vector. The individual data-points were summarized as boxplots. Finally, as an alternative method to obtain a measure of the transcriptional noise of a single cell, we first subsampled the gene count list to 100 000 counts per cell. We then selected a set of invariant genes evenly across the range of mean expression. First we binned the genes in 10 equally sized bins by mean abundance, then we selected the 10% of genes with the lowest CV from each bin, omitting the bins at the high and low extremes. We then used these genes to determine the Euclidean distance from each cell to the average profile across all cells.
To determine the genes whose mRNA abundance were significantly dependent upon transcriptional instability, we used linear rank regression on the CPM values. p values were adjusted for multiple testing using the FDR procedure of Benjamini & Hochberg (with FDR < 1E-15 as significance cutoff), and ordered by their coefficient.
DATA AND SOFTWARE AVAILABILITY
The accession number for the single-cell mRNA-seq data reported in this paper is GEO: GSE81547. All custom scripts will be provided upon request to the Lead Contact.
Supplementary Material
Table.
Summary of sequenced cells Sequencing statistics are median values.
| Passed QC | Failed QC | |
|---|---|---|
|
|
|
|
| Cells | 2544 (94.9%) | 136 (5.1%) |
| Sequencing statistics | ||
|
| ||
| aligned reads | 932172 | 962153 |
|
| ||
| transcripts detected | 3203 | 1392 |
|
| ||
| % aligned | 78.54% | 79.94% |
|
| ||
| % ERCC | 8.06% | 33.20% |
|
| ||
| % exonic (non-ERCC) | 62.85% | 29.03% |
|
| ||
| % mitochondrial | 6.47% | 10.53% |
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| HPx1-Dylight 488 | Novus | NBP1-18951G |
| HPi2-Dylight 650 | Novus | NBP1-18946C |
| CD133/1 – Biotin | Miltenyi Biotec | 130-090-664 |
| CD133/2 – Biotin | Miltenyi Biotec | 130-090-852 |
| Streptavidin-eFluor780 | eBioscience | 47-4317-82 |
| Streptavidin-APC | eBioscience | 17-4317-82 |
| anti human EpCAM-APC, | Biolegend | 324208 |
| Anti human Insulin | DAKO | A0564 |
| Anti human Glucagon | Sigma | G 2654 |
| 8-oxo-dG mouse Ab | MyBioSource | MBS606843 |
| Biological Samples | ||
| Human pancreatic samples | Integrated Islet Distribution Network (IIDP), | N/A |
| Human pancreatic samples | UCSF Islet Isolation Core (San Francisco, CA USA) | N/A |
| Human pancreatic samples | International Institute for the Advancement of Medicine (IIAM) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Antifade gold | Invitrogen | P36930 |
| UNG | Thermo Fisher | N8080096 |
| Critical Commercial Assays | ||
| Nextera XT | Illumina | FC-131-1096 |
| KAPA HiFi HotStart ReadyMix | KAPA Biosystems | KK2601 |
| Deposited Data | ||
| Single cell mRNA-seq data | This paper | GEO:GSE81547 |
| Experimental Models: Cell Lines | ||
| GP5d colon adenocarcinoma cell line | Sigma-Aldrich | 95090715 |
| Oligonucleotides | ||
| GCG primer for staining: G+TC+TC+TC+AA+AT+TC+ATCGTGACGTTT | This paper | N/A |
| INS primer for staining: G+CA+CC+AG+GGC+CCC+CGCCCAGCTCCA | This paper | N/A |
| GCG padlock probe: Phosp-GAATAACATTGCCAAACGTGTGTCTATTTAGTGGATCCCGTGCGCCTGGTAGCAATTAGCTCCACTGTTACTAGATTGGAATACCAAGAGGAACAG | This paper | N/A |
| INS padlock probe: Phosp-AGGTGGGGCAGGTGGAGCCTCAATGCTGCTGCTGTACTCTACGATTTTACCAGTTGCCCTAGATGTTCCGCTATTGTCCGGGAGGCAGAGGACCTGC | This paper | N/A |
| SmartSeq2 OligodT: 5′–AAGCAGTGGTATCAACGCAGAGTACT30VN-3′ | Picelli et al., 2014 | N/A |
| SmartSeq2 TSO: 5′-AAGCAGTGGTATCAACGCAGAGTACATrGrG3′ | Picelli et al., 2014 | N/A |
| SmartSeq2 ISPCR: 5′-AAGCAGTGGTATCAACGCAGAGT-3′ | Picelli et al., 2014 | N/A |
| Detection probes for in situ RNA staining – see Table S8 | This paper | N/A |
| Software and Algorithms | ||
| GATK pipeline | McKenna et al., 2010; Van der Auwera et al., 2013 | https://software.broadinstitute.org/gatk/ |
| HTSeq | Anders et al., 2014 | https://github.com/simon-anders/htseq |
| STAR | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| GSEA | Subramanian et al., 2005 | software.broadinstitute.org/gsea |
| Picard | McKenna et al., 2010 | https://broadinstitute.github.io/picard/ |
| TSNE | van der Maaten and Hinton, 2008 | https://github.com/jdonaldson/rtsne/ |
Highlights.
RNA-seq of single cells from donors allows detection of stochastic age-related errors
Cells from older donors have increased transcriptional noise and signs of fate drift
Endocrine pancreas cells display an oxidative stress-related mutational signature
Cellular stress and metabolic genes are high in cells with accumulation of errors
Acknowledgments
The authors thank Norma Neff and Gary Mantalas for assistance with sequencing and Spyros Darmanis, Geoff Stanley, and Felix Horns for helpful discussions. This study was supported by the California Institute for Regenerative Medicine (GC1R-06673 to S.R.Q.), the Center of Excellence for Stem Cell Genomics and NIH (U01-HL099999 and U01-HL099995 to S.R.Q.), and by the NIH (UC4DK104211, DK10261201, and P30DK116074-01 to S.K.K.), the Helmsley Charitable Trust (to S.K.K.), the H.L. Snyder Foundation (to S.K.K.), the Elser Foundation (to S.K.K.), and the JDRF (to S.K.K.). M.E. was supported by the Wallenberg Research link at Stanford University (KAW 2013.0391). H.E.A. was supported by a postdoctoral fellowship from the JDRF (3-APF-2016-172-A-N) and an NIDDK training grant to the Endocrinology Division, Department of Medicine, Stanford (5T32DK007217-39). M.M. was supported by the Swedish Research Council (grant 2015-00599).
Footnotes
Supplemental Information includes five figures and eight tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2017.09.004.
AUTHOR CONTRIBUTIONS
M.E., H.E.A., S.K.K., and S.R.Q. designed the research. M.E., H.E.A., J.B., and M.M. performed research. R.B. isolated the islets. M.E. and S.R.Q. analyzed the data. M.E., H.E.A., M.M., S.K.K., and S.R.Q. wrote the paper.
References
- Alexander MP, Begins KJ, Crall WC, Holmes MP, Lippert MJ. High levels of transcription stimulate transversions at GC base pairs in yeast. Environ. Mol. Mutagen. 2013;54:44–53. doi: 10.1002/em.21740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013a;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, et al. Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain. Signatures of mutational processes in human cancer. Nature. 2013b;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Jones PH, Wedge DC, Sale JE, Campbell PJ, Nik-Zainal S, Stratton MR. Clock-like mutational processes in human somatic cells. Nat. Genet. 2015;47:1402–1407. doi: 10.1038/ng.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arda HE, Li L, Tsai J, Torre EA, Rosli Y, Peiris H, Spitale RC, Dai C, Gu X, Qu K, et al. Age-dependent pancreatic gene regulation reveals mechanisms governing human βεcell function. Cell Metab. 2016;23:909–920. doi: 10.1016/j.cmet.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dollé MET, Calder RB, Chisholm GB, Pollock BH, Klein CA, Vijg J. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- Banerjee D, Mandal SM, Das A, Hegde ML, Das S, Bhakat KK, Boldogh I, Sarkar PS, Mitra S, Hazra TK. Preferential repair of oxidized base damage in the transcribed genes of mammalian cells. J. Biol. Chem. 2011;286:6006–6016. doi: 10.1074/jbc.M110.198796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blodgett DM, Nowosielska A, Afik S, Pechhold S, Cura AJ, Kennedy NJ, Kim S, Kucukural A, Davis RJ, Kent SC, et al. Novel observations from next-generation RNA sequencing of highly purified human adult and fetal islet cell subsets. Diabetes. 2015;64:3172–3181. doi: 10.2337/db15-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokzijl F, de Ligt J, Jager M, Sasselli V, Roerink S, Sasaki N, Huch M, Boymans S, Kuijk E, Prins P, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538:260–264. doi: 10.1038/nature19768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AM, Halter JB. Aging and insulin secretion. Am. J. Physiol. Endocrinol. Metab. 2003;284:E7–E12. doi: 10.1152/ajpendo.00366.2002. [DOI] [PubMed] [Google Scholar]
- Chen H, Gu X, Liu Y, Wang J, Wirt SE, Bottino R, Schorle H, Sage J, Kim SK. PDGF signalling controls age-dependent proliferation in pancreatic β-cells. Nature. 2011;478:349–355. doi: 10.1038/nature10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Hayden Gephart MG, Barres BA, Quake SR. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. USA. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugaard M, Rohde M, Jäättelä M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007;581:3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- De Tata V. Age-related impairment of pancreatic Beta-cell function: pathophysiological and cellular mechanisms. Front. Endocrinol. (Lausanne) 2014;5:138. doi: 10.3389/fendo.2014.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat. Rev. Genet. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- Gonitel R, Moffitt H, Sathasivam K, Woodman B, Detloff PJ, Faull RLM, Bates GP. DNA instability in postmitotic neurons. Proc. Natl. Acad. Sci. USA. 2008;105:3467–3472. doi: 10.1073/pnas.0800048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman D. The free radical theory of aging: effect of age on serum copper levels. J. Gerontol. 1965;20:151–153. doi: 10.1093/geronj/20.2.151. [DOI] [PubMed] [Google Scholar]
- Higuchi M, Single FN, Köhler M, Sommer B, Sprengel R, Seeburg PH. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- Kamiya H, Suzuki A, Yamaguchi Y, Handa H, Harashima H. Incorporation of 8-hydroxyguanosine (8-oxo-7,8-dihydroguanosine) 50-triphosphate by bacterial and human RNA polymerases. Free Radic. Biol. Med. 2009;46:1703–1707. doi: 10.1016/j.freeradbiomed.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Kasar S, Brown JR. Mutational landscape and underlying mutational processes in chronic lymphocytic leukemia. Mol. Cell. Oncol. 2016;3:e1157667. doi: 10.1080/23723556.2016.1157667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuta H, Akashi T, Katsuta R, Nagaya M, Kim D, Arinobu Y, Hara M, Bonner-Weir S, Sharma AJ, Akashi K, Weir GC. Single pancreatic beta cells co-express multiple islet hormone genes in mice. Diabetologia. 2010;53:128–138. doi: 10.1007/s00125-009-1570-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke R, Mignardi M, Pacureanu A, Svedlund J, Botling J, Wählby C, Nilsson M. In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods. 2013;10:857–860. doi: 10.1038/nmeth.2563. [DOI] [PubMed] [Google Scholar]
- Kino K, Sugiyama H. Possible cause of G-C–>C-G transversion mutation by guanine oxidation product, imidazolone. Chem. Biol. 2001;8:369–378. doi: 10.1016/s1074-5521(01)00019-9. [DOI] [PubMed] [Google Scholar]
- Kino K, Sugiyama H. UVR-induced G-C to C-G transversions from oxidative DNA damage. Mutat. Res. 2005;571:33–42. doi: 10.1016/j.mrfmmm.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Kreutzer DA, Essigmann JM. Oxidized, deaminated cytosines are a source of C–>T transitions in vivo. Proc. Natl. Acad. Sci. USA. 1998;95:3578–3582. doi: 10.1073/pnas.95.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Klughammer J, Farlik M, Penz T, Spittler A, Barbieux C, Berishvili E, Bock C, Kubicek S. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep. 2016;17:178–187. doi: 10.15252/embr.201540946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato MA, Woodworth MB, Lee S, Evrony GD, Mehta BK, Karger A, Lee S, Chittenden TW, D’Gama AM, Cai X, et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science. 2015;350:94–98. doi: 10.1126/science.aab1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j. 2011;17:10–12. [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignardi M, Mezger A, Qian X, La Fleur L, Botling J, Larsson C, Nilsson M. Oligonucleotide gap-fill ligation for mutation detection and sequencing in situ. Nucleic Acids Res. 2015;43:e151. doi: 10.1093/nar/gkv772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya M, Ou C, Bodepudi V, Johnson F, Takeshita M, Grollman AP. Site-specific mutagenesis using a gapped duplex vector: a study of translesion synthesis past 8-oxodeoxyguanosine in E. coli. Mutat. Res. 1991;254:281–288. doi: 10.1016/0921-8777(91)90067-y. [DOI] [PubMed] [Google Scholar]
- Muraro MJ, Dharmadhikari G, Grün D, Groen N, Dielen T, Jansen E, van Gurp L, Engelse MA, Carlotti F, de Koning EJ, van Oudenaarden A. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 2016;3:385–394.e3. doi: 10.1016/j.cels.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Wedge DC, Alexandrov LB, Petljak M, Butler AP, Bolli N, Davies HR, Knappskog S, Martin S, Papaemmanuil E, et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat. Genet. 2014;46:487–491. doi: 10.1038/ng.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB, Martin S, Wedge DC, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paneni F, Osto E, Costantino S, Mateescu B, Briand S, Coppolino G, Perna E, Mocharla P, Akhmedov A, Kubant R, et al. Deletion of the activated protein-1 transcription factor JunD induces oxidative stress and accelerates age-related endothelial dysfunction. Circulation. 2013;127:1229–1240. doi: 10.1161/CIRCULATIONAHA.112.000826. [DOI] [PubMed] [Google Scholar]
- Park C, Qian W, Zhang J. Genomic evidence for elevated mutation rates in highly expressed genes. EMBO Rep. 2012;13:1123–1129. doi: 10.1038/embor.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smartseq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Segerstolpe Å, Palasantza A, Eliasson P, Andersson EM, Andréasson AC, Sun X, Picelli S, Sabirsh A, Clausen M, Bjursell MK, et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 2016;24:593–607. doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin CS, Moon BS, Park KS, Kim SY, Park SJ, Chung MH, Lee HK. Serum 8-hydroxy-guanine levels are increased in diabetic patients. Diabetes Care. 2001;24:733–737. doi: 10.2337/diacare.24.4.733. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Kumar S. Neutral substitutions occur at a faster rate in exons than in noncoding DNA in primate genomes. Genome Res. 2003;13:838–844. doi: 10.1101/gr.1152803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedge M, Lortz S, Drinkgern J, Lenzen S. Relation between antioxidant enzyme gene expression and antioxidative defense status of insulin-producing cells. Diabetes. 1997;46:1733–1742. doi: 10.2337/diab.46.11.1733. [DOI] [PubMed] [Google Scholar]
- Toone WM, Morgan BA, Jones N. Redox control of AP-1-like factors in yeast and beyond. Oncogene. 2001;20:2336–2346. doi: 10.1038/sj.onc.1204384. [DOI] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L, Hinton G. Visualizing data using T-SNE. JMLR. 2008;9:2579–2605. [Google Scholar]
- Vijg J. Impact of genome instability on transcription regulation of aging and senescence. Mech. Ageing Dev. 2004;125:747–753. doi: 10.1016/j.mad.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Wang YJ, Schug J, Won KJ, Liu C, Naji A, Avrahami D, Golson ML, Kaestner KH. Single-cell transcriptomics of the human endocrine pancreas. Diabetes. 2016;65:3028–3038. doi: 10.2337/db16-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren LA, Rossi DJ, Schiebinger GR, Weissman IL, Kim SK, Quake SR. Transcriptional instability is not a universal attribute of aging. Aging Cell. 2007;6:775–782. doi: 10.1111/j.1474-9726.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- Xin Y, Kim J, Ni M, Wei Y, Okamoto H, Lee J, Adler C, Cavino K, Murphy AJ, Yancopoulos GD, et al. Use of the fluidigm C1 platform for RNA sequencing of single mouse pancreatic islet cells. Proc. Natl. Acad. Sci. USA. 2016;113:3293–3298. doi: 10.1073/pnas.1602306113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Cai Z, Cui M, Nie P, Sun Z, Sun S, Chu S, Wang X, Hu L, Yi J, et al. The orphan nuclear receptor Nur77 inhibits low shear stress-induced carotid artery remodeling in mice. Int. J. Mol. Med. 2015;36:1547–1555. doi: 10.3892/ijmm.2015.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.