Dear Editor
Rats are reference laboratory animal models for understanding mechanism of human diseases such as diabetes, hypertension, and neurological disorder1,2. CRISPR/Cas9 system has proved an efficient and flexible tool to generate gene modified rats3–6. Great efforts have been made to reduce the side effects and extend the application of this system7–10. Cytosine base editor (CBE), containing the engineered cytosine deaminase with CRISPR/Cas9 can be used to modify mammal genomic DNA without induction of double-strand DNA break or template. CBE targets sequence by inducing C·G to T·A conversion with a window of approximately five nucleotides11. Recently, the same group developed a new dCas9-guide tRNA adenosine deaminase which was capable of inducing A·T to G·C conversion. Together with CBE, ABE enables introduction of all four nucleotides transitions (C to T, A to G, T to C, and G to A) in target genomic sequence12. These base-editing tools provide a much safer approach compared with wild-type CRISPR/Cas9 system for gene correction of human disease. Here we report the application of this newly developed adenine base editors in rat base editing.
Two experiments were designed to test this system in rats. For the first experiment, we selected one targeting site at Hemgn gene locus (Fig. 1, Supplementary Table S1). For the second experiment, we selected two targeting sites at N-deacetylase and N-sulfotransferase 4 (Ndst4) gene loci (Fig. 1, Supplementary Table S1). The ABE and sgRNAs were prepared in vitro as described4,12. Twenty-five nanograms per microliter of ABE mRNA and 20 ng/ml of sgRNA were prepared for microinjection4. Animals in Sprague Dawley (SD) background were used in all experiments. Tail genomic DNA of born pups was extracted and used as a template for genotyping.
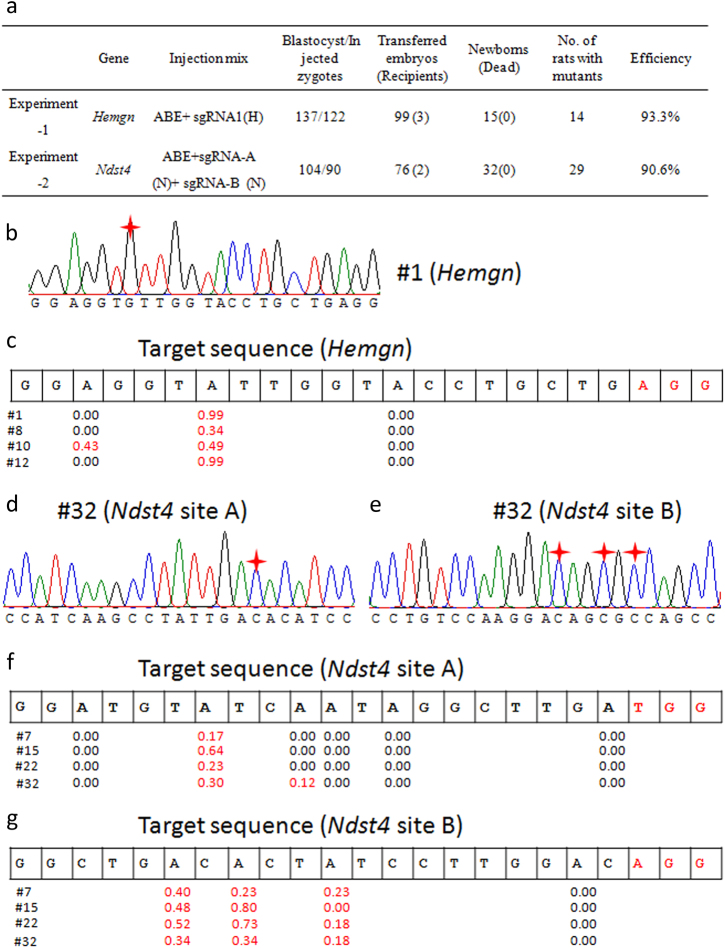
Fig. 1. Highly efficient base editing using ABE system in rats.
a Summary of the base editing experiments in rats. b The representative sequence chromatogram of Hemgn targeting sequence (potential founder #1). The conversion of A to G at target site was marked with red star. c Summary of the targeted deep sequencing of on-target site for Hemgn (potential founders #1, #8, #10, and #12). The conversion efficiency of every A was indicated. The PAM was highlight in red. d The representative sequence chromatogram of Ndst4-A targeting sequence (clone #4 of potential founder #32). The conversion of T to C at target site was marked with red star. e The representative sequence chromatogram of Ndst4-B targeting sequence (clone #4 of potential founder #32). The conversion of T to C at target site was marked with red star. f Summary of the targeted deep sequencing of on-target site for Ndst4-A (potential founders #7, #15, #22, and #32). The conversion efficiency of every A was indicated. The PAM was highlight in red. g Summary of the targeted deep sequencing of on-target site for Ndst4-A (potential founders #7, #15, #22, and #32). The conversion efficiency of every A was indicated. The PAM was highlight in red
For Hemgn targeting, a total of 99 injected zygotes were transferred to 3 pseudopregnant female SD rats and 15 pups were born (Fig. 1a). To test the base editing efficiency, the fragment including the target site was amplified and sequenced (Supplementary Table S2). Fourteen rats (14/15) (potential founders #1–2, #4–15) contained an A to G conversion at the 14th base distal from the PAM (Fig. 1a, b; Supplementary Fig. S1). Two rats (2/15) (potential founders #5, #10) revealed an A to G conversion at the 18th base distal from the PAM (Fig. 1a, b; Supplementary Fig. S1), indicating the high base-editing efficiency.
To further analyze the on-target editing effects, deep sequencing was applied to sample #1, #8, #10, and #12. More than 3 M (1024×1024 bit) clean data for each sample were obtained (Supplementary Table S4). The result showed that 14th base distal from the PAM showed A to G conversion with efficiency as high as 0.99 for #1, 0.34 for #8, 0.49 for #10, and 0.99 for sample #12. The conversion efficiency at 18th base distal from the PAM was 0.43 for #10 (Fig. 1c). No other alteration or indel was detected in the selected samples (data not shown). The results showed ABE is a highly precise and efficient base editor for rat genome editing.
For most of human diseases with more than one mutation. Multiple base editing capability is valuable for gene therapy. To test this possibility, two sgRNAs targeting different exons at Ndst4 gene locus were designed in the second experiment (Supplementary Table S1). Ten nanograms of each sgRNA was used for microinjection to avoid toxic effects. The target sites amplified by the same strategy were analyzed and sequenced. For Ndst4 targeting, a total of 76 injected zygotes were transferred to 2 pseudopregnant female SD rats and 32 pups were born (Fig. 1a). Fifteen rats (15/32) (potential founders #7, #11, #13–15, #17, #19, #21–25, #29–30, and #32) showed an A to G conversion at the 14th base distal from the PAM at targeting site A (Supplementary Fig. S2). Twenty-nine rats (29/32) (potential founders #1–8, #10–25, #27–30, and #32) showed an A to G conversion at targeting site B (Fig. 1a, d, e, Supplementary Fig. S3). In targeting site A, all mutations were occurred at 14th nucleotide distal from PAM. In targeting site B, 5 rats (potential founders #7, #18, #20, #22, and #32) showed triple A to G conversions at the 15th, 13th, 10th nucleotide distal from PAM, 9 rats (potential founders #1, #2, #5, #6, #14, #15, #21, #24, and #27) showed dual A to G conversions at the 15th, 13th nucleotide distal from PAM, 1 rat (potential founder #3) showed dual A to G conversions at the 15th, 10th nucleotide distal from PAM, 13 rats (potential founders #4, #8, #10, #11–13, #16, #17, #19, #23, #25, #28, and #30) showed single A to G conversion at the 15th nucleotide distal from PAM, and 1 rat (potential founder #29) showed single A to G conversion at the 13th nucleotide distal from PAM. Fifteen rats (15/32) (potential founders #7, #11, #13–15, #17, #19, #21–25, #29–30, #32) revealed an A to G conversion at both targeting sites (A and B) simultaneously (Fig. 1a, e; Supplementary Figs. S2 & S3, Table S3).
For further analysis, four samples (potential founders #7, #15, #22, and #32) were selected for on-target (site A and site B) analysis by deep sequencing (Supplementary Table S4). The results showed that conversion rate at site A at the 14th nucleotide was about 0.17 for #7, 0.64 for #15, 0.23 for #22, and 0.30 for #32, which was consistent with the results of PCR products sequencing (Fig. 1f). The conversion rate at site B at the 15th nucleotide was about 0.40 for #7, 0.48 for #15, 0.52 for #22, and 0.34 for #32. The conversion rate at site B at the 13th nucleotide was about 0.23 for #7, 0.80 for #15, 0.73 for #22, and 0.34 for #32. The conversion rate at site B at the 10th nucleotide was about 0.23 for #7, 0.00 for #15, 0.18 for #22, and 0.18 for #32 (Fig. 1g).
Genome-editing functions beyond one-cell stage may induce mosaicism. Therefore, we detected whether ABE system causes mosaicism, and all mutant rats were genotyped by TA clone and subsequent sequencing. Our results showed that more than two mutations were detected for Ndst4-B (potential founders #1–3, #5, #17, #18, #20, #22, and #32) (Supplementary Fig. S4). No evidence of mosaicism was found for Hemgn and Ndst4-A (Supplementary Fig. S4). These results indicate that ABE system does induce genetic mosaicism in rats.
In addition, we also tested the transmission of substitutions by crossing 3 F0 mutants (potential founders #11 and #13 for Hemgn; potential founder #21 for Ndst4-A and -B) with wild-type SD rats and determined the genotypes of the F1 rats. The PCR products of F1 rats were further analyzed by sequencing. Sequencing results showed that the same substitutions as their parent rats appeared in offsprings (Supplementary Fig. S5), demonstrating that ABE-induced substitutions in rats are transmittable.
Testing the off-target effects is very important for evaluating a new genome-editing tool. Here, we examined 11 potential off-target sites (on-target site not included) for Hemgn sgRNA in 4 selected samples (potential founders #1, #8, #10, and #12) and 14 off-target sites (on-target site not included) for Ndst4 sgRNA-A, 14 off-target sites (on-target site not included) for Ndst4 sgRNA-B in 4 selected samples (potential founders #7, #15, #22, and #32) identified using Cas-OFFinder13 (Supplementary Table S4 & S5). All PCR products were sequenced and analyzed. No off-target mutation was found in those samples. To further analyze the off-target effects, we performed targeted deep sequencing on the selected potential off-target sites. As described above, more than 3 M clean reads for each off-target site were obtained. As expected, no off-target effects were detected. We next performed whole-genome sequencing (WGS) with a sequencing depth of ×30 for samples H-8 (Hemgn targeting rat), H-10 (Hemgn targeting rat), N-15 (Ndst4-A and -B targeting rat), and N-32 (Ndst4-A and -B targeting rat) to detect ABE-induced off-target effects. Based on Cas-OFFinder, 33,718 potential off-target sites for Hemgn sgRNA, 20,774 potential off-target sites for Ndst4 sgRNA-A, 22,547 potential off-target sites for Ndst4 sgRNA-B were obtained with up to 2-bp mismatch in seed region and 8-bp mismatch in non-seed region with NRG PAM (Supplementary Fig. S6). Among these potential off-target sites, only several potential off-target sites were obtained from WGS data (Supplementary Fig. S6, Table S6). We further sequenced these potential off-target sites, and no real off-target sites were obtained (data not shown). Taken together, these results showed that ABE system is a reliable genome-editing tool for rat base editing.
We also analyzed the possible indels mediated by ABE at on-target sites and off-target sites. Only very low signals were detected at these sites, this may be caused by noise of deep sequencing (data not shown). Taken together, our results showed that the ABE system induces base conversion in a much safer way compared with traditional CRISPR/Cas9 system.
In summary, our data showed the successful application of ABE in rat base editing. This exciting strategy showed a highly efficient, precise, and safe way to edit the rat genome. Our data demonstrate the potential of ABE in correction of human genetic disease-associated mutations.
Electronic supplementary material
Acknowledgements
We thank all the members of Huang and Zhang for helpful discussions. We also thank Shanghai Institutes of Biological Sciences, Chinese Academy of Sciences for the help of deep sequence analysis. The present work was supported in part by the CAMS Innovation Fund for Medical Sciences (CIFMS) (2017-I2M-3-015, 2016-I2M-1-004), National Natural Science Foundation of China (31501001, 81571222).
Author contributions
L.Z., X.H., and Y.M. designed experiments. Y.M., Y.L. X.Z., L.B., W.C., J.L., S.P., and X.Q. performed the rat experiments. C.X. and S.H. performed targeted deep sequencing and whole-genome sequencing data analysis. Y.M., X.Z., and L.Y. collected the data and performed the data analysis. Y.M. wrote the manuscript. L.Z., X.H., and R.G. revised the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
Supplementary Information accompanies the paper at (10.1038/s41421-018-0047-9).
Contributor Information
Xingxu huang, Email: huangxx@shanghaitech.edu.cn.
Lianfeng Zhang, Email: zhanglf@cnilas.org.
References
- 1.Iannaccone PM, et al. Rats! Dis. Model Mech. 2009;2:206–210. doi: 10.1242/dmm.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smalley E. CRISPR mouse model boom, rat model renaissance. Nat. Biotechnol. 2016;34:893–894. doi: 10.1038/nbt0916-893. [DOI] [PubMed] [Google Scholar]
- 3.Li D, et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:681–683. doi: 10.1038/nbt.2661. [DOI] [PubMed] [Google Scholar]
- 4.Ma Y, et al. Generating rats with conditional alleles using CRISPR/Cas9. Cell Res. 2014;24:122–125. doi: 10.1038/cr.2013.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Y, et al. Generation of eGFP and Cre knockin rats by CRISPR/Cas9. Febs. J. 2014;281:3779–3790. doi: 10.1111/febs.12935. [DOI] [PubMed] [Google Scholar]
- 6.Yoshimi K, et al. Allele-specific genome editing and correction of disease-associated phenotypes in rats using the CRISPR-Cas platform. Nat. Commun. 2014;5:4240. doi: 10.1038/ncomms5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleinstiver BP, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleinstiver BP, et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 2015;33:1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu Y, et al. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleinstiver BP, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komor AC, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaudelli NM, et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bae S, et al. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.