Abstract
Cassia tora is a plant of medicinal importance. Medicinal plants from different localities are believed to differ in their therapeutic potency. In this study, six populations of C. tora with different eco-geographical origins were investigated genotypically (ISSR) and phytochemically (FTIR) to establish an integrated approach for population discrimination and authentication of the origin of this medicinal herb. CHS gene expression analysis and determination of flavonoid content were carried out to substantiate the study. A total of 19 population-specific authentication bands were observed in 11 ISSR fingerprints. Authentication codes were generated using six highly polymorphic bands, including three authentication bands. FTIR spectra revealed that the peaks at wavenumber 1623 cm−1 (carbonyl group) and 1034 cm−1 (>CO- group) were powerful in separating the populations. These peaks are assigned to flavonoids and carbohydrates, respectively, were more intense for Ranchi (highland) population. Variation in the transcript level of CHS gene was observed. The findings of FTIR and RT-PCR analyses were in agreement with the TFC analysis, where, the lowest amount of flavonoids observed for Lucknow (lowland) population. All the populations of C. tora have been authenticated accurately by ISSR analyses and FTIR fingerprinting, and the Ranchi site was observed to be more suitable for the potential harvesting of therapeutic bioactive compounds.
Introduction
The therapeutic potential of plants has been utilized in traditional medicines such as Chinese, Ayurveda, Siddha, and Unani etc. Being relatively nontoxic and easily affordable, there has been resurgence in the demand for medicinal plants1. Cassia tora L. Syn. Senna tora (L.) Roxb. verna. Chakwad, commonly known as sickle senna, belongs to family Caesalpiniaceae (Subfamily: Caesalpinioideae, tribe: Cassieae, sub tribe: Cassiinae). It is the wild annual herbal crop, indigenous to palaeotropical region (Africa and Asia to eastward Polynesia) and distributed throughout the tropical and sub-tropical regions of the world2–4. The plant is widely consumed as a potent source of sennosides (laxative), and enlisted in the World Health Organisation’s ‘List of Essential Medicines’5. It's medicinal potentials have been described in the traditional Chinese medicine (TCM) and Ayurvedic practices with the special reference to cure psoriasis and other skin degenerative disorders6. In addition to this, the plant was also used traditionally to cure diabetes, dermatitis, constipation, cough, cold and fever, etc7. The plant harbors anti-proliferative, hypolipidemic, immunostimulatory, and anticancerous properties8. Earlier, It has been reported that C. tora possessed a large amount of flavonoids, the potent antioxidants9,10. The production of flavonoids in plants is linked with the expression of chalcone synthase (CHS) gene encoding CHS enzyme which is the first committed enzyme in flavonoid biosynthesis11,12. CHS is ubiquitous to higher plants and belongs to the family of polyketide synthase (PKS) enzymes (known as type III PKS). It is believed to act as the central hub for the enzymes involved in the flavonoid pathway12,13. The expression of CHS gene is the important step in the biosynthesis of flavonoids14,15 and CHS transcription is regulated by endogenous programs in response to environmental signals16. Plant samples from different geographical origins have different biochemical compositions due to variations in the environmental conditions and genetic reasons17–19. Therefore, it is crucial to identify the medicinal herb at the locality level.
The general approach to identification is dependent on morphological, anatomical, and chemical features, but such characteristics are often affected by environmental and other developmental factors during plant growth20. Additionally, medicinal plants are processed for use as crude drugs, which affect many morphological and anatomical characteristics, as well as resulting in changes in some chemical constituents21. Therefore, it is difficult to identify the crude herbs through anatomical and chemotaxonomical studies. The established DNA barcoding approaches to authenticate plant species were based on either a short, and standardized DNA sequence region or DNA polymorphism using genetic markers like ISSR, and SNP etc.22–24. In plants, variations among the plastid gene sequences, for example, rbcL, matK and trnL genes, and ITS regions were used in identification, population discrimination and authentication of various plant species25–29. However, the lack in the prior information of genomic regions and low evolutionary rate of change in the coding regions are serious limitations to such analysis. A DNA based polymorphism assay may be the suitable alternative for the population authentication of herbal medicines. Earlier studies based on genetic markers (RAPD, SCAR and ISSR etc.) could significantly identify the plant populations30,31, however ISSRs were found highly reproducible, more variable and efficient than the currently used DNA marker32–34 in being the more robust to even slight changes in DNA concentrations. In addition, they retained the benefits over other PCR-based techniques such as the need for very little template material. Nevertheless, DNA genotyping also has limitations such as within species variation. Furthermore, the technique does not reveal the composition of the active ingredients or chemical constituents. DNA remains the same irrespective of the plant part used, while the phytochemical compositions may vary with the plant parts used, physiology, and the environment19,21. Therefore, proper integration of DNA based techniques like ISSRs and analytical tools like FTIR for chemo-profiling would be more efficient to authenticate the population and will lead to the development of a comprehensive system of characterization that can be conveniently applied at the industry level for quality control of herbal drugs.
Therefore, the aim of this study was focused upon a) development of molecular markers to distinguish the test populations of C. tora and b) discriminate the same populations based on the variability in phytochemical (mainly flavonoid) content. For the aforementioned purposes, we used the ISSRs and FTIR as the rapid and efficient techniques. In addition, we also employed a qPCR based approach to test the expression level of CHS gene and total flavonoid content (TFC) analysis to substantiate the study. To the best of our knowledge, this study is the first attempt of its type.
Results
Amplified products
A total of 130 clear and reproducible bands were amplified from six populations of C. tora which were collected from different geographical locations (Fig. 1, Supplementary Table S1) using the 11 selected ISSR primers, of which, 118 were polymorphic (90.76%). The total number of loci varied from 31 to 54 per primer for all the populations (Table 1), with fragment size ranging from 200–3000 bp (Fig. 2). Among the samples, the CT-5 (Ranchi) population had the highest ISSR polymorphism (85.90%), while the CT-1 (Dehradun) population, the lowest (81.97%). The lowest genetic distance, based on Jaccard's coefficient, was between CT-3 (Varanasi) and CT-4 (Patna) populations, and the highest between CT-2 (Lucknow) and CT-6 (Puri) (Supplementary Table S2). ISSR fingerprinting of six populations using primer ISSR-8 and ISSR-10 is shown in Fig. 2.
Figure 1.
Collection sites of Cassia tora. This image is prepared with the help of an editable map of India (Source: https://yourfreetemplates.com).
Table 1.
Details of markers selected for the study and their amplified products.
| S. No. | ISSR Primers | Sequence (5′-3′) | Annealing temperature (°C) | Size range of Amplicons | No. of amplified bands | Polymorphic loci | Polymorphism (%) |
|---|---|---|---|---|---|---|---|
| 1. | ISSR5 | (GTG)5 | 52 | 200 bp–2.8 kb | 13 | 12 | 92.30 |
| 2. | ISSR7 | (AC)8G′ | 48 | 100 bp–2.9 kb | 17 | 17 | 100 |
| 3. | ISSR8 | (GA)8CT | 52 | 150 bp–2.8 kb | 15 | 15 | 100 |
| 4. | ISSR10 | (CA)7CC | 48 | 200 bp–2.5 kb | 11 | 9 | 81.82 |
| 5. | ISSR11 | (CA)7CG | 48 | 100 bp–2 kb | 10 | 9 | 90 |
| 6. | ISSR13 | (GT)7CG | 48 | 250 bp–2.8 kb | 9 | 8 | 88.89 |
| 7. | ISSR16 | (GTG)4GAC | 52 | 200 bp–2.1 kb | 9 | 9 | 100 |
| 8. | ISSR17 | (AG)8G | 49.5 | 350 bp–2.1 kb | 11 | 8 | 72.73 |
| 9. | ISSR21 | (TC)8C | 49.5 | 250 bp–2 kb | 11 | 11 | 100 |
| 10. | ISSR22 | (TC)8G | 49.5 | 300 bp–1.1 kb | 13 | 10 | 76.92 |
| 11. | ISSR25 | (GA)8GT | 49.5 | 250 bp–3 kb | 11 | 10 | 90.91 |
| Total | 130 | 118 | 90.76 |
Figure 2.
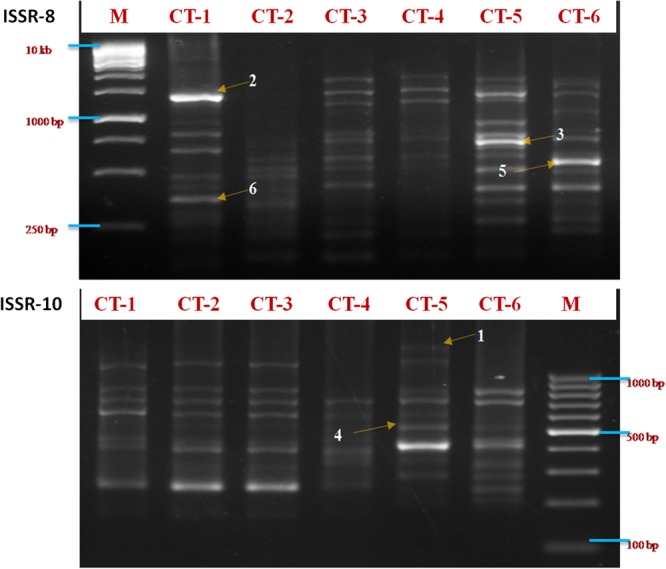
Agarose gel images showing amplification pattern of six C. tora populations obtained by ISSR-8 and ISSR-10 indicating selected polymorphic bands for the authentication of C. tora population.
Development of specific authentication markers for Cassia tora population
From the DNA fingerprints, based on ISSR primers, highly polymorphic bands were selected for the population identification. Total nineteen (14.62%) specific authentication bands observed which were present in one population but absent in other (Table 2). Total six highly polymorphic bands were selected as authentication bands to identify the C. tora population (Fig. 2). These were scored as zero (0) and one (1), based on the absence and presence of the polymorphic bands in the rest of the population (Table 3).
Table 2.
Specific authentication bands (kb) from six C. tora populations.
| S. No. | Primers | Cassia tora population | |||||
|---|---|---|---|---|---|---|---|
| CT-1 | CT-2 | CT-3 | CT-4 | CT-5 | CT-6 | ||
| 1. | ISSR5 | 1 | |||||
| 2. | ISSR7 | 0.7 | 0.52 | ||||
| 3. | ISSR8 | 0.7 | 0.6 | ||||
| 4. | ISSR10 | 2.5 | |||||
| 5. | ISSR11 | 0.9 | 0.7 | ||||
| 6. | ISSR13 | 0.3 | 2.8 | ||||
| 7. | ISSR16 | 0.45 | 0.8 | ||||
| 8. | ISSR17 | 0.75 | |||||
| 9. | ISSR21 | 0.3 | 0.9 | ||||
| 10. | ISSR22 | 0.55 | |||||
| 11. | ISSR25 | 0.25 | 0.35 | 3 | |||
Table 3.
ISSR genotypes of six C. tora populations.
| Population code | ISSR authentication markers | Authentication code | |||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | ||
| CT-1 | 0 | 1 | 1 | 0 | 1 | 1 | 011011 |
| CT-2 | 1 | 0 | 1 | 0 | 0 | 0 | 101000 |
| CT-3 | 1 | 1 | 0 | 1 | 1 | 1 | 110111 |
| CT-4 | 1 | 1 | 1 | 1 | 1 | 0 | 111110 |
| CT-5 | 1 | 0 | 1 | 1 | 0 | 1 | 101101 |
| CT-6 | 0 | 1 | 1 | 0 | 1 | 0 | 011010 |
FTIR analysis
FTIR spectra of six C. tora populations having different geographic origins (Supplementary Table 1) are depicted in Fig. 3A. Though, the repeat measurements from one region showed no significant difference in the spectra hence, only one profile was given for a population (Fig. 3A). The spectra showed broadly similar transmittance patterns for all the tested populations. Several prominent peaks in spectra indicated the presence of specific functional groups in common among all the populations. The result showed high absorbance at wavenumber region of 3400–3200, 3200–2800, 1800–1500 and 1100–950 cm−1. The fingerprint region, 2000–900 cm−1 was chosen for further analysis. Besides the similar transmittance pattern observed in spectra for all the populations, the distinct intensity of prominent peaks were observed between different populations i.e. CT-1 (Deharadun) and CT-5 (Ranchi) populations which showed intense peaks compared to rest of the populations (Fig. 3A,B). According to geographic elevation, we divided the all populations into two groups, highland (<500 m) and lowland (>500 m), and performed student’s t-test to determine the level of significance of difference between their transmittance patterns, and it was highly significant (P>0.01). However, smaller differences in-between populations were most difficult to resolve as many spectra overlapped. Thus secondary derivatives (SD)-IR were used to enhance the resolution and to amplify small differences in the IR spectra. The SD-IR spectra were more idiosyncratic among the different populations. The spectra shown in (Fig. 3C), revealed the three prominent peaks assigned to wavenumbers 1384 and 1034 cm−1. Peaks at 1034 cm−1 were more intense, and is assigned to carbonyl group (carbohydrate region)35 (Supplementary Table S3). However, the fluctuation between peaks intensities could be noticed easily throughout whole spectra.
Figure 3.
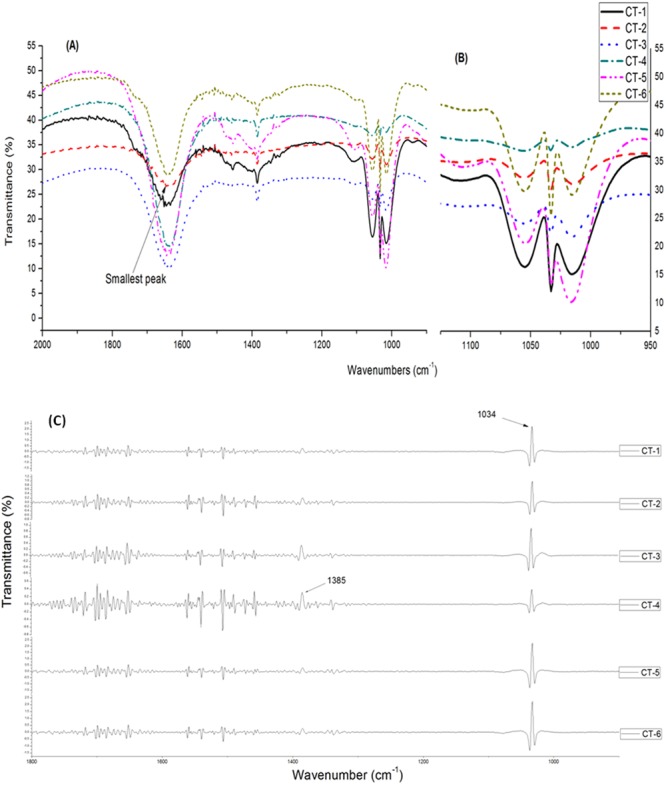
FTIR-spectra of six populations of C. tora collected from Dehradun (CT-1), Lucknow (CT-2), Varanasi (CT-3), Patna (CT-4), Ranchi (CT-5), and Puri (CT-6). (A) A portion of phytochemically important region (1800-900 cm−1); (B) An enhanced view; (C) Secondary derivatives of FTIR-spectra of six population of C. tora.
Multivariate analysis
Cluster analysis was performed using polymorphic data generated by ISSR analyses to observe similarity among the C. tora populations (Fig. 4a). The visual observation of FTIR spectra showed no significant difference in the characteristic transmittance bands among tested populations, and the intensity of peaks at certain wavenumbers did not differ among each other especially at fingerprint region 2000–900 cm−1. Therefore, it is more practical to incorporate statistical method for the aid of interpreting the measurements obtained. Since the authentication of different geographical origins of the herb based on the slight differences among particular absorption bands is too subjective, the results may vary among the analysts as reported earlier. The principal component analysis (PCA) and Pearson’s correlation were carried out between the selected spectral regions (2000–900 cm−1) (Fig. 4b). PCA revealed 62.35% variance for principal component (PC)-1 and 22.39% for the PC-2 (Fig. 5).
Figure 4.
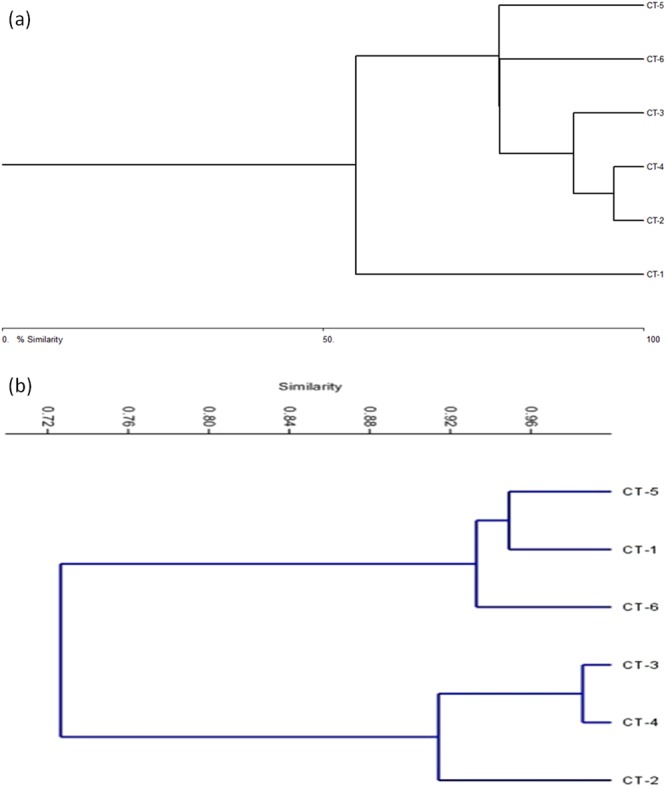
ISSR data (a) and FTIR absorbance (b) based clustering of C. tora populations.
Figure 5.
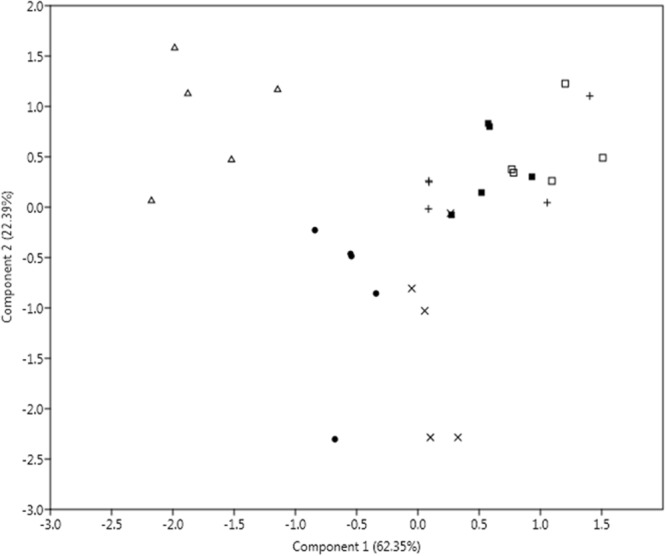
PCA model based on transmission data analysed by IR spectra of the six accessions of C. tora. The percentage of variation of the data explained by each component is provided in the plot. Dot, plus, square, triangle, X and fill square symbols indicated CT-1, CT-2, CT-3, CT-4, CT-5 and CT-6 populations, respectively.
Total flavonoids content
The total flavonoids content (TFC) in extracts, was determined using the formula (y = 0.005x + 0.085), derived from the calibration curve, and expressed as mg/g leaf dry wt (in terms of quercetin equivalent). High yield (21.53 mg/g) of TFC was observed for CT-5 (Ranchi) population. However, lowest yield (13.87 mg/g) was found in CT-2 (Lucknow) population (Supplementary Table S4, Fig. 6).
Figure 6.
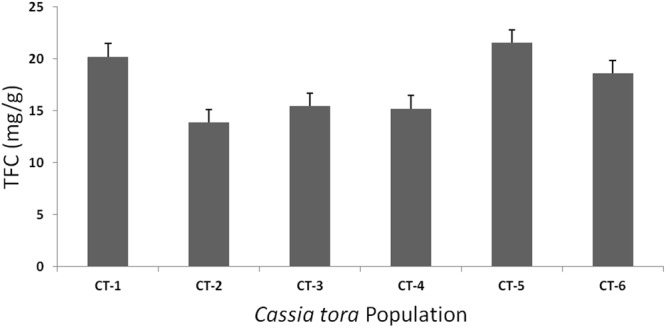
Total flavonoid content (TFC) of six C. tora populations.
CHS gene analysis
CHS1 and CHS2 genes of C. tora from six different geographic origins were analysed by semi quantitative RT-PCR. CHS1 gene showed clear variation in the relative expressions among the populations. The lowest transcript level was observed in the CT-4 (Patna) population and the high quantity of transcripts was observed for the CT-6 (Puri) population. CHS2 analysis disclosed the lowest transcript level for CT-2 (Lucknow) population however, relatively high expression was observed in CT-5 (Ranchi) population (Fig. 7). The result indicated the occurrence of variable transcript level of the CHS gene among C. tora populations of different geographical regions.
Figure 7.
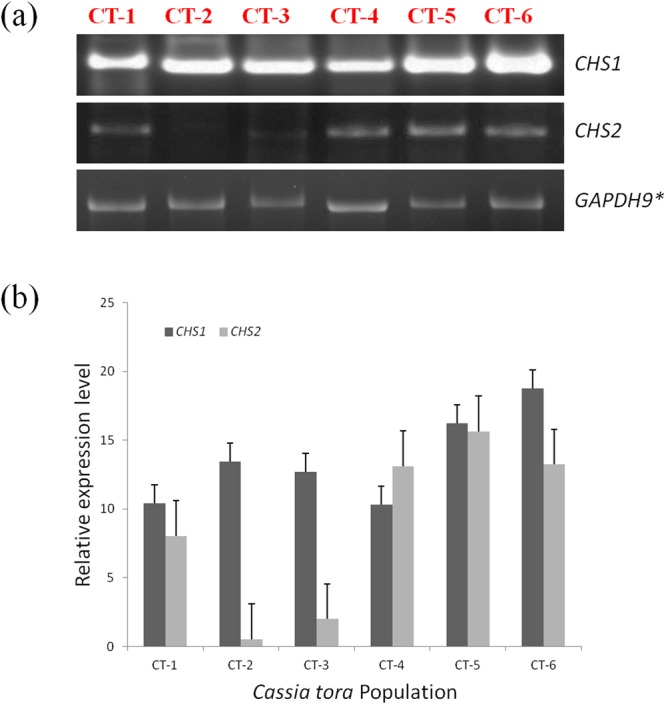
RT-PCR of Chalcone synthase (CHS) gene: (a) visualization of amplified genes (b) Relative transcript expression level in C. tora leaves of the six accessions was determined. Error bars indicate the standard error of the mean ± SE of three replicate measurements. (*control gene).
Discussion
Traditionally, C. tora is claimed to be useful in the treatment of psoriasis and other skin diseases6,7. Earlier, antipsoriatic activity of three flavonoids, namely quercetin-3-O-Dglucuronide, luteolin-7-O-glucopyranoside and formononetin-7-O-D-glucoside from C. tora leaves were investigated using UV-B induced photodermatitis model, revealed the significant (p < 0.01) percentage reduction of relative epidermal thickness compared to positive control6,36. The environment has influences on plant development and metabolism, and can alter the plant’s chemical compositions and therapeutic potency18,19. Therefore, selection of the genuine populations for potent application has become the key issue in the modernization of traditional medicines. It is, however, difficult to authenticate genuine from among the wild populations accurately using the conventional techniques, as they are similar in morphological, anatomical characteristics, and also, sometimes in chemical components too37. Hence, the precise identification and authentication of genuine population is a prerequisite for the chemical and pharmacological investigations of traditional medicines, as well as for their clinical applications.
In this study, we have investigated the qualities of 25 ISSR primers to generate polymorphic DNA fragments among which eleven primers were selected. Total 130 bands were obtained in fingerprints by 11 ISSR markers, among which, 118 bands were polymorphic (90.63%) which indicates that the simple sequences were abundant and highly dispersed throughout the genome of C. tora, and highly polymorphic. The results are consistent with the view point that level of genetic diversity as affected by the species distribution38. At the same time, we have detected 19 population-specific authentication bands, and established ISSR authentication codes involving three authentication bands for the each population of C. tora (Tables 2 and 3), which efficiently enhanced population authentication and validated the ISSR-PCR technique as the efficient marker system to be utilized to construct DNA fingerprints and to authenticate the plant populations. Earlier, ISSR authentication codes had been generated to authenticate the various medicinal plant populations like Dendrobium officinale30 and rhubarb39. The high polymorphism among populations also points out the rich genetic variability of C. tora. The lowest polymorphic bands (50) for Dehradun population proved the declination of genetic functions of a species at higher altitude and low temperatures40,41. Bary-Curtis cluster analysis of all populations favored the above findings and split Dehradun population from the rest (Fig. 4a). However, Ranchi population had highest polymorphism (85.9%) indicating that genetic exchange and differentiation of populations increased slightly at the higher elevation, probably due to extensive gene flow at the altitudes42.
FTIR has been proven to be an accurate, fast and simple method for phytochemical screening43. It provides more information through the fingerprint regions of herbal medicines, rendering the technique direct and simple21,44. Previously, FTIR has been used in identification and population discrimination studie35,45,46. Samples from the different populations can be discriminated based on the functional group absorption. Peak absorbance at the particular wavenumber is presented in (Supplementary Table S3). The IR spectra of wave-region (3400–3200, 3200–2800, 1800–1500 and 1100–950 cm−1) were similar for all the populations with the variable intensity, which implies the presence of similar major chemical components in all the samples obtained from different locations. Previously, the methanolic extract of C. tora had been analyzed using FTIR, and flavonoids were reported as a major phenolic component in the plant10. Carbonyl (>C=O) group constitutes the functional group of flavonoid, in which, stretching vibrations in carbonyl compounds lie between 1750-1600 cm−1 of mid-IR47. The sharp peaks in this region indicated that the all extracts were flavonoid-rich. Nevertheless, the high absorbance of O-H, C=C, and C-O-C functional group in methanol extracts of the leaf indicated that the phenol and flavonoids could be the dominant compounds10. The absorbance by C-O, C-H, C=C, C=O, and C-N functional groups between 1800–900 cm−1 are indicative of benzene, aldehyde and carbohydrate groups (Supplementary Table S3). Stronger absorption peaks in these regions (Fig. 3) for CT-1 (Dehradun) and CT-5 (Ranchi) sample suggests a high amount of such compounds among highland populations21. Chan et al. (2007)48 had also reported the high amount of phenolics in highlands population of ginger. The high flavonoid content in Dehradun and Ranchi populations may be due to ecological stresses like a decrease in soil moisture and nutrients availability or the decreasing temperature49. These stresses could have led to oxidative damages, and as the antagonistic response, plants synthesized abundant antioxidants especially the phenolics50.
The analysis based on SD-IR provides a better way to distinguish the populations when the peaks overlapped as SD-IR spectra enhanced the apparent resolution and amplified the tiny differences in the IR spectrum45. In this study, several peaks overlapped together at a single wavenumber making their appearance incoherent; therefore, SD-IR was used to resolve the peaks and to reveal the weaker spectral features (1800-800 cm−1) for interpreting the components with a low concentration and weak absorption peaks44. Results of SD-IR of the C. tora leaf illustrated the two distinct and sharp peaks in-between the wave region (1500-900 cm−1) assigned to benzene (1384 cm−1) and carbonyl group (1034 cm−1) (Fig. 3C), which clearly exhibited the presence of phenolics and carbohydrates in all the six populations with variations in their constituents. Peaks between wavenumbers (1500- 1200 cm−1) and (1100–900 cm−1) assigned to the carbohydrate region21,35. Patna population had the highest absorption peak at 1384 cm−1, and lowest at 1034 cm−1. Low-intensity peak indicated the presence of low amount of the carbohydrates of this population compared to all the populations. According to the above findings, Patna population could be easily discriminated from rest of the populations. Earlier, similar studies had also been used for population discrimination of Polygonum minus, species discrimination in between Tephrosia tinctoria and Atylosia albicans, and identification of genuine American ginseng population21,45,46.
Cluster analysis based on similarity matrix successfully discriminated all the populations into two separate groups, one with the populations CT-1, CT-5, and CT-6; and another with CT-2, CT-3 and CT-4 (Fig. 4b). Both the groups were highly (28%) dissimilar. Dehradun and Ranchi populations were 94% similar while Varanasi and Patna populations 98% similar, indicating that they shared almost the common phytochemical constituents. However, Puri and Lucknow populations distantly placed in their cluster showed the difference in chemical compositions and could be easily discriminated. PCA analysis also showed a disparity among the populations (Fig. 5) and separated them with the varying eco-geographical features especially in-between Highland (Dehradun and Ranchi) and Lowland populations. Such variations in absorbance were linked to quality and quantity of the phytochemical constituents may also be due to the altitude effect51. Higher altitude like that of Dehradun, exposed the plants to intense solar radiation than the rest (Supplementary Table S1) and can be inversely affected by temperature, and therefore, plant defense system produces excessive phenolics to protect against photo-damage52.
In order to substantiate our study, we have also conducted the quantitative analysis of TFC. Among all the six populations, the highland populations (Ranchi and Dehradun) had highest flavonoid content (21.53 mg/g and 20.20 mg/g, respectively) followed by lowland populations (Fig. 6, Supplementary Table S4). The similar findings were also reported for the highlands population of ginger48. ISSR analysis also corroborated these findings where maximum polymorphism was observed for Ranchi population (85.9%). However, with relatively lower polymorphic DNA (81.9%) compared to other populations, Dehradun population estimated high amounts of flavonoids after Ranchi, and it might be due to the geographical elevation and other physical and physiological stresses49,50. Therefore, it is emphasized that Ranchi population produced secondary metabolites in greater abundance and is more genetically affluent than the others. Hence, this population can be better exploited for the germplasm conservation and breeding purposes. The high TFC content of Ranchi populations along with Dehradun and Puri populations (Fig. 6) could be correlated with the FTIR spectra (Fig. 3) and cluster analysis (Fig. 4b) where, these populations were clustered separately. Varanasi and Patna populations had comparatively larger and intense peaks at wave region 1750–1100 cm−1 than Lucknow populations, indicating these populations to be rich in flavonoids over the Lucknow population and that were also substantiated by TFC analysis (Figs 3 and 6). Dehradun populations showed the lowest DNA polymorphism, and clustered separately from rest of the population in accordance with indices of Bary-Curtis similarity, although contained good amounts of the flavonoids as per the analysis of IR and TFC data (Figs 3, 4a and 6). Based on the above results, this can be suggested that plants growing at relatively low temperatures (Supplementary Table1) might have high phenylalanine ammonia lyase activity, the key enzyme of phenylpropanoid pathway that possibly leads to the accumulation of flavonoids53.
Up regulated transcription of genes encoding enzymes involved in phytochemical biosyntheses, such as CHS, leads to increased phytochemical (i.e. flavonoids) concentrations in plants11,54. Genes encoding CHS constitute a multigene family in which the copy number varies among the plant species, and functional divergence appears to have repeatedly occurred55. CHS gene expression has been studied extensively in relation to flavonoids production in many plant species14,15. However, there are few reports about the CHS gene analysis in sub-tribe Cassiinae. Panigrahi et al. (2013)56 correlate the flavonoid content with the presence of CHS gene in between C. laevigata and C. fistula, and Samappito et al. (2013)57 studied the expression of CHS gene in C. alata roots and correlates their role in the synthesis of flavonoids. In our study, a preliminary attempt was made to access the expression level of CHS among all the tested populations of C. tora because this plant is rich in flavonoids which are a major source of bioactive compounds10. CHS1 analysis showed higher transcript level for Ranchi and Puri populations compare to the rest (Fig. 7b). Earlier, it has been reported that CHS is constitutively expressed in plants but can also be subject to induced expression through light and temperature58. Therefore, higher expression of CHS1 gene in Puri population might be due to the high geographical temperature (Supplementary Table S1), responsible for the larger production of flavonoids as observed in FTIR spectra and TFC analysis (Figs 3A and 6). CHS2 showed lowest expression for Lucknow population (Fig. 3B), might be linked to the lesser production of flavonoids as observed in IR-spectra (Fig. 3A) with the smallest (low intense) peak in the flavonoid zone (1750-950 cm−1) and was also in agreement with TFC analysis (Fig. 6). Nevertheless, the higher transcript level of CHS2 in Ranchi population (Fig. 3) was also in agreement with FTIR analysis and TFC. Thus it may be inferred that the variable transcript level of CHS gene might be responsible for the lopsided distribution of flavonoids56,57, among C. tora populations and proficient to discriminate the populations from different localities.
Conclusions
ISSR fingerprinting was the suitable method for estimating the genetic differences among the populations and the authentication codes developed during analysis, will be helpful in differentiating the C. tora populations. However, FTIR spectrum analysis seemed appropriate to monitor the phytochemical variations among different C. tora populations. Both the techniques, ISSRs and FTIR established a very rapid, efficient and cost-effective technique to characterize the C. tora populations having different eco-geographical origins. C. tora population of Ranchi locality was genetically effluent comparatively rich in bioactive compounds, and hence this site would be most suited for the collection of germplasm and high amount potent bioactive compounds. Furthermore, we can also conclude that highland populations of C. tora produced certain secondary metabolites (flavonoids) in greater quantity than lowlands ones.
Materials and Methods
Plant material
C. tora plants were collected from their natural habitats in August 2014 at six different locations in India (Fig. 1). All the samples were identified using the morphological characters encrypted in the monograph and other relevant literature9. In addition to this, the plants were also authenticated by Prof. N. K. Dubey, taxonomist of the department of Botany, Banaras Hindu University (BHU), Varanasi, India. For the each population, herbarium specimen was prepared and deposited at the ‘Herbarium’ of the above-mentioned institution with the voucher specimen number (Caesal/2014/1). These taxonomically authenticated samples are referred to as Biological Reference Material (BRM)59. The plant collection sites with eco-geographical details are given in (Supplementary Table S1). Three individuals per population were taken with technical replicates for all the experimental analyses.
DNA extraction
The genomic DNA was extracted from lyophilized young leaves using the cetyl trimethyl ammonium bromide (C-TAB) method of Wang (2010)60 with the given modifications. 30 mg of Polyvinylpyrrolidone (PVP) was added to remove polyphenols and extracted sample was treated with RNase (30 µg, 37 °C) for 30 min. DNA concentration and purity were determined by spectrophotometry (ND-2000, NanoDrop, USA) and electrophoresed on 0.8% agarose gels. The final concentration of each DNA sample was diluted to approx 20 ng/ml with Mili-Q water and stored at 20 °C till further use.
PCR amplification
ISSR-PCR
PCR amplification was carried out in a total volume of 25 µl, containing 20 ng of template DNA, 2.5 µl 10X PCR buffer, 2.5 µl 25 mM MgCl2, 2 µl 10 mM dNTPs, 0.32 mM primer, 2.5 unit of Taq polymerase, and Mili-Q water. The reactions were performed in a Mastercycler thermocycler (BioRad, USA). The program consisted of an initial denaturation at 94 °C for 4 min, followed by 35 cycles of 45 s at 94 °C, 45 s at 48–52 °C (depending on the primer), 2 min at 72 °C, and the final extension of 10 min at 72 °C. A negative control, with the template DNA omitted, was included in each PCR. Amplification products were electrophoretically separated at a constant voltage of 60 V for 3 h, in 1.5% agarose gels with 0.5X TAE buffer, stained with ethidium bromide and visualized under UV. The 100 bp and 1 kb DNA ladders were used to estimate the molecular size of the fragments. Twenty-five primers were tested to identify those that produced sharp and reproducible bands. Three individuals from each population of C. tora were randomly chosen for the experiment. The eleven primers selected for this study were used to amplify all the C. tora DNA (Table 1).
CHS gene analysis
Total RNA was isolated from the leaf samples (100 mg) using TRIZOL reagent (GIBCO-BRL) as per instructions given in the manufacturers’ protocol. The total RNA was digested with DNase at 37 °C for 15 min and then reverse transcribed into cDNA using M-MLV Reverse Transcriptase RNaseH (Bio-Rad CFX-96TM system). Primers were designed using the software Primer 3 (CHS1 forward: 5′-AGCCAGTGAAGCAGGTAGCC-3′, CHS1 reverse: 5′-GTGATCCGGAAGTAGTAAT-3′ and CHS2 forward: 5′-AGCCAGTGAAGCAGGTAGCC-3′, CHS2 reverse: 5′- GTGATCCGGAAGTAGTAAT -3′), referring to accessed sequences in the Genbank (Supplementary Table S5). The semi quantitative RT-PCR of selected genes was done according to Goto-Yamamoto et al. (2002)61. Triplet of all sample reactions were carried out and negative control of master mix in addition to primers was performed in all RT-PCR runs. GAPDH was taken as control because of its constitutive expression. The accuracy of primers was tested using genomic DNA of the plant as positive control. The intensities of the PCR products on agarose gels were quantified with the Gel Doc 2000 system and volume tool of the Quantity one software (BioRad, USA).
Data analysis
The amplified products were scored in terms of the binary code as present (1) or absent (0), each treated as the unit character regardless of its intensity. Polymorphism at the population level was calculated as the ratio of polymorphic loci to the total number of loci scored in all accessions of the same population. The pair wise genetic distance matrix was computed using UPGMA, NTSYSpc version 2.02e (Supplementary Table S2).
Sample preparation and extraction
Leaves of C. tora were selected for the preparation of extracts because they are more frequently used for medicinal purposes62. Leaves are generally more sensitive to changes in the environmental factors than other organs, and the difference in their traits has been used to classify plants and to establish the genetic relatedness. Phytochemicals were extracted according to Gomez-Romero et al. (2010)63 with slight modifications. Each dried leaf samples of C. tora (weight 5 g) was lyophilized in liquid nitrogen and ground to fine powder. The powder was then defatted with hexane (50 ml), and extracted using a soxhlet extractor (Quickfit, India) (30 min). Hexane was discarded through rotary evaporation. Finally, extracts were dissolved in 50 ml of methanol, incubated overnight (25 °C), filtered through 0.2 µm Millipore filter and stored (4 °C).
FTIR
FTIR spectra were obtained from potassium bromide (KBr) pellets. The fraction (10 µl) of each extract (100 mg/ml) of C. tora was applied on KBr. The infrared spectra were obtained at the resolution of 1 cm−1 in the mid-IR range of 4,000–400 cm−1 using a FTIR spectrophotometer (System 2000, Perkin Elmer, Wellesley, MD, USA). All determinations were in five technical replicate, and data analyzed using statistical software OriginPro 8.0 (Fig. 3). Since, spectral reproducibility is important for creating the robust classification model; hence, variations between replicate spectra due to baseline effect, were removed by derivatization. We further obtained the FTIR spectra of standard quercetin (Supplementary Figure S1) to compare the presence of flavonoids in the tested populations of C. tora.
Total flavonoid content (TFC)
TFC was determined according to Zengin et al. (2011)64. An aliquot of diluted leaf samples (1 mg/ml) as well as the standard solution of quercetin were added to 75 ml of NaNO2 solution, and mixed (6 min), by adding 0.15 ml AlCl3 (100 g/L). After 5 min, 0.5 ml of NaOH was added and the final volume adjusted to 2.5 ml with distilled water, and thoroughly mixed. Absorbance of the mixture was read at 510 nm against the blank using a UV-VIS spectrophotometer (V-550 model, Jasco, Japan). TFC concentration is expressed (mg/g dry wt) (Supplementary Table S4) based on the standard (Fig. 6). All samples were analysed in triplicate.
Statistical analysis
FTIR-data were plotted using statistical software OriginLab (version 8.0). Differences between combined data of highland and lowland populations were analysed using a Student’s t-test analysis in SPSS software v12.0.1 (Chicago, IL, USA). Changes with P<0.05 were considered to be significant. Calibration curve of quercetin (Supplementary Figure 2) was plotted using Microsoft Office Excel (2007).
Multivariate analysis
Polymorphic data obtained by ISSR analyses were analyzed for Bary-Curtis differentiation in the software BioDiversity Pro (version 2.0). Correlation and PCA analysis were done in PAST software (version 2.1) for clustering the transmittance data of six populations of different origins.
Electronic supplementary material
Acknowledgements
The authors are thankful to the Head, Department of Botany, Banaras Hindu University for providing necessary lab facilities and Prof. S. P. Singh for revising the sentence structure of the manuscript. University Grant Commission is also acknowledged for providing financial assistance to VK (F1-17.1/2011-12/RGNF-SC-UTT-4945/(SA-III/Website)).
Author Contributions
V.K. and B.K.R. designed and conceived the experiment. V.K. performed the experiment, analyzed the data and wrote the article. Both the authors have read and approved the final version of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-29114-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jayashree A, Maneemegalai S. Studies on the antibacterial activity of the extracts from Tridax procumbens L. and Ixora coccinea L. Biomedicine. 2008;28:190–194. [Google Scholar]
- 2.Smith, A. C. F V Nova: A new flora of Fiji. National Tropical Botanical Garden, Lawai, H.I., USA, 110–111 (1985).
- 3.Jain S, Patil UK. Phytochemical and pharmacological profile of Cassia tora L.: An overview. IJNPR. 2010;1(4):430–437. [Google Scholar]
- 4.Kumar V, Kumar A, Pandey KD, Roy BK. Isolation and characterization of bacterial endophytes from the roots of Cassia tora L. Ann Microbiol. 2014;65:1391–1399. doi: 10.1007/s13213-014-0977-x. [DOI] [Google Scholar]
- 5.WHO WHO Model List of Essential Medicines. World Health Organization. (2014).
- 6.Vijayalakshmi A, Madhira G. Anti-psoriatic activity of flavonoids from Cassia tora leaves using the rat ultraviolet B ray photodermatitis model. Revista Brasileira de. Farmacognosia. 2014;24(3):322–329. doi: 10.1016/j.bjp.2014.07.010. [DOI] [Google Scholar]
- 7.Meena AK, et al. Cassia tora Linn.: A review on its ethanobotany, phytochemical and pharmacological profile. J. pharmacy Res. 2010;3(3):557–560. [Google Scholar]
- 8.Abraham A, Rejiya CS, Cibin TR. Leaves of Cassia tora as a novel cancer therapeutic–An in vitro study. Toxicol. in Vitro. 2009;23:1034–1038. doi: 10.1016/j.tiv.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Pawar, H. A., & D’mello, P. M. Cassia tora Linn.: An Overview. IJPSR, 2(9), 2286–2291 (2011).
- 10.Vats S, Kamal R. Identification of flavonoids and antioxidant potential of Cassia tora L. Amer. J. Drug Disc. Devel. 2014;4(1):50–57. doi: 10.3923/ajdd.2014.50.57. [DOI] [Google Scholar]
- 11.Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell Biology, and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tohge T, Yonekura-Sakakibara K, Niida R, Wantanabe-Takahasi A, Saito K. Phytochemical genomics in Arabidopsis thaliana: A case study for functional identification of flavonoid biosynthesis genes. Pure Appl. Chem. 2007;9(4):811–23. doi: 10.1351/pac200779040811. [DOI] [Google Scholar]
- 13.Hrazdina G, Wagner GJ. Metabolic pathways as enzyme complexes: evidence for the synthesis of phenylpropanoids and flavonoids on membrane associated enzyme complexes. Arch. Biochem. Biophys. 1985;237(1):88–100. doi: 10.1016/0003-9861(85)90257-7. [DOI] [PubMed] [Google Scholar]
- 14.Sun W, et al. Molecular and Biochemical Analysis of Chalcone Synthase from Freesia hybrid in flavonoid biosynthetic pathway. PLoS One. 2015;10(3):e0119054. doi: 10.1371/journal.pone.0119054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao H, et al. Deep sequencing of the transcriptome reveals distinct flavonoid metabolism features of black tartary buckwheat (Fagopyrum tataricum Garetn.) Prog Biophys Mol Biol. 2016;124:49–60. doi: 10.1016/j.pbiomolbio.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Thain SC, Murtas G, Lynn JR, McGrath RB, Millar AJ. The circadian clock that controls gene expression in Arabidopsis is tissue specific. Plant Physiol. 2002;130:102–110. doi: 10.1104/pp.005405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou HT, et al. A study on genetic variation between wild and cultivated populations of Paeonia lactiflora Pall. Acta Pharm. Sin. 2002;37:383–388. [PubMed] [Google Scholar]
- 18.Dong TT, et al. Chemical assessment of roots of Panax notoginseng in China: regional and seasonal variations in its active constituents. J. Agr. Food Chem. 2003;51:4617–4623. doi: 10.1021/jf034229k. [DOI] [PubMed] [Google Scholar]
- 19.Said SA, et al. Inter-population variability of terpenoid composition in leaves of Pistacia lentiscus L. from Algeria: A chemoecological approach. Molecules. 2011;16(3):2646–2657. doi: 10.3390/molecules16032646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Y, David B, Tu P, Barbin Y. Recent analytical approaches in quality control of traditional Chinese medicines—a review. Analytica chimica acta. 2010;657(1):9–18. doi: 10.1016/j.aca.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 21.Khairudin K, Sukiran NA, Goh HH, Baharum SN, Noor NM. Direct discrimination of different plant populations and study on temperature effects by Fourier transform infrared spectroscopy. Metabolomics. 2014;10:203–211. doi: 10.1007/s11306-013-0570-5. [DOI] [Google Scholar]
- 22.Kress, W. J. & Erickson, D. L. DNA barcodes: genes, genomics, and bioinformatics. Proc Natl. Acad. Sci. USA, 26, 105(8), 2761 (2008). [DOI] [PMC free article] [PubMed]
- 23.Techen N, Parveen I, Ikhlas ZP, Khan A. DNA barcoding of medicinal plant material for identification. Curr. Opin. Biotech. 2014;25:103–110. doi: 10.1016/j.copbio.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 24.Chen X, et al. Identification of crude drugs in the Japanese pharmacopoeia using a DNA barcoding system. Sci. Rep. 2017;7:42325. doi: 10.1038/srep42325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gigliano GS. Restriction profiles of trnL (UAA) intron as a tool in Cannabis sativa L. identification. Delpinohaa. 1995;37(8):85–95. [Google Scholar]
- 26.Asahina H, Shinozaki J, Masuda K, Morimitsu Y, Satake M. Identification of medicinal Dendrobium species by phylogenetic analyses using matK and rbcL sequences. J. Nat. Med. 2010;64:133–138. doi: 10.1007/s11418-009-0379-8. [DOI] [PubMed] [Google Scholar]
- 27.Chen S, et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE. 2010;5:8613. doi: 10.1371/journal.pone.0008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song M, Li J, Xiong C, Liu H, Liang J. Applying high-resolution melting (HRM) technology to identify five commonly used Artemisia species. Sci. Rep. 2016;6:34133. doi: 10.1038/srep34133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raclariu AC, et al. Comparative authentication of Hypericum perforatum herbal products using DNA metabarcoding, TLC and HPLC-MS. Sci. Rep. 2017;7(1):1291. doi: 10.1038/s41598-017-01389-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen J, et al. Intersimple Sequence Repeats (ISSR) Molecular Fingerprinting Markers for Authenticating Populations of Dendrobium officinale. Biol. Pharm. Bull. 2006;29(3):420–422. doi: 10.1248/bpb.29.420. [DOI] [PubMed] [Google Scholar]
- 31.Ahn CH, Kim YS, Lim S, Yi JS, Choi YE. Random amplified polymorphic DNA (RAPD) analysis and RAPD-derived sequence characterized amplified regions (SCAR) marker development to identify Chinese and Korean ginseng. J. Med. Plant Res. 2011;5:4487–4492. [Google Scholar]
- 32.Esselman EJ, Jianqiang L, Crawford DJ, Windus JL, Wolfe AD. Clonal diversity in the rare Calamagrostis porteri ssp. Insperata (Poaceae): comparative results for allozymes and random amplified polymorphic DNA (RAPD) and intersimple sequence repeat (ISSR) markers. Mole. Ecol. 1999;8:443–451. doi: 10.1046/j.1365-294X.1999.00585.x. [DOI] [Google Scholar]
- 33.Mort ME, et al. Relationships among the Macaronesian members of Tolpis (Asteraceae: Lactuceae) based upon analyses of inter simple sequence repeat (ISSR) markers. Taxon. 2003;52:511–518. doi: 10.2307/3647449. [DOI] [Google Scholar]
- 34.Acharya L, Mukherjee AK, Panda PC. Separation of the genera in the subtribe Cassiinae (Leguminosae: Caesalpinioidae) using molecular markers. Acta. Bot. Bras. 2011;25(1):223–233. doi: 10.1590/S0102-33062011000100026. [DOI] [Google Scholar]
- 35.Carballo-Meilan A, Goodman AM, Baron MG, Gonzalez-Rodriguez J. A specific case in the classification of woods by FTIR and chemometric: discrimination of Fagales from Malpighiales. Cellulose. 2014;21:261–273. doi: 10.1007/s10570-013-0093-2. [DOI] [Google Scholar]
- 36.Vijayalakshmi A, Geetha M, Ravichandiran V. Quantitative evaluation of the antipsoriatic activity of flavonoids from Cassia tora Linn. Leaves. Iran J Sci Technol Trans Sci. 2017;41:307. doi: 10.1007/s40995-017-0219-8. [DOI] [Google Scholar]
- 37.Zhang Y-B, Pang-Chui S, Cho-Wing S, Tong Y. Z-TW. Molecular Authentication of Chinese Herbal Materials. J.Food Drug Anal. 2007;15(1):1–9. [Google Scholar]
- 38.Leffler EM, et al. Revisiting an old riddle: what determines genetic diversity levels within species? PLoS Biol. 2012;10:e1001388. doi: 10.1371/journal.pbio.1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang XM. Inter-simple sequence repeats (ISSR) molecular fingerprinting markers for authenticating the genuine species of rhubarb. J. Med. Plants Res. 2011;5(5):758–764. [Google Scholar]
- 40.Korner, C. Alpine plant life, 2nd edition. Heidelberg: Springer, (2003).
- 41.Byars SG, Parsons Y, Hoffmann AA. Effect of altitude on the genetic structure of an Alpine grass. Poa hiemata. Ann.Bot. 2009;103:885–899. doi: 10.1093/aob/mcp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hahn T, et al. Patterns of Genetic Variation across Altitude in Three Plant Species of Semi-Dry Grasslands. PLoS ONE. 2012;7(8):41608. doi: 10.1371/journal.pone.0041608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allwood JW, Ellis DI, Goodacre R. Metabolomic technologies and their application to the study of plants and plant–host interactions: A review. Physiol. Plantarum. 2008;132:117–135. doi: 10.1111/j.1399-3054.2007.01001.x. [DOI] [PubMed] [Google Scholar]
- 44.Sim, C. O., Hamdani, M. R., Ismail, Z. & Ahmad, M. N. Assessment of herbal medicines by chemometrics-assisted interp retation of FTIR spectra. J. Anal. Chim. Acta.1–14 (2004).
- 45.Li YM, et al. Identification of American ginseng from different regions using FTIR and two-dimensional correlation IR spectroscopy. Vib. Spectrosc. 2004;36:227–232. doi: 10.1016/j.vibspec.2003.12.009. [DOI] [Google Scholar]
- 46.Kumar JK, Devi Prasad AG. Identification and comparison of bio-molecules in medicinal plants of Tephrosia tinctoria and Atylosia albicans by using FTIR. Rom. J. Biophys. 2011;21(1):63–71. [Google Scholar]
- 47.Heneczkowski M, Kopackz M, Nowak D, Kuzniar A. Infrared spectrum analysis of some flavonoids. Acta Pol. Pharm. 2001;58(6):415–420. [PubMed] [Google Scholar]
- 48.Chan EWC, Lim YY, Lim TY. Total Phenolic Content and Antioxidant Activity of Leaves and Rhizomes of Some Ginger Species in Peninsular Malaysia. Gardens’ Bulletin Singapore. 2007;59(1&2):47–56. [Google Scholar]
- 49.Wang G, Cao F, Wang G, Yousry A. & El-Kassaby, Role of Temperature and Soil Moisture Conditions on Flavonoid Production and Biosynthesis-Related Genes in Ginkgo (Ginkgo biloba L.) Leaves. Nat. Prod. Chem. Res. 2015;3:162. doi: 10.4172/2329-6836.1000162. [DOI] [Google Scholar]
- 50.Zidorn C. Altitudinal variation of secondary metabolites in flowering heads of the Asteraceae: trends and causes. Phytochem. Rev. 2010;9:197–203. doi: 10.1007/s11101-009-9143-7. [DOI] [Google Scholar]
- 51.Sundqvist MK, Wardle DA, Olofsson E, Giesler R, Gundale MJ. Chemical properties of plant litter in response to elevation: subarctic vegetation challenges phenolic allocation theories. Funct. Ecol. 2012;26:1090–1099. doi: 10.1111/j.1365-2435.2012.02034.x. [DOI] [Google Scholar]
- 52.Close DC, McArthur C. Rethinking the role of many plant phenolics- protections from photodamage not herbivores? Oikos. 2002;99:166–172. doi: 10.1034/j.1600-0706.2002.990117.x. [DOI] [Google Scholar]
- 53.Janas KM, Cvikrova M, Palagiewicz A, Eder J. Alterations in phenylpropanoid content in soybean roots during low temperature acclimation. Plant Physiol. Biochem. 2000;38:587–593. doi: 10.1016/S0981-9428(00)00778-6. [DOI] [Google Scholar]
- 54.Pandey A, et al. AtMYB12 expression in tomato leads to large scale differential modulation in transcriptome and flavonoid content in leaf and fruit tissues. Sci. Rep. 2015;5:12412. doi: 10.1038/srep12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang J, Huang J, Gu H, Zhong Y, Yang Z. Duplication and adaptive evolution of the chalcone synthase genes of Dendranthema (Asteraceae) Mol. Biol. Evol. 2002;19(10):1752–1759. doi: 10.1093/oxfordjournals.molbev.a003997. [DOI] [PubMed] [Google Scholar]
- 56.Panigrahi GK, et al. Preparative thin-layer chromatographic separation followed by identification of antifungal compound in Cassia laevigata by RP-HPLC and GC-MS. J. Sci. Food Agr. 2013;94(2):308–315. doi: 10.1002/jsfa.6259. [DOI] [PubMed] [Google Scholar]
- 57.Samappito S, Schmidt AJ, Eknamkul WD, Kutchan TM. Molecular characterization of root-specific chalcone synthases from Cassia alata. Planta. 2002;216:64–71. doi: 10.1007/s00425-002-0872-8. [DOI] [PubMed] [Google Scholar]
- 58.Schulze-Lefert P, Becker-Andre M, Schulz W, Hahlbrock K, Dangl JL. Functional architecture of the light-responsive chalcone synthase promoter from parsley. The Plant Cell. 1989;1(7):707–714. doi: 10.1105/tpc.1.7.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seethapathy GS, et al. Assessing product adulteration in natural health products for laxative yielding plants, Cassia, Senna, and Chamaecrista, in Southern India using DNA barcoding. Int. J. Leg. Med. 2015;129(4):693–700. doi: 10.1007/s00414-014-1120-z. [DOI] [PubMed] [Google Scholar]
- 60.Wang XM. Optimization of DNA isolation, ISSR-PCR system and primers screening of genuine species of rhubarb, an important herbal medicine in China. J. Med. Plants Res. 2010;., 4(10):904–908. [Google Scholar]
- 61.Goto-Yamamoto N, Wan GH, Masaki K, Kobayashi S. Structure and transcription of three chalcone synthase genes of grapevine (Vitis vinifera) Plant Science. 2002;162:867–872. doi: 10.1016/S0168-9452(02)00042-0. [DOI] [Google Scholar]
- 62.Sirappuselvi S, Chitra M. In vitro Antioxidant Activity of Cassia tora Lin. Int. Res. J. Biol. Sci. 2012;1(6):57–61. [Google Scholar]
- 63.Gomez-Romero M, Segura-Carretero A. & Fernandez-Gutierrez, Metabolite profiling and quantification of phenolic compounds in methanol extracts of tomato fruit. Phytochem. 2010;71:1848–1864. doi: 10.1016/j.phytochem.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 64.Zengin G, Aktumsek A, Guler GO, Cakmak YS, Yildiztugay E. Antioxidant properties of methanolic extract and fatty acid composition of Centaurea urvillei DC. sub sp. hayekiana Wagenitz. Rec. Nat. Prod. 2011;5(2):123. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.