Abstract
The multi-tissue DNA methylation estimator of chronological age (DNAm-age) has been associated with a wide range of exposures and health outcomes. Still, it is unclear how DNAm-age can have such broad relationships and how it can be best utilized as a biomarker. Understanding DNAm-age’s molecular relationships is a promising approach to address this critical knowledge gap. In this review, we discuss the existing literature regarding DNAm-age’s molecular relationships in six major categories: animal model systems, cancer processes, cellular aging processes, immune system processes, metabolic processes, and nucleic acid processes. We also present perspectives regarding the future of DNAm-age research, including the need to translate a greater number of ongoing research efforts to experimental and animal model systems.
Background
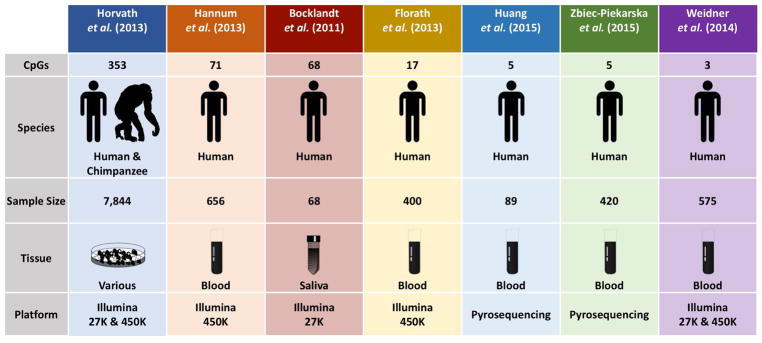
Since its introduction in 2013, the multi-tissue 353-CpG DNA methylation-based estimator of chronological age (DNAm-age) has gained notoriety as the leading molecular measure of human aging. DNAm-age – which is also referred to as “the epigenetic clock” and “epigenetic age” – is not the only DNA methylation-based predictor of chronological age (1–6), but it is the estimator with the greatest organismal and tissue applicability (Figure 1). In contrast to other methylation-based predictors which are based on data from blood cells or one tissue type, DNAm-age was developed using data from 82 Illumina DNA methylation array datasets. These datasets were comprised of 7,844 non-cancer human samples from 51 healthy tissues or cell types. Horvath et al. (2013) began by regressing a transformed version chronological age on 21,369 CpGs – shared between the Illumina 27K and 450K methylation array platforms – using a penalized regression elastic net model. 353 CpGs were selected by the elastic net: 193 were hypermethylated with age and 160 were hypomethylated. The weighted average of the regression coefficients from each of the 353 CpG sites was then used to develop an algorithm to calculate one measure of age prediction, DNAm-age. With only a few exceptions (e.g. breast tissue, dermal fibroblasts, heart tissue, uterine endometrium, and skeletal muscle), DNAm-age has a prediction performance error of ± 3.6 years across cell types. It is still unclear why DNAm-age performs poorly in these tissues, but it is believed to be due to some intrinsic property of the tissues themselves (7).
Figure 1. DNA methylation based predictors of chronological age.
This figure presents seven of the known DNA methylation based predictors of chronological age, the number of CpGs used in each measure, and the species where the measure can be applied. The figure also presents the sample sizes, tissues, and platforms used to generate each measure.
After the release of the seminal DNAm-age paper, researchers began reporting statistically and biologically significant associations of DNAm-age with a host of disease states. Many of these relationships persisted even after accounting for chronological age, and were identified in tissues where DNAm-age was known to have high predictive performance. One such study reported that every 10 unit increase in Body Mass Index (BMI) was significantly associated with a 3.3 year increase in the hepatocyte DNAm-age of their study subjects (8). As time progressed, more evidence suggested that DNAm-age was not simply a predictor of chronological age. Rather, DNAm-age was a novel measure of biological age that could be indicative of disease risk. One of the most compelling studies that supported this biological age theory was a meta-analysis of 13 population-based cohorts amounting to 13,089 individuals. This study found that increases in blood DNAm-age were predictive of mortality even when accounting for chronological age and additional disease states and lifestyle risk factors (9). To date, DNAm-age continues to be associated with a growing number of health-related outcomes and exposures ranging from Parkinson’s disease and human immunodeficiency virus (HIV) infection to cumulative lifetime stress and air pollution (Figure 2) (10–26).
Figure 2. Exposures and Health Outcomes Associated with DNAm-age.
This figure lists the exposures and outcomes that have been reported to have significant associations with DNAm-age in the existing literature.
Although the exposure and outcome associations have been helpful in beginning to understand DNAm-age’s utility as a biomarker, this work has also generated additional questions. For instance: [1] how can DNAm-age be related to so many different exposures and outcomes; and [2] if these relationships are indeed valid, what basic biological process is DNAm-age actually reflecting? To address these questions, researchers will require a greater understanding of DNAm-age’s molecular relationships. In this review, we shift from DNAm-age’s macro-relationships with diseases and exposures and survey what is known about DNAm-age’s molecular relationships. We do this by focusing on experimental research and observational studies that emphasize DNAm-age’s molecular biology. Lastly, we offer some perspectives about the future of DNAm-age research including the need to translate ongoing research efforts to experimental and animal model systems.
Cellular Aging Processes
After DNAm-age demonstrated high accuracy in predicting chronological age and promising utility as a measure of biological aging, researchers wondered how it would perform when compared to other measures of biological age already being used in research. Telomeres, nucleoprotein structures at the ends eukaryotic chromosomes, help facilitate complete chromosomal replication and are one measure of biological age commonly used in biomedical research. Telomere attrition is a hallmark of normal cellular aging and has been associated with a number of aging-related diseases (27). The first published study to report on the relationship of DNAm-age with telomere length examined this relationship in the whole blood of 1,820 male and female elderly participants from the German ESTHER study. Here, a one unit change in age acceleration – DNAm-age minus chronological age – was not significantly associated with telomere length even after adjusting for leukocyte distribution and lifestyle factors (βrange = −0.0020 to 0.0024; pminimum =. 0.45) (17). Three later studies using the Scottish Lothian birth cohorts (N=1,334), the VA Normative Aging Study (N=857), and the New Zealand Dunedin Study (N=964) examined the relationship of DNAm-age with telomere length measured in blood cells of elderly and middle-age individuals and also reported null findings (12, 28, 29).
A study performed by Chen et al. (2017) used blood cell measurements from participants of three cohorts – Framingham Heart Study (FHS; N= 909), Bogalusa Heart Study (BHS; N=826), and the Women’s Health Initiative (WHI; N=804) – and used a variation of DNAm-age: intrinsic age acceleration (IEAA). The IEAA is DNAm-age already adjusted for chronological age and blood cell counts using Horvath’s algorithm. Thus, IEAA represents biological aging unaffected by variation in blood cell composition and chronological age. In this study, IEAA was not significantly correlated with telomere length in the WHI or FHS, but was in the BHS (r = 0.08, p = 0.016) (30). Together, the bulk of the existing evidence demonstrates little if any relationship of DNAm-age with telomere length. Even in the one study where a significant correlation was reported, this was a weak (r = 0.08) relationship not adjusted for potentially important confounders like lifestyle factors or diseases.
Experimental evidence also supports a difference between DNAm-age and telomere biology. Lowe et al. (2016) used primary human endothelial cells to examine the role of senescence in DNAm-age biology (31). Senescence can be generally defined as a state of permanent growth arrest where cells are irreversibly unable to divide but do not undergo cell death. Telomeres are specifically known to play a role in replicative senescence. With each cell division, DNA must be replicated. With each cycle of replication, telomeres gradually shorten (32). However, the enzyme telomerase can extend the ends of telomeres. In embryonic stem cells and young cells, telomerase is very active and enables these cells to continually divide. But, as cells age, telomerase becomes less active and telomeres reach a critical length where they are so short that genomic fidelity would be sacrificed if further cell replications were allowed. At this point, cellular signals are transmitted to prevent further division and cells enter a state of senescence (33). These researchers first cultured endothelial cells and passaged them for 5–6 weeks until they began showing signs of senescence and could no longer proliferate. They then measured the DNAm-age in these cells and found it to be increased. This initial data suggested that telomere length and DNAm-age were inversely related because telomere length shortened as the cells were being passaged. The researchers next immortalized telomerase in cultured endothelial cells and passaged them. After 50 passages, they noted that the cells again had increased DNAm-ages but did not show any signs of replicative senescence. This latter experiment uncoupled telomere and DNAm-age biology and provides the most compelling evidence that the two are indeed distinct forms of biological aging. Importantly, this data also suggests that the positive relationship between cell passaging and DNAm-age is likely due to some other biological process that is independent of telomere length (7, 34). These findings have recently been validated in an independent study, which demonstrated that IEAA continues to increase with passage number in telomerase reverse transcriptase (hTERT) immortalized fibroblasts (35).
Also, relevant to the relationship of DNAm-age with cellular aging are two studies showing how the DNAm-age of cells changes when cells are transformed to stem cells or another cell type. Horvath et al. (2013) noted that induced pluripotent stem cells, stem cells generated from adult cells, have a DNAm-age close to zero (7). Huh et al. (2016) demonstrated that adult fibroblasts directly reprogrammed to neurons retain their DNAm-age (36). Hence, something about converting adult cells to stem cells via transcription factors resets DNAm-age biology, but when an adult cell type is directly converted to another adult cell type using reprogramming miRNAs, DNAm-age remains intact (36). Understanding this difference will inform DNAm-age relationships with molecular aging and cell development.
Nucleic Acid Processes
The central dogma of molecular biology describes the relationships that allow for the translation of nucleotide sequences to protein effectors. These processes can be further regulated by epigenetic modifications and non-coding RNAs to fine-tune nucleotide sequence expression. This fine-tuning, ultimately determines which protein products are functional in the cell (37). Given that DNAm-age is derived from epigenetic modifications on DNA, it is not farfetched to hypothesize that it may have relationships with other nucleic acids or entities involved in nucleic acid processing. Pathway analysis of the genes that co-locate with the 353 CpG sites that make up DNAm-age revealed enrichment for the biological processes of organism tissue development; cellular growth and proliferation; cell death and survival; and cancer (7). Moreover, the 190 hypermethylated component CpGs were more likely to be in poised promoters and were over-represented near Polycomb-group target genes. Polycomb-group target genes are known to play a critical role during embryonic development (38). The 160 hypomethylated CpGs were more likely to be in weak promoters or strong enhancers and were over-represented in CpG shores (7).
With regard to other nucleic acids, a study based on the peripheral blood mononuclear cells of 2,295 participants in the Framingham Heart Study offspring cohort compared DNAm-age to predictors of aging that were developed using miRNAs and mRNAs. DNAm-age had moderate but significant correlations with both miRNA-age (r = 0.34, p = 1.0e-59) and mRNA-age (r = 0.43, p = 6.6e-96) (39). Importantly, these relationships were not corrected for chronological age. Thus, the age-independent relationships of DNAm-age with miRNA-age and mRNA-age still remain unclear. A study in the VA Normative Aging Study cohort further established a connection between miRNA genetics and DNAm-age by focusing on a panel of normally occurring polymorphisms in genes involved in miRNA processing (40). More specifically, this study was focused on understanding a previously reported association of long-term fine particle exposure with DNAm-age, and the authors hypothesized that miRNA processing gene SNPs may play a role in that relationship. Using fully-adjusted linear mixed models, this study first identified two SNPs – rs4961280 (AGO2) and rs784567 (TARBP2) – that had direct associations with DNAm-age. For both SNPs, individuals with at least one copy of the minor allele had on average a DNAm-age that was at least 1.13 years lower than that of individuals who were homozygous for the major allele. Moreover, the rs4961280 (AGO2) SNP significantly modified the association of long-term fine particle exposure with DNAm-age. In the effect modification analysis, having at least one copy of the major allele reduced the magnitude of the fine particle association with DNAm-age. Although, these findings still need to be reproduced in a different and larger cohort, they demonstrate early evidence supporting a relationship between DNAm-age and miRNA processing biology.
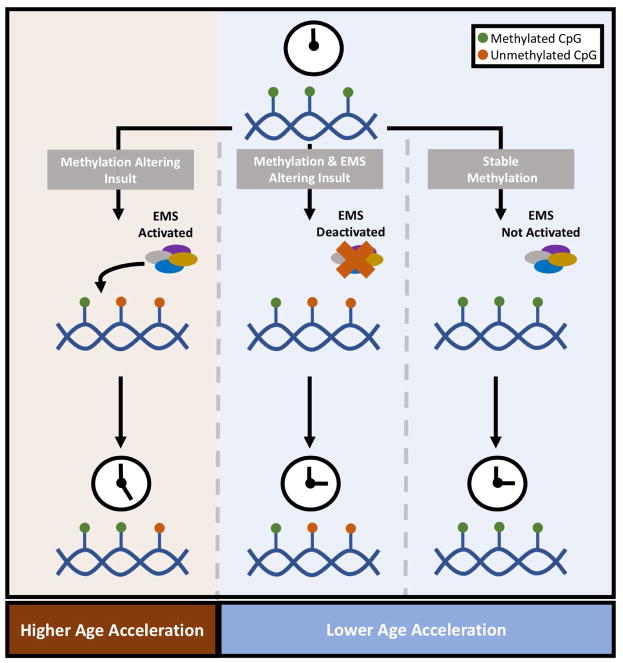
The relationships between DNAm-age and nucleic acids may also be useful for understanding what DNAm-age is believed to represent at its molecular core. It is hypothesized that DNAm-age reflects the sum total of work being performed by a yet to be defined epigenetic maintenance system (EMS). Under the EMS hypothesis, any event or exposure that disrupts epigenetic homeostasis would result in more work being done by the components of the EMS in an attempt to reestablish homeostasis and would manifest itself as a higher DNAm-age. This theory was supported by the fact that DNAm-age only increased with time and that the rate of DNAm-age increases was the lowest in times of genomic inactivity as seen with perfectly young embryonic stem cells (7). However, the idea that lower rates of DNAm-age acceleration are strictly associated with epigenomic inactivity of normal EMS components may be a bit misleading. It is equally plausible under the EMS hypothesis that a lower than expected DNAm-age could occur with an insult that damages or prevents components of the EMS from operating at all. Evidence for this consideration comes from cell culture work where endothelial cells were irradiated, experienced DNA damage, but did not demonstrate any changes in DNAm-age (31). These authors deduced that the DNA damage process was not important for DNAm-aging, but it could also be possible that EMS components were unable to perform any work given this type of damage (Figure 3). There is a great need for studies to further test these hypotheses and begin identifying actual components of the EMS.
Figure 3. The Epigenetic Maintenance System Hypothesis.
This figure describes the Epigenetic Maintenance System (EMS) hypothesis as it is currently understood in the literature. As depicted, a methylation altering insult is hypothesized to result in activation of and work performed by the EMS. In any tissue, at baseline, DNAm-age would accelerate at a particular rate. However, cells experiencing methylation altering insults and subsequent EMS work would display higher than expected age acceleration. Lower age acceleration can result from epigenetic stability/inactivity or some hindrance of the EMS’s ability to perform work. Again, some rate of age acceleration would be expected in any tissue. However, EMS deactivation or epigenetic stability/inactivity in a cell would not hasten the age acceleration further.
It has been hypothesized that DNA methyltransferases (DNMTs), the enzymes that primarily methylate DNA, may themselves be EMS components. Yet, this has not been confirmed (7). Most of the data that has provided some insight into what the EMS components may actually be are findings from genome wide association studies (GWAS). Existing GWAS studies of DNAm-age have been principally performed using brain tissues. One study using post-mortem brain samples from a supercentenarian and younger subjects (N=212) noted that the cerebellum aged slower than other brain regions. In exploring whether differences in gene expression could account for this difference in aging, the researchers found that most of the transcripts differentially expressed between the cerebellum and other brain regions belonged to two superfamilies of helicases (SF1 and SF2). Moreover, when the researchers explored what SNPs were associated with DNAm-aging in the cerebellum, the majority of these SNPs were located near genes from the same helicase superfamilies (41). A second DNAm-age GWAS in post-mortem cerebellar tissue from 555 subjects of European ancestry identified five significant SNPs in two loci: 2p22.1 (inside the gene DHX57) and 16p13.3 (near the gene MLST8) (42). DHX57 is a putative ATP-binding RNA helicase. MLST8 encodes a subunit of the MTORC1 and MTORC2 complexes which play a role in regulating cell growth and survival in response to nutrient and other microenvironment signals (43). A third GWAS study (N=1,163) found 7 SNPS in the 17q11.2 locus that were significantly associated with DNAm-age across brain regions. Of these 7, the leading SNP, rs2054847, is located in the gene SLC6A4 (44). SLC6A4 is a serotonin re-uptake transporter that removes serotonin from the synaptic cleft and the rs2054847 SNP has been associated with risk of schizophrenia in Han Chinese people (45). The most recent and largest of the GWAS studies (N=9907) was performed in blood cells and identified five gene variants/loci that were significantly associated with IEAA: TERT, TRIM59, SCGN, STXBP4, and KIF13A-NHLRC1. Interestingly, the TERT (telomerase reverse transcriptase) locus identified on the GWAS overlapped with a TERT locus known to be associated with telomere length. Nonetheless, this TERT locus was still significantly associated with IEAA when a subsequent GWAS was performed adjusting for leukocyte telomere length (35). It is also important to note that only one of the significant loci – SCGN – identified in all of these GWASs co-located with a DNAm-age component CpG. Hence, the majority of these associations are indeed novel. Although much remains unknown regarding the specific relevance of these loci with DNAm-age biology, they represent some of the top candidates for EMS components. Subsequent analyses will be crucial for determining if the EMS actually does exist, and if so, its characteristics. Moreover, pairing gene expression studies with protein expression studies of DNMTs and other EMS candidates will also be critical in truly understanding these relationships.
Immune System Processes
Collecting white blood cells (WBCs) is usually inexpensive and can be done with little discomfort or risks to study subjects (46). For these reasons, the majority of the observational literature use WBC-derived DNAm-age. Even though DNAm-age is approximately the same across most tissue types, there is still some chance that blood cell composition could confound any relationship of blood DNAm-age. Hence, many observational studies deem it necessary to adjust for blood cell composition in their statistical models (18, 47).
Apart from immune system relationships with WBC composition, there has been a great deal of evidence suggesting that infectious agents can accelerate DNAm-age. While some relationships of DNAm-age and infectious agents like Cytomegalovirus have only been described on a macro scale (16), other studies have started to examine relationships of DNAm-age with the molecular biology of infection. Gao et al. (2017) examined associations of DNAm-age with molecular characteristics related to Helicobacter Pylori (H. pylori) infection using blood samples from 1,477 elderly men and women enrolled in the ESTHER study between 2000 and 2001. These analyses did adjust for blood cell proportions along with age, and revealed that infection with chronic atrophic gastritis virulent H. pylori strains was significantly associated with age acceleration (18). Another study comparing blood and postmortem brain samples of HIV-1 infected individuals and healthy controls observed significant age acceleration (DNAm-age greater than chronological age) of ~7.4 years in some brain regions (i.e. occipital cortex and cerebellum), but not in others (i.e. frontal cortex) (15). They report no significant difference in age acceleration of cerebrospinal fluid (CSF) samples of cases compared to controls. However, in blood, cases demonstrated age acceleration. From regression analysis, it was observed that individuals with a detectable viral load (> 35 copies/mL) on average had an age acceleration of 3.6 years when compared to individuals without a detectable viral load. It is important to note that none of the blood analyses in this study took blood cell composition into account likely because changes in WBC composition are a known effect of HIV-1 infection. It is also interesting that blood viral load (N = 70) and not CSF viral load (N = 48) was associated with age acceleration. These viral load relationships still need to be validated and may be attributable to slightly smaller sample size of CSF viral load, but it could again demonstrate that blood DNAm-age is particularly sensitive to phenomena that impact blood cells.
Studies have also explored associations of blood DNAm-age with less disease-specific immune responses like serum C-reactive protein (CRP) and general cytokine levels. In a pilot study of 23 community-dwelling adults in the Canadian Longitudinal Study of Aging, serum levels of IFN-γ, TNF, IL-1β, IL-6, IL-8, IL-10, and IL-12p70 were not significantly correlated with DNAm-age or age acceleration (48). Of the two studies that have examined CRP relationships, one revealed a null relationship with DNAm-age in 23 nursing home residents and the second revealed a significant positive correlation with IEAA (r = 0.08, p = 2e-5) (49, 50). The study examining IEAA relationships was based on 4,173 women from the Women’s Health Initiative (WHI). In examining these data, it is likely that the two studies of 23 individuals were underpowered to detect statistically significant relationships, but the weak correlation reported from the WHI does create some skepticism regarding the biological significance of CRP relationships with DNAm-age. Nonetheless, even studies that did not explicitly aim to explore immune relationships indicate that this area of research should be further explored. In a sample of 779 observations from men in the VA Normative Aging Study, associations of DNAm-age were explored with three serum endothelial function markers: ICAM, VCAM, and VEGF. After adjusting for blood cell types, chronological age, and diseases, DNAm-age was significantly positively associated with ICAM and VCAM, but not VEGF. Unlike VEGF which is produced by hypoxic cells, ICAM and VCAM are often specifically produced by endothelial cells and are involved with endothelial cell interactions with inflammatory cells. Thus, an association with ICAM and VCAM but not VEGF suggests that the endothelial relationship with DNAm-age may be closely related to immune physiology (47). A longitudinal study of 46 adolescent girls examined associations of DNAm-age with cortisol, a stress hormone and known suppressant of immune function. Here, cortisol was positively associated with DNAm-age (51). This result may suggest that: [1] higher stress is associated with increased DNAm-age or [2] greater risk of immune susceptibility is associated with DNAm-age. Each of these interpretations make additional assumptions that are not necessarily supported by the data. Still, it is known that 85 of the 353 DNAm-age loci are located near glucocorticoid receptor elements, and 110 loci demonstrated significant DNA methylation changes following exposure to dexamethasone, a glucocorticoid receptor agonist (13). The data as a whole suggest that the relationships between general inflammatory mediators and DNAm-age need to be better defined and merit further exploration.
Metabolic Processes
Dietary factors can be considered among the exposures that have been associated with DNAm-age. Many of these relationships were uncovered using serum data from 4,173 women in the WHI and 402 men and women in the European InCHANTI study. Serum levels of insulin, glucose, and triglycerides were weakly but significantly positively correlated with IEAA (rmax = 0.07; pmin = 8e-5). Total cholesterol was positively correlated with IEAA (r = 0.03, p = 0.04), while HDL was negatively correlated with IEAA (r = −0.04, p = 0.01) (50). In fully-adjusted linear models, poultry intake was associated with having a lower IEAA (β = −3.30, p = 1e-3). This study also reported a positive association of BMI with IEAA, which expanded on findings from an earlier study that demonstrated a positive association of BMI with DNAm-age in hepatocytes but not in blood (8, 50). This earlier study was based on 141 liver samples and 274 blood samples. Both of these sample sizes are much smaller than that of the WHI and InCHANTI cohorts; yet, a statistically significant relationship was still observed in hepatocytes. This suggests that certain tissues may be more sensitive for answering particular DNAm-age questions. However, because DNAm-age is largely consistent across tissues, these issues of tissue sensitivity can possibly be overcome with an adequately powered study. This may explain why significant metabolic-relevant findings are observed in a small but metabolically-sensitive liver study sample, but are only observed in blood with a much larger sample size. There is also evidence that the age of study subjects may be relevant when attempting to understand the relationships of DNAm-age and BMI (52).
BMI is not a molecular measure, but we discuss the results in this review because of noteworthy findings from interventional studies with molecular implications. In 21 subjects, researchers examined if bariatric surgery, which is associated with metabolic changes and weight loss, impacted DNAm-age. Although the BMI of the study subjects significantly decreased by an average of 14.6 units following surgery, no significant differences in liver DNAm-age were observed between pre-surgery and 9 months post-surgery samples. Additional studies examining changes in BMI due to exercise also did not report any significant DNAm-age associations (8). Further studies will be necessary to determine if the rate of DNAm-age acceleration truly can not be altered through lifestyle changes or if additional follow-up time is necessary to observe statistically significant changes.
Mitochondria are the primary energy producing organelles in the cell and they have been implicated in aging-related diseases (53). Mitochondrial DNA copy number has been negatively associated with DNAm-age (β = −3.23, p < 0.0001) and decreases in copy number were statistically estimated to mediate 12% of the relationship of one-year ambient fine particle exposure with DNAm-age (54). Copy number is viewed as a measure of the mitochondria’s ability to buffer stressors (55). These findings suggest that there is a greater likelihood of biological aging when buffer capacity is decreased. Associations of copy number with DNAm-age have also been explored in the context of bipolar disorder (56). However, in this study of 58 subjects including bipolar cases, healthy controls and healthy siblings; copy number was not correlated with DNAm-age. It was only when the sample was restricted to individuals over the age of 33 that a statistically significant positive correlation was observed (r = 0.69, p < 0.001). This relationship is the opposite direction of what we expect, but it is a correlation in a smaller group of subjects and does not take into account important confounders. Still, additional observational and experimental studies will be useful to better characterize the relationship of mitochondrial physiology with DNAm-age.
Cancer Processes
Given their ability to continuously proliferate and evade aging-related processes like apoptosis and senescence (57), it was unclear if cancer cells would exhibit younger than expected DNAm-ages. In analyses of cancer methylation datasets, some cancers had higher than expected DNAm-ages while others – like those with TP53 mutations – had DNAm-ages that were lower than expected. Cancer cells with high age acceleration also exhibited fewer somatic mutations (7). Both the TP53 and somatic mutation findings have implications for how we may interpret the EMS hypothesis. As previously mentioned, under the EMS hypothesis higher age acceleration is interpreted as greater epigenetic instability and more work being performed by the EMS to reestablish homeostasis. The finding that cancers with higher age acceleration have fewer somatic mutations supports this hypothesis. Theoretically, work was performed by the EMS (higher DNAm-age) and that work helped reestablish some degree of homeostasis as made evident by fewer somatic mutations. Also in line with the EMS hypothesis, lower age acceleration can be interpreted as greater epigenetic stability/inactivity or an inability of the EMS to perform work. The data from TP53 mutations appears to support the latter of these possibilities, and may indicate that TP53 expression is highly related to the putative EMS. Other groups have replicated the TP53 finding and even shown relationships between other cancers and genes associated with DNAm-age component CpGs (58, 59). Continued work in this area may help in the development of cancer cell type and prognosis biomarkers while providing greater mechanistic insight into DNAm-age biology.
One study has also explored DNAm-age relationships in the context of cancer therapeutics. The researchers analyzed the DNAm-ages of donor hematopoietic stem cells before and following transplantation into a host suffering from de novo acute myeloid leukemia (60). Prior to receiving donor cells, chemotherapy or irradiation is given to the hosts to eradicate their diseased myeloid cells (61). After transplantations, donor cells reconstitute the host’s myeloid system. In this population, DNAm-age was strongly correlated with donor’s chronological age before transplantation (r = 0.96, p = 5.4e-8). However, in the first six months following transplantation, there was a significant decrease in age acceleration. Two years after transplantation, DNAm-age of the transplanted cells was again highly correlated with donor age (r = 0.91, p = 8.7e-31). The researchers deduced that donor cells may experience a sort of rejuvenation following transplantation. However, after about a year, DNAm-age is primarily determined by the age of the donor rather than the age of the recipient and to a lesser extent by the presence of graft versus host disease (60). It remains unclear if this early rejuvenation is in fact due to transplantation itself or due to factors (e.g. granulocyte colony-stimulating factor) that are given around the time of transplantation. Nevertheless, this initial data provides evidence to motivate future studies of DNAm-age as a biomarker for transplantation outcomes, cancer prognosis, or therapeutic success.
Animal Models
It is not clear if DNAm-age relationships from observational studies are causative or correlative. A key step in making this distinction will be exploring these relationships in experimental settings including in vivo animal models. The human DNAm-age metric performs accurately in chimpanzees but has diminished performance in gorillas (7). This difference implies that biomarker performance may be related to evolutionary distances. To date, only one study has attempted to translate the human DNAm-age metric to a more evolutionary distant animal model, mice. However, these researchers found that the human metric was not fully conserved in mice (62). Still, this limitation does not necessarily mean that DNAm-age is useless. Difficulties in cross-species translation are a common issue in molecular biology, and just because a metric is not directly translatable does not mean that important biology can not be learned by the parts that are translatable. It is possible that a non-exact mouse variation of DNAm-age could be capturing the same underlying biology as the human metric (7, 62). A number of similar methylation-based aging measures have also been developed in mice and other animals including dogs, wolves, and whales (63–66). Again, these metrics are not direct translations of the human metric but they could still offer some utility for understanding aging biology. Dogs and whales for instance, have lifespans that are closer to that of humans than mice. Already research groups are beginning to explore the underlying biology of these related age metrics in animals. A different metric but similar to DNAm-age has already been used to demonstrate that aging-related interventions like caloric restriction can reduce methylation aging in rhesus monkeys (67). There will surely need to be cross-talk between animal and human studies, but it appears that the underlying biology of methylation age is robust enough for researchers to really dive into understanding how these metrics mechanistically perform in these model organisms.
Conclusions
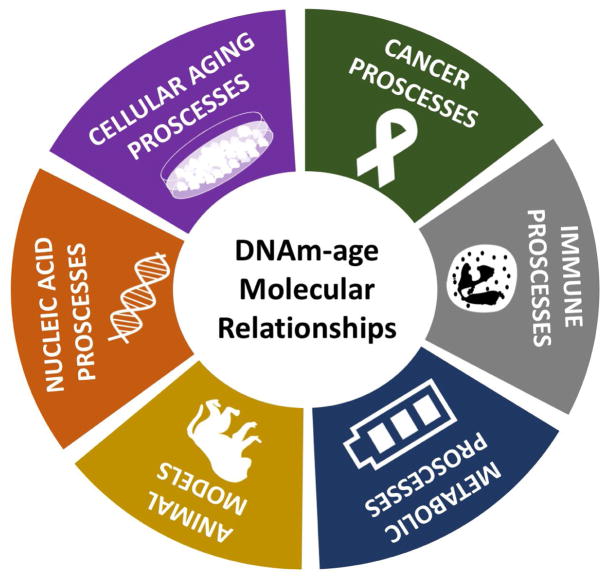
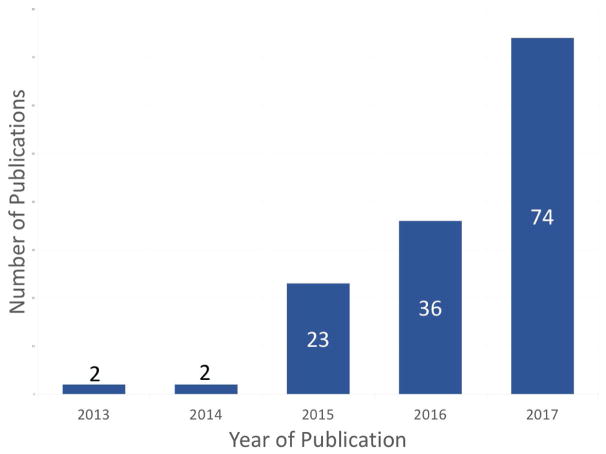
In this review, we have summarized the known molecular relationships of the multi-tissue 353-CpG DNAm-age metric. Although this summary is derived from a number of studies spanning the areas of cellular aging, nucleic acid, immune system, metabolic, and cancer biology research; much of this evidence was still acquired from molecular epidemiologic studies (Figure 4). As the number of published studies examining DNAm-age continues to grow (Figure 5), there is an ever-growing need to perform experimental studies alongside the epidemiologic research. This experimental work will help in elucidating DNAm-age’s molecular relationships and clarifying if these relationships are correlative or causal.
Figure 4. Categories of DNAm-Age Molecular Relationships.
This figure summarizes the six main categories of DNAm-age molecular relationships that are discussed in this review.
Figure 5. Counts of Studies Examining DNAm-Age by Publication Year.
This figure depicts the number of published studies examining DNAm-age by year of publication.
Understanding the molecular biology of DNAm-age is also relevant for determining how to best use DNAm-age as a biomarker in biomedical research, and potentially, clinical medicine. A major question is whether DNAm-age can be used as sensitive and specific marker of disease, or whether it is simply a measure of a broader biological process with disease implications. The bulk of evidence from this present review and the existing literature supports the latter. Analogous to how C-reactive protein (CRP) and erythroid sedimentation rate (ESR) are non-specific markers of general inflammatory processes, DNAm-age may be a non-specific marker of a yet to be defined process. Usually, CRP and ESR are not solely utilized to diagnose a specific disease. Rather, they are used in addition to other diagnostic tools/algorithms and are monitored to track treatment effectiveness and disease progression/resolution (68–70). Similarly, rates of DNAm-age acceleration could one day be monitored to track disease courses and treatment effectiveness. In an effort to increase the quality of studies needed to address this gap in DNAm-age molecular biology and to help realize the full promise of this biomarker, we offer three major summary points with corresponding considerations for future and current researchers:
[A] Cell Types
Summary Point
Although DNAm-age measures display consistency across tissues, emerging research still demonstrates important nuances regarding cell type. One of the best examples from the reviewed studies is that the blood of HIV infected individuals demonstrated significant DNAm-age age acceleration, but their CSF did not.
Important Consideration
It remains important for researchers to scrutinize whether the cell type being used in their study is relevant to the research question being asked. Furthermore, in instances when mixed tissues like blood are being used, cell composition should also be considered. Continuing with the example of HIV infection, changes in blood cell composition will likely be an effect of infection. Thus, studies will be faced with the task of teasing apart direct and indirect relationships of DNAm-age with the independent variable. Molecular epidemiologic and basic science studies can accomplish this by reporting results of models unadjusted and adjusted for cell composition. Studies may also choose to first sort cells with techniques like flow cytometry and then report results for specific cell types.
[B] Study Generalizability and Reproducibility
Summary Point
In this review, we summarize the first studies that have attempted to create a DNAm-age measure in animal models including mice, wolves, dogs, and whales. We also summarize the results from the few existing DNAm-age experimental studies. Conducting rigorous and reproducible studies has been a longstanding issue in other areas of molecular research – especially experimental studies – and it will likely be an issue as the field of DNAm-age research grows.
Important Consideration
Whether a study reports findings using a particular cell line, a specific animal model, or even a particular patient population, it will be critical to first consider the generalizability of these findings before attempting to reproduce them. Moreover, when possible researchers should make efforts to demonstrate that their findings are externally valid. This may involve showing parallel results in two animal models/strains, performing some type of epidemiologic replication, or running the experiments in parallel using different cell lines. If such analyses that attempt to broaden generalizability can not be performed, the researchers should clearly characterize the context of their findings.
[C] DNAm-age Terminology
Summary Point
The studies summarized in this review demonstrate that age acceleration, DNAm-age, and IEAA have all been used as outcomes in DNAm-age research. Age acceleration is at times defined as the difference between DNAm-age and chronological age (or vice versa) or even as the residuals of regression DNAm-age on chronological age. Moreover, in some study designs researchers choose to use DNAm-age and adjust for blood cell counts as covariates while researchers in other studies simply use IEAA, which already accounts for blood cell proportions.
Important Consideration
In a perfect world, each of these variations of studying DNAm-age would result in similar findings; however, to simply believe that they would is a major assumption. A future study showing how each of these variations is related in the context of one outcome or one biological process could be useful for remedying this issue. Establishing more consistent DNAm-age terminology will also be essential for achieving better communication and continuity among research studies. In this review, we use DNAm-age over “epigenetic age” or the “epigenetic clock” because DNA methylation is only one epigenetic process. In the future, groups may be able to develop age predictors based on histone acetylation or other epigenetic modifications. These future measures could also be referred to as a form of “epigenetic age.”
In conclusion, DNAm-age is a novel biomarker that is pertinent to human aging and aging related conditions. Just as DNAm-age has been associated with a range of exposures and health outcomes, it also appears to be related to a range of molecular processes (e.g. nucleic acid, immune system, metabolic, and cancer biology). Establishing a better understanding of DNAm-age’s molecular relationships, via experimental and animal studies, will be critical for actualizing the maximum utility of this biomarker. DNAm-age may prove to be sensitive and specific for a particular disease. It may also prove to simply be a useful measure of a more general biological process with disease implications. Developing consistency in DNAm-age research communications and methodologies will be important for reaching either of these conclusions.
Highlights.
DNAm-age has been associated with a wide range of exposures and health outcomes.
Yet, it is unclear how DNAm-age can have such broad relationships.
We highlight DNAm-age molecular relationships in six major areas.
We offer perspectives for future research, including needed experimental and animal model systems.
Understanding these molecular relationships will be critical for biomarker utility.
Acknowledgments
This work was supported by a NIH/NIA Ruth L. Kirschstein National Research Service Award (1 F31AG056124-01A1).
Footnotes
Competing financial interests related to this research: None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, Deconde R, Chen M, Rajapakse I, Friend S, Ideker T, Zhang K. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weidner CI, Lin Q, Koch CM, Eisele L, Beier F, Ziegler P, Bauerschlag DO, Jockel KH, Erbel R, Muhleisen TW, Zenke M, Brummendorf TH, Wagner W. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 2014;15:R24. doi: 10.1186/gb-2014-15-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bocklandt S, Lin W, Sehl ME, Sanchez FJ, Sinsheimer JS, Horvath S, Vilain E. Epigenetic predictor of age. PLoS One. 2011;6:e14821. doi: 10.1371/journal.pone.0014821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Y, Yan J, Hou J, Fu X, Li L, Hou Y. Developing a DNA methylation assay for human age prediction in blood and bloodstain. Forensic Sci Int Genet. 2015;17:129–136. doi: 10.1016/j.fsigen.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Zbiec-Piekarska R, Spolnicka M, Kupiec T, Parys-Proszek A, Makowska Z, Paleczka A, Kucharczyk K, Ploski R, Branicki W. Development of a forensically useful age prediction method based on DNA methylation analysis. Forensic Sci Int Genet. 2015;17:173–179. doi: 10.1016/j.fsigen.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Florath I, Butterbach K, Muller H, Bewerunge-Hudler M, Brenner H. Cross-sectional and longitudinal changes in DNA methylation with age: an epigenome-wide analysis revealing over 60 novel age-associated CpG sites. Hum Mol Genet. 2014;23:1186–1201. doi: 10.1093/hmg/ddt531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horvath S, Erhart W, Brosch M, Ammerpohl O, von Schonfels W, Ahrens M, Heits N, Bell JT, Tsai PC, Spector TD, Deloukas P, Siebert R, Sipos B, Becker T, Rocken C, Schafmayer C, Hampe J. Obesity accelerates epigenetic aging of human liver. Proceedings of the National Academy of Sciences. 2014;111:15538–15543. doi: 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai PC, Roetker NS, Just AC, Demerath EW, Guan W, Bressler J, Fornage M, Studenski S, Vandiver AR, Moore AZ, Tanaka T, Kiel DP, Liang L, Vokonas P, Schwartz J, Lunetta KL, Murabito JM, Bandinelli S, Hernandez DG, Melzer D, Nalls M, Pilling LC, Price TR, Singleton AB, Gieger C, Holle R, Kretschmer A, Kronenberg F, Kunze S, Linseisen J, Meisinger C, Rathmann W, Waldenberger M, Visscher PM, Shah S, Wray NR, McRae AF, Franco OH, Hofman A, Uitterlinden AG, Absher D, Assimes T, Levine ME, Lu AT, Tsao PS, Hou L, Manson JE, Carty CL, LaCroix AZ, Reiner AP, Spector TD, Feinberg AP, Levy D, Baccarelli A, van Meurs J, Bell JT, Peters A, Deary IJ, Pankow JS, Ferrucci L, Horvath S. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY) 2016;8:1844–1865. doi: 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horvath S, Ritz BR. Increased epigenetic age and granulocyte counts in the blood of Parkinson’s disease patients. Aging (Albany NY) 2015;7:1130–1142. doi: 10.18632/aging.100859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine AJ, Quach A, Moore DJ, Achim CL, Soontornniyomkij V, Masliah E, Singer EJ, Gelman B, Nemanim N, Horvath S. Accelerated epigenetic aging in brain is associated with pre-mortem HIV-associated neurocognitive disorders. J Neurovirol. 2015 doi: 10.1007/s13365-015-0406-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nwanaji-Enwerem JC, Colicino E, Trevisi L, Kloog I, Just AC, Shen J, Brennan K, Dereix A, Hou L, Vokonas P, Schwartz J, Baccarelli AA. Long-term ambient particle exposures and blood DNA methylation age: findings from the VA normative aging study. Environ Epigenet. 2016;2 doi: 10.1093/eep/dvw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zannas AS, Arloth J, Carrillo-Roa T, Iurato S, Roh S, Ressler KJ, Nemeroff CB, Smith AK, Bradley B, Heim C, Menke A, Lange JF, Bruckl T, Ising M, Wray NR, Erhardt A, Binder EB, Mehta D. Lifetime stress accelerates epigenetic aging in an urban, African American cohort: relevance of glucocorticoid signaling. Genome Biol. 2015;16:266. doi: 10.1186/s13059-015-0828-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demanelis K, Virani S, Colacino JA, Basu N, Nishijo M, Ruangyuttikarn W, Swaddiwudhipong W, Nambunmee K, Rozek LS. Cadmium exposure and age-associated DNA methylation changes in non-smoking women from northern Thailand. Environmental Epigenetics. 2017;3:dvx006-dvx006. doi: 10.1093/eep/dvx006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horvath S, Levine AJ. HIV-1 Infection Accelerates Age According to the Epigenetic Clock. J Infect Dis. 2015;212:1563–1573. doi: 10.1093/infdis/jiv277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kananen L, Nevalainen T, Jylhava J, Marttila S, Hervonen A, Jylha M, Hurme M. Cytomegalovirus infection accelerates epigenetic aging. Exp Gerontol. 2015;72:227–229. doi: 10.1016/j.exger.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Breitling LP, Saum KU, Perna L, Schottker B, Holleczek B, Brenner H. Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clin Epigenetics. 2016;8:21. doi: 10.1186/s13148-016-0186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao X, Zhang Y, Brenner H. Associations of Helicobacter pylori infection and chronic atrophic gastritis with accelerated epigenetic ageing in older adults. Br J Cancer. 2017;117:1211–1214. doi: 10.1038/bjc.2017.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horvath S, Langfelder P, Kwak S, Aaronson J, Rosinski J, Vogt TF, Eszes M, Faull RL, Curtis MA, Waldvogel HJ, Choi OW, Tung S, Vinters HV, Coppola G, Yang XW. Huntington’s disease accelerates epigenetic aging of human brain and disrupts DNA methylation levels. Aging (Albany NY) 2016;8:1485–1512. doi: 10.18632/aging.101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maierhofer A, Flunkert J, Oshima J, Martin GM, Haaf T, Horvath S. Accelerated epigenetic aging in Werner syndrome. Aging (Albany NY) 2017;9:1143–1152. doi: 10.18632/aging.101217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll JE, Irwin MR, Levine M, Seeman TE, Absher D, Assimes T, Horvath S. Epigenetic Aging and Immune Senescence in Women With Insomnia Symptoms: Findings From the Women’s Health Initiative Study. Biol Psychiatry. 2017;81:136–144. doi: 10.1016/j.biopsych.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeremian R, Chen YA, De Luca V, Vincent JB, Kennedy JL, Zai CC, Strauss J. Investigation of correlations between DNA methylation, suicidal behavior and aging. Bipolar Disord. 2017;19:32–40. doi: 10.1111/bdi.12466. [DOI] [PubMed] [Google Scholar]
- 23.Levine ME, Hosgood HD, Chen B, Absher D, Assimes T, Horvath S. DNA methylation age of blood predicts future onset of lung cancer in the women’s health initiative. Aging (Albany NY) 2015;7:690–700. doi: 10.18632/aging.100809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Binder AM, Corvalan C, Mericq V, Pereira A, Santos JL, Horvath S, Shepherd J, Michels KB. Faster ticking rate of the epigenetic clock is associated with faster pubertal development in girls. Epigenetics. 2017:1–31. doi: 10.1080/15592294.2017.1414127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lind PM, Salihovic S, Lind L. High plasma organochlorine pesticide levels are related to increased biological age as calculated by DNA methylation analysis. Environ Int. 2018;113:109–113. doi: 10.1016/j.envint.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 26.Roetker NS, Pankow JS, Bressler J, Morrison AC, Boerwinkle E. Prospective Study of Epigenetic Age Acceleration and Incidence of Cardiovascular Disease Outcomes in the ARIC Study (Atherosclerosis Risk in Communities) Circ Genom Precis Med. 2018;11:e001937. doi: 10.1161/CIRCGEN.117.001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montpetit AJ, Alhareeri AA, Montpetit M, Starkweather AR, Elmore LW, Filler K, Mohanraj L, Burton CW, Menzies VS, Lyon DE, Jackson-Cook CK. Telomere length: a review of methods for measurement. Nurs Res. 2014;63:289–299. doi: 10.1097/NNR.0000000000000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marioni RE, Harris SE, Shah S, McRae AF, von Zglinicki T, Martin-Ruiz C, Wray NR, Visscher PM, Deary IJ. The epigenetic clock and telomere length are independently associated with chronological age and mortality. Int J Epidemiol. 2016 doi: 10.1093/ije/dyw041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belsky DW, Moffitt TE, Cohen AA, Corcoran DL, Levine ME, Prinz JA, Schaefer J, Sugden K, Williams B, Poulton R, Caspi A, Telomere Eleven. Epigenetic Clock, and Biomarker-Composite Quantifications of Biological Aging: Do They Measure the Same Thing? Am J Epidemiol. 2017 doi: 10.1093/aje/kwx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen BH, Carty CL, Kimura M, Kark JD, Chen W, Li S, Zhang T, Kooperberg C, Levy D, Assimes T, Absher D, Horvath S, Reiner AP, Aviv A. Leukocyte telomere length, T cell composition and DNA methylation age. Aging (Albany NY) 2017;9:1983–1995. doi: 10.18632/aging.101293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowe D, Horvath S, Raj K. Epigenetic clock analyses of cellular senescence and ageing. Oncotarget. 2016;7:8524–8531. doi: 10.18632/oncotarget.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sosinska P, Mikula-Pietrasik J, Ksiazek K. Molecular bases of cellular senescence: Hayflick phenomenon 50 years later. Postepy Hig Med Dosw (Online) 2016;70:231–242. doi: 10.5604/17322693.1197485. [DOI] [PubMed] [Google Scholar]
- 33.McHugh D, Gil J. Senescence, aging: Causes, consequences, and therapeutic avenues. J Cell Biol. 2017 doi: 10.1083/jcb.201708092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Z, Wong A, Kuh D, Paul DS, Rakyan VK, Leslie RD, Zheng SC, Widschwendter M, Beck S, Teschendorff AE. Correlation of an epigenetic mitotic clock with cancer risk. Genome Biol. 2016;17:205. doi: 10.1186/s13059-016-1064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu AT, Xue L, Salfati EL, Chen BH, Ferrucci L, Levy D, Joehanes R, Murabito JM, Kiel DP, Tsai PC, Yet I, Bell JT, Mangino M, Tanaka T, McRae AF, Marioni RE, Visscher PM, Wray NR, Deary IJ, Levine ME, Quach A, Assimes T, Tsao PS, Absher D, Stewart JD, Li Y, Reiner AP, Hou L, Baccarelli AA, Whitsel EA, Aviv A, Cardona A, Day FR, Wareham NJ, Perry B, JR, Ong KK, Raj K, Lunetta KL, Horvath S. GWAS of epigenetic aging rates in blood reveals a critical role for TERT. Nat Commun. 2018;9:387. doi: 10.1038/s41467-017-02697-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huh CJ, Zhang B, Victor MB, Dahiya S, Batista LF, Horvath S, Yoo AS. Maintenance of age in human neurons generated by microRNA-based neuronal conversion of fibroblasts. Elife. 2016;5 doi: 10.7554/eLife.18648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leenen FA, Muller CP, Turner JD. DNA methylation: conducting the orchestra from exposure to phenotype? Clin Epigenetics. 2016;8:92. doi: 10.1186/s13148-016-0256-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell. 2017;171:34–57. doi: 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 39.Huan T, Chen G, Liu C, Bhattacharya A, Rong J, Chen BH, Seshadri S, Tanriverdi K, Freedman JE, Larson MG, Murabito JM, Levy D. Age-associated microRNA expression in human peripheral blood is associated with all-cause mortality and age-related traits. Aging Cell. 2017 doi: 10.1111/acel.12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nwanaji-Enwerem JC, Colicino E, Dai L, Di Q, Just AC, Hou L, Vokonas P, De Vivo I, Lemos B, Lu Q, Weisskopf MG, Baccarelli AA, Schwartz JD. miRNA processing gene polymorphisms, blood DNA methylation age and long-term ambient PM2.5 exposure in elderly men. Epigenomics. 2017;9:1529–1542. doi: 10.2217/epi-2017-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horvath S, Mah V, Lu AT, Woo JS, Choi O-W, Jasinska AJ, Riancho JA, Tung S, Coles NS, Braun J, Vinters HV, Coles LS. The cerebellum ages slowly according to the epigenetic clock. Aging (Albany NY) 2015;7:294–306. doi: 10.18632/aging.100742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu AT, Hannon E, Levine ME, Hao K, Crimmins EM, Lunnon K, Kozlenkov A, Mill J, Dracheva S, Horvath S. Genetic variants near MLST8 and DHX57 affect the epigenetic age of the cerebellum. Nat Commun. 2016;7:10561. doi: 10.1038/ncomms10561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kakumoto K, Ikeda J, Okada M, Morii E, Oneyama C. mLST8 Promotes mTOR-Mediated Tumor Progression. PLoS One. 2015;10:e0119015. doi: 10.1371/journal.pone.0119015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu AT, Hannon E, Levine ME, Crimmins EM, Lunnon K, Mill J, Geschwind DH, Horvath S. Genetic architecture of epigenetic and neuronal ageing rates in human brain regions. Nat Commun. 2017;8:15353. doi: 10.1038/ncomms15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin H, Lei Y, Zhang B, Dai Z, Lu X. Common variants of HTR1A and SLC6A4 confer the increasing risk of Schizophrenia susceptibility: A population-based association and epistasis analysis. Am J Med Genet B Neuropsychiatr Genet. 2015;168:749–755. doi: 10.1002/ajmg.b.32380. [DOI] [PubMed] [Google Scholar]
- 46.Mayeux R. Biomarkers: potential uses and limitations. NeuroRx. 2004;1:182–188. doi: 10.1602/neurorx.1.2.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nwanaji-Enwerem JC, Bind MA, Dai L, Oulhote Y, Colicino E, Di Q, Just AC, Hou L, Vokonas P, Coull BA, Weisskopf MG, Baccarelli AA, Schwartz JD. Modifying Role of Endothelial Function Gene Variants on the Association of Long-term PM2.5 Exposure with Blood DNA Methylation Age: the VA Normative Aging Study. Toxicol Sci. 2017 doi: 10.1093/toxsci/kfx077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verschoor CP, McEwen LM, Kohli V, Wolfson C, Bowdish DM, Raina P, Kobor MS, Balion C. The relation between DNA methylation patterns and serum cytokine levels in community-dwelling adults: a preliminary study. BMC Genet. 2017;18:57. doi: 10.1186/s12863-017-0525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verschoor CP, McEwen LM, Kobor MS, Loeb MB, Bowdish DME. DNA methylation patterns are related to co-morbidity status and circulating C-reactive protein levels in the nursing home elderly. Exp Gerontol. 2017 doi: 10.1016/j.exger.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 50.Quach A, Levine ME, Tanaka T, Lu AT, Chen BH, Ferrucci L, Ritz B, Bandinelli S, Neuhouser ML, Beasley JM, Snetselaar L, Wallace RB, Tsao PS, Absher D, Assimes TL, Stewart JD, Li Y, Hou L, Baccarelli AA, Whitsel EA, Horvath S. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany NY) 2017;9:419–446. doi: 10.18632/aging.101168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davis EG, Humphreys KL, McEwen LM, Sacchet MD, Camacho MC, MacIsaac JL, Lin DTS, Kobor MS, Gotlib IH. Accelerated DNA methylation age in adolescent girls: associations with elevated diurnal cortisol and reduced hippocampal volume. Transl Psychiatry. 2017;7:e1223. doi: 10.1038/tp.2017.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nevalainen T, Kananen L, Marttila S, Jylhava J, Mononen N, Kahonen M, Raitakari OT, Hervonen A, Jylha M, Lehtimaki T, Hurme M. Obesity accelerates epigenetic aging in middle-aged but not in elderly individuals. Clin Epigenetics. 2017;9:20. doi: 10.1186/s13148-016-0301-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Payne BA, Chinnery PF. Mitochondrial dysfunction in aging: Much progress but many unresolved questions. Biochim Biophys Acta. 2015;1847:1347–1353. doi: 10.1016/j.bbabio.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nwanaji-Enwerem JC, Colicino E, Dai L, Cayir A, Sanchez-Guerra M, Laue HE, Nguyen VT, Di Q, Just AC, Hou L, Vokonas P, Coull BA, Weisskopf MG, Baccarelli AA, Schwartz JD. Impacts of the Mitochondrial Genome on the Relationship of Long-Term Ambient Fine Particle Exposure with Blood DNA Methylation Age. Environ Sci Technol. 2017;51:8185–8195. doi: 10.1021/acs.est.7b02409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peng C, Cayir A, Sanchez-Guerra M, Di Q, Wilson A, Zhong J, Kosheleva A, Trevisi L, Colicino E, Brennan K, Dereix AE, Dai L, Coull BA, Vokonas P, Schwartz J, Baccarelli AA. Associations of Annual Ambient Fine Particulate Matter Mass and Components with Mitochondrial DNA Abundance. Epidemiology. 2017;28:763–770. doi: 10.1097/EDE.0000000000000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fries GR, Bauer IE, Scaini G, Wu MJ, Kazimi IF, Valvassori SS, Zunta-Soares G, Walss-Bass C, Soares JC, Quevedo J. Accelerated epigenetic aging and mitochondrial DNA copy number in bipolar disorder. Transl Psychiatry. 2017;7:1283. doi: 10.1038/s41398-017-0048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ukraintseva SV, Yashina AI. Cancer as “rejuvenescence”. Ann N Y Acad Sci. 2004;1019:200–205. doi: 10.1196/annals.1297.032. [DOI] [PubMed] [Google Scholar]
- 58.Galamb O, Kalmar A, Bartak BK, Patai AV, Leiszter K, Peterfia B, Wichmann B, Valcz G, Veres G, Tulassay Z, Molnar B. Aging related methylation influences the gene expression of key control genes in colorectal cancer and adenoma. World J Gastroenterol. 2016;22:10325–10340. doi: 10.3748/wjg.v22.i47.10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin Q, Wagner W. Epigenetic Aging Signatures Are Coherently Modified in Cancer. PLoS Genet. 2015;11:e1005334. doi: 10.1371/journal.pgen.1005334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stolzel F, Brosch M, Horvath S, Kramer M, Thiede C, von Bonin M, Ammerpohl O, Middeke M, Schetelig J, Ehninger G, Hampe J, Bornhauser M. Dynamics of epigenetic age following hematopoietic stem cell transplantation. Haematologica. 2017;102:e321–e323. doi: 10.3324/haematol.2016.160481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hegde A, Murthy HS. Frailty: the missing piece of the pre- hematopoietic cell transplantation assessment? Bone Marrow Transplant. 2017 doi: 10.1038/bmt.2017.192. [DOI] [PubMed] [Google Scholar]
- 62.Stubbs TM, Bonder MJ, Stark AK, Krueger F, Team BIAC, von Meyenn F, Stegle O, Reik W. Multi-tissue DNA methylation age predictor in mouse. Genome Biol. 2017;18:68. doi: 10.1186/s13059-017-1203-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson MJ, vonHoldt B, Horvath S, Pellegrini M. An epigenetic aging clock for dogs and wolves. Aging (Albany NY) 2017;9:1055–1068. doi: 10.18632/aging.101211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Polanowski AM, Robbins J, Chandler D, Jarman SN. Epigenetic estimation of age in humpback whales. Mol Ecol Resour. 2014;14:976–987. doi: 10.1111/1755-0998.12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petkovich DA, Podolskiy DI, Lobanov AV, Lee SG, Miller RA, Gladyshev VN. Using DNA Methylation Profiling to Evaluate Biological Age and Longevity Interventions. Cell Metab. 2017;25:954–960 e956. doi: 10.1016/j.cmet.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang T, Tsui B, Kreisberg JF, Robertson NA, Gross AM, Yu MK, Carter H, Brown-Borg HM, Adams PD, Ideker T. Epigenetic aging signatures in mice livers are slowed by dwarfism, calorie restriction and rapamycin treatment. Genome Biol. 2017;18:57. doi: 10.1186/s13059-017-1186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maegawa S, Lu Y, Tahara T, Lee JT, Madzo J, Liang S, Jelinek J, Colman RJ, Issa JJ. Caloric restriction delays age-related methylation drift. Nat Commun. 2017;8:539. doi: 10.1038/s41467-017-00607-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Limper M, de Kruif MD, Duits AJ, Brandjes DP, van Gorp EC. The diagnostic role of procalcitonin and other biomarkers in discriminating infectious from non-infectious fever. J Infect. 2010;60:409–416. doi: 10.1016/j.jinf.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 69.Pepys MB, Hirschfield GM. C-reactive protein: a critical update. J Clin Invest. 2003;111:1805–1812. doi: 10.1172/JCI18921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bray C, Bell LN, Liang H, Haykal R, Kaiksow F, Mazza JJ, Yale SH. Erythrocyte Sedimentation Rate and C-reactive Protein Measurements and Their Relevance in Clinical Medicine. WMJ. 2016;115:317–321. [PubMed] [Google Scholar]