Abstract
After injury to the mature central nervous system (CNS), myelin-derived inhibitory ligands bind to the Nogo-66 tripartite receptor complex expressed on axonal growth cones, comprised of LINGO-1 and p75NTR/TROY and induce growth cone collapse through the RhoA pathway. We have also shown that amphoterin-induced gene and open reading frame-3 (AMIGO3) substitutes for LINGO-1 and can signal axon growth cone collapse. Here, we investigated the regeneration of dorsal root ganglion neuron (DRGN) axons/neurites after treatment with a short hairpin RNA (sh) AMIGO3 plasmid delivered with a non-viral in vivo-jetPEI vector, and the pro-survival/axogenic neurotrophin (NT) 3 in vitro and in vivo. A bicistronic plasmid, containing both shAMIGO3 and NT3 knocked down >75% of AMIGO3 mRNA in cultured DRGN and significantly overexpressed NT3 production. In vivo, intra-DRG injection of in vivo-jetPEI plasmids containing shAMIGO3/gfp and shAMIGO3/nt3 both knocked down AMIGO3 expression in DRGN and, in combination with NT3 overexpression, promoted DC axon regeneration, recovery of conduction of compound action potentials across the lesion site and improvements in sensory and locomotor function. These findings demonstrate that in vivo-jetPEI is a potential non-viral, translatable DRGN delivery vehicle in vivo and that suppression of AMIGO3 disinhibits the growth of axotomised DRGN enabling NT3 to stimulate the regeneration of their DC axons and enhances functional recovery.
Introduction
Adult mammalian central nervous system (CNS) axons do not regenerate after injury probably because ontogenetic axogenic intracellular signalling is suppressed during CNS maturation. Axon growth inhibitory ligands also become incorporated into the maturing CNS neuropil, derived from both myelin and the incipient scar, which further limits CNS axon regeneration1. The former comprises Nogo-A, myelin associated glycoprotein (MAG), and oligodendrocyte-derived myelin glycoprotein (OMgp), while the latter comprise chondroitin sulphate proteoglycan (CSPG), NG2, semaphorins and ephrins (secreted by reactive astrocytes and invading meningeal fibroblasts)1–6. After binding to their cognate receptors, myelin- and scar-derived inhibitory ligands activate intracellular signals which converge on the RhoGTPase pathway, mediating axon growth cone collapse1–6. Myelin-derived inhibitors bind to a tripartite receptor complex comprised of NGR1/p75NTR/LINGO-1 in which TROY4,7 substitutes for p75NTR and AMIGO3 (amphoterin-induced gene and open reading frame-3) for LINGO1 in the immediate post-injury period8. Evidence for the latter proposition is derived from experiments in which: in vitro (i), knockdown of AMIGO3 permits retinal ganglion cell (RGC) and dorsal root ganglion neuron (DRGN) neurites to grow on a CNS myelin extract (CME) substrate; (ii), RhoA is activated in response to co-transfection with amigo3/ngr1/p75ntr; and (iii), significant numbers of neurites grow after knockdown of AMIGO3; and (iv), in vivo, AMIGO3, not LINGO-1, becomes elevated in RGC/DRGN for up to 10 days after optic nerve/dorsal column (DC) crush injury8.
Viral vectors are commonly used to transfect CNS cells with shRNA to inhibit target mRNA translation9–11. Viral vectors, such as adeno-associated virus (AAV), transfect multiple genes but are restricted by the insert capacity whilst shRNA delivery delays suppression of targeted mRNA by 14–28 days. Both these disadvantages retard the therapeutic effect and reduce their translational potential12–14. Another significant impediment to the use viral vectors in vivo is that maximum transgene expression requires 7–14 days and hence viral vector transfection is limited in acute conditions. Non-viral gene delivery vectors include cationic lipid agents and a more recently formulated non-lipid polymer, polyethylenimine (in vivo-jetPEI), which transfects cells both in vitro and in vivo15,16. In vivo-jetPEI has high transfection efficiency and yields up to 4× higher transfection rates than naked DNA15. Moreover, in vivo-jetPEI is easy to prepare, stable and safe, and in vivo-jetPEI/shRNA transfection induces faster suppression of mRNA translation than viral vectors, giving immediate therapeutic benefit17. For example, in the CNS, Melanopsin expression is rapidly abolished in RGC after intravitreal injection of in vivo-jetPEI-shMelanopsin18.
In this study, we show that in vivo-jetPEI successfully delivers a bicistronic plasmid DNA to DRGN encoding shAMIGO3 to suppress AMIGO3 and perturb axon growth inhibitory signalling while, at the same time inducing the overexpression of neurotrophin-3 (NT3). We show that suppression of AMIGO3 and concomitant stimulation of DRGN with NT3 enhanced disinhibited DRGN neurite/axon regeneration and improved functional recovery. These results suggest that in vivo-jetPEI PEI delivers genes to DRGN without the disadvantages of viral vectors, rendering in vivo-jetPEI PEI as translationally relevant.
Results
In vitro experiments
In vivo-jetPEI delivered plasmids knock down AMIGO3 mRNA
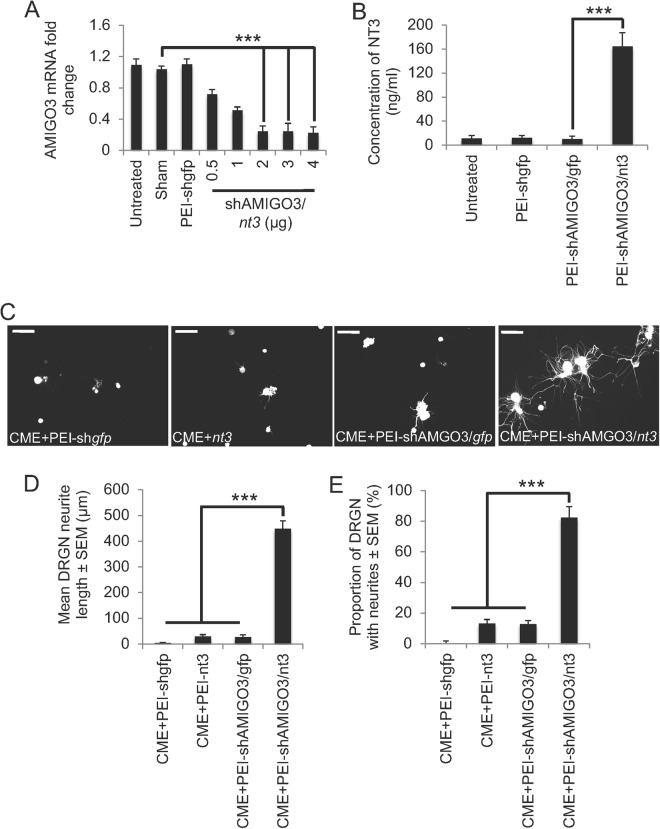
In untreated, sham and non-specific shgfp control transfected DRGN, there was no change in mRNA for AMIGO3 suggesting that none of these treatments had any non-specific effects on amigo3 mRNA (Fig. 1A). Treatment with increasing amounts of shAMIGO3 plasmid delivered by in vivo-jetPEI (referred to as PEI in the results section from herein) caused a dose-dependent decrease in amigo3 mRNA to a minimum at 2 µg of plasmid DNA, correlating with 80% knockdown compared to untreated, sham or shgpf controls (Fig. 1A). Increasing the amount of plasmid DNA above 2 µg did not decrease amigo3 mRNA levels further, confirming that 2 µg of plasmid DNA gave optimal knockdown.
Figure 1.
Knockdown of AMIGO3 and NT3 over-expression by PEI-delivered plasmid DNA disinhibited DRGN neurite outgrowth. (A) Increasing concentrations of plasmid DNA encoding shAMIGO3/nt3 efficiently suppressed AMIGO3 mRNA in cultured DRGN. (B) Plasmids encoding nt3 significantly increased the titres of NT3 in DRGN culture media. (C) Representative images show that in the presence of CME, plasmid DNA encoding gfp, nt3 or shAMIGO3/gfp did not, but that plasmids encoding shAMIGO3 and nt3 did promote DRGN neurite outgrowth. Note: DRGN do not have neurites due to the presence of inhibitory concentrations of CME, which does not affect their survival. (D) Quantification of the mean DRGN neurite length and (E) the proportion of DRGN with neurites showed that AMIGO3 suppression combined with nt3 overexpression promoted significant disinhibited DRGN neurite outgrowth. Scale bars in C = 50 μm. ***P < 0.0001, ANOVA.
PEI delivered shAMIGO3/nt3 plasmids increased NT3 secretion into the culture media
In untreated, sham, non-specific PEI-shgfp and PEI-shAMIGO3/gfp-treated DRGN, little or no NT3 was detected by ELISA in culture supernatant (Fig. 1B). However, in culture media from DRGN, treated with 2 µg of PEI-shAMIGO3/nt3 plasmid DNA significant production and release of NT3 occurred (164 ± 24 ng/ml, P < 0.0001) compared to PEI-shAMIGO3-gfp (Fig. 1B). These results suggest that 2 µg of plasmid DNA was optimal for amigo3 mRNA knockdown and NT3 production.
Knockdown of AMIGO3 and concomitant stimulation of NT3 disinhibited DRGN neurite outgrowth
In PEI-shgfp-, PEI-nt3 or PEI-shAMIGO3/gfp-treated cultures, little or no DRGN neurite outgrowth was observed on CME substrates (Fig. 1C–E). However, treatment of cultures with PEI-shAMIGO3/nt3 significantly increased both neurite length (448 ± 31 µm, P < 0.0001 compared to PEI-shAMIGO3/gfp), and the proportion of DRGN with neurites (82 ± 8%, P < 0.0001 compared to PEI-shAMIGO3/gfp; Fig. 1C–E). Thus, delivery of shAMIGO3/nt3 plasmids by PEI significantly enhanced DRGN neurite outgrowth on a CME substrate.
In vivo experiments
PEI-shAMIGO3 enhanced transduction in all sizes of DRGN
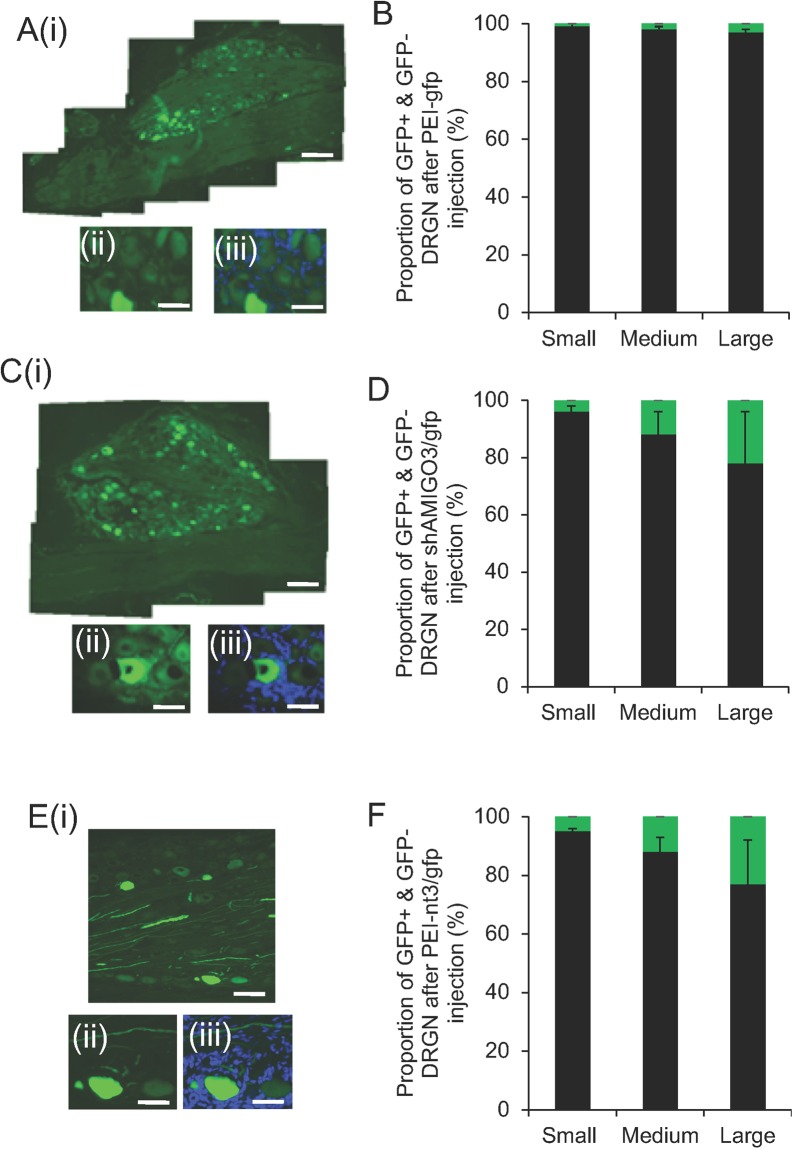
No GFP+ DRGN was observed in either intact controls (IC) or in dorsal column (DC) crush injured animals (not shown). In the DC + PEI-gfp (Fig. 2A(i–iii),B), DC + PEI-nt3/gfp (not shown) and DC + PEI-shAMIGO3/nt3 groups, similar numbers of DRGN were GFP+ (green) (Fig. 2C(i–iii),D). High power insets of GFP (Fig. 2A(ii),C(ii)) and images merged with DAPI counterstain (blue) (Fig. 2A(iii),C(iii)) showed variable weak and high levels of GFP+ DRGN. Approximately 1, 2 and 3% of small, medium and large diameter DRGN were GFP+, respectively, in the DC + PEI-gfp group (Fig. 2B) and, in the DC + PEI-shAMIGO3/gfp group, GFP expression increased significantly to 4, 12 and 22% in small, medium and large diameter DRGN, respectively (Fig. 2D). Similar levels of DRGN transduction were also observed in DC + PEI-nt3/gfp groups (Fig. 2E,F). These results suggested that PEI delivered plasmids encoding shAMIGO3-/gfp or nt3/gfp enhanced transduction in all sizes of DRGN compared to PEI-gfp alone.
Figure 2.
GFP+ (green) DRGN in DRG in the DC + PEI-gfp group (A(i,ii)); high power to show GFP+ DRGN, (iii); high power GFP+ DRGN with DAPI counterstained (blue) nuclei), (B) the proportion of GFP+ small (0–29 μm), medium (30–59 μm) and large (>60 μm) diameter DRGN (green bar); % total GFP+ + GFP− small, medium and large DRGN (black bar). (C) GFP+ DRGN in the DC + PEI-shAMIGO3/gfp group (C(i),(ii)), high power to show GFP+ DRGN, (iii); high power GFP+ DRGN with DAPI counterstained (blue) nuclei), (D) the proportion of GFP+ small (0–29 μm), medium (30–59 μm) and large (>60 μm) diameter DRGN (green bar); % total GFP+/GFP− small, medium and large diameter DRGN (black bar). (E) GFP+ DRGN in the DC + PEI-nt3/gfp group (E(i),(ii)), high power to show GFP+ DRGN, (iii); high power GFP+ DRGN with DAPI counterstained (blue) nuclei), (F) the proportion of GFP+ small (0–29 μm), medium (30–59 μm) and large (>60 μm) diameter DRGN (green bar); % total GFP+/GFP− small, medium and large diameter DRGN (black bar). Scale bar in (A(i)), (C(i)) and (E(i)) = 400 μm, in (A(ii)), (A(iii)), (C(ii)) and (C(iii)), (E(ii)) and E(iii) = 100 μm.
PEI/shAMIGO3 down regulated AMIGO3 in DRGN in vivo
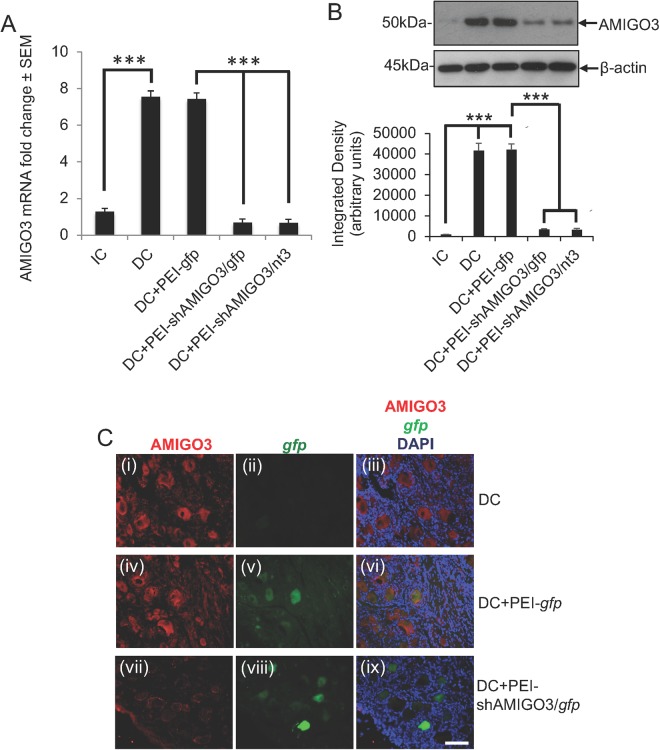
The levels of amigo3 mRNA increased significantly by 7.54 ± 0.33 and 7.42 ± 0.34-fold (P < 0.0001) in both DC and DC + PEI-gfp groups compared to those observed in the IC group (Fig. 3A). However, amigo3 mRNA levels were suppressed by nearly 11-fold in DRG treated with both PEI-shAMIGO3/gfp and PEI-shAMIGO3/nt3 (P < 0.0001 compared to DC + PEI-gfp; Fig. 3A). Western blot and subsequent densitometry reflected these significant increased changes (P < 0.0001) in AMIGO3 protein in DC and DC + PEI-gfp groups whilst a significant reduction (P < 0.0001) was observed in DC + PEI-shAMIGO3/gfp and DC + PEI-shAMIGO3/nt3 groups (Fig. 3B). Immunohistochemistry also detected high levels of AMIGO3 (red) in DC and DC + PEI-gfp-treated DRG in GFP+ (green) and GFP− DRGN (Figs 3C(i)-(iii) and 4C(iv)-(vi), respectively; panels (iii) and (vi) show merged images with DAPI+ nuclei stained in blue). In the DC-PEI-gfp group (Fig. 3C(iv)-(vi)) almost 30% of the DRGN were GFP+/AMIGO3+. In the DC + PEI-shAMIGO3/gfp group, similar numbers of DRGN as in the DC + PEI-gfp group were GFP+ but few DRGN were AMIGO3+ (Fig. 3C(vii)-(ix); panel (ix) = merged with DAPI+ nuclei stained in blue). These results demonstrate that: (1), PEI-delivered plasmids encoding shAMIGO3 efficiently knocked down AMIGO3 protein in DRGN; and (2), knockdown of AMIGO3 occurred in both transfected and un-transfected DRGN.
Figure 3.
AMIGO3 levels are suppressed in DRGN after injection of PEI transduced plasmids encoding shAMIGO3. (A) Low levels of AMIGO3 mRNA in IC increased significantly after DC injury (P < 0.0001) and remained high in the DC + PEI-gfp group. AMIGO3 mRNA levels reduced significantly in both DC + PEI/shAMIGO3/gfp and DC + PEI-shAMIGO3/nt3 groups (P < 0.0001). (B) Western blot and subsequent densitometry for AMIGO3 protein reflected the changes in mRNA. β-actin was used as a loading control (full scans shown). (C) Immunohistochemistry for AMIGO3 showed high levels of AMIGO3 (red) in DRGN after DC (C(i)), which were gfp− (C(ii)). AMIGO3 levels remained high in DC + PEI-gfp groups (C(iv)), with gfp expression (green) in some DRGN (C(v)). AMIGO3 levels were suppressed in DRGN from DC + PEI-shAMIGO3/gfp groups (C(vii)), with gfp expression in some DRGN (C(viii)). (C(iii)), (C(vi)) and (C(ix)) are merged images showing AMIGO3 (red), gfp (green) and DAPI stained nuclei (blue). Scale bar = 100 μm. ***P < 0.0001, ANOVA.
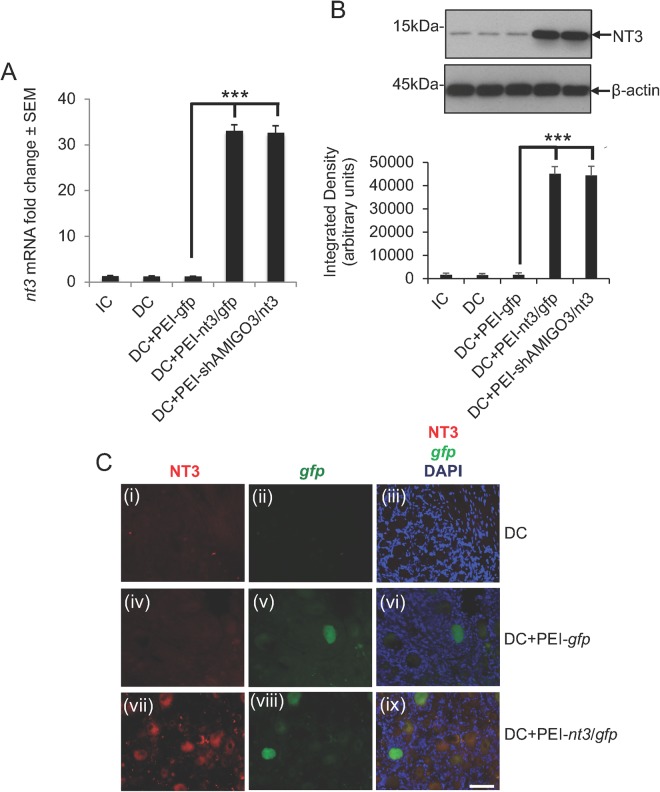
Figure 4.
NT3 levels were overexpressed in DRGN after injection of in vivo-jetPEI transduced plasmids encoding shAMIGO3. (A) Low levels of nt3 mRNA were detected in IC, DC and DC + PEI-gfp groups, whilst significantly higher levels (P < 0.0001) were detected in DC + PEI-nt3/gfp and DC + PEI-shAMIGO3/nt3 groups. (B) Western blot and subsequent densitometry for NT3 protein reflected the changes in mRNA. β-actin was used as a loading control (full scans shown). (C) Immunohistochemistry for NT3 showed low levels of NT3 (red) in DRGN after DC. (C(i)), which were gfp− (C(ii)) and in DC + PEI-gfp groups (C(iv)), with gfp expression (green) in some DRGN (C(v)). NT3 levels (Cvii)) were high in both gfp+ and gfp− DRGN (C(viii)). Scale bar = 100 μm. C(iii), C(vi) and C(ix) are merged images showing NT3 (red), gfp (green) and DAPI stained nuclei (blue). ***P < 0.0001, ANOVA.
NT3 levels are overexpressed in DRGN after PEI-shAMIGO3/nt3 transfection
The levels of nt3 mRNA remained low or unchanged in IC, DC and DC + PEI-gfp-treated DRGN (Fig. 4A). However, in DRGN treated with DC + PEI-nt3/gfp and DC + PEI-shAMIGO3/nt3, nt3 mRNA levels increased significantly and by 33.0 ± 1.35-fold, compared to DC + PEI-gfp-treated DRGN (P < 0.0001; Fig. 4A). Similar changes in the levels of NT3 protein were corroborated by western blot and subsequent densitometry showing high levels of NT3 production in DC + PEI-nt3/gfp and DC + PEI-shAMIGO3/nt3-treated groups (Fig. 4B). Little or no NT3 immunoreactivity (red) was present in GFP+ (green) and GFP− DRGN in IC (not shown), DC (Fig. 4C(i)-(iii)) and DC + PEI-gfp (Fig. 4C(iv)-(vi); panels (iii) and (vi) show merged images with DAPI+ nuclei stained in blue) groups. However, in the DC + PEI-nt3/gfp group (Fig. 4C(vii)-(ix); panel (ix) = merged with DAPI+ nuclei stained in blue), high levels of NT3 immunoreactivity were observed in both GFP+ and GFP− DRGN. These results demonstrate that: (1), PEI successfully transfected DRGN with plasmids encoding nt-3 resulting in NT3 protein expression; and (2), NT3 was detected in both transfected and un-transfected DRGN.
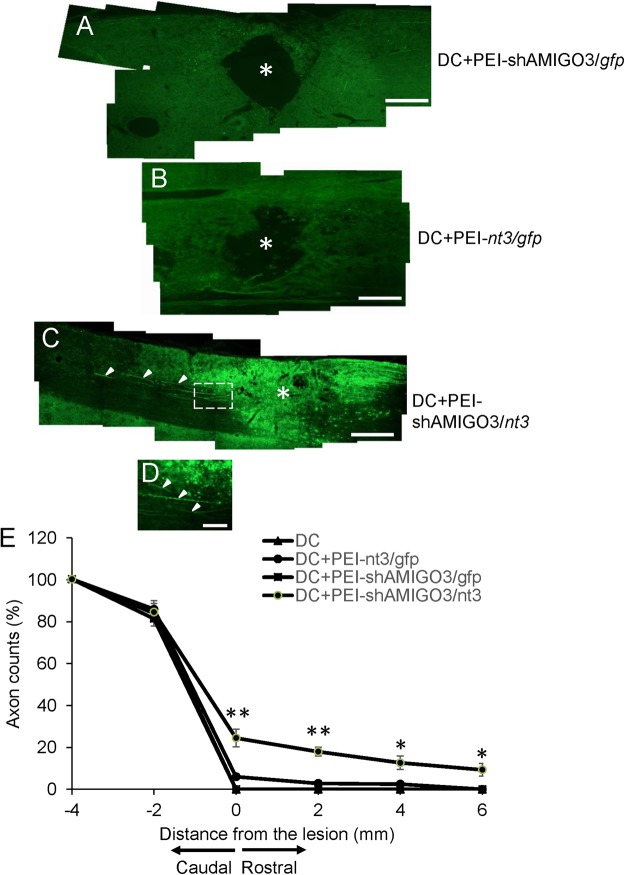
PEI-shAMIGO3/nt3 transfected axotomised DRGN regenerate axons in the DC
DC injury induced a large cavity at the lesion site (*) in the cords of DC and DC + PEI/gfp (not shown), DC + PEI-shAMIGO3/gfp (Fig. 5A) and DC + PEI-nt3/gfp (Fig. 5B) groups; few immunoreactive GAP43+ axons were seen (Fig. 5A,B). By contrast, in the injured cords of the DC + PEI-shAMIGO3/nt3 groups, no cavities developed and many regenerating GAP43+ axons crossed through caudal cord segment and some traversed the lesion site (*), growing for long distances into the rostral segment of the spinal cord (Fig. 5C (arrowheads), Fig. 5D; high power magnification of boxed region in Fig. 5C). Quantification of the number of GAP43+ DC axons regenerating through the lesion site shows that 24.5 ± 4.2, 18.0 ± 2.1, 12.7 ± 3.3 and 9.3 ± 3.1% of axons regenerated rostrally at 0, 2, 4 and 6 mm beyond the lesion site, respectively, in DC + PEI-shAMIGO3/nt3 whilst no axons were present rostrally in either DC, DC + PEI-nt3/gfp or DC + PEI-shAMIGO3/gfp. These results demonstrate that PEI-shAMIGO3/nt3 plasmids promote DC axon regeneration after DC injury.
Figure 5.
Suppression of AMIGO3 in DRGN promoted axon regeneration in the cord rostral to the lesion. There were no GAP43+ regenerating axons in the DC of the (A) PEI + shAMIGO3/gfp and (B) DC + PEI-nt3/gfp groups with a large cavity (*) present at the lesion site. (C) Knockdown of AMIGO3 and co-incident up-regulation of NT3 in the DC + shAMIGO3/nt3 group promoted GAP43+ DRGN axon (arrowheads) regeneration through the DC lesion site. ((D), high power view of axons coursing rostrally from the lesion site (arrowheads)), with the absence of a cavity in the lesion site. Scale bar in A–C = 500 μm; in D = 50 μm. (E) GAP43+ axon fiber counts at specific distances rostral and caudal to the lesion site showed a significant proportion of axons present at 2, 4 and 6 mm rostral to the lesion site. **P < 0.001, ANOVA; * < 0.05, ANOVA.
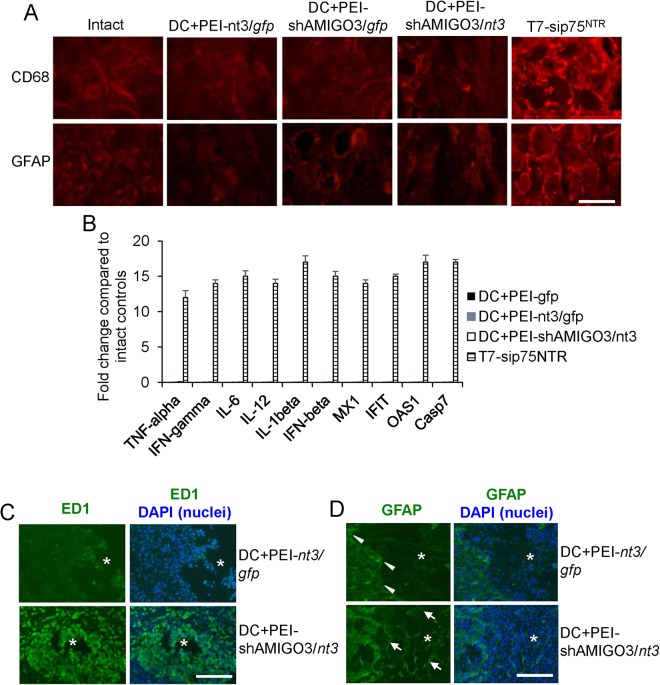
PEI-delivered plasmids did not invoke off-target effects after intra-DRG injection
Intra-DRG injection of any of the plasmids that were delivered by PEI did not invoke changes in the CD68 and GFAP immunoreactivity in DRG (Fig. 6A). In contrast, intra-DRG injection of T7-sip75NTR, a chemically synthesized siRNA that induces off-target effects19, invoked a marked increase in CD68 and GFAP immunoreactivity in DRG (Fig. 6A). Likewise, intra-DRG injection of PEI delivered plasmids did not invoke off-target immune-mediated cytokines or innate immunity genes such as TNF-α, IFN-γ, IL-6, IL-12/IL-23, IL-1β, and IFN-β, MX1, IFIT, OAS1 or Casp7 (Fig. 6B). In contrast, intra-DRG injection of sip75NTR induced 12–17-fold increases in the mRNA of all of these genes (Fig. 6B). These results suggest that PEI-delivered plasmids do not invoke off-target effects in DRGN.
Figure 6.
Response to PEI-delivered plasmids. (A) CD68 and GFAP immunoreactivity in intact and after PEI-delivered plasmids in section of DRG to show a lack of induction of immunological responses to these markers. In contrast, T7-sip75NTR invoked significant changes in CD68 and GFAP+ immunoreactivity in DRG. (B) Lack of activation of off-target cytokines and innate immunity related genes in DRG after PEI-delivered plasmid transduction in DRG. Once again, T7-sip75NTR showed significant upregulation of mRNA for all of these genes. (C) ED1 immunoreactivity was low in DC + PEI-nt3/gfp-treated animals whilst abundant ED1+ cells were present within the lesion site (*) in DC + PEI-shAMIGO3/nt3-treated animals. (D) GFAP+ astrocytes were restricted to the lesion edge in DC + PEI-nt3/gfp-treated animals whilst GFAP+ astrocytes invaded the lesion site in DC + PEI-shAMIGO3/nt3-treated animals. Scale bars in A = 100 µm; C and D = 500 µm.
High numbers of ED1+ cells and astrocyte invasion into the lesion site were observed in DC + PEI-shAMIGO3/nt3 treatment
In DC (not shown) and DC + PEI-nt3/gfp-treated groups, ED1+ immunoreactivity (green) was largely confined to the lesion (*demarcates lesion site) cavity edge (Fig. 6C) whilst in DC + PEI-shAMIGO3/nt3-treated groups, large numbers of ED1+ cells (green) were localized within the lesion core along with other non-ED1+ cells (DAPI stained nuclei; blue) (Fig. 6C). GFAP+ astrocytes (green) were also restricted to the edges of the lesion cavity in DC (not shown) and DC + PEI-nt3/gfp-treated groups (Fig. 6C; arrowheads show GFAP+ astrocytes at the lesion cavity edge). In contrast, GFAP+ (green) astrocytes (arrows) invaded the lesion core in DC + PEI-shAMIGO3/nt3-treated groups and were present amongst other cells (DAPI stained nuclei; blue) (Fig. 6D).
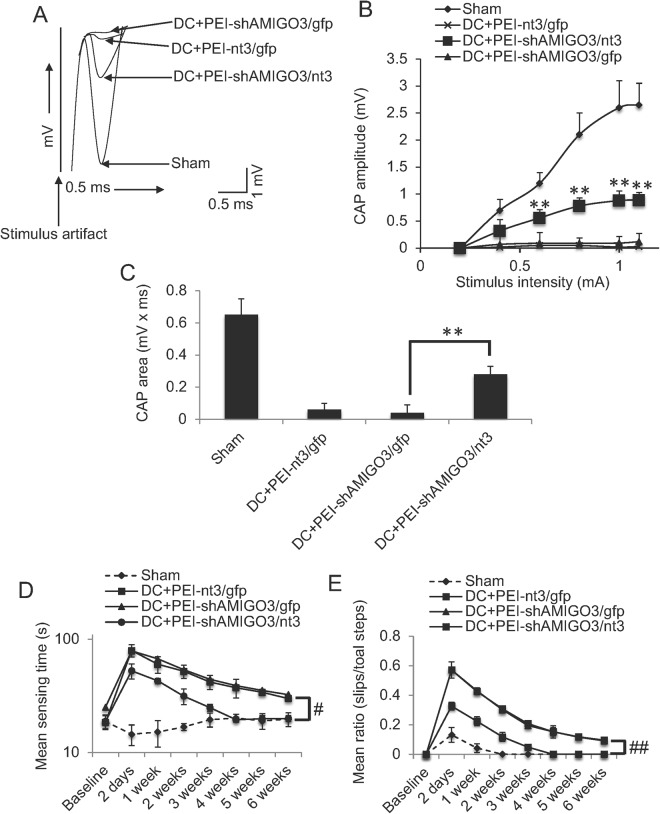
PEI-shAMIGO3/nt3 transfection of axotomised DRGN improved compound action potentials and sensory and locomotor function
Superimposed CAP traces from representative Sham control, DC + PEI-nt3/gfp, DC + PEI-shAMIGO3/gfp and DC + shAMIGO3/nt3 groups show that in the DC + PEI-nt/gfp3 and DC + PEI-shAMIGO3/gfp group, the negative CAP wave was significantly attenuated in amplitude compared to Sham controls, whereas in PEI-shAMIGO3/nt3, the negative amplitude increased markedly (Fig. 7A). The mean CAP amplitude was reduced after DC, compared to Sham controls (Fig. 7B). Significantly larger CAP amplitude was observed in DC + PEI-shAMIGO3/nt3-treated animals at all stimulation intensities, compared to DC + PEI-nt3/gfp and DC + PEI-shAMIGO3/gfp treatment groups (P < 0.001; Fig. 7B). In Sham controls, CAP area (0.51 ± 0.06 mV x ms) was reduced to 7.8 ± 9.8% of the CAP area in Sham controls (0.04 ± 0.05 mV x ms) (Fig. 7C). CAP area in DC + PEI-shAMIGO3/nt3 was significantly larger (P < 0.001) than in DC + PEI-nt/gfp3 and DC + PEI-shAMIGO3/gfp groups and was increased to 54.9 ± 9.8% of Sham control CAP area (Fig. 7C).
Figure 7.
Suppression of AMIGO3 in DRGN preserved the spinal compound action potentials (CAP) across the lesion site. (A) Superimposed CAP traces from representative Sham controls, DC + PEI-shAMIGO3/gfp DC + PEI-nt3/gfp and DC + shAMIGO3/nt3 groups. (B) Compared to Sham controls, negative CAP amplitudes (mV) were highly attenuated in DC + PEI-shAMIGO3/gfp and DC + PEI-nt3/gfp groups but was significantly improved in DC + PEI-shAMIGO3/nt3 groups (P < 0.001, ANOVA). (C) Mean CAP area at different stimulus intensities recorded from Sham controls was significantly attenuated in DC + PEI-shAMIGO3/gfp but improved significantly in DC + PEI-shAMIGO3/nt3 compared to DC + PEI-shAMIGO3/gfp and DC + PEI-nt3/gfp groups (**P < 0.001, ANOVA). (D) Mean sensing time for the tape removal test showed return to baseline Sham control levels by 4-wekks after DC and treatment with DC + PEI-shAMIGO3/nt3 compared to other groups. (E) Mean error ratio to show the number of slips versus total number of steps in the horizontal ladder walking test. Once again DC + PEI-shAMIGO3/nt3-treated animals returned to Sham control levels by 4 weeks after DC injury. ##P < 0.012, generalised linear mixed models; #P < 0.0011, linear mixed models.
The mean sensing time for the tape removal test which assess sensory function20 remained between 12–25 s throughout the 6-week experimental time period in sham treated animals (Fig. 7D). The mean sensing time increased to 80–85 s in DC + PEI-nt3 and DC + PEI-shAMIGO3/gfp-treated groups at 2d after DC lesion (Fig. 7D). However, in DC + PEI-shAMIGO3/nt3-treated animals, the mean sensing time at 2d was significantly lower (P < 0.001), taking 53 ± 8 s to detect the tape (Fig. 7D). The mean sensing time improved over the 6-week time period in all animals, however, in DC + PEI-shAMIGO3/nt3, the mean sensing time returned to sham treated baseline levels by 4 weeks after DC lesion. Over the 6-week time period, DC + PEI-shAMIGO3/nt3-treated animals were significantly better at detecting the tape and removing it compared to all other treatment groups (P < 0.0012, linear mixed model).
The mean ratio (slips/total steps) in the horizontal ladder crossing test, which detects locomotor function20 remained between 0.1 and 0 throughout the 6-week time period in sham treated animals (Fig. 7E). The mean ratio increased to 0.58 ± 0.05 in DC + PEI- in DC + PEI-nt3 and DC + PEI-shAMIGO3/gfp-treated groups at 2d after DC lesion (Fig. 7E). However, in DC + PEI-shAMIGO3/nt3-treated animals, the mean ratio at 2d was significantly lower (P < 0.001), at 0.33 ± 0.02 (Fig. 7D). The mean ratio improved over the 6-week time period in all animals, however, in DC + PEI-shAMIGO3/nt3, the mean ratio returned to sham treated baseline levels by 4 weeks after DC lesion. Over the 6-week time period, DC + PEI-shAMIGO3/nt3-treated animals were significantly better at ladder crossing compared to all other treatment groups (P < 0.01, generalized linear mixed models).
Taken together, these results show that suppression of AMIGO3 and simultaneous NT3 stimulation of DC axon regeneration promotes sensory and locomotor function recovery.
Discussion
In this study, we investigated the use of a non-viral vector for gene delivery to DRGN and showed in vitro and that shAMIGO3 knockdown combined with NT3 over-expression, induced DRGN to grow neurites on a CME substrate. We also showed in vivo that in vivo-jetPEI-delivered shAMIGO3/gfp plasmid DNA: (1), increased transduction in all sizes of DRGN; (2), efficiently knocked down AMIGO3 while simultaneously increasing NT3 production; (3), stimulated DC axon regeneration which conducted action potentials across the lesion site and led to enhanced sensory and locomotor function.
In vitro findings
We tested the efficiency of in vivo-jetPEI-delivered plasmids to suppress AMIGO3 and upregulate NT3 in DRGN cultures. It has been recognised by us and others that a combinatorial strategy will be required to promote optimal CNS axon regeneration due to the complexity of the injury. For example, we have shown that suppression of axon growth inhibitory molecules combined with neurotrophic factor stimulation promotes significant disinhibited DRGN neurite outgrowth in the presence of CME21–24. In this study, 2 µg of plasmid DNA encoding shAMIGO3 was sufficient to significantly knock down AMIGO3, to similar levels that we observed with siAMIGO38. The same amount of DNA was also able to promote over-expression of NT3 in DRGN and significantly increased titres of NT3 secretion into the culture medium.
However, similar to our previous observations with siAMIGO3, knockdown of AMIGO3 using an shAMIGO3 alone did not overcome CME-mediated neurite growth inhibition. Added simultaneous stimulation with NT3 to promote DRGN neurite outgrowth was required, confirming that disinhibition alone in insufficient to promote DRGN neurite outgrowth in the presence of CME but that concomitant stimulation of growth is required to drive regeneration22,25,26.
In vivo findings
We compared the targeting efficiency of non-viral plasmids delivered to DRGN through intra-DRG injection with in vivo-jetPEI. There is a high heterogeneity of DRGN in size distribution27,28 and our plasmids, especially in the presence of shAMIGO3, increased transduction in all sizes of DRGN. In a previous study, we found that preferential targeting of large diameter DRGN was achieved with AAV8 delivered gfp28. It is possible that the large surface area exposed to the plasmids results in an increased incidence of transduction28. It is well established that the majority of the DRGN that project their axons through the DC are large diameter DRGN and hence their increased transduction rates by our plasmids is actually beneficial, after DC injury. It is clear from our study that NT3 production by DRGN through the local production of NT3 may also have paracrine effects on neighbouring DRGN, since both GFP+ (i.e. transduced DRGN) and GFP− DRGN (i.e. non-transduced DRGN) were NT3+.
A common concern with RNA interference (RNAi) is RNAi-associated immune stimulation, which is a major hurdle to the safety and efficacy of RNAi. The induction of innate immunity may arise from sequence-dependent effects, delivery vehicles and the RNAi process itself leading to undesired effects and misinterpretation of the data29,30. However, our study found no evidence of changes in the most common off-target cytokines and innate immunity genes after transduction of DRGN with in vivo-jetPEI-delivered plasmids. Others have also reported that in vivo-jetPEI vector is safe and does not induce off-target immune-mediated cytokines such as TNF-α, IFN-γ, IL-6, IL-12/IL-23, IL-1β, and IFN-β31. Our study therefore suggests that in vivo-jetPEI may be safe for clinical applications.
Not only did the in vivo-jetPEI-delivered shAMIGO3 suppress AMIGO3 levels in >80% DRGN, immunoreactivity for AMIGO3 was reduced in both gfp+ and gfp− DRGN. This suggests that the plasmid efficiently suppressed AMIGO3 levels in both transduced and non-transduced DRGN. At present, we cannot explain why non-transduced DRGN showed reduced AMIGO3 levels but, to our knowledge, this is the first report to show such an effect. It may suggest that suppression of AMIGO3 in DRGN had paracrine effects on neighbouring DRGN, representing an intriguing possibility that not all DRGN require targeting to promote therapeutically effective outcomes. Similarly, over-expression of NT3 was also apparent in both gfp+ and gfp− DRGN suggesting that both transfected and non-transfected DRGN produced high titres of NT3. Our results therefore demonstrate that in vivo-jetPEI efficiently knocked down AMIGO3 and up-regulated NT3 in DRGN.
Knockdown of AMIGO3 and simultaneous stimulation with NT3 promoted significant DRGN axon regeneration after DC injury. This was correlated with significant improvements in CAP amplitudes across the lesion site and sensory and locomotor function. This is the first demonstration that knockdown of AMIGO3 promotes spinal cord axon regeneration in vivo and demonstrates that AMIGO3 may require targeting to promote CNS axon regeneration. We previously showed that AMIGO3 could substitute for LINGO-1 in binding to p75NTR and NgR1 to induce RhoA activation in response to CME. Our current results are consistent with these findings. Axons regenerated for up to 6 mm rostral to the lesion site and conducted action potentials across the lesion site correlating with improved sensory and locomotor function. Other types of behavioural analyses however, have been shown to be inconclusive with the T8 DC lesion model, since measurable functional improvements in mechanical and thermal hyperplasia do not occur32,33.
Our current study demonstrates in vivo proof-of-principle that in DRGN cultures grown in the presence of CNS myelin extracts, knockdown of AMIGO3 promoted signifcant DRGN neurite outgrowth8 and that knockdown of AMIGO3 together with co-incident growth stimulation (here by NT3) promoted signficant DC axon regeneration and improved functional recovery. In addition, spinal cord cavitation was reduced and the lesion site was filled with invading cells, suggesting a further therapeutic use of AMIGO3 and NT-3 in spinal cord injury. It remains to be elucidated why knockdown of AMIGO3 suppressed cavitation that normally results after DC injury in rats33. One possibility is that since AMIGO3 belongs to a family of adhesion molecules, knocking down AMIGO3 may have limited the influx of inflammatory cells after DC injury and hence reduced inflammation34 limiting secondary damage and cavitation35,36.
Although ED1+ cells were upregulated in the lesion site of shAMIGO3/nt3-treated animals, we did not identify whether these cells were of the M1 (classical activation) or M2 (alternatively activated macrophages) lineage37. However, pro-regenerative M2 macrophage lineages are only present for the first week whilst M1 macrophages persist for more than 4 weeks. Therefore, it is unlikely that ED1+ cells were pro-regenerative M2 phenyotypes. However, we are unsure why there are more ED1+ cells after treatment with shAMIGO3/nt3 and are currently investigating why this is so. Astrocytes on the other hand play multifaceted roles after spinal cord injury. Normally astrocytes are the major component of the glial scar and prevent axon regeneration by forming a physical and chemical barrier at the lesion borders. However, it appears that shAMIGO3/nt3 treatment promoted astrocyte invasion into the lesion site, transforming the lesion border from a dense network of tightly woven and thickened processes to a more open meshwork, allowing astrocytes to invade the lesion site. It is possible that astrocyte invasion allowed greater axon regeneration in these animals since they are able to restore the extracellular ionic environment, sequester extracellular glutamate and produce neurotrophic factors38–41. Astrocytes secrete axon growth inhibitory CSPG but also adhesive extracellular molecules such as laminin that are supportive to axon growth42–45. TGFα infusion into the spinal cord lesion site in mouse also allowed similar astrocyte invasion and new cells as was observed by us with shAMIGO3/nt3 treatment. For example, the centre of the lesion site contained greater number of cells and increased astrocyte invasion, which correlated with increased numbers of GAP43+ axons46. These results suggest that astrocytes may provide axon growth supportive roles by migrating into the lesion site.
Conclusion
Our results demonstrated that non-viral plasmid delivery enhanced transduction in all sizes of DRGN, significantly downregulated AMIGO3 and upregulated NT-3 production from DRGN. This reduced cavity formation and promoted axon regeneration and functional recovery in the damaged spinal cord. These results suggest that AMIGO3 might comprise a future therapy to promote axon regeneration and functional recovery after spinal cord injury. Plasmid transfection is likely to be more translational since viruses need at least 2 weeks to start producing effective titres of the shRNA of choice, whereas plasmids begin to express the relevant payload within hours. Moreover, in vivo-jetPEI vector is safe and does not induce off-target immune-mediated cytokines. Therefore, the lack of immunogenic responses after in vivo-jetPEI injection is to be expected and is a further advantage over viral vectors.
Methods
Preparation of CNS myelin extracts
CNS myelin extracts (CME), containing significant titres of Nogo-A, OMgp, MAG and CSPG were prepared according to our previously published methods21.
In vitro experiments
shAMIGO3/NT-3 plasmid construction: Plasmids (control gfp, shgfp, shAMIGO3/gfp, nt3/gfp, shAMIGO3/nt3) were prepared as described previously47. To construct bicistronic plasmids (pH1-shAMIGO3-CMV-gfp (shAMIGO3/gfp) and pH1-shAMIGO3-CMV-nt3 (shAMIGO3/nt3), shAMIGO3 encoding complementary DNA sequences was designed using the online Ambion software and oligonucleotides were hybridized together and cloned into pRNAT-H1.1/shuttle (Addgene, Cambridge, MA, USA). To create the secretable form of NT3, pCMV-nt3-gfp (nt3/gfp), the coding sequence of rat NT3 and the nerve growth factor secretion signal sequence48 were cloned into pIRES-EGFP (Clonetech, Mountain View, CA, USA). The efficiency of plasmids to knock down AMIGO3 gene expression was optimised using quantitative real-time PCR (qRT-PCR).
DRGN cultures: Adult DRGN cultures were prepared as described by us earlier21. DRGN were cultured in Neurobasal-A (Invitrogen, Paisley, UK) at a plating density of 500/well in chamber slides (Beckton Dickinson, Oxford, UK) pre-coated with 100 μg/ml poly-D-lysine (Sigma, Poole, UK).
In vivo-jetPEI (PEI; Polyplus Transfection, New York, USA) was prepared according to the manufacturer’s instructions. DRGN were transfected with 0.5, 1, 2, 3 and 4 μg of plasmid DNA containing either a control shgfp (PEI-shgfp) or shAMIGO3/nt3 (PEI-shAMIGO3/nt3). Additional controls included untreated DRGN (Untreated), DRGN transfected with in vivo-jetPEI only (Sham), and shAMIGO3/gfp (PEI-shAMIGO3/gfp) groups. DRGN were allowed to incubate for 3d before harvesting of cells and extraction of total RNA for validation of shAMIGO3 knockdown using qRT-PCR, as described below.
To monitor NT3 production and disinhibited DRGN neurite outgrowth after shAMIGO3 knockdown and NT3 stimulation, DRGN were transfected with the optimal dose of plasmid DNA (2 μg) and incubated for 3d in the presence of 100 μg/ml CME21, before harvesting culture supernatant to detect NT3 production by ELISA and neurite outgrowth by immunocytochemistry.
Quantitative real-time PCR to confirm shAMIGO3 knockdown: Total RNA from treated DRGN (n = 6 wells/treatment) was prepared using Trizol according to the manufacturer’s instructions (Invitrogen). AMIGO3 knockdown and nt3 over-expression were validated, using qRT–PCR from complementary DNA prepared from extracted mRNA, with appropriate forward and reverse primers (Table 1) using the LightCycler real time qRT-PCR system (Roche, Burgess Hill, UK) according to our previously published methods19. Fold-changes were computed using the ΔΔCt method.
Table 1.
Primers used in this study for qRT-PCR.
| Rat genes | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| AMIGO3 | CGGCTGCGTGCCTTGTACCT | AGCACTTAGGCCCCGCTGGT |
| NT3 | GCCCCCTCCCTTATACCTAATG | CATAGCGTTTCCTCCGTGGT |
| TNF-α | ACCACGCTCTTCTGTCTACTG | CTTGGTGGTTTGCTACGAC |
| IFN-γ | AGGATGCATTCATGAGCATCGCC | CACCGACTCCTTTTCCGCTTCCT |
| IL-6 | TCTCTCCGCAAGAGACTTCCA | ATACTGGTCTGTTGTGGGTGG |
| IL-12 | AATGTTCCAGTGCCTCAACCA | GATCAATCTCTTCGGAAGTGCA |
| IL-1β | GCAATGGTCGGGACATAGTT | AGACCTGACTTGGCAGAGGA |
| IFN-β | CGTTCCTGCTGTGCTTCTC | TGTAACTCTTCTCCATCTGTGAC |
| MX1 | AACCCTGCTACCTTTCAA | AAGCATCGTTTTCTCTATTTC |
| IFIT | CTGAAGGGGAGCGATTGATT | AACGGCACATGACCAAAGAGTAGA |
| OAS1 | TTCTACGCCAATCTCATCAGTG | GGTCCCCCAGCTTCTCCTTAC |
| Casp7 | CAACGACACCGACGCTAATC | GGTCCTTGCCATGCTCATTC |
| GAPDH | AGACAGCCGCATCTTCTTGT | CTTGCCGTGGGTAGAGTCAT |
NT-3 ELISA: NT-3 was assayed in supernatant from n = 6 wells/treatment using the NT-3 DuoSet ELISA kit according to the manufacturer’s instructions (R&D Systems, Abingdon, UK).
Immunocytochemistry: Cells were fixed in 4% paraformaldehyde, washed in 3 changes of PBS before immunocytochemistry as described by us previously21. To visualise neurites, DRGN were stained with monoclonal anti-βIII tubulin (Sigma) detected with Alexa-488 anti-mouse antibodies (Invitrogen). Slides were then viewed with an epi-fluorescent Axioplan 2 microscope, equipped with an AxioCam HRc and running Axiovision Software (all from Carl Zeiss, Hertfordshire, UK). The proportion of DRGN with neurites and mean neurite lengths were calculated using Axiovision Software, as previously described21. All experiments were performed in duplicate and repeated on 3 independent occasions (i.e. n = 6 wells/condition).
In vivo experiments
Surgical procedures: All animal surgeries were carried out in strict accordance to the guidelines of the UK Animals Scientific Procedures Act, 1986 and the Revised European Directive 1010/63/EU and conformed to the guidelines and recommendation of the use of animals by the Federation of the European Laboratory Animal Science Associations. Experiments were licensed by the UK Home Office and all experimental protocols were approved by the University of Birmingham’s Animal Welfare and Ethical Review Board. Adult female Sprague-Dawley rats weighing 170–220 g (Charles River, Margate, UK) were used in all experiments. The experiments comprised 8 groups (n = 12 rats/group), including 4 control groups comprising: Intact controls (Group 1: IC; to detect baseline levels), partial laminectomy but no DC lesion; Group 2: sham; to detect surgery-induced changes), DC transected controls (Group 3: DC; to detect injury-mediated changes), and DC + intra-DRG injection of in vivo-jetPEI-gfp (Group 4: DC + PEI-gfp; to monitor DRGN transduction efficiencies) and 4 experimental groups in which DC were transected and DRG injected with plasmid-containing in vivo-jetPEI complexes, comprised of: DC + intra-DRG injection of in vivo-jetPEI-gfp (Group 1: DC + PEI-gfp); DC + intra-DRG injection of in vivo-jetPEI-nt3/gfp (Group 2: DC + PEI-nt3/gfp; to monitor DRGN transduction and NT3 over-expression); DC + in vivo-jetPEI-shAMIGO3/gfp (Group 3: PEI-shAMIGO3/gfp; to monitor AMIGO3 knockdown in DRGN and to establish if AMIGO3 knockdown without concomitant stimulation with NT3 promoted DRGN axon regeneration); and DC + intra-DRG injection of in vivo-jetPEI-shAMIGO3/nt3 (Group 4: DC + PEI-shAMIGO3/nt3; to determine if AMIGO3 knockdown and simultaneous stimulation with NT3 was required for DRGN axon regeneration).
Before surgery, rats were injected subcutaneously with 0.05 ml Buprenorphine and anaesthetised using 5% of Isoflurane in 1.8 ml/l of O2 with body temperature and heart rate monitored throughout surgery. After partial T8 laminectomy, DC were crushed bilaterally using calibrated watchmaker’s forceps and, after injection of in vivo jetPEI/plasmids into L4/L5 DRG28,33, rats were killed at either 28d (western blot, immunohistochemistry, qRT-PCR and ELISA) or 6 weeks (electrophysiology and functional tests) in rising concentrations of CO2.
Analysis of off-target cytokines and innate immune response genes: Total RNA from n = DRG/treatment were extracted using Trizol reagent as described above. A selection off-target cytokines and innate immune response genes (Table 1) were analysed by qRT-PCR from complementary DNA prepared from extracted mRNA as described above19. Fold-changes were computed using the ΔΔCt method.
Tissue preparation and cryo-sectioning: Rats were intracardially perfused with 4% formaldehyde and DRG and segments of T8 cord containing the DC injury sites were post-fixed for 2 h at RT, prepared for cryoprotection and sectioned using a cryostat as described by us previously33.
Immunohistochemistry: Immunohistochemistry was performed on sections from the middle of DC lesion sites or middle of the DRG as described previously8,21,33. Primary antibodies included: anti-AMIGO3 (1:200 dilution; Santa Cruz Biotechnology, San Diego, CA, USA), anti-GAP43 (1:400 dilution; Zymed, San Francisco, CA, USA); anti-NT3 (1:400 dilution; Abcam, Cambridge, UK); anti-GFAP (1:400 dilution, Sigma); anti-CD68 (1:500 dilution, Abcam) and anti-rat ED1 (1:400 dilution; Serotec, Oxford, UK).
Protein extraction, western blot and densitometry: Total protein from DRG was extracted and western blot followed by densitometry was performed according to our previously published methods49,50. Briefly, 40 µg of total protein extract was resolved on 12% SDS gels, transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Watford, UK) and probed with relevant primary antibodies: anti-AMIGO3 (1:400 dilution, Santa Cruz); anti-NT-3 (1:1000 dilution, Abcam) and β-actin (1:1000 dilution, Sigma). Membranes were then incubated with relevant HRP-labelled secondary antibodies and bands were detected using the enhanced chemiluminescence kit (GE Healthcare, Buckinghamshire, UK). Anti β-actin antibodies were used as a protein loading control.
For densitometry, western blots were scanned into Adobe Photoshop (Adobe Systems Inc, San Jose, CA, USA) keeping all scanning parameters the same between blots and the integrated density of bands analysed using the built-in-macros for gel analysis in ImageJ (NIH, USA, http://imagej.nih.gov/ij)21,50–52. Means ± SEM were plotted in Microsoft Excel (Microsoft Corporation, CA, USA).
Analysis of GFP expression: Images of sections (n = 160) of L4/L5 DRG from each animal (n = 10) were captured at x10 magnification using the Zeiss Axioplan 200 (Carl Zeiss). Images were merged in Adobe Photoshop (Adobe Systems Incorporated) using Photomerge and the total number of GFP+ DRGN and their diameters recorded, as described previously28.
Quantification of axons: Axon regeneration in the spinal cord was quantified according to previously published methods53. Briefly, serial parasagittal sections of cords were reconstructed by collecting all serial 50 μm-thick sections (∼70–80 sections/animal; n = 10 rats/treatment). In each section, the numbers of intersections of GAP43+ fibers through a dorsoventral orientated line was counted from 4 mm rostral to 4 mm caudal to the lesion site. Axon number was calculated as a percentage of the fibers seen 4 mm above the lesion, where the DC was intact.
Electrophysiology: Four weeks after surgery or treatment, compound action potentials (CAP) were recorded after shAMIGO3/nt3 treatment as previously described54,55. The experimenter was blinded to the treatment status of the animals. The amplitude of CAP was calculated between the negative deflection after the stimulus artifact and the next peak of the wave. The area of the CAP was calculated by rectifying the CAP component (full-wave rectification) and measuring its area. At the end of each experiment, the dorsal half of the spinal cord was transected between stimulating and recording electrodes to confirm that a CAP could not be detected.
Functional tests
Functional testing after DC lesions was carried out as described by others20. Animals (n = 18/group) were trained to master traversing the rope and ladder for 1w before functional testing. All functional tests were performed 2-3d before injury to establish baseline parameters. Animals were then tested 2d after DC lesion + treatment and then weekly for 6 weeks. Experiments were performed by 2 observers (from which the treatment conditions were masked) in the same order, the same time of day and each test performed for 3 individual trials.
Horizontal ladder test: This tests the animals locomoter function and is performed on a 0.9-meter-long horizontal ladder with a diameter of 15.5 cm and randomly adjusted rungs with variable gaps of 3.5–5.0 cm. Animals were assessed traversing the ladder and the left and right rear paw slips were recorded along with the total number of steps. To calculate the mean error rate: the number of slips were divided by the total number of steps.
Tape removal test (sensory function): The tape removal test determines touch perception from the left hind paw. Animals were held with both hind-paws extended and a piece of tape of 15 × 15 mm (Kip Hochkrepp, Bocholt, Germany) was affixed to the palm of the left hind-paw. The time it took for the animals to detect and remove the tape was recorded and used to calculate the mean sensing time.
Statistical analysis
Data are presented as means ± SEM. Significant differences were calculated using SPSS Version 22 (IBM, NJ, USA) software by one-way analysis of variance (ANOVA), with Bonferroni post hoc tests, set at P < 0.05.
For the horizontal ladder crossing functional tests, data was analysed according to Fagoe et al.20 using R package (www.r-project.org). Whole time-course of lesioned and sham-treated animals were compared using binomial generalized linear mixed models (GLMM), with: lesion/sham set to true in lesioned animals post-surgery (set to false otherwise); operated/unoperated set to false before surgery, true after surgery as fixed factors; animals as random factors and time as a continuous covariate. Binomial GLMMs were fitted in R using package lme4 with the glmer function. P values were then calculated using parametric bootstrap. For the tape removal test, linear mixed models (LMM) were calculated by model comparison in R using the package pbkrtest, with the Kenward-Roger method20.
Data availability
All data generated or analysed during this study are included in this article.
Acknowledgements
The authors would like to thank the Saudi Education Ministry in the UK and the Wellcome Trust (grant no. 092539/Z/10/Z) for funding this work. The authors would also like to thank University of Birmingham’s Biomedical Services Unit staff who were involved in masking/unmasking animal ID’s for all in vivo experiments.
Author Contributions
S.A., Z.A. and M.B. drafted manuscript; S.A. and Z.A. performed experiments; A.L. and M.B. revised the manuscript; Z.A. and A.L. designed the research and approved final version of manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sandvig A, Berry M, Barrett LB, Butt A, Logan A. Myelin-, reactive glia-, and scar-derived CNS axon growth inhibitors: expression, receptor signaling, and correlation with axon regeneration. Glia. 2004;46:225–251. doi: 10.1002/glia.10315. [DOI] [PubMed] [Google Scholar]
- 2.Hunt D, Coffin RS, Anderson PN. The Nogo receptor, its ligands and axonal regeneration in the spinal cord; a review. J Neurocytol. 2002;31:93–120. doi: 10.1023/A:1023941421781. [DOI] [PubMed] [Google Scholar]
- 3.Liu BP, Fournier A, GrandPre T, Strittmatter SM. Myelin-associated glycoprotein as a functional ligand for the Nogo-66 receptor. Science. 2002;297:1190–1193. doi: 10.1126/science.1073031. [DOI] [PubMed] [Google Scholar]
- 4.Shao Z, et al. TAJ/TROY, an orphan TNF receptor family member, binds Nogo-66 receptor 1 and regulates axonal regeneration. Neuron. 2005;45:353–359. doi: 10.1016/j.neuron.2004.12.050. [DOI] [PubMed] [Google Scholar]
- 5.Wang KC, Kim JA, Sivasankaran R, Segal R, He Z. P75 interacts with the Nogo receptor as a co-receptor for Nogo, MAG and OMgp. Nature. 2002;420:74–78. doi: 10.1038/nature01176. [DOI] [PubMed] [Google Scholar]
- 6.Berry M, Ahmed Z, Lorber B, Douglas M, Logan A. Regeneration of axons in the visual system. Restor Neurol Neurosci. 2008;26:147–174. [PubMed] [Google Scholar]
- 7.Park JB, et al. A TNF receptor family member, TROY, is a coreceptor with Nogo receptor in mediating the inhibitory activity of myelin inhibitors. Neuron. 2005;45:345–351. doi: 10.1016/j.neuron.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 8.Ahmed Z, Douglas MR, John G, Berry M, Logan A. AMIGO3 is an NgR1/p75 co-receptor signalling axon growth inhibition in the acute phase of adult central nervous system injury. PLoS One. 2013;8:e61878. doi: 10.1371/journal.pone.0061878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson BL, Breakefield XO. Viral vectors for gene delivery to the nervous system. Nat Rev Neurosci. 2003;4:353–364. doi: 10.1038/nrn1104. [DOI] [PubMed] [Google Scholar]
- 10.Raoul C, Barker SD, Aebischer P. Viral-based modelling and correction of neurodegenerative diseases by RNA interference. Gene Ther. 2006;13:487–495. doi: 10.1038/sj.gt.3302690. [DOI] [PubMed] [Google Scholar]
- 11.Reynolds A, et al. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 12.Miyagoe-Suzuki Y, Takeda S. Gene therapy for muscle disease. Exp Cell Res. 2010;316:3087–3092. doi: 10.1016/j.yexcr.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 13.Mason MR, et al. Comparison of AAV serotypes for gene delivery to dorsal root ganglion neurons. Mol Ther. 2010;18:715–724. doi: 10.1038/mt.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarty DM, Monahan PE, Samulski RJ. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 2001;8:1248–1254. doi: 10.1038/sj.gt.3301514. [DOI] [PubMed] [Google Scholar]
- 15.Wiseman JW, Goddard CA, McLelland D, Colledge WH. A comparison of linear and branched polyethylenimine (PEI) with DCChol/DOPE liposomes for gene delivery to epithelial cells in vitro and in vivo. Gene Ther. 2003;10:1654–1662. doi: 10.1038/sj.gt.3302050. [DOI] [PubMed] [Google Scholar]
- 16.Boussif O, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lungwitz U, Breunig M, Blunk T, Gopferich A. Polyethylenimine-based non-viral gene delivery systems. Eur J Pharm Biopharm. 2005;60:247–266. doi: 10.1016/j.ejpb.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 18.Liao, H. W. & Yau, K. W. In vivo gene delivery in the retina using polyethylenimine. Biotechniques42, 285–286, 288 (2007). [DOI] [PMC free article] [PubMed]
- 19.Read ML, et al. Profiling RNA interference (RNAi)-mediated toxicity in neural cultures for effective short interfering RNA design. J Gene Med. 2009;11:523–534. doi: 10.1002/jgm.1321. [DOI] [PubMed] [Google Scholar]
- 20.Fagoe ND, et al. Evaluation of Five Tests for Sensitivity to Functional Deficits following Cervical or Thoracic Dorsal Column Transection in the Rat. PLoS One. 2016;11:e0150141. doi: 10.1371/journal.pone.0150141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmed Z, et al. Disinhibition of neurotrophin-induced dorsal root ganglion cell neurite outgrowth on CNS myelin by siRNA-mediated knockdown of NgR, p75NTR and Rho-A. Mol Cell Neurosci. 2005;28:509–523. doi: 10.1016/j.mcn.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 22.Ahmed Z, et al. Schwann cell-derived factor-induced modulation of the NgR/p75NTR/EGFR axis disinhibits axon growth through CNS myelin in vivo and in vitro. Brain. 2006;129:1517–1533. doi: 10.1093/brain/awl080. [DOI] [PubMed] [Google Scholar]
- 23.Ahmed Z, Jacques SJ, Berry M, Logan A. Epidermal growth factor receptor inhibitors promote CNS axon growth through off-target effects on glia. Neurobiol Dis. 2009;36:142–150. doi: 10.1016/j.nbd.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 24.Ahmed Z, Douglas MR, Read ML, Berry M, Logan A. Citron kinase regulates axon growth through a pathway that converges on cofilin downstream of RhoA. Neurobiol Dis. 2011;41:421–429. doi: 10.1016/j.nbd.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 25.Ahmed Z, et al. Optic nerve and vitreal inflammation are both RGC neuroprotective but only the latter is RGC axogenic. Neurobiol Dis. 2010;37:441–454. doi: 10.1016/j.nbd.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 26.Vigneswara V, et al. Combined suppression of CASP2 and CASP6 protects retinal ganglion cells from apoptosis and promotes axon regeneration through CNTF-mediated JAK/STAT signalling. Brain. 2014;137:1656–1675. doi: 10.1093/brain/awu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmalbruch H. The number of neurons in dorsal root ganglia L4-L6 of the rat. Anat Rec. 1987;219:315–322. doi: 10.1002/ar.1092190313. [DOI] [PubMed] [Google Scholar]
- 28.Jacques SJ, et al. AAV8(gfp) preferentially targets large diameter dorsal root ganglion neurones after both intra-dorsal root ganglion and intrathecal injection. Mol Cell Neurosci. 2012;49:464–474. doi: 10.1016/j.mcn.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 29.Olejniczak M, Polak K, Galka-Marciniak P, Krzyzosiak WJ. Recent advances in understanding of the immunological off-target effects of siRNA. Curr Gene Ther. 2011;11:532–543. doi: 10.2174/156652311798192770. [DOI] [PubMed] [Google Scholar]
- 30.Meng Z, Lu M. RNA Interference-Induced Innate Immunity, Off-Target Effect, or Immune Adjuvant? Front Immunol. 2017;8:331. doi: 10.3389/fimmu.2017.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonnet ME, Erbacher P, Bolcato-Bellemin AL. Systemic delivery of DNA or siRNA mediated by linear polyethylenimine (L-PEI) does not induce an inflammatory response. Pharm Res. 2008;25:2972–2982. doi: 10.1007/s11095-008-9693-1. [DOI] [PubMed] [Google Scholar]
- 32.Kanagal SG, Muir GD. The differential effects of cervical and thoracic dorsal funiculus lesions in rats. Behav Brain Res. 2008;187:379–386. doi: 10.1016/j.bbr.2007.09.035. [DOI] [PubMed] [Google Scholar]
- 33.Surey S, Berry M, Logan A, Bicknell R, Ahmed Z. Differential cavitation, angiogenesis and wound-healing responses in injured mouse and rat spinal cords. Neuroscience. 2014;275:62–80. doi: 10.1016/j.neuroscience.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Kuja-Panula J, Kiiltomaki M, Yamashiro T, Rouhiainen A, Rauvala H. AMIGO, a transmembrane protein implicated in axon tract development, defines a novel protein family with leucine-rich repeats. J Cell Biol. 2003;160:963–973. doi: 10.1083/jcb.200209074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorio A, et al. Recombinant human erythropoietin counteracts secondary injury and markedly enhances neurological recovery from experimental spinal cord trauma. Proc Natl Acad Sci USA. 2002;99:9450–9455. doi: 10.1073/pnas.142287899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popovich PG, et al. Depletion of hematogenous macrophages promotes partial hindlimb recovery and neuroanatomical repair after experimental spinal cord injury. Exp Neurol. 1999;158:351–365. doi: 10.1006/exnr.1999.7118. [DOI] [PubMed] [Google Scholar]
- 37.Kong X, Gao J. Macrophage polarization: a key event in the secondary phase of acute spinal cord injury. J Cell Mol Med. 2017;21:941–954. doi: 10.1111/jcmm.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sykova E, Svoboda J, Simonova Z, Jendelova P. Role of astrocytes in ionic and volume homeostasis in spinal cord during development and injury. Prog Brain Res. 1992;94:47–56. doi: 10.1016/S0079-6123(08)61738-0. [DOI] [PubMed] [Google Scholar]
- 39.Rothstein JD, et al. Knockout of glutamate transporters reveals a major role for astroglial transport in excitotoxicity and clearance of glutamate. Neuron. 1996;16:675–686. doi: 10.1016/S0896-6273(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 40.Lee MY, Kim CJ, Shin SL, Moon SH, Chun MH. Increased ciliary neurotrophic factor expression in reactive astrocytes following spinal cord injury in the rat. Neurosci Lett. 1998;255:79–82. doi: 10.1016/S0304-3940(98)00710-1. [DOI] [PubMed] [Google Scholar]
- 41.Ikeda O, et al. Acute up-regulation of brain-derived neurotrophic factor expression resulting from experimentally induced injury in the rat spinal cord. Acta Neuropathol. 2001;102:239–245. doi: 10.1007/s004010000357. [DOI] [PubMed] [Google Scholar]
- 42.Fitch MT, Silver J. CNS injury, glial scars, and inflammation: Inhibitory extracellular matrices and regeneration failure. Exp Neurol. 2008;209:294–301. doi: 10.1016/j.expneurol.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frisen J, et al. Spinal axons in central nervous system scar tissue are closely related to laminin-immunoreactive astrocytes. Neuroscience. 1995;65:293–304. doi: 10.1016/0306-4522(94)00467-J. [DOI] [PubMed] [Google Scholar]
- 44.Morgenstern DA, Asher RA, Fawcett JW. Chondroitin sulphate proteoglycans in the CNS injury response. Prog Brain Res. 2002;137:313–332. doi: 10.1016/S0079-6123(02)37024-9. [DOI] [PubMed] [Google Scholar]
- 45.Costa S, et al. Astroglial permissivity for neuritic outgrowth in neuron-astrocyte cocultures depends on regulation of laminin bioavailability. Glia. 2002;37:105–113. doi: 10.1002/glia.10015. [DOI] [PubMed] [Google Scholar]
- 46.White RE, Yin FQ, Jakeman LB. TGF-alpha increases astrocyte invasion and promotes axonal growth into the lesion following spinal cord injury in mice. Exp Neurol. 2008;214:10–24. doi: 10.1016/j.expneurol.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng L, et al. A convenient plasmid-based system containing three reporter genes for real-time and quantitative analysis of messenger RNA silencing. Anal Biochem. 2009;394:284–286. doi: 10.1016/j.ab.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 48.Zhou L, Baumgartner BJ, Hill-Felberg SJ, McGowen LR, Shine HD. Neurotrophin-3 expressed in situ induces axonal plasticity in the adult injured spinal cord. J Neurosci. 2003;23:1424–1431. doi: 10.1523/JNEUROSCI.23-04-01424.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahmed Z, et al. Matrix metalloproteases: degradation of the inhibitory environment of the transected optic nerve and the scar by regenerating axons. Mol Cell Neurosci. 2005;28:64–78. doi: 10.1016/j.mcn.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 50.Ahmed Z, et al. TACE-induced cleavage of NgR and p75NTR in dorsal root ganglion cultures disinhibits outgrowth and promotes branching of neurites in the presence of inhibitory CNS myelin. Faseb J. 2006;20:1939–1941. doi: 10.1096/fj.05-5339fje. [DOI] [PubMed] [Google Scholar]
- 51.Douglas MR, et al. Off-target effects of epidermal growth factor receptor antagonists mediate retinal ganglion cell disinhibited axon growth. Brain. 2009;132:3102–3121. doi: 10.1093/brain/awp240. [DOI] [PubMed] [Google Scholar]
- 52.Ahmed Z, et al. Ocular neuroprotection by siRNA targeting caspase-2. Cell Death Dis. 2011;2:e173. doi: 10.1038/cddis.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hata K, et al. RGMa inhibition promotes axonal growth and recovery after spinal cord injury. J Cell Biol. 2006;173:47–58. doi: 10.1083/jcb.200508143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hains BC, Saab CY, Lo AC, Waxman SG. Sodium channel blockade with phenytoin protects spinal cord axons, enhances axonal conduction, and improves functional motor recovery after contusion SCI. Exp Neurol. 2004;188:365–377. doi: 10.1016/j.expneurol.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 55.Lo AC, Saab CY, Black JA, Waxman SG. Phenytoin protects spinal cord axons and preserves axonal conduction and neurological function in a model of neuroinflammation in vivo. J Neurophysiol. 2003;90:3566–3571. doi: 10.1152/jn.00434.2003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this article.