Abstract
Macrofungi belonging to the phylum Basidiomycota are mostly used as medicinal mushrooms in many countries. In the present study, hundred basidiocarp of macrofungi were collected from Tamilnadu during rainy season. The basidiocarp was found in association with root/trunk of living trees, wood log and decayed matter. Among the hundred basidiocarp, 49 were grown into axenic cultures. Notable variations in the macroscopic characteristics of the basidiome and culture morphology were observed. To study the genetic diversity, the molecular taxonomy of the isolates was carried out using internal transcribed spacer (ITS) and 5.8S rRNA gene sequence marker. Thirty-two strains belonging to the order Polyporales, Hymenochataeles and Russuales under the division Basidiomycota were classified based on phylogeny analysis. This study provides first evidence for the occurrence of species Fulvifomes fastuosus (LDCMY39 and LDCMY43) and Ganoderma wiiroense (LDCMY02, LDCMY08, LDCMY11, LDCMY17 and LDCMY19) from southern India. Molecular evidence for the existence of Phellinus badius was given for the first time as well. These data enhance our understanding on the diversity of macrofungi in India, which could be further exploited for biomedical applications.
Introduction
The kingdom fungi are a distinct group of eukaryotic organisms encompassing about 1.5 M species1,2, where 77,000 fungal species are identified by ITS sequence and been reported in GenBank repository3. They are identified by filamentous mycelium, absence of motile cells and chlorophyll, presence of chitin-rich cell walls and secretion of external digestive enzymes to degrade the food. Their mode of reproduction is via asexual and sexual spores4. These are considered to be the key decomposers of terrestrial ecosystems and known to play crucial ecological role5–7. Wild mushrooms from the natural habitat have profound biological and economic impact due to their major role in ecosystem maintenance8–10. Destruction of environment is the major threat for fungal diversity; exploration of diversity of macrofungi and their taxonomy are acquired importance for reforestation programmes11.
The phylum Basidiomycota includes largely of fleshy fungi (e.g., mushrooms, toadstools, rusts) and ranked second with approximately 23,000 species4. Abundant growth of Basidiomycetes are prevalent in the rainy seasons where the environmental conditions such as temperature, relative humidity and sunshine are favourable, which aids them in the breakdown of dead organic tissue12. These are the potential indicators of environmental quality13. Many fleshy fungi are edible and harmless, but few are poisonous14. However, approximately 700 species of Basidiomycetes were reported to exhibit notable pharmacological activities15,16. These mainly aids in immune system enhancement, regulation of biorhythm, maintenance of homeostasis and are considered to be the biofactor of effective compounds to cure various diseases as anti-fungal, anti-inflammatory, anti-tumor, anti-viral, anti-bacterial, hepatoprotective, anti-diabetic, hypolipedemic, anti-thrombotic and hypotensive activities17,18. Though countless number of macrofungi demonstrates an array of medicinal values only a small fraction has been subjected to scientific examination.
India is rich in fungal biodiversity and consists of one-third of global fungal diversity in which only 50% is characterized and explored19. Until 1975, study on mushrooms was neglected in states such as Tamil Nadu, Kerala, Karnataka, and Andhra Pradesh in South India. Natarajan and colleagues20 worked on the prospection of mushrooms from southern and south-western region excluding Kerala and, listed 230 agaric and bolete species belonged to 67 genera.
The diversity of Basidiomycetes is studied by classical and molecular methods. It involved collection of basidiome, in vitro culture, molecular identification, and preservation of the macrofungi. Classical taxonomy of macrofungi involves description of macro- and micro-morphological characters such as attachment of basidiocarp, types of basidiocarp, pileus surface, margin, pore surface, hyphal system, setae, basidia, basidiospore and reaction to KOH, Meltzer’s reagent etc.21–23. Traditional survey alone cannot detect many species of fungi, as they do not produce visible basidicarp or species-specific characteristics. Those can be studied using molecular methods24–26. The focus of the present study was to explore the diversity of ethnomycologically important Basidiomycetes in Southern Tamil Nadu, India and we have employed molecular methods for the identification of macrofungi.
Many methods have been used in molecular systematics of macrofungi namely DNA-DNA hybridization; restriction enzyme analysis - RFLP (restriction fragment length polymorphism), rDNA (nuclear ribosomal DNA), mtDNA (mitochondrial DNA); and sequencing analysis – spacers (ITS-internal transcribed spacer), 5S nuclear rRNA, mitochondrial rRNA27. The universal primer for fungal phylogenetics comprised of fungal ribosomal operon: large subunit (26S or 28S), small subunit (18S) and the ITS comprising of ITS1 and ITS2 containing the conserved 5.8S28–30. The ITS1 and ITS4 primers amplify the highly variable ITS1 and ITS2 sequences surrounding coding sequence of 5.8S and it’s exclusively specific for basidiomycetes31,32. This study focussed on sequencing the entire ITS1, 5.8S rRNA and ITS2 for identification of isolated macrofungi. Based on phylogenetic analysis, thirty-two strains belonging to the division Basidiomycota were classified. This study provided additional information to the present knowledge on the data of diversity of fungi in Tamilnadu and also to understand their bioprospects.
Results
This study is the first report on the occurrence of species Fulvifomes fastuosus and Ganoderma wiiroense from India. In addition, molecular evidence for the existence of Phellinus badius in southern Tamilnadu is also provided. In the present study, hundred basidiomata were collected from different locations: Lady Doak College Campus (Fig. 1), Nagamalai (Fig. 2), Pudhupatti (Fig. 3), Ayyanar falls and Kovai Kutralam (Fig. 4), and Tirunelveli (Fig. 5). The collection details such as habitat, host, attachment pattern and position of basidiome on the tree are mentioned in Table 1. The species richness was found in the following order: Lady Doak College Campus (22%), Pudhupatti (21%), Nagamalai (19%), Ayyanar falls (23%), Tirunelveli (13%), Kovai Kutralam (1%), and Thenkasi (1%). The host of the isolates are as follows: Albizzia sp., Azadirachta sp., Canthium dicoccum, Cocos nucifera, Nerium sp., Tamarindus sp., wood log and decayed leaf litters. In this study, Albizzia sp. (58%) was found to be the predominant host. Nearly 56% of the basidiome were associated with tree roots, 36% with tree trunks and 8% with decayed matter. The attachment pattern with the host varied among the isolates: sessile (67%) and stipitate (33%).
Figure 1.
Field photographs of Basidiomata collected from Lady Doak Campus, Madurai District. The macrofungi grown on the host species: Albizzia sp., - LDCBIF01, LDCBIF82, LDCBIF83, LDCBIF84; Azadirachta sp., - LDCBIF09; Araccaceae sp., - LDCBIF101. Few isolates were collected from the decayed matter (LDCBIF02, LDCBIF10 & LDCBIF104) and wood log (LDCBIF03 - LDCBIF07, LDCBIF11 - LDCBIF13, LDCBIF86 & LDCBIF87).
Figure 2.
Field photographs of Basidiomata collected from Nagamalai, Madurai District. The macrofungi grown on the host species: Albizzia sp., - LDCBIF32, LDCBIF33, LDCBIF35, LDCBIF72, LDCBIF76; Azadirachta sp., - LDCBIF30; Cocos sp., - LDCBIF24 and Tamarindus sp., - LDCBIF15, LDCBIF36. Few isolates were collected from the decayed matter (LDCBIF23, LDCBIF25, LDCBIF26, LDCBIF28, LDCBIF29, LDCBIF31) and wood log (LDCBIF14, LDCBIF27, LDCBIF34).
Figure 3.
Field photographs of Basidiomata collected from Pudhupatti, Madurai District. The macrofungi grown on the host species: Albizzia sp., - LDCBIF16 - LDCBIF22, LDCBIF39, LDCBIF40 - LDCBIF50 and LDCBIF77; Tamarindus sp., - LDCBIF08.
Figure 4.
Field photographs of Basidiomata collected from Ayyanar Falls, Dindigul and Kovai kutralam, Coimbatore District. The macrofungi grown on the host species: Ayyanar Falls - Albizzia sp., - LDCBIF51, LDCBIF52, LDCBIF58, LDCBIF59, LDCBIF60, LDCBIF66. Few isolates were collected from the decayed matter (LDCBIF79 - LDCBIF81) and wood log (LDCBIF37 & LDCBIF38). Kovai Kutralam - wood log (LDCBIF85)
Figure 5.
Field photographs of Basidiomata collected from Tirunelveli District. The macrofungi grown on the host species: Nerium sp., - LDCBIF88; Canthium sp., -LDCBIF89; Albizzia sp., - LDCBIF90 - LDCBIF98 and Tamarindus sp., - LDCBIF99 & LDCBIF100.
Table 1.
Basidiomata collected.
| S. No. | Basidiome Id | Host | Attachment to the Host | Position of basidiome on the tree | Size L * W (in cm) | Xanthochroic |
|---|---|---|---|---|---|---|
| 1. | LDCBIF01* | Albizzia sp | Stipitate | Root | 15, 10.5 | − |
| 2. | LDCBIF02* | Decayed material | Stipitate | — | 10.5, 7.5 | − |
| 3. | LDCBIF03* | Wood Log | Sessile | Root | 11, 9 | − |
| 4. | LDCBIF04* | Wood Log | Sessile | Root | 7.5, 4.5 | − |
| 5. | LDCBIF05* | Wood Log | Sessile | Root | NA | − |
| 6. | LDCBIF06* | Wood Log | Sessile | Root | NA | − |
| 7. | LDCBIF07* | Wood Log | Sessile | Root | 6, 5 | − |
| 8. | LDCBIF08* | Tamarindus sp. | Stipitate | Root | NA | − |
| 9. | LDCBIF09* | Azadirachta sp. | Stipitate | Root | 16, 13.5 | − |
| 10. | LDCBIF10* | Decayed material | Sessile | — | 4, 5 | − |
| 11. | LDCBIF11* | Wood Log | Stipitate | Root | 12, 5 | − |
| 12. | LDCBIF12* | Wood Log | Stipitate | Root | 15, 7 | − |
| 13. | LDCBIF13* | Wood Log | Stipitate | Root | 9.5, 7 | − |
| 14. | LDCBIF14# | Wood Log | Sessile | Root | 15.2, 8 | − |
| 15. | LDCBIF15# | Tamarindus sp. | Sessile | Root | 19, 10.5 | − |
| 16. | LDCBIF16@ | Albizzia sp. | Stipitate | Root | 4, 2 | − |
| 17. | LDCBIF17@ | Albizzia sp. | Stipitate | Root | 9, 5 | − |
| 18. | LDCBIF18@ | Albizzia sp. | Stipitate | Root | 7, 4.5 | − |
| 19. | LDCBIF19@ | Albizzia sp. | Stipitate | Root | 9, 7 | − |
| 20. | LDCBIF20@ | Albizzia sp. | Stipitate | Root | 5.5, 3 | − |
| 21. | LDCBIF21@ | Albizzia sp. | Stipitate | Root | 5,3 | − |
| 22. | LDCBIF22@ | Albizzia sp. | Sessile | Root | 12,7.5 | − |
| 23. | LDCBIF23# | Decayed material | Stipitate | Root | 7.5, 6 | − |
| 24. | LDCBIF24# | Cocos sp. | Sessile | Root | 39, 20 | − |
| 25. | LDCBIF25# | Decayed material | Stipitate | Root | 6, 3 | − |
| 26. | LDCBIF26# | Decayed material | Stipitate | Root | 7.8, 6 | − |
| 27. | LDCBIF27# | Wood Log | Sessile | Root | NA | − |
| 28. | LDCBIF28# | Decayed material | Stipitate | Root | 8.5, 7 | − |
| 29. | LDCBIF29# | Decayed material | Stipitate | Root | 7,6 | − |
| 30. | LDCBIF30# | Azadirachta sp. | Stipitate | Root | 5.8, 3.5 | − |
| 31. | LDCBIF31# | Decayed material | Sessile | Root | 5, 3.5 | − |
| 32. | LDCBIF32# | Albizzia sp. | Sessile | Root | 25, 16.5 | − |
| 33. | LDCBIF33# | Albizzia sp. | sessile | Root | 11.5, 7 | − |
| 34. | LDCBIF34# | Wood Log | Sessile | Root | 4.5, 2.5 | − |
| 35. | LDCBIF35# | Albizzia sp. | Sessile | Root | 14, 6 | − |
| 36. | LDCBIF36# | Tamarindus sp. | Sessile | Trunk | 15.5, 10 | − |
| 37. | LDCBIF37$ | Wood Log | Sessile | Root | 25, 18 | − |
| 38. | LDCBIF38$ | Wood Log | Sessile | Root | 12, 10.5 | − |
| 39. | LDCBIF39@ | Albizzia sp. | Sessile | Trunk | 4.5, 3 | + |
| 40. | LDCBIF40@ | Albizzia sp. | Sessile | Trunk | 4.8, 2.8 | + |
| 41. | LDCBIF41@ | Albizzia sp. | Sessile | Trunk | 4.5, 3.5 | + |
| 42. | LDCBIF42@ | Albizzia sp. | Sessile | Trunk | 5.5, 4 | + |
| 43. | LDCBIF43@ | Albizzia sp. | Sessile | Trunk | 5, 3.5 | + |
| 44. | LDCBIF44@ | Albizzia sp. | Sessile | Trunk | 10, 6 | + |
| 45. | LDCBIF45@ | Albizzia sp. | Sessile | Trunk | 7, 3.5 | + |
| 46. | LDCBIF46@ | Albizzia sp. | Sessile | Trunk | 6, 4.5 | + |
| 47. | LDCBIF47@ | Albizzia sp. | Sessile | Trunk | 12, 5.6 | + |
| 48. | LDCBIF48@ | Albizzia sp. | Sessile | Trunk | 15, 6.5 | + |
| 49. | LDCBIF49@ | Albizzia sp. | Sessile | Trunk | 19.5, 9 | + |
| 50. | LDCBIF50@ | Albizzia sp. | Sessile | Trunk | 10.5, 6 | + |
| 51. | LDCBIF51$ | Albizzia sp. | Sessile | Trunk | 12, 9.5 | + |
| 52. | LDCBIF52$ | Albizzia sp. | Sessile | Trunk | 10, 5.5 | + |
| 53. | LDCBIF53$ | Albizzia sp. | Sessile | Trunk | 14, 8 | + |
| 54. | LDCBIF54$ | Albizzia sp. | Sessile | Trunk | 11.5,7 | + |
| 55. | LDCBIF55$ | Albizzia sp. | Sessile | Trunk | 5.5, 4.5 | + |
| 56. | LDCBIF56$ | Albizzia sp. | Sessile | Trunk | 7, 4 | + |
| 57. | LDCBIF57$ | Albizzia sp. | Sessile | Trunk | 9.5, 6 | + |
| 58. | LDCBIF58$ | Albizzia sp. | Sessile | Trunk | 13, 5.5 | + |
| 59. | LDCBIF59$ | Albizzia sp. | Sessile | Trunk | 9, 6 | + |
| 60. | LDCBIF60$ | Albizzia sp. | Sessile | Trunk | 6, 4.5 | + |
| 61. | LDCBIF61$ | Albizzia sp. | Sessile | Trunk | 4, 2 | + |
| 62. | LDCBIF62$ | Albizzia sp. | Sessile | Trunk | 7.5, 4 | + |
| 63. | LDCBIF63$ | Albizzia sp. | Sessile | Trunk | 5, 2.5 | + |
| 64. | LDCBIF64$ | Albizzia sp. | Sessile | Trunk | 6.5, 3 | + |
| 65. | LDCBIF65$ | Albizzia sp. | Sessile | Trunk | 7, 5 | + |
| 66. | LDCBIF66$ | Albizzia sp. | Sessile | Trunk | 5, 5 | + |
| 67. | LDCBIF67$ | Albizzia sp. | Sessile | Trunk | 11, 7 | + |
| 68. | LDCBIF68$ | Albizzia sp. | Sessile | Trunk | 11, 4.8 | + |
| 69. | LDCBIF71# | Albizzia sp. | Sessile | Trunk | 8.5, 5 | + |
| 70. | LDCBIF72# | Albizzia sp. | Sessile | Trunk | 5.5, 3.5 | + |
| 71. | LDCBIF73* | Albizzia sp. | Sessile | Root | 6, 5 | + |
| 72. | LDCBIF74* | Albizzia sp. | Sessile | Trunk | 3, 2 | + |
| 73. | LDCBIF75* | Albizzia sp. | Sessile | Root | 5.5, 3 | + |
| 74. | LDCBIF76# | Albizzia sp. | Sessile | Trunk | 6, 4.5 | + |
| 75. | LDCBIF77@ | Albizzia sp. | Sessile | Trunk | NA | + |
| 76. | LDCBIF78≠ | Wood Log | Sessile | — | 11.5, 7 | − |
| 77. | LDCBIF79# | Decayed material | Stipitate | — | 3, 3 | − |
| 78. | LDCBIF80# | Decayed material | Stipitate | — | 7, 5 | − |
| 79. | LDCBIF81# | Decayed material | Stipitate | — | 6, 5.8 | − |
| 80. | LDCBIF82* | Albizzia a sp. | Sessile | Root | 4, 2.5 | + |
| 81. | LDCBIF83* | Albizzia sp. | Sessile | Root | NA | + |
| 82. | LDCBIF84* | Albizzia sp. | Sessile | Root | NA | + |
| 83. | LDCBIF85€ | Wood Log | Sessile | — | NA | − |
| 84. | LDCBIF86* | Wood Log | Stipitate | Root | NA | − |
| 85. | LDCBIF87* | Wood Log | Sessile | Trunk | NA | − |
| 86. | LDCBIF88® | Nerium sp. | Sessile | Root | 16, 8.4 | − |
| 87. | LDCBIF89® | Canthium sp. | Sessile | Root | 11, 8 | − |
| 88. | LDCBIF90® | Cocos sp. | Stipitate | Root | 9.1, 8 | − |
| 89. | LDCBIF91® | Cocos sp. | Stipitate | Root | 3, 3.5 | − |
| 90. | LDCBIF92® | Albizzia sp. | Stipitate | Root | 8, 5 | − |
| 91. | LDCBIF93® | Albizzia sp. | Stipitate | Root | 5, 4.3 | − |
| 92. | LDCBIF94® | Albizzia sp. | Stipitate | Root | 7.2, 5.1 | − |
| 93. | LDCBIF95® | Albizzia sp. | Sessile | Root | 3, 2.8 | − |
| 94. | LDCBIF96® | Albizzia sp. | Stipitate | Root | 4, 3.8 | − |
| 95. | LDCBIF97® | Albizzia sp. | Stipitate | Root | 7, 5.2 | − |
| 96. | LDCBIF98® | Albizzia sp. | Stipitate | Root | 4.8, 3.4 | − |
| 97. | LDCBIF99® | Tamarindus sp. | Sessile | Root | 10.8, 6 | − |
| 98. | LDCBIF100® | Tamarindus sp. | Stipitate | Root | 4.4, 4 | − |
| 99. | LDCBIF101* | Araccaceae sp. | Sessile | Root | 7.8, 6.8 | − |
| 100. | LDCBIF104* | Decayed Material | Stipitate | — | NA | − |
$, €, *, #, ≠, @, ®used to denote the strains collected from different places. $Ayyanar falls; €Coimbatore; *Lady Doak College Campus; #Nagamalai; ≠Thenkasi; @Pudhupatti; ®Tirunelveli.
Among the hundred basidiome collected only forty-nine isolates (49%) could be grown in axenic cultures. The mycelial growth significantly varied from 7 days to 30 days. The colour of the mycelia varies for each strain: white, orange white, yellowish white, pale yellow, greyish orange, light yellow, pale orange and brownish orange (Fig. 6, Table 2). The pure cultures of all isolates were stored in mineral oil till further use.
Figure 6.
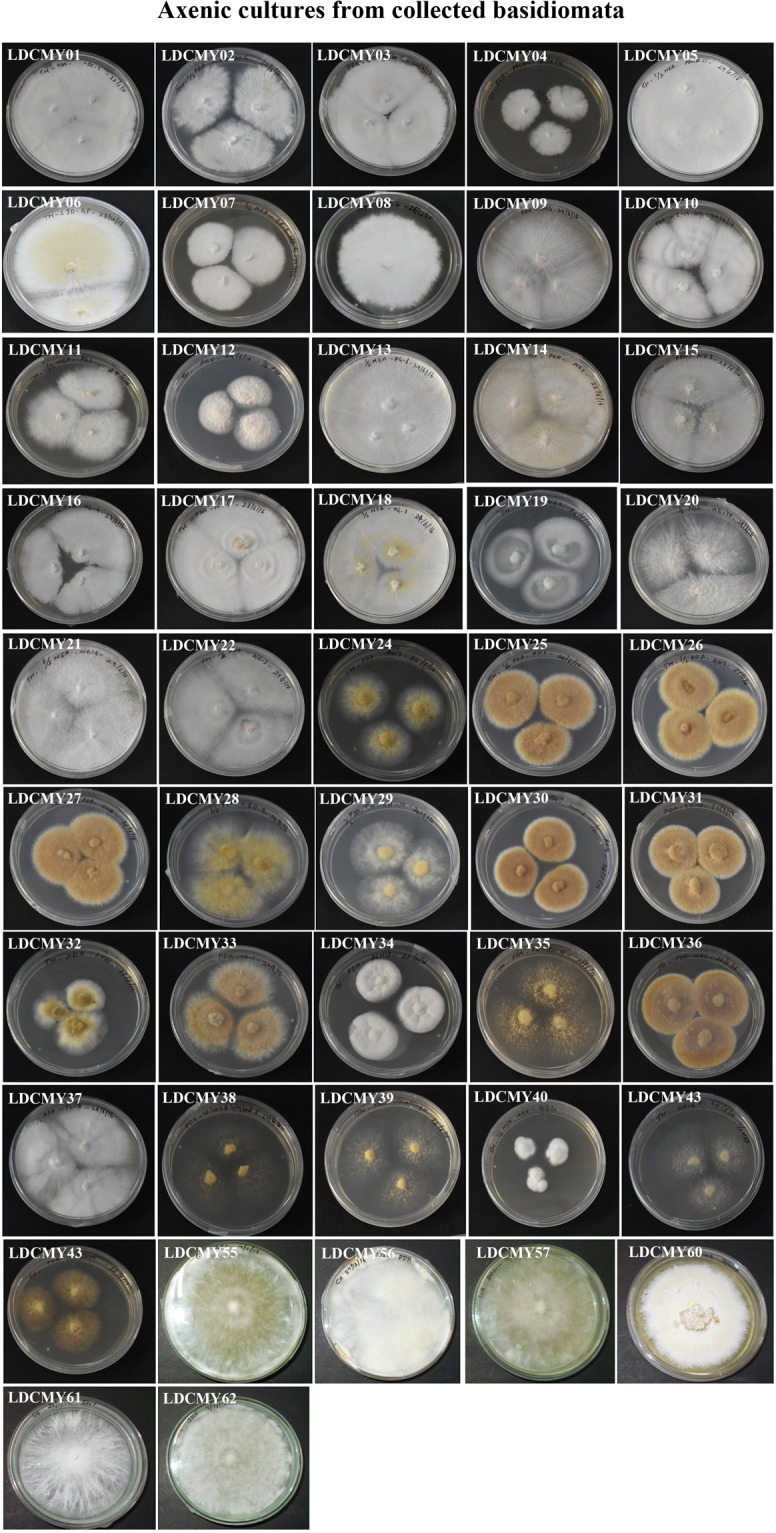
Axenic culture of collected basidiomata. The mycelium culture on PDA plates. Variations in growth and the color of the mycelium was observed (See Table 2). The identified strains by sequencing; Amylosporous sp. - LDCMY57 & LDCMY58; Coriolopsis caperata - LDCMY42; Fomitopsis ostreiformis - LDCMY21; Fulvifomes fastuosus - LDCMY39, LDCMY43; Ganoderma resinaceum - LDCMY01; Ganoderma sp. - LDCMY04, LDCMY05, LDCMY06; LDCMY12, LDCMY14, LDCMY16, LDCMY18, LDCMY22, LDCMY41. Ganoderma wiiroense - LDCMY19, LDCMY08, LDCMY11, LDCMY17 and LDCMY02; Inonotus rickii - LDCMY52; Phellinus badius - LDCMY36; Phellinus sp. - LDCMY23, LDCMY24, LDCMY27, LDCMY28, LDCMY29, LDCMY31, LDCMY34, LDCMY45; Trametes elegans - LDCMY37.
Table 2.
Growth and characteristics of mycelium culture.
| S. No. | Basidiome Id | Strain Id | Mycelial growth in PDA plates | |||
|---|---|---|---|---|---|---|
| Initial radial expansion (in mm) | Complete colonization (in days) | Front Color | Reverse Color | |||
| 1. | LDCBIF01 | LDCMY01* | 23.44 ± 0.24 | 7 | White | Pale Yellow |
| 2. | LDCBIF02 | LDCMY02* | 22.00 ± 0.15 | 7 | Orange White | Pale Yellow |
| 3. | LDCBIF03 | LDCMY03* | 37.66 ± 0.20 | 7 | White | Pale Yellow |
| 4. | LDCBIF04 | LDCMY04* | 18.00 ± 0.26 | 7 | White | White |
| 5. | LDCBIF06 | LDCMY41* | 25.33 ± 0.15 | 7 | White | Pale Yellow |
| 6. | LDCBIF08 | LDCMY05* | 19.33 ± 0.05 | 7 | Orange White | Pale Yellow |
| 7. | LDCBIF09 | LDCMY06* | 25.00 ± 0.15 | 7 | White | Pale Yellow |
| 8. | LDCBIF10 | LDCMY07* | 22.33 ± 0.25 | 7 | White | Light Yellow |
| 9. | LDCBIF11 | LDCMY08* | 22.00 ± 0.55 | 7 | White | Light Yellow |
| 10. | LDCBIF12 | LDCMY09* | 36.66 ± 0.55 | 7 | White | Pale Yellow |
| 11. | LDCBIF13 | LDCMY10* | 32.66 ± 0.15 | 7 | White | Light Yellow |
| 12. | LDCBIF16 | LDCMY11@ | 25.66 ± 0.15 | 7 | White | Light Yellow |
| 13. | LDCBIF19 | LDCMY12@ | 21.33 ± 0.11 | 7 | White | Light Yellow |
| 14. | LDCBIF21 | LDCMY13@ | 41.33 ± 0.92 | 7 | White | Pale Yellow |
| 15. | LDCBIF23 | LDCMY14# | 29.66 ± 0.20 | 7 | White | Light Orange |
| 16. | LDCBIF25 | LDCMY15# | 24.00 ± 0.30 | 7 | White | Pale Yellow |
| 17. | LDCBIF26 | LDCMY16# | 19.33 ± 0.23 | 7 | Yellowish White | Greyish Yellow |
| 18. | LDCBIF28 | LDCMY17# | 23.00 ± 0.26 | 7 | White | Pale Yellow |
| 19. | LDCBIF29 | LDCMY18# | 32.00 ± 0.32 | 7 | Pale Yellow | Light Orange |
| 20. | LDCBIF31 | LDCMY19# | 37.33 ± 0.25 | 7 | White | White |
| 21. | LDCBIF32 | LDCMY20# | 34.00 ± 0.36 | 7 | White | Pale Yellow |
| 22. | LDCBIF34 | LDCMY21# | 33.00 ± 0.75 | 7 | White | Pale Yellow |
| 23. | LDCBIF35 | LDCMY22# | 28.33 ± 0.05 | 7 | White | White |
| 24. | LDCBIF39 | LDCMY23@ | 18.00 ± 0.10 | 30 | Greyish Orange | Greyish Orange |
| 25. | LDCBIF43 | LDCMY24@ | 18.00 ± 0.15 | 27 | Light Yellow | Greyish Yellow |
| 26. | LDCBIF44 | LDCMY25@ | 23.00 ± 0.20 | 14 | Light Yellow | Greyish Yellow |
| 27. | LDCBIF55 | LDCMY26$ | 26.66 ± 0.05 | 17 | Greyish Orange | Greyish Orange |
| 28. | LDCBIF58 | LDCMY27$ | 22.66 ± 0.11 | 17 | Greyish Orange | Greyish Orange |
| 29. | LDCBIF59 | LDCMY28$ | 27.00 ± 0.51 | 17 | Greyish Orange | Greyish Orange |
| 30. | LDCBIF60 | LDCMY29$ | 22.33 ± 0.47 | 17 | Light Yellow | Light Yellow |
| 31. | LDCBIF62 | LDCMY30$ | 22.66 ± 0.32 | 17 | Greyish Orange | Greyish Orange |
| 32. | LDCBIF66 | LDCMY31$ | 22.00 ± 0.17 | 30 | Pale Orange | Light Orange |
| 33. | LDCBIF68 | LDCMY32$ | 34.00 ± 0.45 | 27 | Light Yellow | Greyish Yellow |
| 34. | LDCBIF71 | LDCMY44# | 27.33 ± 0.15 | 20 | Brownish Orange | Deep Orange |
| 35. | LDCBIF72 | LDCMY34# | 21.66 ± 0.25 | 19 | Light Yellow | Brownish Yellow |
| 36. | LDCBIF73 | LDCMY35* | 37.66 ± 0.40 | 17 | Greyish Orange | Greyish Orange |
| 37. | LDCBIF74 | LDCMY43* | 28.00 ± 0.10 | 20 | Brownish Orange | Deep Orange |
| 38. | LDCBIF77 | LDCMY36@ | 23.33 ± 0.05 | 15 | Greyish Orange | Greyish Orange |
| 39. | LDCBIF78 | LDCMY37≠ | 17.00 ± 0.26 | 5 | White | Pale Yellow |
| 40. | LDCBIF82 | LDCMY38* | 27.33 ± 0.45 | 17 | Greyish Yellow | Deep Orange |
| 41. | LDCBIF84 | LDCMY39* | 25.33 ± 0.20 | 20 | Brownish Orange | Deep Orange |
| 42. | LDCBIF85 | LDCMY40* | 18.00 ± 0.43 | 5 | White | Light Yellow |
| 43. | LDCBIF86 | LDCMY41* | 21.00 ± 0.39 | 7 | White | Pale Yellow |
| 44. | LDCBIF87 | LDCMY42£ | 32.00 ± 0.21 | 5 | White | Light Yellow |
| 45. | LDCBIF88 | LDCMY57® | 27.00 ± 0.10 | 5 | White | Pale Yellow |
| 46. | LDCBIF96 | LDCMY58® | 17.33 ± 0.15 | 5 | White | Pale Yellow |
| 47. | LDCBIF100 | LDCMY60® | 18.00 ± 0.43 | 7 | White | Pale Yellow |
| 48. | LDCBIF101 | LDCMY61® | 26.66 ± 0.25 | 7 | White | Pale Yellow |
| 49. | LDCBIF104 | LDCMY62* | 21.66 ± 0.11 | 7 | White | Pale Yellow |
$,€,*,#,≠,@,®Used to denote the strains collected from different places. $Ayyanar falls; €Coimbatore; *Lady Doak College Campus; #Nagamalai; ≠Thenkasi; @Pudhupatti; ®Tirunelveli.
The radial expansion was measured on the 3rd day (shown in bold) and 7th day. The measurements are given in mean ± SD. The total number of days taken for complete colonization (80 mm) in PDA medium varied among the isolates and ranged from 5–30 days for different strains.
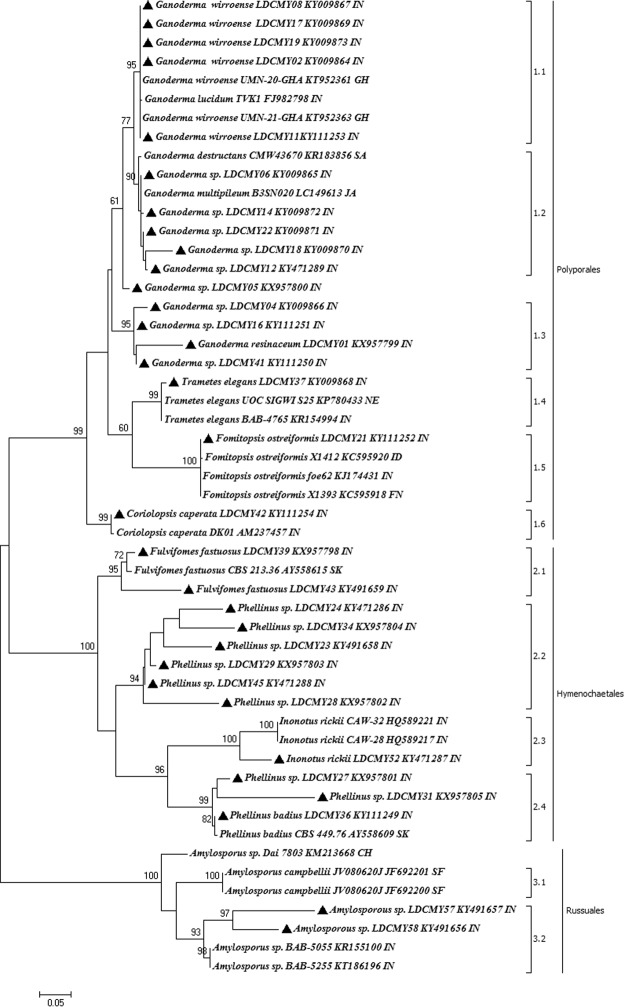
Genomic DNA was obtained and 5.8S ribosomal RNA gene segment was amplified using sequence specific primers. Thirty-two isolates were successfully sequenced and the size of the amplicon ranged from 599 bp to 902 bp. The sequences were deposited in GenBank and accession numbers were obtained (Table 3). Variation in genetic makeup was observed among the isolates from the same environment. Molecular phylogentic analysis was carried out using 52 ITS sequences in which 20 reference sequences were retrieved from GenBank, NCBI to clarify the variation among the sequences. The phylogenetic tree constructed using maximum likelihood (ML) method (Fig. 7). The basidiomycete species were clustered into three clades: Clade 1 - Polyporales, Clade 2 - Hymenochaetales and Clade 3 - Russuales. The three clades are detailed below:
Table 3.
Species and their GenBank accession number used for constructing molecular phylogeny.
| S.No | Organism Name | Strain/Isolate Name | Source of DNA | Geographical Origin | Sequence Length (ITS1/ITS4) | Accession No |
|---|---|---|---|---|---|---|
| 1. | Amylosporous sp. | LDCMY58® | Mycelium | Tirunelveli, South India | 741 | KY491656 |
| 2. | Amylosporous sp. | LDCMY57® | Mycelium | Tirunelveli, South India | 774 | KY491657 |
| 3. | Amylosporus sp. | BAB-5055 | — | India | 897 | KR155100 |
| 4. | Amylosporus sp. | BAB-5255 | — | India | 775 | KT186196 |
| 5. | Amylosporus sp. | Dai 7803 | — | China | 748 | KM213668 |
| 6. | Amylosporus campbellii | JV080620J | — | Southern Florida | 807 | JF692201 |
| 7. | Amylosporus campbellii | JV080620J | — | Southern Florida | 810 | JF692200 |
| 8. | Coriolopsis caperata | LDCMY42* | Mycelium | Lady Doak College Campus, Madurai, South India | 614 | KY111254 |
| 9. | Coriolopsis caperata | DK01 | — | New Delhi | 585 | AM237457 |
| 10. | Fomitopsis ostreiformis | LDCMY21# | Mycelium | Nagamalai, Madurai, South India | 599 | KY111252 |
| 11. | Fomitopsis ostreiformis | X1412 | — | Indonesia | 1600 | KC595920 |
| 12. | Fomitopsis ostreiformis | foe62 | — | Karnataka- India | 636 | KJ174431 |
| 13. | Fomitopsis ostreiformis | X1393 | — | Finland | 1600 | KC595918 |
| 14. | Fulvifomes fastuosus | LDCMY39* | Mycelium | Lady Doak College Campus, Madurai, South India | 756 | KX957798 |
| 15. | Fulvifomes fastuosus | LDCMY43* | Mycelium | Lady Doak College Campus, Madurai, South India | 738 | KY491659 |
| 16. | Fulvifomes fastuosus | CBS 213.36 | — | South Korea | 768 | AY558615 |
| 17. | Ganoderma destructans | CMW43670 | — | South Africa | 640 | KR183856 |
| 18. | Ganoderma lucidum | TVK1 | — | India | 603 | FJ982798 |
| 19. | Ganoderma multipileum | B3SN020 | — | Japan | 832 | LC149613 |
| 20. | Ganoderma resinaceum | LDCMY01* | Mycelium | Lady Doak College Campus, Madurai, South India | 614 | KX957799 |
| 21. | Ganoderma sp. | LDCMY04* | Mycelium | Lady Doak College Campus, Madurai, South India | 610 | KY009866 |
| 22. | Ganoderma sp. | LDCMY05* | Mycelium | Lady Doak College Campus, Madurai, South India | 620 | KX957800 |
| 23. | Ganoderma sp. | LDCMY06* | Mycelium | Lady Doak College Campus, Madurai, South India | 608 | KY009865 |
| 24. | Ganoderma sp. | LDCMY12@ | Mycelium | Pudhupatti, South India | 606 | KY471289 |
| 25. | Ganoderma sp. | LDCMY16# | Mycelium | Nagamalai, Madurai, South India | 607 | KY111251 |
| 26. | Ganoderma sp. | LDCMY18# | Mycelium | Nagamalai, Madurai, South India | 722 | KY009870 |
| 27. | Ganoderma sp. | LDCMY22# | Mycelium | Nagamalai, Madurai, South India | 619 | KY009871 |
| 28. | Ganoderma sp. | LDCMY14# | Mycelium | Nagamalai, Madurai, South India | 614 | KY009872 |
| 29. | Ganoderma sp. | LDCMY41* | Mycelium | Lady Doak College Campus, Madurai, South India | 642 | KY111250 |
| 30. | Ganoderma wiiroense | LDCMY02* | Mycelium | Lady Doak College Campus, Madurai, South India | 608 | KY009864 |
| 31. | Ganoderma wiiroense | LDCMY08* | Mycelium | Lady Doak College Campus, Madurai, South India | 618 | KY009867 |
| 32. | Ganoderma wiiroense | LDCMY11@ | Mycelium | Pudhupatti, South India | 611 | KY111253 |
| 33. | Ganoderma wiiroense | LDCMY17# | Mycelium | Nagamalai, Madurai, South India | 612 | KY009869 |
| 34. | Ganoderma wiiroense | LDCMY19# | Mycelium | Nagamalai, Madurai, South India | 647 | KY009873 |
| 35. | Ganoderma wiiroense | UMN-20-GHA | — | USA | 769 | KT952361 |
| 36. | Ganoderma wiiroense | UMN-21-GHA | — | USA | 722 | KT952363 |
| 37. | Inonotus rickii | LDCMY52$ | Basidiome | Ayyanar falls, Dindigul, South India | 902 | KY471287 |
| 38. | Inonotus rickii | CAW-32 | — | Rajasthan- India | 747 | HQ589221 |
| 39. | Inonotus rickii | CAW-28 | — | Rajasthan - India | 750 | HQ589217 |
| 40. | Phellinus badius | LDCMY36@ | Mycelium | Pudhupatti, South India | 688 | KY111249 |
| 41. | Phellinus badius | CBS 449.76 | — | South Korea | 714 | AY558609 |
| 42. | Phellinus sp. | LDCMY23@ | Mycelium | Pudhupatti, South India | 709 | KY491658 |
| 43. | Phellinus sp. | LDCMY 24@ | Mycelium | Pudhupatti, South India | 668 | KY471286 |
| 44. | Phellinus sp. | LDCMY27$ | Mycelium | Ayyanar falls, Dindigul, South India | 662 | KX957801 |
| 45. | Phellinus sp. | LDCMY28$ | Mycelium | Ayyanar falls, Dindigul, South India | 693 | KX957802 |
| 46. | Phellinus sp. | LDCMY29$ | Mycelium | Ayyanar falls, Dindigul, South India | 683 | KX957803 |
| 47. | Phellinus sp. | LDCMY31$ | Mycelium | Ayyanar falls, Dindigul, South India | 685 | KX957805 |
| 48. | Phellinus sp. | LDCMY34# | Mycelium | Nagamalai, Madurai, South India | 681 | KX957804 |
| 49. | Phellinus sp. | LDCMY45$ | Basidiome | Ayyanar falls, Dindigul, South India | 677 | KY471288 |
| 50. | Trametes elegans | LDCMY37≠ | Mycelium | Thenkasi, South India | 606 | KY009868 |
| 51. | Trametes elegans | UOC SIGWI S25 | — | Nepal | 655 | KP780433 |
| 52. | Trametes elegans | BAB-4765 | — | India | 637 | KR154994 |
$,€,*,#,≠,@,®Used to denote the sequence data generated from the strains collected from different places. $Ayyanar falls; €Kovai kutralam *Lady Doak College Campus; #Nagamalai; ≠Thenkasi; @Pudhupatti; ®Tirunelveli.
Figure 7.
The evolutionary relationship was inferred using the maximum Likelihood method in MEGA6. The analysis involved 52 nucleotide sequences; thirty two sequences generated in this study are highlighted. The initial trees were obtained with the random addition of sequences. All positions containing gaps and missing data were eliminated. Numerical values above the internodes are the percentage of 1000 bootstrap replications. Bootstrap values higher than 60% are indicated. Scale bar 0.05 represents nucleotide substitutions per position. Three clades were predicted Clade 1: Polyporales; Clade 2: Hymenochaetales; Clade 3: Russuales. The abbreviated letters next to accession number indicates the localities from which the sample is collected: IN - India, GH - Ghana, CH- China, ID - Indonesia, FL - Finland, NE - Nepal, SA - South Africa, SF - South Florida, SK - South Korea, SL - Sri Lanka. The diversity within subpopulation was predicted as 0.1, the diversity within entire population - 0.3 with a Mean inter population Diversity - 0.3 and Coefficient of differentiation - 0.8.
Clade 1: Polyporales - Found in all study sites except Ayyanar falls. Eighteen strains were grouped under this clade and fifteen sequences were further categorised under the family Ganodermataceae, two under Polyporaceae and one in Fomitopsidaceae. The isolated strains belong to the Polyporales were Coriolopsis caperata, Fomitopsis ostreiformis, Ganoderma resinaceum, Ganoderma sp., Ganoderma wiiroense and Trametes elegans. Coriolopsis caperata LDCMY42 collected from Nagamalai showed 99% similarity with the strain Coriolopsis caperata DK01 (AM237457). Monophyletic origin of Fomitopsis ostreiformis was determined with 100% bootstrap support. Five strains were identified as Ganoderma wiiroense (LDCMY19, LDCMY08, LDCMY11, LDCMY17 and LDCMY02) and showed highest similarity with the strains reported from United States of America (KT952361 and KT952363). Variations in the genetic makeup as well in the morphology of the Ganoderma wiiroense strains were observed. Majority of the Ganoderma strains were found to be stipitate. Based on molecular analysis, this is the first evidence for the occurrence of Ganoderma wiiroense from India.
The Clade 1 was supported by 99% bootstrap value and it was further categorized into 6 groups (1.1–1.6). Three groups (1.1–1.3) in this clade consisted of strains from Ganoderma sp. Five strains of Ganoderma wiiroense were grouped in 1.1 and supported by 95% bootstrap value. The mean difference between the sequences in this group was very low (0.000878851). The group 1.2 included Ganoderma sp., which is supported by 90% bootstrap with the mean difference of 0.019876893. The group 1.3 included Ganoderma sp. from different places, which was supported by 95% bootstrap value with the mean difference of 0.049142826. The group 1.4 included Trametes elegans LDCMY37, Thenkasi showed similarity with two strains reported from Nepal and India, and supported by 99% bootstrap value with the mean difference of 0.004707472. The group 1.5 included Fomitopsis ostreiformis LDCMY21 isolated from Nagamalai supported by 100% bootstrap value with the mean difference of 0.001759814. The group 1.6 included Coriolopsis caperata LDCMY42 from LDC Campus and it was supported by 99% bootstrap with the mean difference of 0.003519628.
Clade 2: Hymenochaetales - the isolates categorized in this clade were found in all study sites except Thenkasi. Twelve isolates belonging to the genus Fulvifomes, Phellinus and Inonotus were categorised in this clade. They are Fulvifomes fastuosus (LDCMY39 and LDCMY43), Inonotus rickii (LDCMY52), Phellinus badius (LDCMY36) and Phellinus sp. (LDCMY23, LDCMY24, LDCMY28, LDCMY34 and LDCMY45). Molecular phylogeny analysis confirmed that two strains (LDCMY39 and LDCMY43) obtained from Lady Doak College campus as Fulvifomes fastuosus. The isolates showed highest similarity with the strains reported from Sri Lanka (KR867653) and South Korea (AY558615) and supported with 95% bootstrapping. The host for both the strains were Albizzia sp. We further provided the first significant report on more precise identification of Fulvifomes fastuosus on the basis of the genetic information. A strain collected from Ayyanar falls was identified as Inonotus rickki (LDCMY52) that shared 100% similarity with the strains previously reported from India. The genus Phellinus was found to be present in all study sites. Phellinus badius LDCMY36 shared 93% relatedness with the strain CBS 449.76 from South Korea. This was the first molecular evidence of the species Phellinus badius from India.
This Clade 2 was supported by 100% bootstrap value and consisted of 4 groups (2.1–2.4). The Group 2.1 includes Fulvifomes fastuosus (95% bootstrap) with the mean difference of 0.082737938; Group 2.2 was supported by 94% bootstrap and includes Phellinus sp. (0.100297219); Group 2.3 has only Inonotus rickki and supported by 100% bootstrap value and the mean difference was 0.27677544. Phellinus badius (99% bootstrap) along with few strains of Phellinus sp. were categorised in Group 2.4. The mean difference within the group was 0.096520676.
Clade 3: Russales - This group consisted of samples collected only from Tirunelveli and supported by 100% bootstrap value and consisted of 2 groups (3.1 & 3.2). Two strains (LDCMY57 and LDCMY58) supported with 93% bootstrap value and identified as Amylosporus sp. belonging to the family Bondarzewiaceae and grouped in 3.2. The mean difference among the isolates in this group was 0.134112602. These isolates showed similarity with the strains reported from India (BAB-5055 and BAB-5255), China (Dai 7803) and USA (JV080620J).
The morphological and culture characteristics of first time reported strains from India Ganoderma wiiroense and Fulvifomes fastuosus along with Phellinus badius are given below.
Ganoderma wiiroense
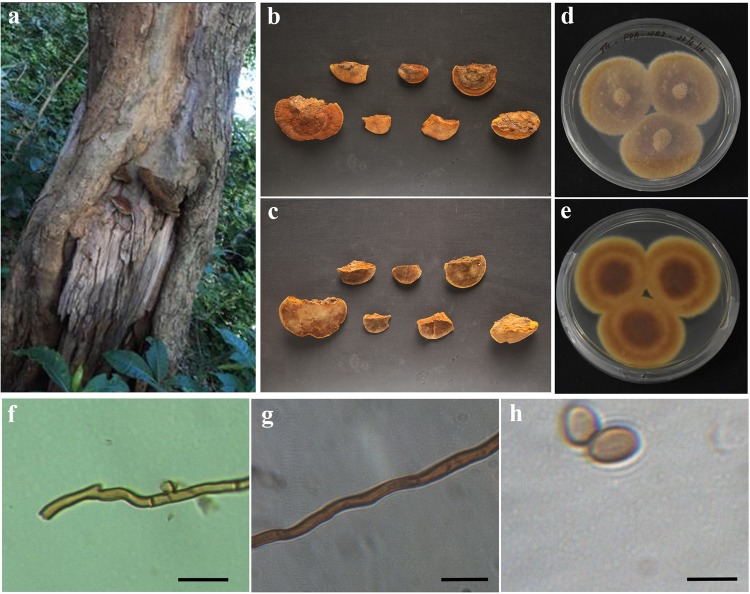
Annual, pileate, basidiocarp, sessile, woody hard, white to creamy yellow when dry. Size of the pileus 10.5 cm × 7.5 cm; Hymenophore poroid, Hyphal system trimitic, generative hyphae with clamp connections, hyaline, thin-walled, branched, 2–4 µm in diameter; skeletal hyphae occasionally branched, 2.5–7.5 μm thick; binding and skeleton-binding hyphae hyaline. Spores ellipsoid (Fig. 8). Colonies of G. wiiroense on PDA was fast growing, 22–37 mm diameter after 3 days and took 7 days to completely colonize 80 mm diameter plates.
Figure 8.
Morphology of Ganoderma wiiroense: (a) Basidiomata; (b) Pileal surface; (c) Hymenial surface; (d and e) Pure culture; (f) Skeletal Hyphae; (g) Generative hyphae; (h) Binding hyphae; (i) Basidiospores. (Scale: 20X – h; 40X – f and G; 100X – i).
Fulvifomes fastuosus
Perennial, pileate, basidiocarp, sessile, woody hard and without odour or taste when dry. Size of the pileus 4.5 cm × 2 cm; Hymenophore poroid, hyphal system Dimitic; generative hyphae without clamp connections, hyaline, thin-walled, simple septate, occasionally branched, 2–3 µm in diameter; skeletal hyphae thick-walled with broad lumen, unbranched, 3–5 µm in diameter. Tissue darkening in KOH. Hymenial setae absent. Spores: subglobose, yellowish, thick-walled, smooth 3.4–5.7 × 3.1–4.2 μm. Yellowish brown, dark reddish brown in KOH (Fig. 9). Colonies of Fulvifomes fastuosus on PDA plate was slow compared to Ganoderma strains, 25–28 mm diameter after 7 days and took 20 days to completely colonize 80 mm diameter plates.
Figure 9.
Morphology of Fulvifomes fastuosus: (a) Basidiomata attached to the host; (b) Pileal surface; (c) Hymenial surface; (d,e) Pure culture; (f) Skeletal Hyphae; (g) Generative hyphae; (h) Basidiospores. (Scale: 40X– g; 100X– f & h).
Phellinus badius
Perennial, pileate, basidiocarp, sessile, woody hard, easily detachable from the host. Hymenophore poroid, hyphal system dimitic; generative hyphae thin walled, simple septate, clampless, moderately branched, hyaline to pale yellow, 3.47 µm; skeletal hyphae thick walled (4.35 µm); Hymenial setae absent. Spores: ellipsoid, moderately thick walled, 4.21–5.54 × 2.83–4.13 μm. Yellowish brown, dark reddish brown in KOH (Fig. 10). The growth of Phellinus badius on PDA was slow, 23–24 mm diameter after 7 days and took 15 days to completely colonize 80 mm diameter plates.
Figure 10.
Morphology of Phellinus badius. (a) Basidiomata attached to the host; (b) Pileal surface; (c) Hymenial surface; (d,e) Pure culture; (f) Skeletal Hyphae; (g) Generative hyphae; (h) Basidiospores. (Scale: 40X– f & g; 100X– h).
Discussion
Fungi are ubiquitous in nature and distributed in all ecosystem. It can survive in diversified habitats such as air, water, soil, litter etc. It contains 1.5 million species, of which 74,000 species are named4. The phylum basidiomycota consist of 37% of all described fungal species33. Threats to fungi due to habitat destruction are a global concern as they play an important role in human welfare19. To understand the distribution and diversity of macrofungi in South India, the basidiomata were collected from living trees, wood log and leaf litters during the rainy season (November to January).
The Basidiomycetes were usually classified based on phenotypic traits; however, classification based on morphological characteristic features alone will be flawed and misleading and the use of molecular classification was found to be more reliable34,35. So far, only 5% of fungal strains were isolated as pure cultures and several described species were acknowledged only as herbarium specimens19. In the present study, pure culture (Fig. 6) was raised from 49% of the isolates and the molecular data were obtained for 65% of the isolates. These molecular data helped in identification of the isolates and was used for construction of genetic diversity among the macrofungal isolates.
Molecular phylogeny of the macrofungal isolates
The molecular systematics of macrofungi has been studied by various methods using DNA-DNA hybridization, restriction enzyme analysis - RFLP, rDNA, mtDNA and sequencing analysis of ITS27. Pectinase isoenzyme36, manganese superoxide dismutase37,38, ITS and 25S ribosomal sequences34,35,39 were used to construct molecular phylogeny in macrofungal species. Later, ITS was used as a DNA barcode for fungal identification32,40,41. In this study, amplification of nuclear ribosomal ITS was used to identify the isolates. The identified isolates belong to three families namely Polyporales, Hymenochaetales and Russuales. The representative strains of the Polyporales from this study were Coriolopsis caperata, Fomitopsis ostreiformis, Ganoderma resinaceum, Ganoderma sp., Ganoderma wiiroense and Trametes elegans. The isolated strains belonging to Hymenochaetales were Fulvifomes fastuosus, Inonotus rickii, Phellinus sp. and Phellinus badius. Amylosporous sp. was the only strain found in our study from the family Russuales. We are the first to report the occurrence of Ganoderma wiiroense and Fulvifomes fastuosus with morphological and molecular evidence; and also provided the molecular evidence for Phellinus badius from India.
G. wiiroense belonging to the Family Polyporales was first reported from Upper Western region of Ghana42. There were only 8 strains available in the GenBank for G. wiiroense, where two from Ghana42 and the rest from this study. Crous et al.42 reported that G. lucidum (TVK1, India; GenBank FJ982798) was closer to G. wiiroense. In our study, we also found that the G. lucidum FJ982798 was closer to G. wiiroense than any other Ganoderma strains reported in this study.
The genus Phellinus belonging to the Family Hymenochaetaceae were important owing to their medicinal values18,43. Three hundred and sixty-seven Phellinus has been reported in the CBS (http://www.punenvis.nic.in/bd_list.htm). In India, eighteen Phellinus species have been reported from Kerala44,45, P. nilgheriensis (Mont.) Cunn., P. shaferi from Gujarat46,47 and P. badius was described morphologically from Punjab48. This study provides the first report on molecular evidence for P. badius from India.
The genus Fulvifomes Murrill was segregated from Phellinus Quél., Murrill49 and typified with F. robiniae (Murrill). It was not accepted as a separate genus and treated as a subgenus of Phellinus till 199950. Later, comprehensive evidences based on molecular phylogenetic analyses proved that it as an independent genus closely associated with Aurificaria Reid and Phylloporia Murrill51,52. The key characteristics of Fulvifomes are pileate basidiocarps, a dimitic hyphal system, coloured basidiospores and absence of setae51. Species with resupinate basidiocarps and/or hymenial setae were included into Fulvifomes based on morphological studies43. Recently, species with monomitic hyphal system were included in Fulvifomes by Zhou53.
Fulvifomes fastuosus was described by Bondartseva and Herrera54. There are 162 reports available in GenBank on the genus Fulvifomes based on molecular data and among them only 18 sequences were on F. fastuosus. The species F. fastuosus was described from China43, Thailand55 and Sri Lanka56. In this study based on molecular phylogeny, two strains collected from Lady Doak College, Tamilnadu, India were identified as Fulvifomes fastuosus.
Macro and micromorphological characteristic features of G. wiiroense, P. badius and F. fastusosus
The identification based on molecular means has been checked with the macro- and micro-morphological characteristic features and were found to be similar with the reported strains. However, the observation on basidiospores was different from the other reports for P. badius and F. fastuosus. The basidiospores of P. badius are ovoid to subglobose to globose and 4–6 × 4–5.5 μm44. Singh and colleagues48 reported that basidiospores were broadly ellipsoid to subglobose. Our observation shows the P. badius basidiospores were ellipsoid and 4.21–5.54 × 2.83–4.13 μm. The basidiospores of F. fastuosus were subglobose, thick-walled, smooth 4.49 × 4.01 μm56. According to Dai43, the basidiospores are 5–6.1 × 4.2–5.6 μm. Our observations shows the basidiospores were 3.4–5.7 × 3.1–4.2 μm, which was smaller than Dai43, but similar to Ediriweera et al.56. However, the variation in the ratio (Q) was the same as previously reported of F. fastuosus strains. The variation in the size of basidiospores might be due to their geographical niche as well as depending on their nutrients from the host species.
Host preference by the macrofungal isolates
There are several factors that influence the distribution of fungi namely ecological niche, climatic conditions, host/substrate type, distribution of fauna and flora19. To study host preference, basidiomata were collected from the living trees, wood log, and leaf litters. Later, the basidiomata was identified by molecular classification.
In India, the information on Ganoderma was first published in the early 1900s57. Nearly 144 hosts were recorded in India58. Among them coconut, betelnut, Casuarina, Areca catechu, Dalbergia sissoo and Toona ciliata59,60 was observed as obvious host of Ganoderma sp. In India and Sri Lanka, Cocus nucifera showed high incidence as a host for Ganoderma species58,61–63. From this study, it was observed that Ganoderma sp. grown on the following host species: Albizzia sp., Tamarindus sp., Azadirachta sp. and Coccus nucifera. Fomitopsis ostreiformis belonging to Ganodermataceae has the host species Albizzia sp., and Coriolopsis caperata from wood log. The newly reported Ganoderma wiiroense has been collected from the trees of Albizzia sp., (Table 1).
The species Fulvifomes fastuosus belongs to the family Hymenochaetaceae and reported to have medicinal properties43. The F. fastuosus has been reported in the trees of Xylocarpus granatum55. In this study, F. fastuosus were found in the host trees of Albizzia sp.
The genera Phellinus have wide host range. Globally Quercus sp. is the more susceptible host and, in India Mangifera sp. followed by Acacia, Artocarpus and Albizzia are the predominant host of Phellinus64,65. It was observed that Albizzia sp. is the host preferred by the genera Phellinus.
The genera Amylosporus was first reported in India among the Asian countries66 with bamboo as their host67. In this study, the Amylosporus sp. was found in the host Nerium sp. and Albizzia sp. Interestingly from this study, Albizzia sp. is found to be the host preferred by most of the macrofungal isolates. This might be due to the abundance of this species in the vicinity of the collected macrofungi.
To conclude, we have identified and report two new macrofungal species G. wiiroense and F. fulvifomes and molecular evidence for P. badius from India. It was observed that Albizzia sp., as the host preferred by most of the macrofungal isolates. Our data provide the existence of G. wiiroense in India; however, we were unable to trace of out the origin of how G. wiiroense might have cross boundaries. We can only speculate G. wiiroense already exists in India; because of the lack of intense mycological study prior, this is the first report on it. These data gains us insight on macrofugal diversity in India, which can be used for the prospection of macrofungi in biomedical and industrial applications.
Methodology
Sample Collection and culture of isolates
Fresh basidiomata of the wild mushrooms belonging to the division basidiomycota were collected from different locations in Dindigul (Ayyanar falls), Madurai (Lady Doak College Campus, Nagamalai, Pudhupatti), Coimbatore (Kovai Kutralam), Thenkasi and Tirunelveli, Tamilnadu (India) during 2013–2017 on rainy seasons i.e., November to January. The basidiomata were cleaned and aseptically transferred to the lab. After surface sterilization with 70% ethanol, small pieces from the contextual layer of basidiomata68 were transferred to sterile potato dextrose agar (PDA) medium supplemented with streptomycin. The plates were incubated at 37 °C for 5–7 days. The pure culture was obtained by continuous sub culturing and used for further analysis. The isolates were stored in PDA plates and slants. The basidiomata were then dehydrated with naphthalene balls for future studies.
The radial growth of the mycelium of all the isolates on the PDA medium was measured using a ruler. Five-millimetre mycelial plugs were removed from the growing edge of the 7-day-old pure culture and inoculated on to the centre of the 80 mm petriplates containing PDA. According to Tomkin69 and our observation, the growth is not constant in the early stage. The lag phase was shorter (1 day) in some strains and longer (5 days) in some strains. The radial/lateral expansion was measured after three days (i.e., 3rd day for strains with shorter lag phase and 7th day for strains with longer lag phase) in diameter (in mm), and the number of days taken to completely colonize 80 mm petridish was recorded. All the measurements were made in triplicates. The representative voucher specimens were deposited in the Department of Biotechnology, Lady Doak College, Madurai, Tamilnadu, India. Taxonomical identification of the isolates was carried out based on molecular identification methods.
After identification, the macromorphological characteristic features such as shape, color, hymenial surface of the basidiomata were studied according to published description70. Microscopical observations (hyphal system, presence/absence of setae and basidiospores) were carried out using brightfield microscope (Olympus system microscope model CX41). Slides were prepared using 5% KOH and cotton blue71.
Molecular characterization of the isolates
Genomic DNA Isolation, PCR amplification and sequencing
Genomic DNA of all the isolates were extracted as described by Moncalvo et al.35. 10 mg of mycelial biomass was homogenized with 3% SDS extraction buffer (3 g SDS, 50 mM Tris, 150 mM NaCl and 80 mM Na2EDTA) and incubated at 60 °C for 20–30 min. The 5.8S nuclear ribosomal RNA gene was amplified using ITS1 (CTTGGTCAT TTAGAGGAAGTAA) and ITS4 (CAGGAGACTTGTACACGGTCCAG) primers30. PCR amplification was carried out using the following condition: initial denaturation (95 °C, 2 min), denaturation (94 °C, 45 sec), annealing (50 °C, 45 sec), extension (72 °C, 1.30 min), final extension (72 °C, 5 min). The PCR products were purified and sequenced (Chromous Biotech Pvt. Ltd, Bangalore). The sequences were read bidirectionally for both strands of the entire ITS1, 5.8S rDNA and ITS2 region. The DNA sequence obtained from both the strands was edited and contig assembly was carried out using DNA Baser sequence assembly software (V.4.36.0). The assembled sequences were submitted to GenBank Database.
Phylogenetic analysis
Additional ITS sequences of Basidiomycetes were downloaded from GenBank to clarify the interspecies relationship. The phylogenetic tree was constructed by maximum likelihood (ML) analysis in MEGA 6 software72. The tree inference options were set as follows: Heuristic Method Nearest-Neighbor-Interchange (NNI) with the very strong branch swap filter with 1000 bootstrap replicates, gaps were treated as missing.
Acknowledgements
This study was funded by Department of Biotechnology – Bioinformatics Infrastructure Facility (reference 102/IFD/SAN/1125/2006-07) to R. Shenbagarathai. The author T. Mowna Sundari would like to acknowledge Dr. W. Isabel, PG and Research Department of Zoology, LDC, Madurai; Dr. V. Mohan, Forest Pathology Division, IFGTB, Coimbatore; Dr. K. Malarvizhi and Dr. Mathivanan, CAS in Botany, University of Madras for their encouragement and support. The help extended by G. Agasta and S. Dhyana is acknowledged. We thank Dr. James Premdoss Clement, JNCASR, Bangalore for proofreading the manuscript.
Author Contributions
T.M. conceived the study, designed and executed the wet lab experiment, designed the evolutionary study, produced figures, analyzed the data and prepared the manuscript. P.J. helped with the experiments. A.A.P.A. produced figures, analyzed the data, reviewed and helped with the manuscript. A.A.P.A. and R.S. made critical revisions and approved final version. All authors reviewed and approved of the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hawksworth DL. The magnitude of fungal diversity: the 1.5 million species estimate revisited** Paper presented at the Asian Mycological Congress 2000 (AMC 2000), incorporating the 2nd Asia-Pacific Mycological Congress on Biodiversity and Biotechnology, and held at the University of Hong Kong on 9–13 July 2000. Mycological research. 2001;105:1422–1432. doi: 10.1017/S0953756201004725. [DOI] [Google Scholar]
- 2.Hawksworth DL. The fungal dimension of biodiversity: magnitude, significance, and conservation. Mycological research. 1991;95:641–655. doi: 10.1016/S0953-7562(09)80810-1. [DOI] [Google Scholar]
- 3.Porras-Alfaro A, et al. Novel root fungal consortium associated with a dominant desert grass. Applied and environmental microbiology. 2008;74:2805–2813. doi: 10.1128/AEM.02769-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hawksworth, D., Kirk, P., Sutton, B. & Pegler, D. Ainsworth and Bisby’s Dictionary of the fungi. 8th edn, xii + 616 (CAB International, 1995).
- 5.Singh SP, Pande K, Upadhyay VP, Singh JS. Fungal communities associated with the decomposition of a common leaf litter (Quercus leucotrichophora A. Camus) along an elevational transect in the Central Himalaya. Biology and Fertility of Soils. 1990;9:245–251. doi: 10.1007/BF00336234. [DOI] [Google Scholar]
- 6.Watling R. Pulling the threads together: habitat diversity. Biodiversity & Conservation. 1997;6:753–763. doi: 10.1023/A:1018374404998. [DOI] [Google Scholar]
- 7.Lodge, D. J., Hawksworth, D. L. & Ritchie, B. J. In Biodiversity and Ecosystem Processes in Tropical Forests (eds Orians, G. H., Dirzo, R. & Cushman, J. H.) 69–100 (Springer Berlin Heidelberg, 1996).
- 8.Wilkins WH, Ellis EM, Harley JL. The ecology of the larger fungi: Constancy and frequency of fungal species in relation to certain vegetation communities, particularly Oak and Beech. Annals of Applied Biology. 1937;24:703–732. doi: 10.1111/j.1744-7348.1937.tb05051.x. [DOI] [Google Scholar]
- 9.Tóth BB, Barta Z. Ecological studies of ectomycorrhizal fungi: an analysis of survey methods. Fungal Diversity. 2010;45:3–19. doi: 10.1007/s13225-010-0052-2. [DOI] [Google Scholar]
- 10.Wiensczyk, A. M., Gamiet, S., Durall, D. M., Jones, M. D. & Simard, S. W. Ectomycorrhizae and forestry in British Columbia: A summary of current research and conservation strategies. Journal of Ecosystems and Management2 (2002).
- 11.Avis PG, Gaswick WC, Tonkovich GS, Leacock PR. Monitoring fungi in ecological restorations of coastal Indiana, USA. Restoration Ecology. 2017;25:92–100. doi: 10.1111/rec.12397. [DOI] [Google Scholar]
- 12.van Dijk H, Onguene NA, Kuyper TW. Knowledge and Utilization of Edible Mushrooms by Local Populations of the Rain Forest of South Cameroon. AMBIO: A Journal of the Human Environment. 2003;32:19–23. doi: 10.1579/0044-7447-32.1.19. [DOI] [PubMed] [Google Scholar]
- 13.Enow E, Kinge TR, Tabi EM, Thiobal N, Mih AM. Diversity and distribution of macrofungi (mushrooms) in the Mount Cameroon Region. Journal of Ecology and The Natural Environment. 2013;5:318–334. doi: 10.5897/JENE2013.0397. [DOI] [Google Scholar]
- 14.Ramsbottom, J. Mushrooms and toadstools: A study of the activities of fungi (Collins 1953).
- 15.Mizuno T. Bioactive biomolecules of mushrooms: Food function and medicinal effect of mushroom fungi. Food Reviews International. 1995;11:5–21. doi: 10.1080/87559129509541017. [DOI] [Google Scholar]
- 16.Wasser S. Medicinal mushrooms as a source of antitumor and immunomodulating polysaccharides. Applied microbiology and biotechnology. 2002;60:258–274. doi: 10.1007/s00253-002-1076-7. [DOI] [PubMed] [Google Scholar]
- 17.Wasser, S. P. & Weis, A. L. Medicinal properties of substances occurring in higher basidiomycetes mushrooms: current perspectives. International Journal of medicinal mushrooms1 (1999). [PubMed]
- 18.Hwang BS, Lee IK, Choi HJ, Yun BS. Anti-influenza activities of polyphenols from the medicinal mushroom Phellinus baumii. Bioorganic & medicinal chemistry letters. 2015;25:3256–3260. doi: 10.1016/j.bmcl.2015.05.081. [DOI] [PubMed] [Google Scholar]
- 19.Manoharachary, C. et al. Fungal biodiversity: distribution, conservation and prospecting of fungi from India. Current Science, 58–71 (2005).
- 20.Natarajan K. Mushroom flora of south India (except Kerala) Advances in Horticulture. 1995;13:387–397. [Google Scholar]
- 21.Gilbertson, R. L. & Ryvarden, L. North American Polypores: Megasporoporia-Wrightoporia (Fungiflora, 1987).
- 22.Schmit JP, Lodge DJ. Classical methods and modern analysis for studying fungal diversity. Mycology Series. 2005;23:193. doi: 10.1201/9781420027891.ch10. [DOI] [Google Scholar]
- 23.Lodge, D. J. et al. Terrestrial and lignicolous macrofungi (Elsevier Inc., 2004).
- 24.Genej GJ, Stchigel AM. Developments in fungal taxonomy. Clinical microbiology reviews. 1999;12:454–500. doi: 10.1128/cmr.12.3.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugiyama J. Relatedness, phylogeny, and evolution of the fungi. Mycoscience. 1998;39:487–511. doi: 10.1007/BF02460912. [DOI] [Google Scholar]
- 26.Bridge P, Hawksworth D. New horizons in the biosystematics of filamentous fungi. Genetic Engineer and Biotechnologist. 1990;10:9–12. [Google Scholar]
- 27.Bruns TD, White TJ. & W., T. J. Fungal Molecular Systematics. Annual Review of Ecology and Systematics. 1991;22:525–564. doi: 10.1146/annurev.es.22.110191.002521. [DOI] [Google Scholar]
- 28.Seifert KA. Progress towards DNA barcoding of fungi. Molecular Ecology Resources. 2009;9:83–89. doi: 10.1111/j.1755-0998.2009.02635.x. [DOI] [PubMed] [Google Scholar]
- 29.Hillis DM, Dixon MT. Ribosomal DNA: molecular evolution and phylogenetic inference. The Quarterly review of biology. 1991;66:411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- 30.White, T. J., Bruns, T., Lee, S. & Taylor, J. In PCR Protocols: A Guide to Methods and Applications (eds Innis, M. A., Gelfand, D. H., Shinsky, J. J. & White, T. J.) 315–322 (Academic Press, 1990).
- 31.Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Molecular ecology. 1993;2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 32.Schoch CL, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirk, P. M., Cannon, P. F., David, J. & Stalpers, J. A. Ainsworth and Bisby’s dictionary of the fungi. (CABI publishing, 2001).
- 34.Moncalvo JM, Wang HF, Hseu RS. Gene phylogeny of the Ganoderma lucidum complex based on ribosomal DNA sequences. Comparison with traditional taxonomic characters. Mycological Research. 1995;99:1489–1499. doi: 10.1016/S0953-7562(09)80798-3. [DOI] [Google Scholar]
- 35.Moncalvo JM, Wang HH, Hseu RS. Phylogenetic Relationships in Ganoderma Inferred from the Internal Transcribed Spacers and 25S Ribosomal DNA Sequences. Mycologia. 1995;87:223–238. doi: 10.2307/3760908. [DOI] [Google Scholar]
- 36.Miller RNG, et al. Isozyme analysis for characterization of Ganoderma strains from south-eastAsia 1. EPPO Bulletin. 1995;25:81–87. doi: 10.1111/j.1365-2338.1995.tb01441.x. [DOI] [Google Scholar]
- 37.Pan SM, Ye JS, Hseu RS. Purification and characterization of manganese superoxide dismutase from Ganoderma microsporum. Biochemistry and molecular biology international. 1997;42:1035–1043. doi: 10.1080/15216549700203491. [DOI] [PubMed] [Google Scholar]
- 38.Frealle E, et al. Manganese superoxide dismutase in pathogenic fungi: an issue with pathophysiological and phylogenetic involvements. FEMS immunology and medical microbiology. 2005;45:411–422. doi: 10.1016/j.femsim.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 39.Moncalvo, J. M. In Ganoderma. Diseases of perennial crops (eds Flood, J., Bridge, P. D. & Holderness, M.) Ch. 2, 23–45 (CABI, 2000).
- 40.Vancov T, Keen B. Amplification of soil fungal community DNA using the ITS86F and ITS4 primers. FEMS microbiology letters. 2009;296:91–96. doi: 10.1111/j.1574-6968.2009.01621.x. [DOI] [PubMed] [Google Scholar]
- 41.Iwen PC, Hinrichs SH, Rupp ME. Utilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. Medical mycology. 2002;40:87–109. doi: 10.1080/mmy.40.1.87.109. [DOI] [PubMed] [Google Scholar]
- 42.Crous PW, et al. Fungal Planet description sheets: 371–399. Persoonia. 2015;35:264–327. doi: 10.3767/003158515X690269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai Y-C. Hymenochaetaceae (Basidiomycota) in China. Fungal Diversity. 2010;45:131–343. doi: 10.1007/s13225-010-0066-9. [DOI] [Google Scholar]
- 44.Leelavathy, K. & Ganesh, P. Polypores of Kerala (Daya Publishing House, 2000).
- 45.Ganesh P, Leelavathy K. New records of Phellinus from India. Current Science. 1986;55:727–728. [Google Scholar]
- 46.Nagadesi P, Arya A. New records of lignicolous fungi deteriorating wood in India. Mycosphere. 2012;3:997–1004. doi: 10.5943/mycosphere/3/6/13. [DOI] [Google Scholar]
- 47.Arya, A. In Vistas in Palaeobotany and Plant Morphology: Evolutionary and Environmental Perspectives (eds Pant, D. D. & Srivastava, P. C.) 321–327 (UP Offset, 2004).
- 48.Singh, A. P., Kaur, G. & Dhingra, G. S. In 8th International Conference on Mushroom Biology and Mushroom Products. 83–91 (World Society for Mushroom Biology and Mushroom Products).
- 49.Murrill, W. Northern polypores. (The Author, 1914).
- 50.Dai, Y.-C. Phellinus sensu lato (Aphyllophorales, Hymenochaetaceae) in East Asia. 115 (Finnish Zoological and Botanical Publishing Board 1999).
- 51.Wagner T, Fischer M. Proceedings towards a natural classification of the worldwide taxa Phellinus s.l. and Inonotus s.l., and phylogenetic relationships of allied genera. Mycologia. 2002;94:998–1016. doi: 10.1080/15572536.2003.11833156. [DOI] [PubMed] [Google Scholar]
- 52.Larsson KH, et al. Hymenochaetales: a molecular phylogeny for the hymenochaetoid clade. Mycologia. 2006;98:926–936. doi: 10.1080/15572536.2006.11832622. [DOI] [PubMed] [Google Scholar]
- 53.Zhou L-W. Notes on the taxonomic positions of some Hymenochaetaceae (Basidiomycota) species with colored basidiospores. Phytotaxa. 2014;177:183–187. doi: 10.11646/phytotaxa.177.3.7. [DOI] [Google Scholar]
- 54.Bondartseva MA, Herrera S, Sandoval D, Cejas F. Taxonomical problems of the Cuban Hymenochaetaceous fungi. Mikol Fitopatol. 1992;26:3–14. [Google Scholar]
- 55.Sakayaroj J, et al. Molecular characterization of basidiomycetes associated with the decayed mangrove tree Xylocarpus granatum in Thailand. Fungal Diversity. 2012;56:145–156. doi: 10.1007/s13225-012-0195-4. [DOI] [Google Scholar]
- 56.Ediriweera, S., Wijesundera, R., Nanayakkara, C. & Weerasena, O. A new record of Fulvifomes fastuosus from Sri Lanka. Journal of the National Science Foundation of Sri Lanka42 (2014).
- 57.Lloyd, C. G. Mycological notes 1–75. Vol. 1–1364 (CG Lloyd, 1895–1925).
- 58.Sankaran KV, Bridge PD, Gokulapalan C. Ganoderma diseases of perennial crops in India – an overview. Mycopathologia. 2005;159:143–152. doi: 10.1007/s11046-004-4437-1. [DOI] [PubMed] [Google Scholar]
- 59.Butler, E. J. Some Disease of Palms. Vol. 1, 299–310 (Thacker, Spink & Company, 1906).
- 60.Butler E. Fomes lucidus (Leys) Fr., a suspected parasite. Indian Forester. 1909;35:514–518. [Google Scholar]
- 61.Snehalatharani A, Maheswarappa H, Devappa V, Malhotra S. Status of coconut basal stem rot disease in India–A review. Indian Journal of Agricultural Sciences. 2016;86:1519–1529. [Google Scholar]
- 62.Peries O. Ganoderma basal stem rot of coconut: a new record of the disease in Sri Lanka. Plant Disease Reporter. 1974;58:293–295. [Google Scholar]
- 63.Petch, T. & Bisby, G. R. The fungi of Ceylon Peradeniya Manual. 6 (1950).
- 64.Ranadive K, Jagtap N, Vaidya J. Host diversity of genus Phellinus from world. Elixir Appl. Botany. 2012;52:11402–11408. [Google Scholar]
- 65.Ranadive K, et al. Host Distribution of Phellinus from India. Indian Journal of Forestry. 2012;35:67–72. [Google Scholar]
- 66.Roy, A. & De, A. B. Polyporaceae Of India. (International Book Distributors, 1996).
- 67.Tarafder E, et al. Contribution to the Macromycetes of West Bengal, India: 13–17. Research Journal of Pharmacy and Technology. 2017;10:1123–1130. doi: 10.5958/0974-360X.2017.00203.7. [DOI] [Google Scholar]
- 68.Lodge, D. J., Ammirati, J. F., O’Dell, T. E. & Mueller, G. M. In Biodiversity of fungi: inventory and monitoring methods (eds Mueller, G. M., Bills, G. & Foster, M. S.) 128–158 (Elsevier Academic. San Diego, California, 2004).
- 69.Tomkins RG. Measuring growth: The petri dish method. Transactions of the British Mycological Society. 1932;17:150–153. doi: 10.1016/S0007-1536(32)80033-1. [DOI] [Google Scholar]
- 70.Kornerup, A. & Wanscher, J. H. Methuen Handbook of Colour. 3 edn, (E. Methuen, 1978).
- 71.Ryvarden, L. Genera of polypores: nomenclature and taxonomy. 1–373 (Lubrecht & Cramer Ltd, 1991).
- 72.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]