Abstract
The immune system exerts antitumor activity via T cell–dependent recognition of tumor-specific antigens. Although the number of tumor neopeptides—peptides derived from somatic mutations—often correlates with immune activity and survival, most classically defined high-affinity neopeptides (CDNs) are not immunogenic, and only rare CDNs have been linked to tumor rejection. Thus, the rules of tumor antigen recognition remain incompletely understood. Here, we analyzed neopeptides, immune activity, and clinical outcome from 6,324 patients across 27 tumor types. We characterized a class of “alternatively defined neopeptides” (ADNs), which are mutant peptides predicted to bind MHC (class I or II) with improved affinity relative to their nonmutated counterpart. ADNs are abundant and molecularly distinct from CDNs. The load of ADNs correlated with intratumoral T-cell responses and immune suppression, and ADNs were also strong predictors of patient survival across tumor types. These results expand the spectrum of mutation-derived tumor antigens with potential clinical relevance.
Introduction
Tumor neopeptides, defined as peptides derived from somatic mutations in cancer cells, are seen as foreign by the adaptive immune system and are critical mediators of tumor immunity (1). Classically defined neopeptides (CDNs) can be bioinformatically identified by predicting the ability of peptides containing somatic mutations to bind MHC class I or II with high affinity. Higher predicted CDN load correlates with several favorable clinical features, including greater lymphocytic infiltration and survival in untreated colorectal tumors (2) and improved response to CTLA-4 blockade in melanoma (3) and PD1 blockade in non–small cell lung carcinoma (4). Strong associations exist between response to PD1 blockade and mismatch repair deficiency (which results in high neopeptide load; ref. 5), mutation burden in melanoma (6), and MHC class I CDN load in urothelial carcinoma (7).
Despite these associations, only a small percentage of predicted CDNs are immunogenic. In an analysis of 3 patients with metastatic melanoma, in aggregate, only 2 out of 57 selected CDNs elicited endogenous T-cell responses (8). Studies also indicate that CDNs are no more predictive of response to checkpoint blockade in patients with metastatic melanoma than overall somatic mutation load (9). Finally, no difference is observed in CDN binding scores for HLA-A2 or immunogenicity between T cell–inflamed and non–T cell-inflamed metastatic melanomas (10), suggesting that the presence of CDNs does not necessarily correlate with T-cell response. These findings represent a gap in the understanding of immune recognition of tumor antigens.
To address this gap, we analyzed predicted neopeptides and immune activity in 6,324 human cancer samples from The Cancer Genome Atlas (TCGA), using an open-source discovery pipeline to evaluate over 228 million mutant peptide predictions from more than 1 million nonsynonymous missense mutations (NSMs) within this dataset. This analysis revealed the existence of a previously underappreciated class of tumor antigens, for which tumor-associated mutations significantly increased peptide affinity for class I or class II MHC compared with nonmutated counterparts. This class of neopeptides—which we refer to as “alternatively defined neopeptides” (ADNs)—predicted immune phenotypes and patient survival.
Materials and Methods
Tumor and normal sample datasets
DNA variant, gene expression, and clinical data from TCGA (11) were obtained in June 2016 from the Genome Data Analysis Center Firehouse (Spring 2016 run; https://gdac.broadinstitute.org/). DNA variants that were manually curated by TCGA analysis working groups were obtained in mutation annotation format (MAF). Gene expression data were normalized counts from the TCGA RNASeq Version 2 pipeline and included all available “Level 3” samples. Raw tumor and normal paired whole exome sequencing reads were obtained through dbGaP (accession phs000178.v9.p8) and the NCI Cancer Genomics Hub. Samples selected for analysis were those with DNA variant, gene expression, and raw exome reads available for download (Supplementary Fig. S1). Tumor types included in analyses were those with at least 15 such complete cases. Seven tumor types contained < 15 samples for these required data and were excluded: cholangiocarcinoma, kidney chromophobe, mesothelioma, ovarian serous cystadenocarcinoma, rectal adenocarcinoma, thymoma, and uterine carcinosarcoma. Within the remaining included tumor types, samples with no identified tumor variants resulting in amino acid changes that passed quality-control filters were excluded from analysis. Tumor cellularity estimates were determined using Sequenza and ABSOLUTE (12, 13) and were similar to those previously reported (Supplementary Fig. S2; refs. 14, 15). For the non–TCGA-independent datasets (3, 4, 16), variants were assembled from provided Supplementary Tables.
HLA determination
Normal tissue whole-exome sequencing was used for 4-digit HLA class I and II determination. In cases with multiple normal tissue samples, the sample with the greatest read depth was used. HLA class I determination was performed using OptiType (version 1.3.1; ref. 17). This method improves on the accuracy of first-generation HLA determination tools and has been independently validated on whole-exome sequencing (18). OptiType was used with default settings. HLA class II determination was performed using HLA reporter (version 1.0.3; ref. 19). Reads were aligned to assembly GRCh38. HLA class I and II determination was validated to have approximately 95% 4-digit accuracy compared with standard clinical typing using sequence-specific oligonucleotide probe and primer techniques, in agreement with published estimates (18). For class I and II determinations, samples with inadequate read-depth or low-certainty results were excluded (<5%). For the non–TCGA-independent datasets, the provided HLA alleles were used (3, 4, 16).
MHC binding affinity predictions
Variants predicted to yield NSMs present in tumors, but not matched normal tissue, were selected for neopeptide prediction. Validation set variants (3, 4, 16) were re-annotated using SnpEff (version 4.3k) for consistency across datasets (20). Using TCGA MAF files, Human Genome Organization, RefSeq, and Entrez identifiers were converted to Enembl transcript IDs using the R/Bioconductor package biomaRt (version 2.30.0). Variants were then filtered for those in genes with RSEM-normalized count expression of greater than 1. After filtering, a sliding window of 8- to 15-mer peptides centered on each variant site was generated. Peptides were truncated if the window length extended beyond a start or stop codon. Estimated binding affinity for each peptide was then calculated using the Immune Epitope Database and Analysis Resource (IEDB) MHC class I and II prediction tools (version 2.15; FASTA input; 8- to 14-mers for MHC class I and 15-mers only for MHC class II; ref. 21). The “IEDB-recommended” method was chosen for predictions because it considers algorithm benchmarks in large-scale evaluations, as well as availability across HLA alleles (http://www.iedb.org/). For HLA alleles with multiple available prediction methods, the median of all prediction values was used. For MHC class I predictions, the established median half-maximum inhibitory concentrations (IC50) of <5,000 nmol/L or <50 nmol/L were used to classify peptides as potential MHC binders or as CDNs, respectively. These cutoffs correspond to experimentally confirmed peptide–MHC binding and immunogenic CDN thresholds, respectively (22, 23). For MHC class II, peptides with a percentile rank of <4 or <1 were classified as potential MHC binders and potential CDNs, respectively. Percentile rank is recommended for MHC class II due to the greater variability in peptide–MHC binding across MHC types for class II compared with class I (24, 25).
ADN criteria and peptide characteristics
We identified ADNs as mutant peptides predicted to bind MHC class I or II with improved affinity relative to nonmutated peptides, a quality known as differential agretopicity (26). Differential agretopicity index (DAI) is the ratio of MHC binding affinity between mutant and normal peptide. To identify ADNs, we first selected mutant peptides that were at least minimally able to bind MHC (median IC50 < 5,000 nmol/L for MHC class I and percentile rank <4 for MHC class II). DAI then was calculated as the fold change in binding affinity (MHC class I) or percentile rank (MHC class II) between mutant and normal peptides. Based on the distribution of resulting DAI scores, ADNs were identified as having a DAI > 10 for MHC class I or >4 for MHC class II, which corresponds to the first percentile by rank. Shannon entropy was calculated as a diversity index that quantifies the difficulty of class prediction across a dataset, similar to geometric mean of abundance.
Random forest survival analysis
Random forest analysis is a multivariable, nonparametric ensemble partitioning tree method for modeling classification, regression, or survival problems and was performed as previously described (27–30). This approach is used to model the effect of multiple input variables and their interactions on a response variable of interest. Random forest analysis was used for two purposes: (i) to determine if a predictive model could be constructed for response variables (suppressive index, dysfunction index, or cumulative survival hazard function) and (ii) to rank input variables based on their relative contribution to model predictiveness. Advantages of this approach compared with other learning methods include excellent discriminatory ability in high dimensional space, few assumptions about underlying data, resistance to noise, missing data and overfitting, as well as built-in error estimates (31).
Random forest analysis was conducted using the random forest SRC R package (version 2.3.0) as previously described (32, 33). Data ranked for classification into high and low cohorts for prediction were scaled from 0 to 100 using an empirical cumulative distribution function within each tumor type. During bootstrapping, a randomly chosen two-thirds of samples were used to train each tree and remaining (out-of-bag, OOB) samples were used for cross-validation and forest-related estimates. No training was conducted on OOB samples, and no iterative parameter optimization was performed. Missing values were imputed, and classification models were built using the following parameters: ntree = 1500; nodesize = 2; nsplit = 10; mtry = (input variable number3/4). A Gini index splitting rule was used for classification, and undersampling of the majority class was used when the number of samples in each class differed by greater than 5-fold.
Cumulative hazard function is the integrated hazard function over time and, thus, represents the total amount of risk accumulated until time t (i.e., the number of times one would expect to observe an event over a given time period). Prediction error was calculated using the Harrell concordance index and estimates the probability that, in two out-of-bag samples, the case with the worse predicted outcome had an event first.
Variables were then ranked by minimal depth (MD), a dimensionless statistic that measures variable predictiveness in tree-based models (34). MD is defined as the shortest distance between the root node of a tree and the parent node of a maximal subtree, which is the largest subtree whose root node splits on the variable. Smaller MD values indicate greater predictiveness, and a tree-averaged threshold MD was used to classify variables as predictive. After predictive variable identification, models were refit using only predictive variables and tested against out-of-bag samples. If the number of input variables exceeded the number of samples by >10-fold, variable hunting was used to select variables prior to refitting the forest. Variable hunting is a regularized algorithm that exploits maximal subtrees for more effective variable selection under conditions where a large number of noisy variables exist, described previously in detail (34). With variable hunting, the high dimensional feature space is divided into multiple smaller subspaces to better estimate MD, which is returned as the average for each variable across subspaces. Model performance was quantified using geometric mean accuracy (1-mean OOB misclassification error rate across replications) and F-score to integrate predictiveness of models for all response variable categories. Relative stability was determined using the normalized Brier score for each model, a proper score function that measures the mean squared difference between the predicted outcome probabilities from a random forest model and the actual outcome tested using OOB samples. Each analysis was externally cross-validated over 20 iterations using resampling without replacement (35), and model performance was determined by averaging OOB error rate, geometric mean accuracy, and normalized Brier score across bootstrap replicates. MD was similarly averaged for each input variable. Additionally, to guard against the possibility of overfitting or bias due to differences in input data size across models, overall accuracy was then compared with bootstrapped-simulated control data with matched dimensions. No models of simulated control data were predictive. Final model significance was determined using the following criteria: normalized Brier score <90, geometric mean accuracy >0.55, normalized Brier score relative to control <0.8, and significantly increased overall/geometric mean accuracy relative to control by FDR. Final input variable MD importance was determined by averaging normalized MD across significant models.
Gene expression analysis
Gene set enrichment scores were generated using the gene set variation analysis (GSVA) R/Bioconductor package (36). The GSVA package provides robust enrichment scores because it implements a nonparametric, unsupervised method of estimating set expression using a Kolmogorov–Smirnov (KS)-like random walk statistic. This method captures subtle or relative changes in gene expression from RNA-seq data. Cytolytic index was calculated using normalized count expression of GZMA and PRF1. The suppressive index consists of 11 well-characterized, highly expressed T-cell checkpoint molecules: ADORA2A (A2AR), CD274 (PD-L1), PDCD1 (PD1), CTLA4, HAVCR2 (TIM3), IDO1, IDO2, PDCD1LG2 (PD-L2), TIGIT, VISTA (C10orf54), and VTCN1 (B7-H4; ref. 14). T-cell dysfunction index consists of 567 genes identified by Schietinger and colleagues (geneset F; ref. 37).
Statistics and software
The neopeptide pipeline for generating peptides, predicting MHC class I/II binding affinity, and interpreting predictions for normal and mutant peptides (antigen.garnish R package) is available for download: https://github.com/andrewrech/antigen.garnish. Survival used in the Kaplan–Meier estimates was determined as the number of days from diagnosis until death or last contact. Analyses were conducted using all samples or by tumor type. Age, gender, and tumor stage were assessed as potential confounders and were not significantly imbalanced between groups. Tumor types with ≤50 total samples with neopeptide predictions were excluded (cholangiocarcinoma, kidney renal clear cell carcinoma, uterine corpus endometrial carcinoma, colon adenocarcinoma, rectum adenocarcinoma, cervical squamous cell carcinoma and endocervical adenocarcinoma, lymphoid neoplasm diffuse large B-cell lymphoma DLBC, Rizvi and colleagues dataset; ref. 4). Kaplan–Meier estimates, P values, and hazard ratios were generated using the R package survival (version 2.3.0). Adjusted P values less than 0.05 were considered significant. The following additional R (https://www.r-project.org, version 4.0) packages were used: cowplot (https://CRAN.R-project.org/package=cowplot, version 0.6.3): figure layouts; data. table (http://r-datatable.com, version 1.9.6): general data analysis; gglogo (https://CRAN.R-project.org/package=gglogo, version 0.1.2): peptide sequence diagrams; ggplot2 (version 2.1.0; ref. 38): general plots; HiveR (https://CRAN.R-project.org/package=HiveR, version 0.2.55): hive plots; pheatmap (https://CRAN.R-project.org/package=pheatmap, version 1.0.8): heatmap generation; stats (version 3.3.2): tests for significance.
Results
Neopeptide landscape in human cancer
To investigate the landscape of neopeptides in human cancer, we created a simple, performant analytical pipeline for predicting neopeptides that integrates TCGA DNA variants, gene expression, and raw exome-sequencing reads (available as an R package; Fig. 1A and Supplementary Fig. S1). We estimated the tumor DNA fraction for all samples using ABSOLUTE (13) and confirmed results using Sequenza (12). The pan-cancer tumor DNA fraction was 0.79 ± 0.08 (mean ± SD), similar to previously published estimates (14), and no correlation existed between the tumor DNA fraction and nonsynonymous mutation (NSM) load (Supplementary Fig. S2). HLA class I and II alleles were determined using OptiType and HLA reporter, respectively (17, 19). We validated these methods to have 95% 4-digit accuracy compared with standard clinical typing, consistent with other analyses (17–19), and TCGA sample HLA allele frequencies paralleled those reported for the general population in the United States (39).
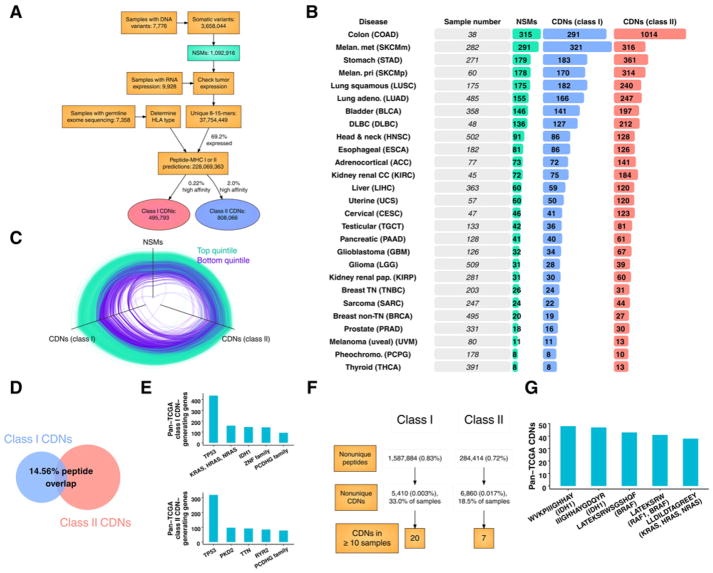
Figure 1.
CDNs across human cancer. A, Pipeline of data types and numbers (see Supplementary Fig. S1 for a detailed pipeline). B, Summary of NSMs, predicted class I (<50 nmol/L IC50) and II (<1% rank) CDNs. Tumor types are ordered from top to bottom by mean number of NSMs. Cohort sizes are shown for MHC class I predictions. C, Parallel coordinate plot showing frequency of NSMs and MHC class I and II CDNs across all tumor types. Edges are samples in the top (green) or bottom (purple) quintile by MHC class I CDN load. Each vertex is normalized from 0 to 1. D, Venn diagram of mean percent MHC class I CDNs (left) contained in class II CDNs (right). Overlapping class I neopeptides are those whose peptide sequence is contained in a class II CDN from the same sample. E, Most frequent pan-TCGA common CDN-generating genes. Y-axis: total number of neopeptides derived from the indicated gene for MHC class I (top) and II (bottom). F, Shared MHC class I and II CDNs across all samples. Percentages: total peptides. Neopeptides were considered duplicates across samples if the same peptide sequence met CDN criteria for sample-specific HLA. G, Most frequent pan-TCGA neopeptides.
We then analyzed all peptides generated from NSMs in expressed genes for the ability to bind MHC class I and II. Peptides with high predicted affinity were identified as CDNs (<50 nmol/L for MHC class I or percentile rank < 1 for MHC class II). Out of a total of 9,928 tumor samples with whole exome sequencing, paired normal tissue and tumor variant calling required to determine tumor variants was available for 7,776 samples. Out of these, raw normal exome sequences (required to determine HLA alleles) were available for 7,358 samples. These inclusion criteria resulted in a total of 3,658,044 high-confidence tumor variants, of which 29.8% were NSMs, resulting in 37,754,449 unique 8- to 15-mer peptides (Fig. 1A and Supplementary Fig. S1). Filtering by tumor RNA expression, 69.2% of NSMs occurred in genes that were at least minimally expressed (>1 RNA sequencing read count) and, therefore, potentially presented by MHC. Overall, MHC binding affinity selection resulted in 495,793 predicted class I and 808,066 predicted class II CDNs from 6,324 samples across 27 tumor types. This higher prevalence of MHC class II CDNs compared with class I CDNs is consistent with previous reports and the greater flexibility of peptide binding in the groove of MHC class II (24, 25).
CDN load differed substantially across tumor types, with wide variability observed within each tumor type (Fig. 1B). The highest mean number of MHC class I CDNs per tumor sample occurred in colon (291 ± 556), metastatic (321 ± 848) and primary (170 ± 222) melanoma, and stomach (183 ± 296) types. Results for MHC class II CDNs were similar (colon: 1,014 ± 3,288; metastatic melanoma: 316 ± 513; primary melanoma: 314 ± 531; and stomach: 361 ± 864). A close correlation existed between the total number of NSMs and CDNs, with greater variability in the case of MHC class II (Fig. 1C; R2 = 0.852 and 0.452, respectively). Despite this correlation in total number, relatively little homology between MHC class I and II CDNs was seen—fewer than 15% of MHC class I CDNs were contained in a class II CDN from the same sample (Fig. 1D). Thus, tumor heterogeneity results in a wide range in CDN load across and within all tumor types. Only a small fraction of missense mutations or derived mutant peptides was predicted to be CDNs for both MHC class I and II. Therefore, MHC class I and class II neopeptides are drawn from distinct sets of genetic lesions.
Common driver mutations critical to oncogenesis may generate common neopeptides amenable to therapeutic targeting. To assess the feasibility of targeting these neopeptides, we determined the number of shared CDNs in TCGA. Genes frequently mutated in cancer often generated neopeptides, as expected (Fig. 1E). The most common genes with mutations generating predicted CDNs across all samples were TP53, Ras family members, and IDH1 for MHC class I and TP53 for MHC class II. Despite the substantial number of neopeptides derived from commonly mutated genes—more than 700 unique CDNs from TP53 alone—few neopeptides were widely shared across samples due to variant and HLA allele diversity. A total of 1,872,298 peptides generated from NSMs were shared between ≥2 samples across all tumor types (<1% of total), resulting in 5,410 shared MHC class I and 6,860 shared MHC class II CDNs (Fig. 1F). Although a sizable fraction of samples analyzed contained at least one such CDN (17% of samples for MHC class I; 70% of samples for MHC class II), most were shared by only 2 to 3 patients. As a result, only 27 MHC class I or II CDNs were shared between ≥10 samples. Commonly shared CDNs were derived from known abundant NSMs in IDH1 (p.R132H), BRAF (p.V600E), and a conserved RAS family mutation (p.Q61R; Fig. 1G). The significant outlier in this analysis was overlapping CDNs derived from IDH1 p.R132H, which occurred in 90 samples. Thus, variant heterogeneity, even among common driver mutations, and HLA allele diversity likely preclude shared neopeptides at frequencies required for widely applicable therapeutic targeting.
ADNs are a distinct class of tumor antigens
DAI expresses the degree to which peptide binding to MHC class I or II differs due to the presence of an NSM (26). Across TCGA, the median DAI of CDNs was 1.183, meaning that most CDNs have nonmutated counterparts that also bind MHC with high affinity. In contrast, we define a class of “alternatively defined neopeptides” (ADN), which are mutant peptides predicted to bind MHC class I or II with greater affinity relative to nonmutated counterparts (i.e., peptides with high DAI). In mice, selection of peptides with high DAI results in a substantially improved rate of experimentally validated epitopes that mediate protection from tumor growth (26). To determine DAI across TCGA, mutant peptides that at least minimally bound to MHC class I or II were selected, and DAI was calculated as the fold change in binding affinity between nonmutant and mutant peptides (Fig. 2A). Based on the distribution of the resulting scores, ADNs were identified as those with DAI > 10 for MHC class I and >4 for class II.
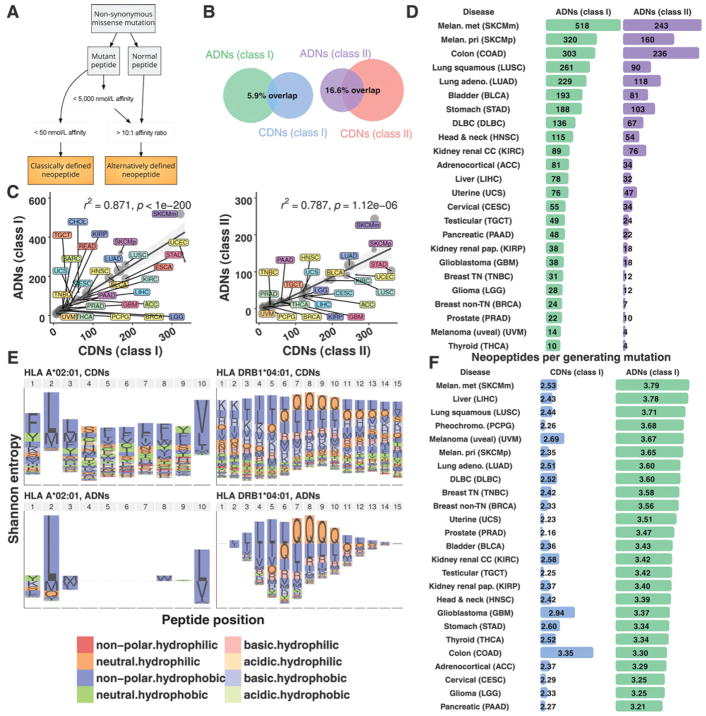
Figure 2.
ADNs are largely distinct from CDNs. A, CDNs were identified based solely on high mutant peptide MHC-binding affinity. ADNs were identified based on ≥10-fold improvement in MHC-binding affinity of mutant peptide vs. nonmutant counterparts, quantified as DAI. B, Venn diagrams of overlap between class I (left) and II (right) neopeptide categories. Denominator for percent overlap: total number of ADNs + CDNs. C, Predicted MHC class I (left) and II (right) ADNs and CDNs. R2 values: linear regression; P values: Spearman rho. Dot size: sample number. Shaded gray region: confidence interval. D, Summary of ADNs. Tumor types are ordered from top to bottom by mean number of predicted MHC class I ADNs. E, Shannon entropy index of mutated amino acids by peptide position for the MHC class I and II alleles for CDNs (top) and ADNs (bottom). Two HLA alleles are shown: HLA A*02:01 and DRB1*04:01. F, Summary of generator rate by neopeptide category and tumor type (see Supplementary Fig. S3 for common ADN-generating genes and shared ADNs).
Applying ADN selection criteria led to the identification of peptides that were distinct from CDNs. For MHC class I, 20% of all MHC binding (defined as IC50 < 5,000 nmol/L) peptides were ADNs, and 28% were CDNs. Yet only a 5.9% overlap between these neopeptide categories was observed (Fig. 2B). For MHC class II, a greater proportion of all MHC binders were ADNs (ADN: 26% vs. CDN: 13%), with only modest overlap between neopeptide categories (16.6% overlap). A close correlation existed between the number of NSMs and ADNs (R2 = 0.810 and 0.827 for MHC class I and II, respectively), as well as between load of ADNs and CDNs across tumor types (Fig. 2C; R2 = 0.87 and 0.79 for MHC class I and II, respectively). An outlier was melanoma, that, for both MHC class I and II, was characterized by a higher ADN to CDN ratio versus all other tumor types (FDR < 0.01). Rates of common ADN-generating genes and shared ADNs paralleled those for CDNs (Supplementary Fig. S3). Thus, MHC class I and II ADNs are abundant, distinct from CDNs, and enriched in certain tumor types, including melanoma.
We observed that a wide distribution of ADN load existed across tumor types and samples (Fig. 2D). For MHC class I, the highest median ADN load occurred in metastatic (518 ± 960) and primary melanoma (320 ± 392), colon adenocarcinoma (303 ± 714), and lung squamous tumor types (261 ± 266). Predicted MHC class II ADN results were similar: metastatic (243 ± 765) and primary melanoma (160 ± 197), colon adenocarcinoma (236 ± 632), and lung squamous (90 ± 81). Therefore, overall number of ADNs, like CDNs, is variable and closely related to the number of NSMs.
To determine whether ADNs and CDNs are distinct at the molecular level, we investigated amino acid composition by HLA allele and position across all samples (Fig. 2E). Overall, an enrichment of nonpolar, hydrophobic and neutral, hydrophilic mutant amino acids existed for all neopeptide categories. ADNs and CDNs differed in the position and identity of the mutant amino acid. For common HLA alleles A*02:01 and DRB1*04:01, frequent amino acid substitutions were valine, glutamine, and leucine. Yet these three amino acids represented a larger proportion of total mutant amino acid substitutions for ADNs compared with CDNs (71% vs. 31%; P <0.001). Although the mutant amino acid position was distributed evenly along the length of CDNs in the binding groove of MHC, mutant amino acids in ADNs were concentrated at anchor positions, an enrichment that logically follows from our selection criteria (Fig. 2E; ref. 26). These results demonstrate that ADNs have a conserved molecular profile distinct from CDNs.
Given this substantial difference in peptide composition between ADNs and CDNs, we hypothesized that ADNs may be generated differently from NSMs. To determine this, we calculated the neopeptide generator rate as:
This metric measures the propensity of NSMs to generate multiple immunogenic peptides, which may correlate with contribution to tumor immunogenicity. The generator rate differed substantially between ADNs and CDNs, as well as across tumor types (Fig. 2F) and was higher for MHC class I ADNs compared with CDNs (FDR < 1 × 10−10). The ADN generator rate was also significantly higher in melanoma, liver, and lung squamous tumor types (FDR < 0.01 vs. all other tumor types). Compared with CDNs, these results show: (i) ADNs arise at higher rates from a smaller pool of NSMs compared with CDNs and (ii) the ADN generator rate is elevated in certain tumor types, likely due to differences in underlying NSM composition. These results are consistent with the constrained profile of mutant amino acid location and identity in ADNs. In contrast, for CDNs, amino acid substitutions were nearly random across the length of peptides, and no significant differences in the generator rate existed across tumor types. Thus, somatic mutations that generate ADNs differ in quality and prevalence across tumor types.
Distinct tumor microenvironment immune associations
To quantify immune dynamics, we measured cytolytic, suppressive, and T-cell dysfunction immune indices and compared expression across TCGA samples and tumor types. Rooney and colleagues developed a metric to assess cytolytic T-cell activity based on expression of granzyme A (GZMA) and perforin-1 (PRF1; cytolytic index; refs. 40, 41). Cytolytic index reflects the presence of activated CD8+ cytolytic T cells and correlates with other metrics of antitumor T-cell immunity. We have previously identified a gene expression index to quantify activation of immune suppressive molecules (suppressive index; ref. 14). Across all TCGA tumor types, cytolytic T-cell activity was correlated with expression of immune suppressive pathways as measured by these indices (Fig. 3A and B top; R2 = 0.66, P < 1 × 10−200). This correlation suggests that cytolytic and suppressive indices are tightly coupled regardless of mutation or neopeptide load. Identified outliers included renal clear cell carcinoma (higher cytolytic index) and low-grade glioma (higher suppressive index; ratio FDR < 0.01 vs. all other tumor types). In contrast to the strong correlation between cytolytic and suppressive indices, a weaker correlation existed between these indices and neopeptide load (Fig. 3A top vs. bottom; P < 9.423 × 10−58 for pan-TCGA immune indices vs. each neoepitope category).
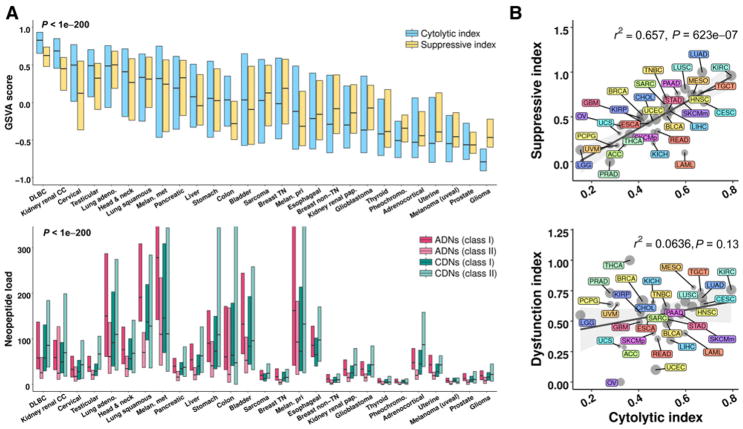
Figure 3.
Immune activity and neopeptide load correlate across tumor types. A, Cytolytic index and suppressive index across tumor types (top) and corresponding neopeptide load for the four indicated categories (bottom). Cytolytic index: expressed as the GSVA score of normalized GZMA and PRF1 expression (40, 41). Suppressive index: the GSVA score of normalized expression for ADORA2A (A2AR), CD274 (PD-L1), PDCD1 (PD1), CTLA4, HAVCR2 (TIM3), IDO1, IDO2, PDCD1LG2 (PD-L2), TIGIT, VISTA (C10orf54), and VTCN1 (B7-H4). Box and line, the 75th to 25th percentiles and median, respectively. Tumor types are ordered from left to right by median cytolytic index. P value: ANOVA across tumor types. B, Cytolytic index (x-axis) and suppressive index (top y-axis) or dysfunction index (bottom y-axis). Dysfunction index: GSVA score of tumor dysfunction genes identified by (37). R2 values: linear regression. P values: Spearman rho. Dot size, sample number. Shaded gray region, confidence interval.
To further assess immune dynamics across tumor types, we also measured T-cell dysfunction, using an index characterized by Schietinger and colleagues (ref. 37; dysfunction index). This index marks a dysfunctional state of tumor-specific human T cells that is distinct from cytolytic index, suppressive index, and virally induced T-cell exhaustion. In contrast to the suppressive index, no significant correlation existed between cytolytic index and dysfunction index, suggesting that tumor-specific T-cell dysfunction, as measured by this index, is either not well-defined by tumor type or that these immune characteristics are decoupled (Fig. 3B, bottom).
Analysis of individual genes comprising the suppressive index revealed that VISTA, IDO1, A2AR, and TIM3 had similarly high expression across tumor types by hierarchical clustering (Supplementary Fig. S4A) and were significantly correlated to cytolytic index (Supplementary Fig. S4B). These checkpoint molecules contribute to potential mechanisms of immune evasion (42), and our data suggest they are coordinately expressed. TIGIT, PD-1, and CTLA-4 expression were also correlated (Supplementary Fig. S4A) and were the most tightly coupled to cytolytic index (Supplementary Fig. S4B), consistent with upregulation of these molecules in association with CD8+ T-cell activation (42). TIGIT is a coinhibitory receptor expressed on T cells and NK cells that regulates T-cell activity, in part, through interaction with its binding partner PVR on antigen-presenting cells (43).
We next assessed relationships between ADNs and immune indices. Across tumor types, mean ADN or CDN load correlated with mean CD8A expression, cytolytic index, and suppressive index (Supplementary Fig. S5). An inverse correlation between mean ADN and dysfunction index approached statistical significance (P = 0.081). In contrast, despite adequate power to detect small effects, few meaningful correlations were detectable between ADN load and the same immune indices case-by-case across samples within each tumor type (Supplementary Fig. S6). These data suggest that, in parallel to the relationship between cytolytic index and suppressive index, the relationship between ADN (or CDN) load and T-cell response, as measured by these immune indices, is defined at the tumor type level, not case-by-case within a tumor type. Thus, the relationship between ADN load and T-cell response predominantly depends on tumor histotype.
Correlation of overall survival and ADNs versus CDNs
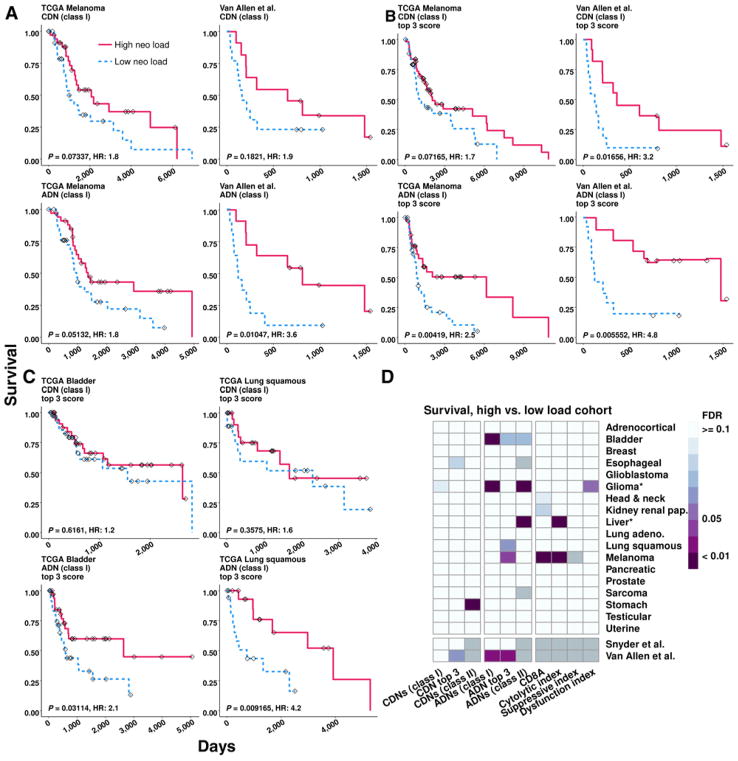
To assess the impact of neopeptide load on patient outcome, we evaluated whether an association between overall survival and ADN or CDN load existed for each TCGA tumor type. Increased load of predicted immunogenic mutations has been associated with significantly improved survival in a study of 512 patients across six tumor types (44). This association with improved survival was found with neopeptides (classified as any peptide with an IC50 < 500 nmol/L for MHC class I) but not with high NSM load or when controling for tumor type. Extending this analysis, we stratified samples in each tumor type by neopeptide load into high- and low-load cohorts (top vs. bottom decile or quartile). We hypothesized that significant differences in immunobiology between these cohorts may allow us to uncover associations with survival. In many tumor types, a minority of tumors respond to checkpoint blockade (45, 46). Similarly, many tumor types have a small, transcriptionally identified “immunogenic” subtype (47–49), and high intratumoral T-cell activity or infiltration is a strong predictor of patient survival (50–52). Stratification by top and bottom deciles corresponds roughly to the minimal size of these identified subtypes. To better model the potential for oligoclonal antitumor responses directed against neopeptides, samples were additionally stratified by a “top three neopeptide score,” which was defined as the sum of the top three affinity scores ( ) for CDNs or the sum of top three DAIs for ADNs. The top three were chosen in each case because this is the minimum number that captures the potential for an oligoclonal T-cell response and mirrors experimentally confirmed oligoclonality of T-cell responses against human tumors (53, 54). The top three score was the least correlated to total neopeptide load (vs. the top 4 through the top 15; R2 = 0.0495) and, therefore, not purely a derivative of total neopeptide load.
We focused first on melanoma because neopeptide load has been shown to correlate with response in this tumor type (3, 9). In both TCGA melanoma and a second, independent melanoma dataset from Van Allen and colleagues (3), we found that high MHC class I ADN load correlated with improved survival (Fig. 4A; P =0.05132 and P =0.01047, respectively). A high top three score also correlated strongly with improved survival in TCGA melanoma (ADNs only) and the Van Allen and colleagues dataset (ADNs and CDNs; Fig. 4B). Using response to checkpoint blockade, the CDN top 3 score differentiated lung adenocarcinoma patients who responded from those who did not (Supplementary Fig. S7; P = 0.0313). In TCGA bladder and lung squamous tumor types, a high top three score for ADNs, but not CDNs, also correlated strongly with improved survival (Fig. 4C). Overall, significantly improved survival was observed in high ADN, but not CDN, load samples in multiple TCGA tumor types after adjustment for multiple hypothesis testing, with the exception of class II CDN load in stomach adenocarcinoma (Fig. 4D, left; Supplementary Table S1). Few significant correlations between survival and cytolytic, suppressive, or dysfunction indices were detected, with the exception of melanoma (Fig. 4D, right). Similar results were obtained by using a 500 nmol/L IC50 cutoff for MHC class I CDNs, a 1,000 nmol/L IC50 cutoff for class I ADNs, or by comparing patients in the high and low neopeptide-load quartiles (Supplementary Table S1).
Figure 4.
ADN load is associated with survival. A–C, Kaplan–Meier curves for the indicated neopeptide category and tumor type showing survival difference between samples with high (red) vs. low (blue) neopeptide load, defined as the top and bottom deciles. Tick marks, time of last known survival status. P value is unadjusted log rank. D, Summary of log-rank FDR values. Comparison is between high vs. low neopeptide load samples. Asterisk, improved survival in the low-neopeptide load cohort; gray box: data not available. Tumor types with <50 samples with neopeptide predictions were excluded. Number of observations for each tumor is listed in Supplementary Table S1.
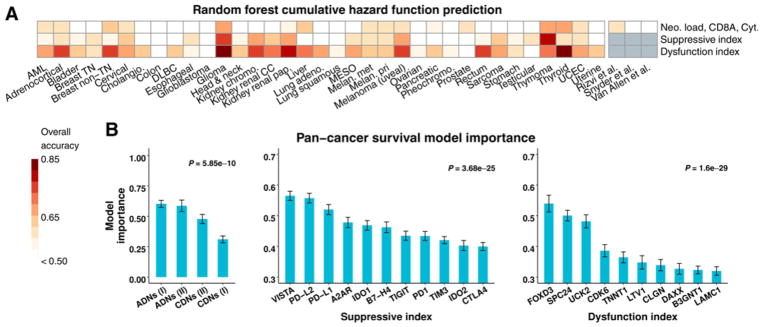
We next confirmed and extended these associations between neopeptide load and survival using random forest (RF) machine learning, an unbiased, robust analytical method (described in Materials and Methods). This analysis measured the ability of ADN load, CDN load, and immune indices to predict cumulative hazard function (CHF) for each tumor type, as well as independent melanoma and lung adenocarcinoma datasets (3, 4, 16). CHF is the integrated hazard function over time, and CHF prediction error estimates the probability that, in two out-of-bag samples, the case with the worse predicted outcome had an event first. RF analysis confirmed our Kaplan–Meier estimate findings, showing ability for ADN load, CDN load, CD8A expression, and cytolytic index to jointly predict CHF in TCGA melanoma, when considered as independent input variables (Fig. 5A; ref. 3). Thus, similar to Kaplan–Meier analysis of the high and low neopeptide load cohorts, RF analysis demonstrated that neopeptide load contributed to the ability to predict survival in some tumor types.
Figure 5.
ADNs are strong predictors of survival. A, Overall cumulative hazard function random forest model prediction accuracy for each TCGA tumor type. Model input variables were ADN and CDN load, CD8A expression, and cytolytic index (top), suppressive index genes (middle) or dysfunction index genes (bottom). An accuracy of 0.5 is equivalent to random guessing. B, Pan-cancer survival model importance. X-axis: model input variables from predictive models, ordered from left to right by importance. P values determined by ANOVA. Error bars, SEM in cross-validation replicates. Only top variables for the dysfunction index are shown.
Suppressive and dysfunction indices were, in general, the best predictors of survival compared with neopeptide load, resulting in accurate prediction of CHF in many tumor types (Fig. 5A). Assessing input variable importance to prediction, class I ADN load was the most important to prediction accuracy and class I CDN load was the least important in predictive models, including only ADN and CDN load (Fig. 5B). For immunosuppressive index, the most important individual genes for survival prediction were VISTA, PD-L1, and PD-L2. VISTA is an inhibitory checkpoint molecule expressed on antigen-presenting cells that is increased after therapy with CTLA-4 blockade in patients (55). In summary, ADNs outperformed CDNs for predicting survival, and immune suppressive molecules—VISTA in particular—and T-cell dysfunction index are strongly predictive of survival across TCGA tumor types.
Discussion
Large-scale studies continue to provide insight into the profound influence of the tumor genome on tumor-immune interactions. We investigated the landscape of neopeptides across human cancer using TCGA, exploring relationships in 27 solid tumor types and evaluating more than 228 million mutant peptide predictions. Our analyses characterized a class of prevalent neopeptides—both MHC class I and II—which we termed ADNs and defined based on improved MHC binding of mutant peptides compared with their nonmutant counterparts. ADNs are molecularly distinct from CDNs and strongly predict patient survival, indicating their biological significance and their potential as an underappreciated source of therapeutic antigens. Nevertheless, suppressive and dysfunction gene signature indices were, in general, superior predictors of survival compared with load of ADN neopeptides or load of CDN neopeptides, emphasizing the importance of tumor cell–extrinsic immune parameters in reflecting cancer immunobiology.
ADNs are generated from selective mutations in the peptide–MHC anchor position (i.e., the agretope) rather than mutations randomly occurring across the peptide sequence. This feature leads to two potentially important and unique immunological characteristics of ADNs. First, unlike CDNs, the TCR-facing peptide sequence in ADNs is likely the same as the corresponding nonmutant peptide. Second, MHC binding ability of the corresponding nonmutant peptide may be so low that its presentation in the thymus is minimal and central tolerance may be bypassed. Experimental evidence provided in previous studies suggests that neopeptides derived from ADNs can be immunogenic. Peptides containing anchor residue mutations, which in retrospect satisfy our ADN selection criteria, are overrepresented among murine and human tumor antigens that had been experimentally confirmed to be immunogenic (23, 26). Mutations in known peptide–MHC anchor residues are very unlikely to alter TCR recognition, providing explicit evidence of the potential immunogenicity of ADNs that escape central tolerance due to poor thymic presentation. An extensive analysis of tumor immunity in a patient with ovarian carcinoma showed that the top six reactive mutant peptides meet our definition of ADNs (56). Our reanalysis of experimentally validated neopeptides from non–small cell lung carcinoma or melanoma patients (54, 57) showed that one third of these were ADNs (resulting from an anchor position substitution that improved MHC affinity > 10-fold). Thus, our findings provide selection criteria to expand the spectrum of mutation-derived tumor antigens with therapeutic potential.
ADNs also exhibit unique associations with patient outcome. High ADN load was associated with improved overall survival in multiple TCGA tumor types, in contrast to CDN load, for which, only a single significant association with survival existed after adjustment for multiple hypothesis testing. Our result differs from that reported by Van Allen and colleagues (3) due to this adjustment. RF analysis of survival provided confirmation of these findings, demonstrating that ADN load was more important to survival prediction than CDN load across all tumor types. Work in pancreatic adenocarcinoma, lung adenocarcinoma, and melanoma has also identified differential agretopicity as a key neopeptide selection criteria, in conjunction with homology to infectious disease-derived peptides, for correlating neopeptide load with survival (58, 59). Our analysis is a comprehensive investigation of the landscape of human neopeptides and provides a timely and critical comparison of CDNs and ADNs for multiple tumor types, peptide lengths, and MHC class I and II. Evaluation of the ability of neopeptide selection criteria that incorporate differential agretopicity to predict immunogenicity and response will be needed in future prospective human studies.
We extended our analyses beyond total load of ADNs or CDNs and also evaluated survival as a function of the predicted highest quality peptides for each patient (i.e., our top three neopeptide score analysis). Using this, we found an even stronger association of survival and the top three score, potentially reflecting the fact that most potent antitumor T-cell responses are oligoclonal (53). A straightforward top three score is immediately translationally and clinically useful because it does not require reconstruction of tumor clonal structure or quantification of tumor evolutionary dynamics. The top three score is not tightly correlated with total neopeptide load, and focusing on the few highest quality ADNs may also provide a strategy to narrow the choice of peptides to be used in patient vaccination from among dozens or hundreds of ADNs (or CDNs) that are otherwise bioinformatically indistinguishable with regard to antigenicity. Finally, the significant associations we identified using a CDN top three score demonstrated that refined methods of quantifying classic, high-affinity neopeptides may yet hold promise, especially given that some high-affinity clones are immunogenic regardless of differential agretopicity (23, 54).
Neither ADNs nor CDNs were widely shared across samples in TCGA. One potential exception is IDH1 p.R132H, which was the source of shared CDNs across 90 samples (1.5% of total samples), with an average predicted IC50 of 33 nmol/L. Peptides generated from this site were predicted to bind HLA alleles, including A*31:01, A*33:03, A*68:01, A*68:03, A*29:01, A*29:02, B*35:01, B*35:14, and B*35:15 (approximate combined U.S. allele frequency: 27.3%; ref. 39). If these predictions are experimentally confirmed, IDH p.R132H may be an immune target many times more frequent than other suggested shared neopeptides such as KRAS p.G12D in C*08:02+ patients, which has an HLA allele frequency of only 4% to 8% in non-Asian, non-Ashkenazi Jewish individuals in the United States (39, 60, 61). Other than IDH p.R132H, even among commonly mutated oncogenic drivers, tumor variant and HLA allele diversity likely precludes shared neopeptides at frequencies amenable to therapeutic targeting.
The overall landscape of tumor neopeptides we revealed here suggests that T-cell immunity to checkpoint blockade or other strategies is not limited to high mutation load patients or tumor types. Our data indicate that a higher quantity of tumor-specific neopeptides alone—ADNs, CDNs, or both—is not necessarily sufficient to elicit a positive immune response discernable by the transcriptome or survival. Many tumor types with high mutation rates were not associated with improved survival in patients belonging to the top decile by mutation or neopeptide load. In low-grade glioma and liver adenocarcinoma, survival and neo-peptide load were negatively correlated. Studies have suggested a cutoff of 192 NSMs per tumor—based on published checkpoint inhibitor response rates—to discriminate patients likely to respond to checkpoint blockade (62). These likely responders include bladder, colon, gastric, and endometrial cancers. Although these patients may be more likely to respond, only a small number of neopeptides are required to drive therapeutically effective antitumor T-cell responses (63). Landmark studies have shown that patients with renal clear-cell carcinoma commonly respond to immune checkpoint therapy despite a low median neopeptide load relative to other tumor types with poorer rate of response, such as colorectal adenocarcinoma (45, 46). We found that neopeptide-immune associations primarily correlated by tumor type, not case-by-case within a tumor type, indicating that tumor histotype plays a dominant role in dictating the nature of the immune response. Thus, our findings support the hypothesis that T-cell immunity in solid tumors is not primarily determined by the abundance of T-cell antigens derived from somatic missense mutations.
Efforts to enhance immunomodulatory strategies such as CTLA-4 and PD-1 blockade may need prioritized consideration of tumor microenvironmental cues on a case-by-case basis. This notion is supported by our ability to model survival more accurately using T-cell suppressive and dysfunction genes, such as VISTA (ref. 55; was highly expressed, was an important predictor of survival, and did not correlate with total neopeptide load; P = 0.49). In summary, MHC class I and II ADNs are prevalent across human cancer and correlate with immunogenicity and survival. Our results demonstrate that the potential pool of existing immune targets is more diverse than currently appreciated. These ADN selection criteria can be used in vaccine studies to evaluate the importance of ADNs as tumor rejection antigens alongside CDNs.
Supplementary Material
Acknowledgments
This work was supported by the NIH (R01-CA169123 to B.Z. Stanger and R.H. Vonderheide) and the Parker Institute for Cancer Immunotherapy (A.J. Rech, K.L. Nathanson, B.Z. Stanger, R.H. Vonderheide).
We gratefully acknowledge Katelyn T. Byrne, Beatriz M. Carreno, Gerald P. Linette, and Mingen Liu for helpful discussions; Lee P. Richman for software development; and The Cancer Genome Atlas/Genome Data Commons (https://portal.gdc.cancer.gov).
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ Contributions
Conception and design: A.J. Rech, D. Balli, B.Z. Stanger, R.H. Vonderheide
Development of methodology: A.J. Rech, D. Balli, H. Ishwaran, K.L. Nathanson, R.H. Vonderheide
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): A.J. Rech, D. Balli, R.H. Vonderheide
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): A.J. Rech, D. Balli, A. Mantero, H. Ishwaran, K.L. Nathanson, B.Z. Stanger, R.H. Vonderheide
Writing, review, and/or revision of the manuscript: A.J. Rech, D. Balli, A. Mantero, K.L. Nathanson, B.Z. Stanger, R.H. Vonderheide
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): A.J. Rech, R.H. Vonderheide
Study supervision: R.H. Vonderheide
References
- 1.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 2.Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016;15:857–65. doi: 10.1016/j.celrep.2016.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–11. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–20. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strønen E, Toebes M, Kelderman S, van Buuren MM, Yang W, van Rooij N, et al. Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science. 2016;352:1337–41. doi: 10.1126/science.aaf2288. [DOI] [PubMed] [Google Scholar]
- 9.Nathanson T, Ahuja A, Rubinsteyn A, Aksoy BA, Hellmann MD, Miao D, et al. Somatic mutations and neoepitope homology in melanomas treated with CTLA-4 blockade. Cancer Immunol Res. 2017;5:84–91. doi: 10.1158/2326-6066.CIR-16-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spranger S, Luke JJ, Bao R, Zha Y, Hernandez KM, Li Y, et al. Density of immunogenic antigens does not explain the presence or absence of the T-cell-inflamed tumor microenvironment in melanoma. Proc Natl Acad Sci USA. 2016;113:E7759–68. doi: 10.1073/pnas.1609376113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Favero F, Joshi T, Marquard AM, Birkbak NJ, Krzystanek M, Li Q, et al. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann Oncol. 2014;26:64–70. doi: 10.1093/annonc/mdu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–21. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balli D, Rech AJ, Stanger BZ, Vonderheide RH. Immune cytolytic activity stratifies molecular subsets of human pancreatic cancer. Clin Cancer Res. 2017;23:3129–38. doi: 10.1158/1078-0432.CCR-16-2128. [DOI] [PubMed] [Google Scholar]
- 15.Şenbabaoğlu Y, Gejman RS, Winer AG, Liu M, Van Allen EM, de Velasco G, et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol. 2016;17:231. doi: 10.1186/s13059-016-1092-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snyder A, Makarov V, Hellmann M, Rizvi N, Merghoub T, Wolchok JD, et al. Genetics and immunology: reinvigorated. OncoImmunology. 2015;4:e1029705. doi: 10.1080/2162402X.2015.1029705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szolek A, Schubert B, Mohr C, Sturm M, Feldhahn M, Kohlbacher O. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics. 2014;30:3310–6. doi: 10.1093/bioinformatics/btu548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shukla SA, Rooney MS, Rajasagi M, Tiao G, Dixon PM, Lawrence MS, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol. 2015;33:1152–8. doi: 10.1038/nbt.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y, Yang J, Ying D, Zhang Y, Shotelersuk V, Hirankarn N, et al. HLA reporter: a tool for HLA typing from next generation sequencing data. Genome Med. 2015;7:25. doi: 10.1186/s13073-015-0145-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly. 2014;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim Y, Ponomarenko J, Zhu Z, Tamang D, Wang P, Greenbaum J, et al. Immune epitope database analysis resource. Nucleic Acids Res. 2012;40:W525–30. doi: 10.1093/nar/gks438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paul S, Weiskopf D, Angelo MA, Sidney J, Peters B, Sette A. HLA class I alleles are associated with peptide-binding repertoires of different size, affinity, and immunogenicity. J Immunol. 2013;191:5831–9. doi: 10.4049/jimmunol.1302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fritsch EF, Rajasagi M, Ott PA, Brusic V, Hacohen N, Wu CJ. HLA-binding properties of tumor neoepitopes in humans. Cancer Immunol Res. 2014;2:522–9. doi: 10.1158/2326-6066.CIR-13-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang P, Sidney J, Dow C, Mothé B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. 2008;4:e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kreiter S, Vormehr M, van de Roemer N, Diken M, Löwer M, Diekmann J, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520:692–6. doi: 10.1038/nature14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duan F, Duitama J, Al Seesi S, Ayres CM, Corcelli SA, Pawashe AP, et al. Genomicandbioinformaticprofilingofmutationalneoepitopesrevealsnew rules to predict anticancer immunogenicity. J Exp Med. 2014;211:2231–48. doi: 10.1084/jem.20141308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breiman L. Machine learning. Berkeley, CA: Statistics Department, University of California; 2001. Random forests. [Google Scholar]
- 28.Chen X, Ishwaran H. Random forests for genomic data analysis. Genomics. 2012;99:323–9. doi: 10.1016/j.ygeno.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS. Random survival forests. Ann Appl Stat. 2008;2:841–60. [Google Scholar]
- 30.Ishwaran H, Kogalur UB. Consistency of random survival forests. Stat Probab Lett. 2010;80:1056–64. doi: 10.1016/j.spl.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boulesteix A-L, Janitza S, Kruppa J, König IR. Overview of random forest methodology and practical guidance with emphasis on computational biology and bioinformatics. Wiley Interdiscip Rev Data Min Knowl Discov. 2012;2:493–507. [Google Scholar]
- 32.Twyman-Saint VC, Rech AJ, Maity A, Rengan R, Pauken KE, Stelekati E, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 2015;520:373–7. doi: 10.1038/nature14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishwaran H, Gerds TA, Kogalur UB, Moore RD, Gange SJ, Lau BM. Random survival forests for competing risks. Biostatistics. 2014;15:757–73. doi: 10.1093/biostatistics/kxu010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishwaran H, Kogalur UB, Gorodeski EZ, Minn AJ, Lauer MS. High-dimensional variable selection for survival data. J Am Sta tAssoc. 2010;105:205–17. [Google Scholar]
- 35.Strobl C, Boulesteix A-L, Kneib T, Augustin T, Zeileis A. Conditional variable importance for random forests. BMC Bioinformatics. 2008;9:307. doi: 10.1186/1471-2105-9-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schietinger A, Philip M, Krisnawan VE, Chiu EY. Tumor-specific T cell dysfunction is a dynamic antigen-driven differentiation program initiated early during tumorigenesis. Immunity. 2016;45:389–401. doi: 10.1016/j.immuni.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer; 2016. [Google Scholar]
- 39.González-Galarza FF, Takeshita LYC, Santos EJM, Kempson F, Maia MHT, da Silva ALS, et al. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 2015;43:D784–8. doi: 10.1093/nar/gku1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson BJ, Costelloe EO, Fitzpatrick DR, Haanen JBAG, Schumacher TNM, Brown LE, et al. Single-cell perforin and granzyme expression reveals the anatomical localization of effector CD8+ T cells in influenza virus-infected mice. Proc Natl Acad Sci USA. 2003;100:2657–62. doi: 10.1073/pnas.0538056100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–61. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu X, Harden K, Gonzalez LC, Francesco M, Chiang E, Irving B, et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol. 2009;10:48–57. doi: 10.1038/ni.1674. [DOI] [PubMed] [Google Scholar]
- 44.Brown SD, Warren RL, Gibb EA, Martin SD, Spinelli JJ, Nelson BH, et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 2014;24:743–50. doi: 10.1101/gr.165985.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brahmer JR, Tykodi SS, Chow LQM, Hwu W-J, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–65. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of antiPD-1 antibody in cancer. N Engl J Med. 2012;366:2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–96. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bailey P, Chang DK, Nones K, Johns AL, Patch A-M, Gingras M-C, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 50.Mlecnik B, Bindea G, Angell HK, Maby P, Angelova M, Tougeron D, et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity. 2016;44:698–711. doi: 10.1016/j.immuni.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 51.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 52.Schumacher K, Haensch W, Röefzaad C, Schlag PM. Prognostic significance of activated CD8(+) T cell infiltrations within esophageal carcinomas. Cancer Res. 2001;61:3932–6. [PubMed] [Google Scholar]
- 53.Gros A, Parkhurst MR, Tran E, Pasetto A, Robbins PF, Ilyas S, et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med. 2016;22:433–8. doi: 10.1038/nm.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carreno BM, Magrini V, Becker-Hapak M, Kaabinejadian S, Hundal J, Petti AA, et al. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 2015;348:803–8. doi: 10.1126/science.aaa3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao J, Ward JF, Pettaway CA, Shi LZ, Subudhi SK, Vence LM, et al. VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer. Nat Med. 2017;57:3325–34. doi: 10.1038/nm.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiménez-Sánchez A, Memon D, Pourpe S, Veeraraghavan H, Li Y, Vargas HA, et al. Heterogeneous tumor-immune microenvironments among differentially growing metastases in an ovarian cancer patient. Cell. 2017;170:927–938. e20. doi: 10.1016/j.cell.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McGranahan N, Furness AJS, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–9. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Balachandran VP, èuksza M, Zhao JN, Makarov V, Moral JA, Remark R, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551:512–6. doi: 10.1038/nature24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.èuksza M, Riaz N, Makarov V, Balachandran VP, Hellmann MD, Solovyov A, et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature. 2017;551:517–20. doi: 10.1038/nature24473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tran E, Robbins PF, Lu Y-C, Prickett TD, Gartner JJ, Jia L, et al. T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med. 2016;375:2255–62. doi: 10.1056/NEJMoa1609279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rech AJ, Vonderheide RH. T-cell transfer therapy targeting mutant KRAS. N Engl J Med. 2017;376:e11. doi: 10.1056/NEJMc1616637. [DOI] [PubMed] [Google Scholar]
- 62.Colli LM, Machiela MJ, Myers TA, Jessop L, Yu K, Chanock SJ. Burden of nonsynonymous mutations among TCGA cancers and candidate immune checkpoint inhibitor responses. Cancer Res. 2016;76:3767–72. doi: 10.1158/0008-5472.CAN-16-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu YC, Yao X, Li YF, El-Gamil M, Dudley ME, Yang JC, et al. Mutated PPP1R3B is recognized by T cells used to treat a melanoma patient who experienced a durable complete tumor regression. J Immunol. 2013;190:6034–42. doi: 10.4049/jimmunol.1202830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.