Abstract
Fatal human cases of avian-origin H10N8 influenza virus infections have raised concern about their potential for human-to-human transmission. H10 subtype avian influenza viruses (AIVs) have been isolated from wild and domestic aquatic birds across Eurasia and North America. We isolated eight H10 AIVs (four H10N7, two H10N9, one H10N1, and one H10N6) from live poultry markets in Bangladesh. Genetic analyses demonstrated that all eight isolates belong to the Eurasian lineage. HA phylogenetic and antigenic analyses indicated that two antigenically distinct groups of H10 AIVs are circulating in Bangladeshi live poultry markets. We evaluated the virulence of four representative H10 AIV strains in DBA/2J mice and found that they replicated efficiently in mice without prior adaptation. Moreover, H10N6 and H10N1 AIVs caused high mortality with systemic dissemination. These results indicate that H10 AIVs pose a potential threat to human health and the mechanisms of their transmissibility should be elucidated.
Introduction
Avian influenza viruses (AIVs) pose continual challenges to human and animal health worldwide. AIVs belong to the family Orthomyxoviridae and are classified on the basis of their surface glycoproteins hemagglutinin (HA) and neuraminidase (NA). Currently, 18 HA subtypes (H1–H18) and 11 NA subtypes (N1–N11) have been identified and, with exception of the H17N10 and H18N11 subtypes found in bats, all AIV subtypes are found in aquatic birds1–4. Only subtypes H1, H2, and H3 are known to be transmissible between humans or associated with human pandemics. However, subtypes H5, H6, H7, H9, and H10 can cross the species barrier from birds to mammals, including humans, and cause sporadic infections but have not yet acquired the ability to transmit between humans5–8. The reasons for this remain unclear. In 2013, three subtypes of AIVs were able to cross the species barrier and infect humans. In February 2013, an H7N9 AIV emerged in China and 1,566 laboratory-confirmed cases of human infection resulted in 613 deaths9. In May 2013, H6N1 AIV infections in humans occurred in Taiwan10. In December 2013, three human cases of H10N8 AIV infection were reported in China5.
A subtype H10 influenza virus was first isolated from a chicken in Germany in 194911. Since then, H10 AIVs have been isolated with increasing frequency from wild and domestic aquatic and terrestrial avian species. H10 AIVs are now divided into two lineages: Eurasian and North American12. The first human infections with H10N7 AIV were reported in 2004 in Egypt13. An additional two cases were then confirmed in Australia in 201014. Each of these human cases was associated with either interactions with, or proximity to, live poultry. H10N7 AIVs were isolated from hunted wild ducks in a live bird market in Egypt and from chickens processed on a farm in Australia that was associated with the 2010 H10N7 poultry outbreak. Those who acquired H10N8 infections were known to have a history of visiting live poultry markets (LPMs) or exposure to live poultry5. Currently, sustained human-to-human transmission has not been reported for H10 AIVs.
H10 AIVs have been isolated from various mammalian hosts, including harbor seals in northwestern Europe15,16. H10N8 AIV was infectious among feral dogs in LPMs in Guangdong Province, China17. Several Eurasian and North American–origin H10 AIVs replicate and cause weight loss in ferrets18. Therefore, H10 AIVs can cause disease in mammals and have the potential to pose a potential public health threat.
LPMs play a critical role in maintaining, amplifying, and disseminating AIVs among poultry species and from poultry to humans. After the first outbreak of highly pathogenic H5N1 AIV in 2007, ongoing active surveillance of poultry in Bangladesh has been conducted. A variety of AIV subtypes, including H5 and H9, have been isolated in LPMs in Bangladesh19. The circulation of H5N1 with other AIVs subtypes may increase the likelihood for emerging pandemic strains through reassortment. Reassortment between the polymerase basic 1 (PB1) gene from the H5N1 and H9N2 AIVs has been observed20,21. Recently, we described the emergence of a novel genotype H5N1 AIV containing the HA, M, and NA genes of circulating Bangladeshi H5N1 AIVs and five genes from low pathogenic Eurasian-lineage AIVs22.
During our active surveillance in Bangladesh, we isolated H10 AIVs from LPMs from 2008 to 2017. To better understand the evolution of these H10 AIVs, we sequenced their full genomes and analyzed their genetic and antigenic characteristics and receptor-binding properties. We determined the growth kinetics and antiviral susceptibility for these viruses and evaluated their pathogenic potential in mice.
Results
Isolation of AIVs from Bangladeshi LPMs
We collected 29,305 samples from LPMs through active surveillance in Bangladesh from 2008 to 2017. We screened all samples and confirmed the H5 subtype with quantitative reverse transcription PCR (qRT-PCR). We then subtyped all samples that tested negative for the H5 subtype by HA and NA sequencing. We detected eight different HA subtypes of AIVs, including H1 (5 isolates), H3 (12 isolates), H4 (4 isolates), H5 (182 isolates), H6 (3 isolates), H7 (1 isolate), H9 (1165 isolates), and H10 (8 isolates)19. We noted that eight H10 subtype AIVs were isolated from samples collected from three different LPMs in Bangladesh. All of the AIVs were isolated from ducks, with the exception of one H10N7 AIV that was isolated from a chicken (Table 1).
Table 1.
H10 AIVs isolated from LPMs in Bangladesh.
| Isolate | Subtype | Abbreviation | Host | GenBank Accession Numbers |
|---|---|---|---|---|
| A/duck/Bangladesh/821/2009 | H10N7 | DK/BD821/09 H10N7 | Duck | MH071472, MH071458, MH071474, MH071475, MH071482, MH071500, MH071501, MH071464 |
| A/duck/Bangladesh/822/2009 | H10N7 | DK/BD822/09 H10N7 | Duck | MH071454, MH071477, MH071463, MH071479, MH071498, MH071481, MH071467, MH071499 |
| A/duck/Bangladesh/824/2009 | H10N7 | DK/BD824/09 H10N7 | Duck | MH071486, MH071502, MH071462, MH071452, MH071506, MH071496, MH071460, MH071473 |
| A/chicken/Bangladesh/842/2009 | H10N7 | CK/BD842/09 H10N7 | Chicken | MH071457, MH071493, MH071497, MH071465, MH071495, MH071470, MH071456, MH071491 |
| A/duck/Bangladesh/8987/2010 | H10N9 | DK/BD8987/10 H10N9 | Duck | MH071453, MH071461, MH071503, MH071490, MH071494, MH071484, MH071455, MH071488 |
| A/duck/Bangladesh/8988/2010 | H10N9 | DK/BD8988/10 H10N9 | Duck | MH071476, MH071492, MH071505, MH071468, MH071471, MH071478, MH071469, MH071483 |
| A/duck/Bangladesh/24035/2014 | H10N1 | DK/BD24035/14 H10N1 | Duck | KY616777, KY616773, KY616772, KY616787, KY616784, KY616756, KY616755, KY616753 |
| A/duck/Bangladesh/24268/2015 | H10N6 | Dk/BD24268/15 H10N6 | Duck | MH071485, MH071487, MH071451, MH071504, MH071459, MH071489, MH071480, MH071466 |
Molecular and phylogenetic analysis of H10 AIVs
To better understand the genetic relationship of the H10 viruses, we sequenced the complete genomes of eight H10 AIVs isolated from LPMs in Bangladesh. Phylogenetic analysis of the HA sequences revealed that they belonged to the Eurasian lineage. We classified the HA genes into two groups based on branch clustering of the phylogenetic tree (Fig. 1). Group 1 contained H10N7 and H10N9 AIVs in branches that also contained viruses from Europe and Asia. The HA gene of four H10N7 AIVs were most closely related to those of H10N7 AIVs isolated from Jiangxi, China in 2009. Group 2 contained H10N1 and H10N6 AIVs along with Asian AIVs and human H10N8 AIVs (Fig. 1).
Figure 1.
Phylogenetic trees for the HA gene of AIVs isolated from LPMs in Bangladesh. Phylogenetic analysis was performed with the neighbor-joining algorithm and the Kimura 2-parameter model. The reliability of the phylogenetic inference at each branch node was estimated by the bootstrap method with 1,000 replications. Evolutionary analyses were conducted with MEGA6 software. The H10 AIVs isolated during the surveillance period are denoted in red font.
The amino acid sequence of the HA protein of all H10 AIVs included a single arginine residue at the cleavage site of HA1 and HA2 (PELMQGR↓GLF), indicating that they were low pathogenic strains. We examined the HA proteins in these AIVs for the presence of N-linked glycosylation motifs by interrogating the proteins for N-(P-[S/T]-P) motifs, which indicate potential N-linked glycosylation sites. We identified four potential glycosylation sites in HA1 at positions 22, 38, 242, and 296, which are highly conserved in all H10 AIVs.
Phylogenetic analysis of the NA genes revealed that all NA subtypes (i.e., N1, N6, N7, and N9) belonged to the Eurasian lineage (Supplementary Fig. S1). The NA genes of four H10N7 AIVs were most closely related to those of H10N7 AIVs isolated from Jiangxi, China in 2009. The NA genes of the H10N9 and H10N6 AIVs were most closely related to those of AIVs isolated from free-range ducks in the Tanguar haor area in Bangladesh in 2015. This indicates that the NA genes may circulate in Bangladesh between poultry sold in LPMs and free-range ducks. The NA gene of the H10N1 AIV was most closely related to those of H6N1 AIVs that were previously isolated fromChina. In addition, we found that some North American–lineage AIVs from Alaska were related to those from the Eurasian lineage (Supplementary Fig. S1).
Phylogenetic analysis of the polymerase basic 2 (PB2) genes revealed that they formed two distinct groups within the Eurasian lineage, all belonging to the Eurasian lineage (Supplementary Fig. S2a). The PB2 gene of H10N7 and H10N9 AIVs were located in one group and shared 99% identity with the closest PB2 gene of A/ruddy shelduck/Mongolia/598C2/2009 (H7N3). The H10N6 AIV PB2 gene was most closely related to AIVs isolated from free-range ducks in the Tanguar haor area in Bangladesh. The PB2 gene of the H10N1 AIV shared 98% nucleotide identity with that of A/tufted duck/Mongolia/1409/2010 (H1N1).
The polymerase basic 1 (PB1) genes of eight H10 AIVs clustered into two groups and belonged to the Eurasian lineage (Supplementary Fig. S2b). All of our isolates expressed a PB1-F2 protein consisting of 90 amino acids. Furthermore, the H10N1 and H10N9 AIVs had an N66S substitution in PB1-F2 (Supplementary Table S1), which increases virulence and replication efficiency in mammalian hosts23. The polymerase acidic (PA) and nucleoprotein (NP) genes of the H10 AIVs clustered into two groups, all of which were of the Eurasian lineage (Supplementary Fig. S2c,d). The PA and NP genes of the H10N7 and H10N9 AIVs were related to those of AIVs isolated from Europe and Mongolia, whereas the NP gene of the H10N1 and H10N6 AIVs were most closely related to viruses isolated from Eastern China and free-range ducks in the Tanguar haor area in Bangladesh. The matrix (M) and nonstructural (NS) genes of all eight H10 AIVs segregated into two groups (Supplementary Fig. S2e,f). The amino acid substitutions at positions 26, 27, 30, 31, and 34 in the M2 protein facilitate resistance to some anti-influenza drugs (i.e., M2 ion channel blockers). We did not observe any of these substitutions in the H10 AIVs we isolated. However, we found that all H10 AIVs contained P42S and V149A substitutions in the NS1 protein, which are associated with virulence and pathogenicity in mammals24,25. We did not detect any deletions in the NS1 proteins in any of the H10 AIVs, and all were of allele A and Eurasian lineage.
Antigenic analysis
To determine the antigenic relations among H10 AIVs isolated from LPMs in Bangladesh, we used a panel of polyclonal and monoclonal antibodies against Eurasian and North American viruses. The H10 AIVs were homogeneous and similar to those of the Eurasian lineage (Table 2). Four H10 AIVs reacted with antisera against A/laughing gull/Delaware Bay/209/2013 (H10N8) and A/glaucous gull/Iceland/CDX16-4552/2015 (H10N7) from North America, although hemagglutination inhibition (HI) assay titers with the H10N1 and H10N6 AIVs were one- to two-fold higher than were those of the other AIVs (Table 2). Two monoclonal antibodies against A/chicken/Jiangxi/34609/2013 (H10N8) cross-reacted with the H10 AIVs at higher titers and one monoclonal antibody to H10 influenza virus (JX13-3) failed to react with any of the Bangladeshi H10 viruses (Table 2). We found that the H10N7 and H10N9 AIVs from group 1, as classified by their HA gene phylogenetic trees, exhibited low cross-reactivity with antisera against group 2 AIVs (e.g., H10N1). These results indicate that the H10 AIVs circulating in LPMs in Bangladesh during the surveillance period were undergoing antigenic drift.
Table 2.
Antigenic characterization by the hemagglutination inhibition assay of H10 AIVs isolated from LPMs in Bangladesh.
| Virus | Monoclonal antibodies | Polyclonal antibodies* | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| JX13-3 | JX13-4 | JX13-5 | N/49 | HK-562 | NL-1 | NL-p14-221 | JX13 | DE-209 | CDX16-4552 | BD-24035 | |
| Reference virus | |||||||||||
| A/chicken/Germany N/49 H10N7 | <100 | 1600 | <100 | 1280 | 80 | 160 | 80 | 320 | 320 | 160 | 80 |
| A/duck/Hong Kong/562/79 H10N9 | <100 | 3200 | 200 | 80 | 80 | 160 | 40 | 160 | 80 | 40 | 20 |
| A/mallard/Netherlands/1/2014 H10N7 | <100 | 3200 | 1600 | 80 | 160 | 160 | 80 | 160 | 80 | 80 | 20 |
| A/chicken/Jiangxi/34609/2013 H10N8 | 3200 | 3200 | 400 | 1280 | 320 | 640 | 320 | 1280 | 640 | 320 | 80 |
| Test virus | |||||||||||
| A/duck/Bangladesh/821/2009 H10N7 | <100 | 800 | 800 | 80 | 40 | 40 | 20 | 40 | 20 | 20 | 20 |
| A/duck/Bangladesh/8988/2010 H10N9 | <100 | 3200 | 400 | 80 | 160 | 160 | 80 | 160 | 80 | 40 | 40 |
| A/duck/Bangladesh/24035/2014 H10N1 | <100 | 3200 | 400 | 640 | 320 | 640 | 160 | 640 | 640 | 160 | 80 |
| A/duck/Bangladesh/24268/2015 H10N6 | <100 | 3200 | 400 | 320 | 320 | 320 | 160 | 320 | 320 | 160 | 80 |
*Polyclonal antibodies were produced in the ferret. Homologous titers are underlined. NL-p14-221, A/seal/Netherlands/p1/2014 (H10N7). DE-209, A/laughing gull/Delaware Bay/209/2013 (H10N8). CDX16-4552, A/glaucous gull/Iceland/CDX16-4552/2015 (H10N7).
Replication of H10 AIVs in mice
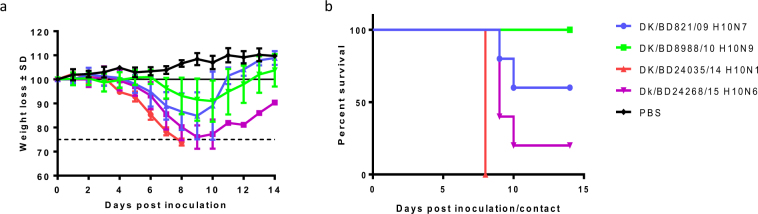
To investigate the replication and virulence of H10 AIVs in mammals, we inoculated groups of 17 DBA/2J mice with 106 50% egg infective dose (EID50) of each AIV. Three mice in each group were euthanized on 2, 4, 6, and 8 days post inoculation (dpi) to determine viral titers in the nasal turbinate, lung, spleen, heart, intestine, and brain. We observed the remaining five mice in each group for 14 days to determine their susceptibility to these AIVs. Mice infected with DK/BD24035/14 (H10N1), Dk/BD24268/15 (H10N6), and DK/BD821/09 (H10N7) experienced significant weight loss (>25%) (Fig. 2a). Four viruses exhibited differing degrees of virulence in mice, The DK/BD24035/14 (H10N1) caused 100% mortality by 8 dpi; Dk/BD24268/15 (H10N6) caused 80% mortality by 10 dpi; DK/BD821/09 (H10N7) caused 40% mortality by day 10 dpi; and DK/BD8988/10 (H10N9) did not cause mortality (Fig. 2b).
Figure 2.
Morbidity and mortality of H10 AIVs in mice. DBA/2J mice (n = 5) were inoculated intranasally with 30 μL of 106 EID50 DK/BD821/09 H10N7, DK/BD8988/10 H10N9, DK/BD24035/14 H10N1, or Dk/BD24268/15 H10N6. (a) Morbidity was examined by recording the body weights of inoculated mice daily, which is shown as the mean percent body weight from the day of inoculation (day 0). (b) Mortality was recorded as actual death or euthanasia at ≥25% weight loss, which was pre-established in our animal protocol.
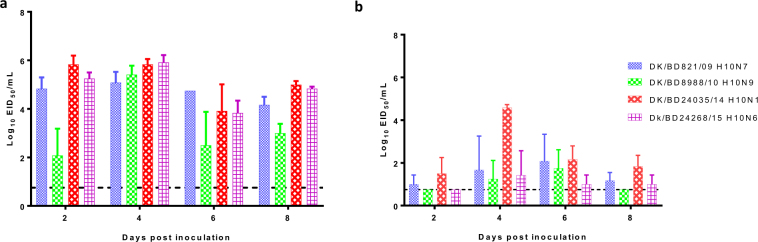
All viruses were detected in the lungs, with mean titers ranging from 3.1 to 5.8 log10 EID50 (Fig. 3a), and in nasal turbinates at all time points, with mean titers ranging from 1.5 to 4.75 log10 EID50 (Fig. 3b). The DK/BD24035/14 (H10N1) AIV titers were markedly higher in nasal turbinates than were the other isolates at 4 dpi (Fig. 3b). These data indicate that all the H10 isolates replicated in mice without prior adaptation.
Figure 3.
Replication of H10 AIVs in the lungs and nasal turbinates of inoculated mice. DBA/2J mice (n = 3) were infected with 106 EID50 of DK/BD821/09 H10N7, DK/BD8988/10 H10N9, DK/BD24035/14 H10N1, or Dk/BD24268/15 H10N6. Lungs (a) and nasal turbinates (b) were collected at 2, 4, 6, and 8 dpi, and viral titers were determined by EID50 assays. The results are expressed as log10 EID50 of tissue; error bars depict standard deviations; and the dotted line indicates the lower limit of detection of infectious virus.
We detected the two DK/BD24035/14 (H10N1) and Dk/BD24268/15 (H10N6) AIVs that caused high mortality in mice and were detected in the heart, liver, and brain tissues. In addition, we detected the DK/BD24035/14 (H10N1) AIV in the spleen at 2 and 4 dpi (Table 3). In contrast, we detected the DK/BD821/09 (H10N7) and DK/BD8988/10 (H10N9) AIVs in the heart and the DK/BD821/09 (H10N7) AIV in the liver at 2 and 4 dpi (Table 3). We did not detect any of the AIVs in the intestines of any of the mice.
Table 3.
Replication kinetics of H10 AIVs in extrapulmonary tissues in mice.
| Virus | Subtype | dpi | Viral titers (log10 EID50/mL) | ||||
|---|---|---|---|---|---|---|---|
| Heart | Spleen | Liver | Intestine | Brain | |||
| A/duck/Bangladesh/821/2009 | H10N7 | 2 | 2/3a (2.5 ± 1.4)b | 0/3 | 1/3 (2.5) | 0/3 | 0/3 |
| 4 | 1/3 (2.5) | 0/3 | 1/3 (2.5) | 0/3 | 0/3 | ||
| 6 | 1/3 (2.5) | 0/3 | 0/3 | 0/3 | 0/3 | ||
| 8 | 2/3 (1.75 ± 0.35) | 0/3 | 0/3 | 0/3 | 0/3 | ||
| A/duck/Bangladesh/8988/2010 | H10N9 | 2 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 |
| 4 | 1/3 (3.25) | 0/3 | 0/3 | 0/3 | 0/3 | ||
| 6 | 1/3 (3.5) | 0/3 | 0/3 | 0/3 | 0/3 | ||
| 8 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | ||
| A/duck/Bangladesh/24035/2014 | H10N1 | 2 | 2/3 (3 ± 0.35) | 1/3 (1.5) | 3/3 (2.92 ± 1.18) | 0/3 | 3/3 (2.25 ± 0.43) |
| 4 | 1/3 (1.5) | 1/3 (1.75) | 0/3 | 0/3 | 1/3 (1.5) | ||
| 6 | 3/3 (2.08 ± 0.57) | 0/3 | 2/3 (1.88 ± 0.53) | 0/3 | 2/3 (1.88 ± 0.53) | ||
| 8 | 3/3 (1.92 ± 0.72) | 0/3 | 2/3 (2.88 ± 1.94) | 0/3 | 2/3 (2 ± 0.35) | ||
| A/duck/Bangladesh/24268/2015 | H10N6 | 2 | 1/3 (1.5) | 0/3 | 1/3 (3.25) | 0/3 | 2/3 (2.13 ± 0.88) |
| 4 | 1/3 (1.5) | 0/3 | 0/3 | 0/3 | 1/3 (2.75) | ||
| 6 | 1/3 (1.5) | 0/3 | 0/3 | 0/3 | 1/3 (1.75) | ||
| 8 | 1/3 (1.5) | 0/3 | 0/3 | 0/3 | 0/3 | ||
aInfected mice were positive for virus detection /Total infected mice.
bThe titers are shown as the mean ± SD.
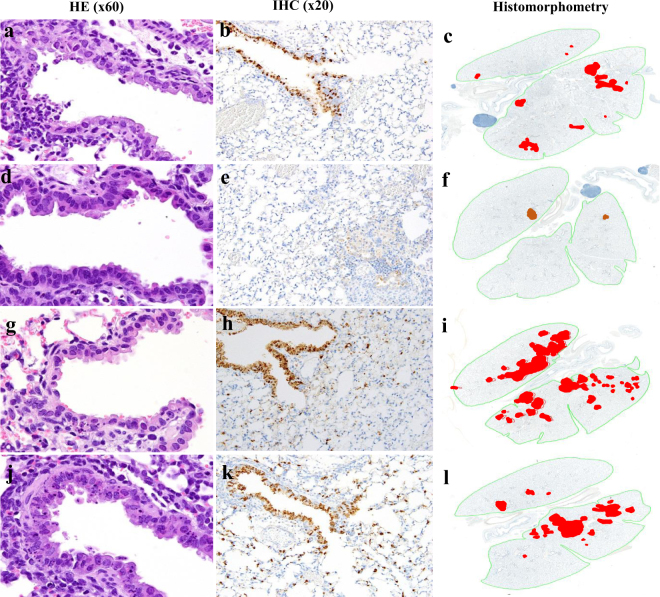
Histopathologic analysis of the lungs of the mice infected with the DK/BD8988/10 (H10N9) AIV revealed minimal viral antigens in bronchiolar epithelial cells and none in alveolar cells. Infection with the DK/BD821/09 (H10N7) AIV caused intermittent infection of bronchiolar epithelial cells and minimal spread to alveoli. Both the DK/BD24035/14 (H10N1) and Dk/BD24268/15 (H10N6) AIVs caused diffuse infection of bronchiolar epithelial cells and spread to surrounding alveolar epithelial cells (Fig. 4).
Figure 4.
Pulmonary lesions and viral spread in the lungs of AIV-infected mice. Lungs infected with DK/BD821/09 H10N7 (a–c) showed scattered degenerating bronchiolar epithelial cells in terminal airways (a) but positive immunohistochemical (IHC) staining for viral antigen was mostly limited to bronchiolar epithelium (b) with only minimal spread to surrounding alveoli (c). Following infections with DK/BD8988/10 H10N9 (d–f) there were no notable lesions in bronchiolar epithelium (d) and minimal viral antigen (e) was detected in the few inflammatory foci present (f). In contrast, in lungs infected with either DK/BD24035/14 H10N1 (g–i) or Dk/BD24268/15 H10N6 (j–l), numerous apoptotic bronchiolar epithelial cells were evident in widely distributed terminal airways (g,j), and IHC showed abundant virus-infected cells in both the terminal airways and extending into surrounding alveoli (h,k). The virus-infection involved a much larger percentage of the total lung field (I,l) than the other two viruses (H10N7 and H10N9). Mouse lungs were fixed in 10% neutral buffered formalin and stained with hematoxylin and eosin (HE), subjected to IHC staining with anti–NP antiserum and analyzed by histomorphometry. Magnifications: 60× (HE), 20× (IHC), and 2× (histomorphometry). For histomorphometry, total lung areas are outlined in green, and areas with antigen-positive cells are shown in red.
Replication of H10 AIVs in A549 cells
To determine the replication of H10 AIVs in human epithelial cells, we compared the multicycle growth kinetics of four viruses in the human A549 epithelial cell line. All four viruses grew efficiently in A549 cells; however, the viral titers of DK/BD24035/14 H10N1-infected cells were significantly higher than those of DK/BD8988/10 H10N9-infected cells at 12, 24, and 48 h post infection (P < 0.01 at 12 and 48 h post infection and P < 0.05 at 25 h, Fig. 5). The viral titers of DK/BD24035/14 H10N1-infected cells were also significantly higher than those of DK/BD821/09 H10N7-infected cells at 48 h postinfection (P < 0.01, Fig. 5).
Figure 5.
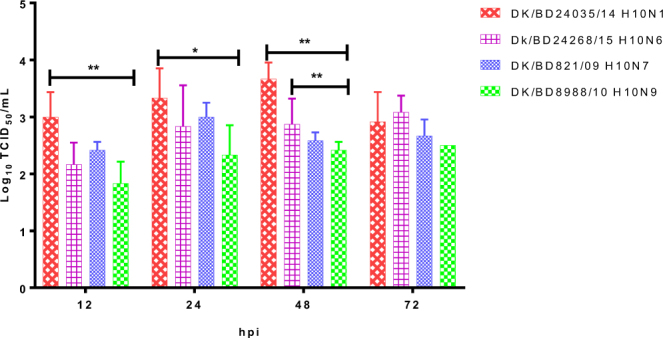
Growth characteristics of H10 AIVs in A549 cells. A549 cells were inoculated at a multiplicity of infection of 0.01 TCID50/cell with the DK/BD821/09 H10N7, DK/BD8988/10 H10N9, DK/BD24035/14 H10N1, or Dk/BD24268/15 H10N6 AIVs. Supernatants were collected at the indicated time points and titrated in MDCK cells by TCID50. Error bars depict standard deviations. *P < 0.05, **P < 0.01.
Receptor-binding assay
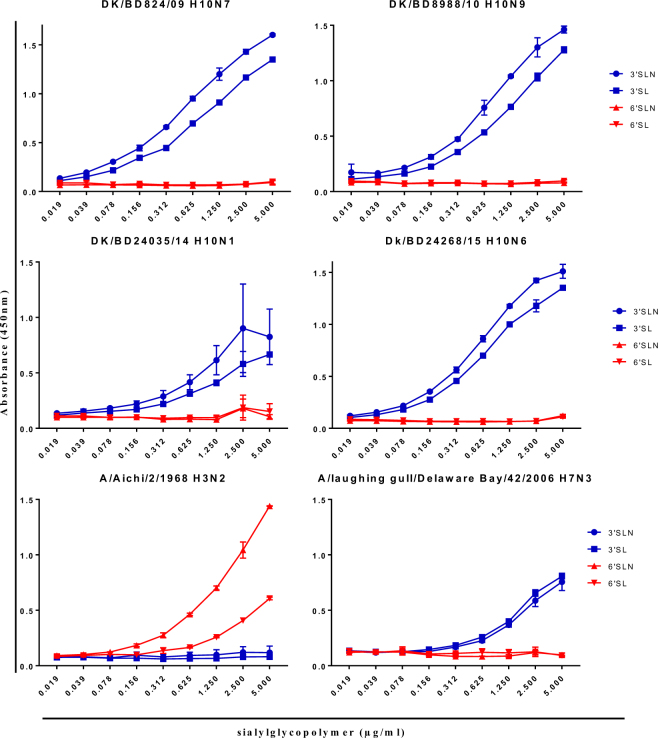
A switch in receptor-binding preference from avian-type receptors to human-type receptors is important for AIV transmission among humans. We tested the receptor-binding preferences of the H10 AIVs and found that all H10 AIVs had higher binding preferences for α2,3 avian-like receptors than for α2,6 human-like receptors (Fig. 6). Expectedly, we found the control A/Aichi/2/1968 (H3N2) human influenza A virus isolate bound to only the human-like receptor, whereas the avian derived virus A/laughing gull/Delaware Bay/42/2006 (H7N3) bound to only the avian-type receptor (Fig. 6).
Figure 6.
Receptor-binding specificity of H10 AIVs. Binding of H10 AIVs to the biotinylated sialylglycopolymers 3′ sialyllactosamine (3′SLN), 3′ sialyllactose (3′SL), 6′ sialyllactosamine (6′SLN), and 6′ sialyllactose (6′SL) were measured. A/Aichi/2/1968 (H3N2) and A/laughing gull/Delaware Bay/42/2006 (H7N3) were used as species-specific controls.
Antiviral susceptibility
To determine whether the Bangladeshi H10 AIVs were sensitive to currently approved antiviral drugs, we examined the susceptibility of these H10 AIVs to the NA inhibitors oseltamivir, zanamivir, and peramivir. All four H10 AIVs that we tested were fully susceptible to all three NA inhibitors, with mean 50% inhibitory concentration (IC50) values ranging from 0.12 to 3.06 nM (Table 4).
Table 4.
Susceptibility of H10 AIVs from Bangladeshi LPMs to NA inhibitors.
| Virus | Subtype | Inhibitory activity (IC50 ± SD, nM)* | ||
|---|---|---|---|---|
| Oseltamivir | Zanamivir | Peramivir | ||
| A/Mississippi/03/2001(WT) | H1N1 | 0.65 ± 0.06 | 0.28 ± 0.02 | 0.10 ± 0.01 |
| A/Mississippi/03/2001(Res) | H1N1, H275Y | 318.95 ± 22.00 | 0.32 ± 0.00 | 48.44 ± 0.97 |
| B/Perth/211/2001 (WT) | 22.96 ± 3.79 | 1.18 ± 0.05 | 0.37 ± 0.02 | |
| B/Perth/211/2001 (Res) | D187E | 186.13 ± 39.83 | 5.25 ± 0.77 | 9.89 ± 0.96 |
| A/duck/Bangladesh/821/2009 | H10N7 | 1.10 ± 0.11 | 2.26 ± 0.32 | 0.54 ± 0.13 |
| A/duck/Bangladesh/8988/2010 | H10N9 | 0.76 ± 0.19 | 0.84 ± 0.10 | 0.14 ± 0.02 |
| A/duck/Bangladesh/24035/2014 | H10N1 | 3.06 ± 0.44 | 0.30 ± 0.03 | 0.12 ± 0.00 |
| A/duck/Bangladesh/24268/2015 | H10N6 | 0.74 ± 0.11 | 3.01 ± 0.58 | 0.43 ± 0.05 |
WT, wild-type virus; Res, control virus resistant to NA inhibitors.
*Inhibitory concentration (IC50) value is the concentration that inhibits viral NA activity by 50%. IC50 values are expressed as the mean ± SD (nM).
Discussion
Fatal human cases of avian-origin H10N8 viral infections have caused increased interest in H10 subtype AIVs5. Extensive surveillance studies have identified H10 AIVs in waterfowl and LPMs26–28. Here, we isolated eight H10 AIVs from LPMs in Bangladesh through active surveillance from 2008 to 2017. Our findings demonstrate that these isolates belong to the Eurasian lineage and replicate efficiently in the respiratory system of mice, without adaptation and with some spread beyond the respiratory tract. This enhanced virulence in mice highlights the need for continued monitoring of AIVs in LPMs, and evaluation of the resultant isolates to identify and understand the pathogenic mechanisms of emerging strains with the potential to infect and cause disease in humans.
Our phylogenetic analysis of the H10 HA gene revealed that two different groups of Eurasian H10 AIVs were introduced into Bangladesh. The group 2 AIVs H10N1 and H10N6 were similar to H10N8 AIVs that formed a stable lineage in Jiangxi Province, China26, suggesting that H10 AIVs may have reassorted with other subtypes in Bangladesh. Our phylogenetic analysis of the NA gene of the H10N1 AIV revealed branching with the NA genes detected in wild and domestic ducks in China. In contrast, the NA genes of H10N6 AIVs branched with those from AIVs recently detected in the Tanguar haor area of Bangladesh and showed similarities to those of Mongolian AIVs. The NA and HA genes of the H10N7 AIVs were similar to those of H10N7 from Jiangxi, China. This suggests that some viruses were introduced to LPMs and others reassorted with viruses circulating in the LPMs or free-range ducks in Bangladesh. The internal genes of the H10 AIVs were more complex and shared high homology with other subtypes of AIVs. Therefore, epidemiologic monitoring of AIVs in migratory birds is essential to better understand their transmission, reassortment and evolution. Antigenic analysis of HA of the H10 AIVs indicated that they share common epitopes. We detected antigenic differences between the A/chicken/Jiangxi/34609/2013 (H10N8) AIV and the other H10 AIVs that correlated with substitutions in the HA at residues 137 and 83 at antigenic sites A and E, respectively. Overall, we observed limited antigenic drift among the North American and Eurasian lineages.
Migratory birds are important in the evolution, maintenance, and spread of AIVs. Bangladesh is located in the Central Asian flyway and is near the Eastern Asian-Australian and Black Sea-Mediterranean flyways, and is a stopover site for migratory birds. The interaction between wild birds and domestic and/or free-range ducks, which introduces AIVs into poultry systems, was responsible for emerging new strains that infected humans. This was observed in the emergence of novel H7N9 and H10N8 strains in China in 201328,29. We previously showed that the viruses isolated from Tanguar haor area played a role in the development of a new genotype of H5N1 clade 2.3.2.1a through reassortment22. Therefore, further study is needed to address the link between AIVs isolated from free-range ducks in the Tanguar haor area and the diversity and evolution of AIVs circulating in LPMs in Bangladesh.
The mouse model has been widely used to evaluate the virulence of AIVs in mammals. We used DBA/2J mice to evaluate the disease potential of H10 AIVs because DBA/2J mice are more sensitive to influenza A infections than are other strains30–32. We found that all H10 AIVs infected and replicated in mice without prior adaptation, some of the AIVs caused some degree of weight loss, and some were lethal. Furthermore, these AIVs replicated to higher titers in the lungs than in the nasal turbinates, which is consistent with previous findings showing that H10N8 AIVs isolated from LPMs in China replicated well in the respiratory tracts of mice, with higher viral loads in the lungs than in nasal turbinates33.
Mouse studies can be useful to investigate the potential of low pathogenic AIVs to replicate and cause disease and also can be used to discover the molecular markers which are important for adaptation to a mammalian host34. Recent studies demonstrated that many low pathogenic H1 and H7 AIVs isolated from wild birds in North America caused mortality in mice prior to adaptation31,35. Recently, we described the replication and pathogenic potential of H3, H7, and H15 viruses from free-range ducks in Bangladesh using mice. We observed that H7N1 and H7N9 viruses caused 100% and 60% mortality respectively, and expanded their tissue tropism beyond the pulmonary tissues to heart, liver, and brain36. Our current study showed this for both H10N1 and H10N6 AIVs as well as high mortality in mice.
The PB2 E627K substitution has been shown to confer increased virulence of AIVs in mammals37,38. All of the H10 AIVs we isolated did not contain the PB2 E627K substitution. The E627K substitution is present in viruses obtained from human samples infected with H10N8 AIVs but not in poultry isolates26,27, suggesting that this substitution is required to improve replication in mammalian hosts. The NS gene of H10N4 AIV has also been shown to contribute to virulence39. In 2014, an H10N7 AIV was isolated from seals in northeastern Europe that caused severe mortality40,41. An H10N4 AIV was also detected in mink farms in Sweden42. We observed that all H10 AIVs had mutations in their NS1 genes that resulted in a P42S substitution, which is associated with virulence and pathogenicity in mammals24,25.
AIV mutations conferring resistance to oseltamivir emerged after the antiviral was used to treat patients with H7N9 and H10N8 AIV infections26,43. All of the H10 AIVs we tested in this study were susceptible to NA inhibitors. However, monitoring influenza viruses for the presence of drug-resistance markers is important to predict the emergence of strains with such markers.
The H9N2 AIV is endemic in Bangladesh21,44–46 and has acquired mammalian host–specific mutations in its internal genes, which have been shown to facilitate transmission from avian species to humans44. H9N2 AIVs are significant donors of genetic material to emerging zoonotic viruses such as H5Nx, H7N9, and H10N8 AIVs posing an enormous threat to both human health and poultry industry. The wide circulation of H9N2 AIVs in Bangladeshi LPMs affords H9N2 with more opportunities for reassortment with other AIV subtypes, such as H10 AIVs. Transmission of H10 AIVs to humans has resulted in some fatal cases, and serologic evidence indicates that H10 AIVs were previously transmitted among turkey farmers in the United States47. This should raise concern about the potential for human-to-human transmission of H10 AIVs. Evaluation of the biologic properties of H10 and other subtypes of AIVs circulating in LPMs is essential for understanding the emergence and evolution of these viruses and to reduce their potential pandemic threat to public health.
Materials and Methods
Ethics statement
All mouse experiments were approved and performed according to the guidelines set by the Animal Care and Use Committee of St. Jude Children’s Research Hospital in an enhanced Animal Biosafety Level 3 containment facility.
Virus isolation
The H10 AIVs used in this study were isolated from LPMs between 2008 and 2017 in Bangladesh during routine surveillance. The HA and NA subtypes of the AIVs were identified via qRT-PCR. All viruses were propagated and titrated for EID50 in the allantoic cavities of 10-day-old embryonated chicken eggs at 35 °C for 48 h. Virus titers were determined by injecting 100 μL of serial 10-fold dilutions of virus into the allantoic cavities of 10-day-old embryonated chicken eggs and then calculating the EID50 according to the method of Reed and Muench48.
Deep amplicon sequencing and genetic analysis
Viral RNA was extracted by using an RNeasy kit (QIAGEN); conventional RT-PCR was then performed by using a SuperScript III first-strand synthesis kit (Invitrogen) with the Uni12 influenza primer. Multiplex PCR of all eight gene segments was conducted by using PCR Supermix HiFi (Invitrogen) with the Uni12/13 primers. PCR products were purified on a spin column (QIAGEN). DNA libraries were prepared by using NEXTera XT DNA-Seq library prep kits (Illumina) with 96 dual-index barcodes, according to manufacturer instructions. Pooled libraries were sequenced with an Illumina MiSeq Personal Genome Sequencer and 150 bp paired-end reads. The closest relatives of the viral genes sequenced of the eight isolated viruses were identified in the Global Initiative on Sharing Avian Influenza Data (GISAID) database and the Influenza Sequence Database (ISD)49,50. For phylogenetic analysis, multiple-sequence alignments for each data set were conducted using the MAFFT program51. MEGA6 was used to construct phylogenetic trees by applying the neighbor-joining method with the Kimura two-parameter distance model and 1,000 bootstrap replicates52. Several virus sequences of the North American lineage were also used as an out group in phylogenetic trees.
Antigenic characterization
The HI assay was used to antigenically characterize the viruses. The four H10 AIVs were tested by using reference antisera against H10 AIVs selected from the North American and Eurasian lineages. The antisera against DK/BD24035/14 (H10N1) was generated for this study. Briefly, ferrets were intranasally infected with 1 mL of 106 EID50/mL viruses and then boosted after 3 weeks by intramuscular injection of virus with adjuvant. Blood was collected 1 week later for serum isolation. A panel of three mouse monoclonal antibodies (JX13-3, JX13-4, and JX13-4) prepared against the H10 HA of A/chicken/Jiangxi/34609/2013 (H10N8) virus. Monoclonal antibodies were prepared at St. Jude Children’s Research Hospital by ClonaCell-HY Hybridoma Kit (StemCell Technologies) according to manufacturer instructions. Anti-H10 monoclonal antibodies were submitted to BEI Research Resources https://www.beiresources.org/ under catalog no. NR-50408, NR-50409, and NR-50410. The HI test was performed according to WHO protocols53.
Pathogenicity in mice
Four H10 isolates were selected to investigate H10 AIVs pathogenicity in DBA/2J mice. Groups of seventeen mice were anesthetized with isoflurane and intranasally inoculated with 30 µL of 106 EID50/mL virus, and an uninfected group was anesthetized and intranasally inoculated with 30 µL of phosphate-buffered saline, pH 7.2 (PBS) as controls. Upon virus challenge, five mice per group were monitored for 14 dpi for disease signs, weight loss, and mortality. Mortality was recorded as actual death or loss of ≥25% body weight, which is the threshold in which animals are required to be euthanized according to our animal protocol. Three mice per group were euthanized via CO2 asphyxiation at 2, 4, 6, and 8 dpi. Lung, intestine, liver, brain, spleen, and heart were collected to determine viral titers. Whole organs of lung, liver, brain, spleen, heart, and parts from intestine were homogenized in 1 ml PBS containing antibiotics with a Qiagen Tissue Lyser II (Qiagen, Gaithersburg, MD). Organ homogenates were centrifuged at 2000g for 10 min, and the supernatants were transferred to clean tubes from which 10-fold dilutions (10−1 to 10−10) were made in a solution of sterile PBS and antibiotics. For each dilution, three 10- to 11-day-old chicken embryos were inoculated via the allantoic cavity to determine the viral titers for each organ in terms of EID50.
Histology and immunohistochemistry
The lungs of mice (n = 3) were collected at 3 dpi and fixed via intratracheal infusion and immersion in a 10% neutral buffered formalin solution. Tissues were embedded in paraffin, sectioned, and stained with hematoxylin and eosin. Immunohistochemical staining was performed with serial histologic sections to determine the distribution of AIV antigens. A goat primary polyclonal antibody (US Biological, Swampscott, MA) against anti-influenza A, USSR (H1N1) was used (1:1,000) on tissue sections that were previously subjected to antigen retrieval for 30 min at 98 °C (Target Retrieval Solution, pH 9; Dako Corp., Carpinteria, CA). To quantify the extent of viral infection in the lungs, digital images of whole lung sections stained for viral antigens were acquired with an Aperio ScanScope XT Slide Scanner (Leica Biosystems, Buffalo Grove, IL). Both uninfected and virus-positive regions were then manually outlined, and the areas of the outlined regions were determined with ImageScope software (Leica Biosystems).
Viral replication in human lung cells
Monolayers of A549 cells were inoculated at a multiplicity of infection of 0.01 50% tissue culture infectious dose (TCID50) with DK/BD821/09 (H10N7), DK/BD8988/10 (H10N9), DK/BD24035/14 (H10N1), and Dk/BD24268/15 (H10N6). Cells were incubated at 37 °C in RPMI containing 0.2 µg/mL of L-1-tosylamido-2-phenylethyl chloromethyl ketone–treated trypsin (Sigma-Aldrich, St. Louis, MO). Supernatants were collected at different time points and titrated in MDCK cells by TCID50.
Receptor specificity assay
Receptor specificity of H10 AIVs was analyzed by using a solid-phase direct-binding assay, as described previously54. Four different glycopolymers were used: biotinylated glycans α2,3 [Neu5Acα2-3Galβ1-4GlcNAcβ-PAA-biotin (3′SLN) and Neu5Acα2-3Galβ 1,4Glc β-PAA-biotin (3′SL)] and α2,6 [Neu5Aca2-6Galβ1-4Glcβ-PAA-biotin (6′SLN) and Neu5Acα2-3Galβ 1,4Glc β-PAA-biotin(6′SL)]. A/Aichi/2/1968 (H3N2) was used as a specificity control for α2,6 binding and A/laughing gull/Delaware Bay/42/2006 (H7N3) was used as a specificity control for α2,3 binding. Binding was detected by measuring absorbance at 450 nm.
Susceptibility of H10 AIVs to NA inhibitors
The NA inhibitors oseltamivir carboxylate (oseltamivir; Hoffmann-La Roche, Basel, Switzerland), zanamivir (GlaxoSmithKline, Research Triangle Park, NC, USA), and peramivir (BioCryst Pharmaceuticals, Birmingham, AL, USA) were used to test the sensitivity of the H10 AIVs to NA inhibitors. A fluorescence-based NA inhibition assay with the fluorogenic substrate 2′-(4-methylumbelliferyl)-α-D-N-acetylneuraminic acid (MUNANA; Sigma-Aldrich, St Louis, MO) was used to determine IC50 values55,56.
Statistical analysis
All statistical analyses were performed by two-way ANOVA in combination with Bonferroni multiple comparison tests by using GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA). P values < 0.05 were considered statistically significant.
Nucleotide sequence accession numbers
The nucleotide sequences of the eight H10 AIVs described in this study were deposited in the GenBank database with the accession numbers shown in Table 1.
Electronic supplementary material
Acknowledgements
We thank Mark Zanin (Department of Infectious Diseases, St. Jude Children’s Research Hospital, Memphis, USA) for reviewing the manuscript. We thank Jon Seiler, Jasmine Turner, David Walker, Kimberly Friedman, Sharmin Akhtar and Subrata Barman for experimental assistance and/or helpful discussion. We also thank James Knowles for administrative assistance and Nisha Badders, PhD, ELS for scientific editing. This work was funded in part by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (HHSN266200700005C and HHSN272201400006C), and by ALSAC.
Author Contributions
Conceived and designed research: R.E., R.J.W., and R.G.W. Performed research: R.E., J.F., B.M., Analyzed the data: R.E., S.K., P.V., and R.G.W. Contributed reagents/materials/analysis tools: M.K.H., M.M.F., S.K., P.V., and P.M. Wrote the paper: R.E., S.K., and R.G.W.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-29079-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tong S, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tong S, et al. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci USA. 2012;109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Y, Wu Y, Tefsen B, Shi Y, Gao GF. Bat-derived influenza-like viruses H17N10 and H18N11. Trends Microbiol. 2014;22:183–191. doi: 10.1016/j.tim.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, et al. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet. 2014;383:714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- 6.Parry J. H7N9 avian flu infects humans for the first time. BMJ. 2013;346:f2151. doi: 10.1136/bmj.f2151. [DOI] [PubMed] [Google Scholar]
- 7.Shi W, Shi Y, Wu Y, Liu D, Gao GF. Origin and molecular characterization of the human-infecting H6N1 influenza virus in Taiwan. Protein Cell. 2013;4:846–853. doi: 10.1007/s13238-013-3083-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuen KY, et al. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998;351:467–471. doi: 10.1016/S0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
- 9.WHO. Influenza at the human-animal interface. 2018.
- 10.Yuan J, et al. Origin and molecular characteristics of a novel 2013 avian influenza A(H6N1) virus causing human infection in Taiwan. Clin Infect Dis. 2013;57:1367–1368. doi: 10.1093/cid/cit479. [DOI] [PubMed] [Google Scholar]
- 11.Feldmann H, et al. The structure of serotype H10 hemagglutinin of influenza A virus: comparison of an apathogenic avian and a mammalian strain pathogenic for mink. Virology. 1988;165:428–437. doi: 10.1016/0042-6822(88)90586-7. [DOI] [PubMed] [Google Scholar]
- 12.Liu S, et al. Panorama phylogenetic diversity and distribution of Type A influenza virus. PLoS One. 2009;4:e5022. doi: 10.1371/journal.pone.0005022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan American Health Organization. Avian influenza virus A (H10N7) circulating among humans in Egypt. 2004 [cited]Available from: http://www1.paho.org/hq/dmdocuments/2010/Avian_Influenza_Egypt_070503.pdf.
- 14.Arzey GG, et al. Influenza virus A (H10N7) in chickens and poultry abattoir workers, Australia. Emerg Infect Dis. 2012;18:814–816. doi: 10.3201/eid1805.111852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodewes R, et al. Seroprevalence of Antibodies against Seal Influenza A(H10N7) Virus in Harbor Seals and Gray Seals from the Netherlands. PLoS One. 2015;10:e0144899. doi: 10.1371/journal.pone.0144899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zohari, S., Neimanis, A., Harkonen, T., Moraeus, C. & Valarcher, J. F. Avian influenza A(H10N7) virus involvement in mass mortality of harbour seals (Phoca vitulina) in Sweden, March through October 2014. Euro Surveill19 (2014). [DOI] [PubMed]
- 17.Su S, et al. First evidence of H10N8 Avian influenza virus infections among feral dogs in live poultry markets in Guangdong province, China. Clin Infect Dis. 2014;59:748–750. doi: 10.1093/cid/ciu345. [DOI] [PubMed] [Google Scholar]
- 18.Sutton, T. C., Lamirande, E. W., Czako, R. & Subbarao, K. Evaluation of the biological properties and cross-reactive antibody response to H10 influenza viruses in ferrets. J Virol (2017). [DOI] [PMC free article] [PubMed]
- 19.Turner JC, et al. Insight into live bird markets of Bangladesh: an overview of the dynamics of transmission of H5N1 and H9N2 avian influenza viruses. Emerg Microbes Infect. 2017;6:e12. doi: 10.1038/emi.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monne I, et al. Reassortant avian influenza A(H5N1) viruses with H9N2-PB1 gene in poultry, Bangladesh. Emerg Infect Dis. 2013;19:1630–1634. doi: 10.3201/eid1910.130534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marinova-Petkova A, et al. The Continuing Evolution of H5N1 and H9N2 Influenza Viruses in Bangladesh Between 2013 and 2014. Avian Dis. 2016;60:108–117. doi: 10.1637/11136-050815-Reg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barman S, et al. Role of domestic ducks in the emergence of a new genotype of highly pathogenic H5N1 avian influenza A viruses in Bangladesh. Emerging microbes & infections. 2017;6:e72. doi: 10.1038/emi.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmolke M, et al. Differential contribution of PB1-F2 to the virulence of highly pathogenic H5N1 influenza A virus in mammalian and avian species. PLoS Pathog. 2011;7:e1002186. doi: 10.1371/journal.ppat.1002186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seo SH, Hoffmann E, Webster RG. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat Med. 2002;8:950–954. doi: 10.1038/nm757. [DOI] [PubMed] [Google Scholar]
- 25.Jiao P, et al. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J Virol. 2008;82:1146–1154. doi: 10.1128/JVI.01698-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi, W. et al. Genesis of the novel human-infecting influenza A(H10N8) virus and potential genetic diversity of the virus in poultry, China. Euro Surveill19 (2014). [DOI] [PubMed]
- 27.Zhang T, et al. Human infection with influenza virus A(H10N8) from live poultry markets, China, 2014. Emerg Infect Dis. 2014;20:2076–2079. doi: 10.3201/eid2012.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma C, et al. Emergence and evolution of H10 subtype influenza viruses in poultry in China. J Virol. 2015;89:3534–3541. doi: 10.1128/JVI.03167-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam TT, et al. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature. 2013;502:241–244. doi: 10.1038/nature12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boon AC, et al. Cross-reactive neutralizing antibodies directed against pandemic H1N1 2009 virus are protective in a highly sensitive DBA/2 mouse influenza model. J Virol. 2010;84:7662–7667. doi: 10.1128/JVI.02444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kocer ZA, Krauss S, Stallknecht DE, Rehg JE, Webster RG. The potential of avian H1N1 influenza A viruses to replicate and cause disease in mammalian models. PLoS One. 2012;7:e41609. doi: 10.1371/journal.pone.0041609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pica N, et al. The DBA.2 mouse is susceptible to disease following infection with a broad, but limited, range of influenza A and B viruses. J Virol. 2011;85:12825–12829. doi: 10.1128/JVI.05930-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng G, et al. Genetics, Receptor Binding, and Virulence in Mice of H10N8 Influenza Viruses Isolated from Ducks and Chickens in Live Poultry Markets in China. J Virol. 2015;89:6506–6510. doi: 10.1128/JVI.00017-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamal RP, Katz JM, York IA. Molecular determinants of influenza virus pathogenesis in mice. Curr Top Microbiol Immunol. 2014;385:243–274. doi: 10.1007/82_2014_388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zanin, M. et al. Potential for Low-Pathogenic Avian H7 Influenza A Viruses To Replicate and Cause Disease in a Mammalian Model. J Virol91 (2017). [DOI] [PMC free article] [PubMed]
- 36.El-Shesheny R, et al. Replication and pathogenic potential of influenza A virus subtypes H3, H7, and H15 from free-range ducks in Bangladesh in mammals. Emerg Microbes Infect. 2018;7:70. doi: 10.1038/s41426-018-0072-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Q, et al. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science. 2013;341:410–414. doi: 10.1126/science.1240532. [DOI] [PubMed] [Google Scholar]
- 38.Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- 39.Zohari S, Metreveli G, Kiss I, Belak S, Berg M. Full genome comparison and characterization of avian H10 viruses with different pathogenicity in Mink (Mustela vison) reveals genetic and functional differences in the non-structural gene. Virol J. 2010;7:145. doi: 10.1186/1743-422X-7-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bodewes R, et al. Avian Influenza A(H10N7) virus-associated mass deaths among harbor seals. Emerg Infect Dis. 2015;21:720–722. doi: 10.3201/eid2104.141675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krog JS, et al. Influenza A(H10N7) virus in dead harbor seals, Denmark. Emerg Infect Dis. 2015;21:684–687. doi: 10.3201/eid2104.141484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klingeborn B, Englund L, Rott R, Juntti N, Rockborn G. An avian influenza A virus killing a mammalian species–the mink. Brief report. Arch Virol. 1985;86:347–351. doi: 10.1007/BF01309839. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y, et al. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet. 2013;381:1916–1925. doi: 10.1016/S0140-6736(13)60903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shanmuganatham K, et al. Genesis of avian influenza H9N2 in Bangladesh. Emerg Microbes Infect. 2014;3:e88. doi: 10.1038/emi.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Negovetich NJ, et al. Live bird markets of Bangladesh: H9N2 viruses and the near absence of highly pathogenic H5N1 influenza. PLoS One. 2011;6:e19311. doi: 10.1371/journal.pone.0019311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biswas, P. K. et al. Incidence of contamination of live bird markets in Bangladesh with influenza A virus and subtypes H5, H7 and H9. Transbound Emerg Dis (2017). [DOI] [PubMed]
- 47.Kayali G, Ortiz EJ, Chorazy ML, Gray GC. Evidence of previous avian influenza infection among US turkey workers. Zoonoses and public health. 2010;57:265–272. doi: 10.1111/j.1863-2378.2009.01231.x. [DOI] [PubMed] [Google Scholar]
- 48.Reed LJ, Muench H. A Simple Method of Estimating Fifty Per Cent Endpoints12. American Journal of Epidemiology. 1938;27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 49.Zhang Y, et al. Influenza Research Database: An integrated bioinformatics resource for influenza virus research. Nucleic Acids Res. 2017;45:D466–D474. doi: 10.1093/nar/gkw857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bogner P, Capua I, Lipman DJ, Cox NJ. & others. A global initiative on sharing avian flu data. Nature. 2006;442:981. doi: 10.1038/442981a. [DOI] [Google Scholar]
- 51.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.WHO. WHO Manual on Animal Influenza Diagnosis and Surveillance. 2002 [cited December 12, 2011]; 2nd Edition:[Available from: http://whqlibdoc.who.int/hq/2002/WHO_CDS_CSR_NCS_2002.5.pdf.
- 54.Matrosovich MN, Gambaryan AS. Solid-phase assays of receptor-binding specificity. Methods Mol Biol. 2012;865:71–94. doi: 10.1007/978-1-61779-621-0_5. [DOI] [PubMed] [Google Scholar]
- 55.Potier M, Mameli L, Belisle M, Dallaire L, Melancon SB. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-D-N-acetylneuraminate) substrate. Anal Biochem. 1979;94:287–296. doi: 10.1016/0003-2697(79)90362-2. [DOI] [PubMed] [Google Scholar]
- 56.Govorkova EA, et al. Antiviral resistance among highly pathogenic influenza A (H5N1) viruses isolated worldwide in 2002–2012 shows need for continued monitoring. Antiviral Res. 2013;98:297–304. doi: 10.1016/j.antiviral.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.