Abstract
Plants interact with diverse microbes including those that result in nutrient-acquiring symbioses. In order to balance the energy cost with the benefit gained, plants employ a systemic negative feedback loop to control the formation of these symbioses. This is particularly well-understood in nodulation, the symbiosis between legumes and nitrogen-fixing rhizobia, and is known as autoregulation of nodulation (AON). However, much less is understood about the autoregulation of the ancient arbuscular mycorrhizal symbioses that form between Glomeromycota fungi and the majority of land plants. Elegant physiological studies in legumes have indicated there is at least some overlap in the genes and signals that regulate these two symbioses but there are major gaps in our understanding. In this paper we examine the hypothesis that the autoregulation of mycorrhizae (AOM) pathway shares some elements with AON but that there are also some important differences. By reviewing the current knowledge of the AON pathway, we have identified important directions for future AOM studies. We also provide the first genetic evidence that CLV2 (an important element of the AON pathway) influences mycorrhizal development in a non-legume, tomato and review the interaction of the autoregulation pathway with plant hormones and nutrient status. Finally, we discuss whether autoregulation may play a role in the relationships plants form with other microbes.
Keywords: autoregulation, nodulation, arbuscular mycorrhizae, CLAVATA, CLE peptide, tomato
Introduction
Mycorrhizal symbioses between plants and fungi are widespread and ancient, with evidence from fossils and extant basal plants indicating that such interactions evolved during the colonization of land by plants between 450 and 475 mya (Field et al., 2015; Martin et al., 2017). The mycorrhizal fungi invade and extend the host plant’s root system, enabling enhanced nutrient uptake in exchange for fixed carbon. The most widespread are the arbuscular mycorrhizal (AM) associations that are characterized by the presence of a unique nutrient exchange unit, the fungal arbuscule. A significant amount of plant-derived carbon is invested, with estimates of between 4 and 20% of the carbon fixed by the plant transferred to the AM fungi (Bago et al., 2000; Douds et al., 2000). To prevent the energy cost to the plant outweighing the benefits of the interaction, plants might be expected to regulate AM colonization. Indeed, elegant physiological studies in flowering plants have revealed powerful systemic regulation of AM colonization. In split root studies, pre-colonization of one side of the root system can suppress subsequent colonization of the other side of the root system, providing evidence for a root–shoot feedback system termed autoregulation of mycorrhiza (AOM) (Vierheilig et al., 2000a, 2008; Meixner et al., 2005). Although there is no direct evidence that such control mechanisms also occur in more basal plant lineages, it is fair to assume that autoregulation would be an important element in the evolution of mutually beneficial plant–mycorrhizal interactions to prevent a potentially parasitic relationship developing.
Significant progress has been made in the past few decades in our understanding of the plant genes and signals that regulate AM symbioses, assisted greatly by the cross-over with the more recently evolved nodulation symbioses that occur between nitrogen-fixing rhizobial bacteria and (almost exclusively) legumes. In particular, the identification of a common symbiotic signaling pathway, required for the formation of both nodules and AM, has revealed that elements of the ancient AM pathway were co-opted into the more recently evolved nodulation pathway (Delaux et al., 2013; Martin et al., 2017). In particular, the common symbiotic pathway includes genes essential for initial communication between the plant host and rhizobia or AM fungi. However, a missing element in these comparisons has been the autoregulation of nodulation (AON) pathway. Like AM, nodulation is under powerful systemic control and the identity of AON signals and transduction pathways are now becoming clear (for review see Reid et al., 2011b; Soyano and Kawaguchi, 2014). Studies in legumes suggest some key cross-overs in the autoregulation pathways, as plant mutants disrupted in the AON pathway display not only hypernodulation but also hypermycorrhizal colonization (Morandi et al., 2000; Shrihari et al., 2000) and split root studies indicate nodulation can systemically suppress AM and vice versa (Catford et al., 2003). In this article, we examine our current understanding of AOM and begin to extend this beyond legumes by providing evidence of a role for the key AON gene in legumes, CLAVATA2 (CLV2), in the AOM pathway of tomato. We also consider the role of plant hormones in autoregulation and examine the potential for the autoregulation of other beneficial plant–microbe interactions.
Genes and Signals of the Autoregulation Pathway(S)
The systemic regulation of AM colonization in split root studies has been observed in both legumes and non-legumes (Vierheilig et al., 2000a; Meixner et al., 2007). This negative regulation appears to require a certain threshold of the amount of root colonized by AM (Vierheilig, 2004). AOM does not appear to be due simply to the strength of the carbon sink (Vierheilig et al., 2000b, 2008), but is rather regulated by a more specific pathway. Nodulation can systemically suppress AM and vice versa and even the application of Nod factors, the rhizobial-derived signaling molecules, can suppress AM (Catford et al., 2003), suggesting a clear overlap between the AON and AOM pathways. Similar conclusions were drawn from studies using a dual inoculation system in which both rhizobia and AM fungi were applied to the one root system (Sakamoto et al., 2013). This has also been explored in the non-legume barley, where it was found that inoculation with Rhizobium sp. could systemically inhibit AM formation. However, this inhibitory effect did not rely on Nod-factor production but was linked instead to the type 3 effector proteins of this rhizobial strain (Khaosaad et al., 2010).
A member of the Leucine-rich repeat (LRR) receptor kinase family that shares similarities with the CLAVATA1 (CLV1) protein (outlined below) has a central role in the AON in legumes (Figure 1). This CLV1-like gene is known as NODULE AUTOREGULATION RECEPTOR KINASE (NARK) in soybean, HYPERNODULATION ABERRANT ROOT FORMATION 1 (HAR1) in Lotus japonicus, SUPER NUMERIC NODULES (SUNN) in Medicago truncatula and SYM29 in pea, and mutations in these genes result in hypernodulation (reviewed by Reid et al., 2011b). The CLV1-like protein appears to function as a shoot receptor for root-derived CLAVATA/ESR-related (CLE) peptides. In the root, events associated with nodulation generate specific rhizobial induced CLE peptides (Rh-CLE Figure 1) which in some cases appear to be arabinosylated via action of the enzyme ROOT DETERMINED NODULATION1 (RDN1) (e.g., MtCLE12, GmRIC1a, and GmRIC1b) (Schnabel et al., 2011; Okamoto et al., 2013; Kassaw et al., 2017; Hastwell et al., 2018; Imin et al., 2018). The CLE peptides are then translocated to the shoot where they are perceived by a receptor complex that includes the CLV1-like protein (e.g., Okamoto et al., 2013). The perception of the root-derived signal(s) in the plant shoot generates a shoot to root signal that inhibits further nodulation. The shoot to root inhibitor is predicted to be a small molecular weight heat-stable molecule (Lin et al., 2010) that has been suggested to be cytokinin (Sasaki et al., 2014). Other elements of the AON pathway include the shoot acting CLV2 (Krusell et al., 2011), CORYNE (CRN) (Crook et al., 2016), and KLAVIER (KLV) (Miyazawa et al., 2010), all three of which encode LRR receptors that may also play a role in CLE perception, and TOO MUCH LOVE (TML), a root-acting F-Box protein that appears to act downstream of the shoot to root signal (Takahara et al., 2013).
FIGURE 1.
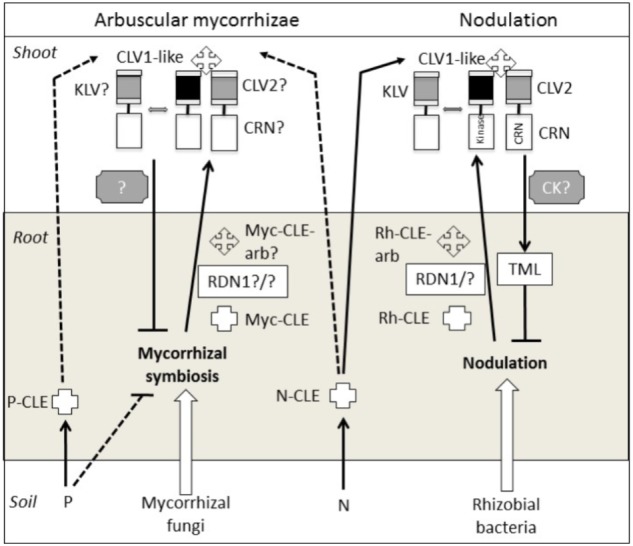
Proteins and signals that act in the shoot and/or root to autoregulate nodulation and mycorrhizal symbioses. Flat-ended lines indicate a negative influence, while arrows indicate a positive influence. Question marks and dotted lines indicate untested elements. CK, cytokinin; N, nitrogen; P, phosphorous; Rh, rhizobial; Myc, mycorrhizal; arb, arabinosylated.
In addition to its role in nodulation, the CLV1-like protein is also essential in the AOM pathway since clv1-like mutants across legume species (sym29, sunn, nark, and har1) also display hypermycorrhizal colonization (Morandi et al., 2000; Solaiman et al., 2000; Sakamoto and Nohara, 2009). It is important to note that the relatively small increase in AM in these mutant plants compared to wild type (WT) (between 20 and 50%) contrasts with the large (many fold) increase in nodule numbers of these lines compared with WT lines. Some have speculated that this relatively small increase may be due to the ability of the AM fungus to spread laterally in the root, meaning it is sometimes difficult to distinguish the effect of clv1-like mutations on subsequent AM infection (Schaarschmidt et al., 2013). One interpretation is that the AOM pathway may inhibit new AM infections at the epidermis but may not limit the spread of AM between cortical cells, although this has not been tested empirically. Such a hypothesis is consistent with the activation of AON during early stages of rhizobial infection (Li et al., 2009). Another way to examine the role of CLV1-like genes in AOM has been through the use of split root studies with mutant plants, which have shown that the CLV1-like gene in soybean (NARK) influences the suppression of subsequent mycorrhizal colonization (Meixner et al., 2005, 2007; Schaarschmidt et al., 2013). A role for NARK in the shoot control of AM was also observed in reciprocal grafts between the soybean nark mutant En6500 and WT plants (Sakamoto and Nohara, 2009) but not in a split root system with the same En6500 mutant (Meixner et al., 2007). Studies of orthologous mutants in other legumes species (e.g., har1, sym29) may be useful to clarify this inconsistency.
Apart from the requirement for the CLV1-like protein in AOM it is not yet clear if other AON genes encoding proteins that act in the root (RDN1, TML), shoot (CLV2, KLV, CRN) or as mobile signals (CLE) are also employed by the AOM pathway (Figure 1). One study examining the effect of a pea clv2 mutant (sym28) found only a small but not significant increase in AM colonization compared with WT, although grafting and split root studies that would reveal if CLV2 plays a role in the systemic regulation of AM were not attempted (Morandi et al., 2000). This and other mutants disrupted in root and shoot acting elements of the AON pathway outlined above are available in a range of legumes, but to date their AM phenotypes and possible role in systemic AOM regulation has not been examined. Recent work in the legume Lotus japonicas indicated that mycorrhizal colonization and phosphate starvation generates CLE peptides distinct from those induced by nodulation or nitrogen (Funayama-Noguchi et al., 2011; Handa et al., 2015), but the function of these CLE peptides has yet to be tested. In an attempt to understand downstream elements of the AOM pathway, Schaarschmidt et al. (2013) analyzed the soybean transcriptome in split root mycorrhizal studies using WT and nark lines. Two putative CCAAT-binding transcription factor genes were identified, GmNF-YA1a and GmNF-YA1b, that are down-regulated by AM in a NARK-dependent manner. Hairy root RNAi lines with reduced expression of GmNF-YA1a/b displayed reduced AM colonization and this occurred in both WT and nark backgrounds, consistent with GmNF-YA1a/b acting downstream of NARK to suppress AM.
The evolutionary origin of the AON/AOM genes is still emerging but is informed by the CLAVATA-WUSCHEL (CLV-WUS) shoot meristem pathway. This pathway is best understood in Arabidopsis and involves several LRR receptors that act locally in the shoot, including CLV1, CLV2 and CRN, to perceive a CLE peptide, CLV3, which in turn activates a feedback loop to maintain a defined stem cell population in the shoot apical meristem (Hazak and Hardtke, 2016). Arabidopsis lines disrupted in these genes displayed altered shoot meristem formation. In pea and Lotus japonicas, mutant studies have revealed CLV2 plays a dual role, acting in both shoot development and AON, as clv2 mutants display hypernodulation and shoot fasciation (Krusell et al., 2011). In contrast, there appear to be specific CLE genes that act in the AON pathway (Mortier et al., 2010). Similarly, disruption of CLV1-like genes closely related to CLV1 in pea, Lotus, Medicago and soybean (SYM29, HAR1, SUNN, and NARK, respectively) result in hypernodulation but not shoot fasciation (reviewed by Reid et al., 2011b). A possible explanation in soybean for divergence between NARK and CLV1, is that CLV1 appears to have undergone a duplication, resulting in NARK and CLV1A (Yamamoto et al., 2000) but this does not appear to be the case in the other legumes examined (see Schnabel et al., 2005). CLV1A is more closely related to AtCLV1 and recent studies have shown it influences shoot architecture but not nodulation (Mirzaei et al., 2017). Although many phylogenetic studies have examined the CLV1, CLV2 and CLE gene families, these studies have been almost exclusively limited to angiosperms (e.g., Sun and Wang, 2011; Zan et al., 2013; Wei et al., 2015; Xu et al., 2015; Hastwell et al., 2017), preventing a more comprehensive understanding of the evolutionary history and possible functional diversification of these genes. One recent transcriptomic study failed to find evidence for the CLAVATA-WUSCHEL (CLV-WUS) pathway in the moss Physcomitrella and liverwort Marchantia (Frank and Scanlon, 2015). However, a more comprehensive examination of these gene families in basal plant lineages and in mycorrhizal vs. non-mycorrhizal species (see approach of Favre et al., 2014; Delaux et al., 2015) might provide an insight into their possible role in the AM program.
Role of Clv2 in Aom of the Non-Legume Tomato
To fully understand the genes, signals and evolutionary history of AOM, we must go beyond legumes. Indeed, a non-legume system removes the possible complication of cross-talk between the AON and AOM pathways to allow us to identify AOM components. Therefore, we employed the tomato clv2-2 line, a CRISPR-Cas9 knock out line that targets the CLV2 gene (Xu et al., 2015). As outlined above, this gene is essential for AON in legumes and also acts to control shoot apical meristem formation in legumes (Krusell et al., 2011) and non-legumes, including clv2 tomato lines that display a weakly fasciated shoot and an increase in the number of floral organs (Xu et al., 2015).
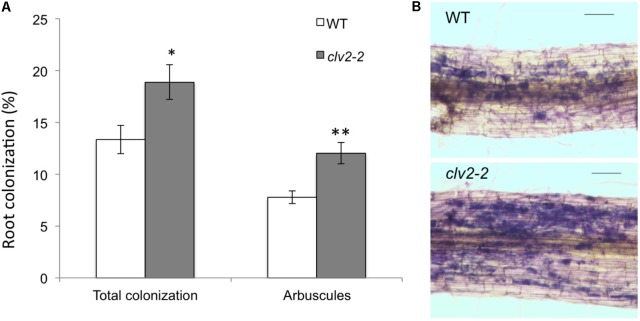
We tested if the CLV2 gene plays a role in AM development by examining the AM phenotype of the tomato clv2-2 line. Compared with WT, clv2 plants displayed a significant increase in AM colonization, including arbuscule frequency, compared with WT plants (Figure 2A). Although more frequent, the mycorrhizal structures observed in clv2 mutants including arbuscules were similar in appearance to WT (Figure 2B). Therefore, we provide the first evidence that CLV2 in tomato, known to be important for AON in legumes, also acts as a negative regulator of AM. This provides the first genetic evidence for the AOM pathway in non-legumes.
FIGURE 2.
Mycorrhizal colonization in tomato wild type (WT, M82) and clv2-2 lines. (A) Percentage of the root colonized by all fungal structures and arbuscules. n = 12, values are means + SE t-tests were performed for each parameter, ∗P < 0.05, ∗∗P < 0.01. (B) Photograph of a typical length of colonized root (scale bar = 100 μm) after staining fungal structures (blue). Tomato seeds were germinated in potting mix for 2 weeks and 12 equal sized seedlings of each genotype were selected and transplanted to 2 L pots. The pots were premixed with vermiculite and gravel (1:1) plus mycorrhizal inoculum (1/5 volume of corn roots colonized with Rhizophagus irregularis and associated potting medium) and topped with vermiculite. The seedlings were grown in a glasshouse with the following condition: 18 h photoperiod, 25°C day/20°C night. Plants were nutriented three times a week with modified Long Ashton solution (5 mM N and 0.5 mM P) (Hewitt, 1966). Plants were harvested 6 weeks after transplanting. The root was cut into 1 to 1.5 cm segments and stained using the ink and vinegar method (Vierheilig et al., 1998). 25 root segments were selected per plant and mycorrhizal colonization scored using the intersect scoring method (McGonigle et al., 1990). Blind labeling was used to avoid any potential bias during the scoring process.
Role of Plant Hormones in Aom
Autoregulation of mycorrhizae and AON are regulated by systemic signals and in addition to mobile CLE peptides, a range of studies have examined the role of plant hormones in AON and in some cases AOM. Double mutant studies in pea indicate gibberellins, brassinosteroids, and strigolactones are not required for the supernodulation phenotype and thus do not appear to act downstream of AON elements, CLV1-like, CLV2 or RDN1 (Ferguson et al., 2011; Foo et al., 2014b). In contrast, transcriptional studies showed that either jasmonic acid (JA) biosynthesis genes or JA regulated genes were systemically regulated by rhizobial colonization, and this was mediated by GmNARK in soybean. These results suggest the AON pathway influences JA signaling (Kinkema and Gresshoff, 2008). Several studies have also examined a role for auxin in autoregulation. In split root studies in soybean, significant auxin accumulation was observed in AM colonized roots but not uncolonized roots. However, this increase was not as great in the roots of a nark mutant (Meixner et al., 2005). This contrasts with nodulation studies with the orthologous mutant in Medicago, sunn, that suggest SUNN may be required to suppress auxin accumulation in the root following rhizobial challenge (van Noorden et al., 2006). In WT Medicago, challenge with rhizobia leads to the downregulation of auxin transport from the shoot to the root. In contrast, sunn mutants displayed elevated auxin levels in the infection zone of the root following inoculation with rhizobia. These studies suggest the AON pathway may modulate shoot to root auxin transport but this has yet to be investigated directly in AOM.
Cytokinin is an interesting case as it has been suggested to be a candidate for the shoot-derived inhibitor in AON, based on several lines of evidence from plants with altered cytokinin or CLE peptide biosynthesis, and measurement of cytokinin levels and transport (Sasaki et al., 2014). However, a key finding of this paper, that the LORE ipt3-2 mutant allele, which is disrupted in a key cytokinin biosynthesis gene, has increased nodulation could not be repeated in an independent study (Reid et al., 2017). In addition, another study indicated cytokinin may promote nodulation via the AON pathway. In Medicago, application of cytokinin directly to roots could induce the expression of the MtCLE13 gene (Mortier et al., 2012) believed to encode the root to shoot AON signal. Given the ability for nodulation to suppress AM and vice versa outlined above, it is likely that the shoot-derived inhibitor is a common signal between the AON and AOM pathways. Studies with grafts between legume species certainly suggest that the shoot-derived inhibitor of nodulation is conserved across species (e.g., Lohar and VandenBosch, 2005; Ferguson et al., 2014; Foo et al., 2014a). However, unlike its clear role in nodulation there is less evidence to suggest cytokinin has an influence on AM. For example, the cytokinin receptor mutant in M. truncatula, cre1, did not display any alteration in AM development (Laffont et al., 2015). However, pharmacological studies suggest that cytokinin may promote AM development in pea (F. Guinel, personal communication). Clearly, questions remain around the role of CK in AON, in particular as the shoot-derived inhibitor, and studies directly testing its endogenous role in AOM are required.
Do Nutrient Status and Other Beneficial Plant–Microbe Symbioses Interact With the Autoregulation Pathway(S)?
Forming symbioses with rhizobial or AM partners may only be beneficial to the plant under conditions of low mineral nutrient availability. In particular, legumes severely reduce nodulation when roots are exposed to elevated nitrogen levels and there are important roles for elements of the AON pathway in this nitrate-response (see Reid et al., 2011b). For example, the clv1-like mutants across legumes display a reduced ability to suppress nodulation in response to nitrate (Schnabel et al., 2005; Lim et al., 2011; Okamoto and Kawaguchi, 2015). This reduced response to nitrogen is also seen in klv and rdn1 mutants (e.g., Jacobsen and Feenstra, 1984; Oka-Kira et al., 2005). However, this has not been comprehensively examined for all AON mutants across species. In addition, nitrate treatment induces the expression of specific CLE peptides, which in some cases are the same as those that are induced by rhizobia (Okamoto et al., 2009; Reid et al., 2011a). Whether the AOM pathway plays any role in nitrogen-regulation of AM symbioses has not been explored and, unlike the clearly suppressive effects on nodulation, it is not even clear if nitrogen is a promoter or inhibitor of AM (Correa et al., 2015). In contrast, phosphate has a strong suppressive influence on AM and this influence is systemic and does not require strigolactones (Breuillin et al., 2010; Foo et al., 2013b). Although phosphate induces expression of specific CLE peptides (Funayama-Noguchi et al., 2011), there is no direct evidence that the AOM pathway mediates the phosphate response of AM. Indeed phosphate regulation of AM is maintained in soybean and pea mutants disrupted in the clv1-like mutants, nark and sym29 (Wyss et al., 1990; Foo et al., 2013a). However, phosphate positively regulates nodule number and studies in pea have found this is disrupted in the sym29 mutant (Foo et al., 2013a), suggesting a cross-over in the AON and phosphate response pathways. MicroRNAs of the 399 family have also been shown to play a role in the phosphate response and some were shown to be induced by phosphate-starvation in AM colonized Medicago (Branscheid et al., 2010). However, as overexpression of these miR399 genes in tobacco did not influence AM colonization, no clear role for these mobile microRNAs were established in the AM phosphate response (Branscheid et al., 2010).
In addition to nodulation and AM, plants form a range of other beneficial interactions with soil microbes. These include actinorhizal symbioses between members of the fabid clade and Frankia bacteria, ectomycorrhizal symbioses and interactions with fungal and bacterial endophytes. Systemic regulation of colonization has been demonstrated for actinorhizal associations (Wall and Huss-Danell, 1997), and for the interaction between Arabidopsis and the fungal enodphyte Piriformospora indica (Pedrotti et al., 2013). Indeed, it has been shown in some split root studies that plants infected with endophytes have a decreased level of AM colonization, although this was not found in all studies (Müller, 2003; Omacini et al., 2006; Mack and Rudgers, 2008). Phylogenetic studies have suggested that the common symbiotic pathway is conserved in AM, rhizobial and actinorhizal associations, although the role of these genes in ectomycorrhizae and endophyte relationships is not known (Martin et al., 2017). Arabidopsis is a particularly interesting case as it appears to have lost the majority of the common symbiotic pathway (Delaux et al., 2014), consistent with the lack of AM colonization and suggesting that this pathway is not important for hosting fungal endophtyes. However, as these studies did not include AON/AOM genes the evolutionary origin of these pathways and their potential role across species is still not clear.
Conclusion and Future Perspectives
The similarities between the AOM and AON pathways and their shared genetic components is consistent with the AON pathway evolving at least in part from a pre-existing AOM pathway in early land plants. However, a lack of phylogenetic, genetic and physiological studies in non-legumes, including basal land plants, has hampered our understanding of the origin and diversification of the autoregulation pathway. In this paper, we show that in the non-legume tomato, the CLV2 gene suppresses AM development, providing the first genetic evidence for an AOM gene in a non-legume. As found in legumes, this gene also plays a role in shoot apical meristem maintenance. However, the precise delineation in function of other AON/AOM elements such as the CLV1 and CLE genes in shoot apical meristem maintenance is still not clear. Furthermore, it is likely that multiple systemic pathways regulate symbioses (Kassaw et al., 2015). For example, novel peptides and accompanying perception pathways with roles in nodulation and root development are now emerging (e.g., CEP1 and CRA2, Mohd-Radzman et al., 2016). Future studies could more systematically examine the role of AON genes and peptide signals in AM development and take a phylogenetic approach to examine the evolutionary origin of symbiotic autoregulation.
Author Contributions
EF conceived the project. CW carried out experiments. CW, JR, and EF wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor Zachary Lippman (Cold Spring Harbor Laboratory) for the kind gift of clv2 tomato seed, Shelley Urquhart and Michelle Lang for assistance with plant husbandry and the Australian Research Council for financial support to EF and JR (FF140100770 and DP140101709).
References
- Bago B., Pfeffer P. E., Shachar-Hill Y. (2000). Carbon metabolism and transport in arbuscular mycorrhizas. Plant Physiol. 124 949 10.1104/pp.124.3.949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branscheid A., Sieh D., Pant B. D., May P., Devers E. A., Elkrog A., et al. (2010). Expression pattern suggests a role of MiR399 in the regulation of the cellular response to local Pi increase during arbuscular mycorrhizal symbiosis. Mol. Plant Microbe Interact. 23 915–926. 10.1094/MPMI-23-7-0915 [DOI] [PubMed] [Google Scholar]
- Breuillin F., Schramm J., Hajirezaei M., Ahkami A., Favre P., Druege U., et al. (2010). Phosphate systemically inhibits development of arbuscular mycorrhiza in Petunia hybrida and represses genes involved in mycorrhizal functioning. Plant J. 64 1002–1017. 10.1111/j.1365-313X.2010.04385.x [DOI] [PubMed] [Google Scholar]
- Catford J. G., Staehelin C., Lerat S., Piché Y., Vierheilig H. (2003). Suppression of arbuscular mycorrhizal colonization and nodulation in split-root systems of alfalfa after pre-inoculation and treatment with Nod factors. J. Exp. Bot. 54 1481–1487. 10.1093/jxb/erg156 [DOI] [PubMed] [Google Scholar]
- Correa A., Cruz C., Ferrol N. (2015). Nitrogen and carbon/nitrogen dynamics in arbuscular mycorrhiza: the great unknown. Mycorrhiza 25 499–515. 10.1007/s00572-015-0627-6 [DOI] [PubMed] [Google Scholar]
- Crook A. D., Schnabel E. L., Frugoli J. A. (2016). The systemic nodule number regulation kinase SUNN in Medicago truncatula interacts with MtCLV2 and MtCRN. Plant J. 88 108–119. 10.1111/tpj.13234 [DOI] [PubMed] [Google Scholar]
- Delaux P.-M., Radhakrishnan G. V., Jayaraman D., Cheema J., Malbreil M., Volkening J. D., et al. (2015). Algal ancestor of land plants was preadapted for symbiosis. Proc. Natl. Acad. Sci. U.S.A. 112 13390–13395. 10.1073/pnas.1515426112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaux P.-M., Séjalon-Delmas N., Bécard G., Ané J.-M. (2013). Evolution of the plant–microbe symbiotic ‘toolkit’. Trends Plant Sci. 18 298–304. 10.1016/j.tplants.2013.01.008 [DOI] [PubMed] [Google Scholar]
- Delaux P. M., Varala K., Edger P. P., Coruzzi G. M., Pires J. C., Ane J. M. (2014). Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLoS Genet. 10:e1004487. 10.1371/journal.pgen.1004487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douds D. D., Pfeffer P. E., Shachar-Hill Y. (2000). “Carbon partitioning, cost, and metabolism of arbuscular mycorrhizas,” in Arbuscular Mycorrhizas: Physiology and Function, eds Kapulnik Y., Douds D. D. (Dordrecht: Springer; ), 107–129. 10.1007/978-94-017-0776-3_6 [DOI] [Google Scholar]
- Favre P., Bapaume L., Bossolini E., Delorenzi M., Falquet L., Reinhardt D. (2014). A novel bioinformatics pipeline to discover genes related to arbuscular mycorrhizal symbiosis based on their evolutionary conservation pattern among higher plants. BMC Plant Biol. 14:333. 10.1186/s12870-014-0333-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B. J., Foo E., Ross J. J., Reid J. B. (2011). Relationship between gibberellin, ethylene and nodulation in Pisum sativum. New Phytol. 189 829–842. 10.1111/j.1469-8137.2010.03542.x [DOI] [PubMed] [Google Scholar]
- Ferguson B. J., Li D., Hastwell A. H., Reid D. E., Li Y., Jackson S. A., et al. (2014). The soybean (Glycine max) nodulation-suppressive CLE peptide, GmRIC1, functions interspecifically in common white bean (Phaseolus vulgaris), but not in a supernodulating line mutated in the receptor PvNARK. Plant Biotechnol. J. 12 1085–1097. 10.1111/pbi.12216 [DOI] [PubMed] [Google Scholar]
- Field K. J., Pressel S., Duckett J. G., Rimington W. R., Bidartondo M. I. (2015). Symbiotic options for the conquest of land. Trends Ecol. Evol. 30 477–486. 10.1016/j.tree.2015.05.007 [DOI] [PubMed] [Google Scholar]
- Foo E., Ferguson B. J., Reid J. B. (2014a). Common and divergent roles of plant hormones in nodulation and arbuscular mycorrhizal symbioses. Plant Signal. Behav. 9 1037–1045. 10.4161/psb.29593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E., Ferguson B. J., Reid J. B. (2014b). The potential roles of strigolactones and brassinosteroids in the autoregulation of nodulation pathway. Ann. Bot. 113 1037–1045. 10.1093/aob/mcu030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E., Yoneyama K., Hugill C., Quittenden L. J., Reid J. B. (2013a). Strigolactones: internal and external signals in plant symbioses? Plant Signal. Behav. 8:e23168. 10.4161/psb.23168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E., Yoneyama K., Hugill C. J., Quittenden L. J., Reid J. B. (2013b). Strigolactones and the regulation of pea symbioses in response to nitrate and phosphate deficiency. Mol. Plant. 6 76–87. 10.1093/mp/sss115 [DOI] [PubMed] [Google Scholar]
- Frank M. H., Scanlon M. J. (2015). Transcriptomic evidence for the evolution of shoot meristem function in sporophyte-dominant land plants through concerted selection of ancestral gametophytic and sporophytic genetic programs. Mol. Biol. Evol. 32 355–367. 10.1093/molbev/msu303 [DOI] [PubMed] [Google Scholar]
- Funayama-Noguchi S., Noguchi K., Yoshida C., Kawaguchi M. (2011). Two CLE genes are induced by phosphate in roots of Lotus japonicus. J. Plant Res. 124 155–163. 10.1007/s10265-010-0342-5 [DOI] [PubMed] [Google Scholar]
- Handa Y., Nishide H., Takeda N., Suzuki Y., Kawaguchi M., Saito K. (2015). RNA-seq transcriptional profiling of an arbuscular mycorrhiza provides insights into regulated and coordinated gene expression in Lotus japonicus and Rhizophagus irregularis. Plant Cell Physiol. 56 1490–1511. 10.1093/pcp/pcv071 [DOI] [PubMed] [Google Scholar]
- Hastwell A. H., Corcilius L., Williams J., Gresshoff P. M., Payne R. J., Ferguson B. J. (2018). Triarabinosylation is required for nodulation-suppressive CLE peptides to systemically inhibit nodulation in Pisum sativum. Plant Cell Environ. 10.1111/pce.13325 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Hastwell A. H., De Bang T. C., Gresshoff P. M., Ferguson B. J. (2017). CLE peptide-encoding gene families in Medicago truncatula and Lotus japonicus, compared with those of soybean, common bean and Arabidopsis. Sci. Rep. 7:9384. 10.1038/s41598-017-09296-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazak O., Hardtke C. S. (2016). CLAVATA 1-type receptors in plant development. J. Exp. Bot. 67 4827–4833. 10.1093/jxb/erw247 [DOI] [PubMed] [Google Scholar]
- Hewitt E. J. (1966). Sand and Water Culture Methods Used in the Study of Plant Nutrition. Wallingford: Centre for Agriculture and Bioscience International. [Google Scholar]
- Imin N., Patel N., Corcilius L., Payne R. J., Djordjevic M. A. (2018). CLE peptide tri-arabinosylation and peptide domain sequence composition are essential for SUNN-dependent autoregulation of nodulation in Medicago truncatula. New Phytol. 218 73–80. 10.1111/nph.15019 [DOI] [PubMed] [Google Scholar]
- Jacobsen E., Feenstra W. J. (1984). A new pea mutant with efficient nodulation in the presence of nitrate. Plant Sci. Lett. 33 337–344. 10.1016/0304-4211(84)90025-7 [DOI] [Google Scholar]
- Kassaw T., Bridges W., Jr., Frugoli J. (2015). Multiple autoregulation of nodulation (AON) signals identified through split root analysis of Medicago truncatula sunn and rdn1 Mutants. Plants 4 209–224. 10.3390/plants4020209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassaw T., Nowak S., Schnabel E., Frugoli J. A. (2017). ROOT DETERMINED NODULATION1 is required for M. truncatula CLE12, but not CLE13 peptide signaling through the SUNN receptor kinase. Plant Physiol. 174 2445–2456. 10.1104/pp.17.00278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaosaad T., Staehelin C., Steinkellner S., Hage-Ahmed K., Ocampo J. A., Garcia-Garrido J. M., et al. (2010). The Rhizobium sp. strain NGR234 systemically suppresses arbuscular mycorrhizal root colonization in a split-root system of barley (Hordeum vulgare). Physiol. Plant. 140 238–245. 10.1111/j.1399-3054.2010.01396.x [DOI] [PubMed] [Google Scholar]
- Kinkema M., Gresshoff P. M. (2008). Investigation of downstream signals of the soybean autoregulation of nodulation receptor kinase GmNARK. Mol. Plant Microbe Interact. 21 1337–1348. 10.1094/MPMI-21-10-1337 [DOI] [PubMed] [Google Scholar]
- Krusell L., Sato N., Fukuhara I., Koch B. E. V., Grossmann C., Okamoto S., et al. (2011). The Clavata2 genes of pea and Lotus japonicus affect autoregulation of nodulation. Plant J. 65 861–871. 10.1111/j.1365-313X.2010.04474.x [DOI] [PubMed] [Google Scholar]
- Laffont C., Rey T., André O., Novero M., Kazmierczak T., Debellé F., et al. (2015). The CRE1 cytokinin pathway is differentially recruited depending on Medicago truncatula root environments and negatively regulates resistance to a pathogen. PLoS One 10:e0116819. 10.1371/journal.pone.0116819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Kinkema M., Gresshoff P. M. (2009). Autoregulation of nodulation (AON) in Pisum sativum (pea) involves signalling events associated with both nodule primordia development and nitrogen fixation. J. Plant Physiol. 166 955–967. 10.1016/j.jplph.2009.03.004 [DOI] [PubMed] [Google Scholar]
- Lim C. W., Lee Y. W., Hwang C. H. (2011). Soybean nodule-enhanced CLE peptides in roots act as signals in GmNARK-mediated nodulation suppression. Plant Cell Physiol. 52 1613–1627. 10.1093/pcp/pcr091 [DOI] [PubMed] [Google Scholar]
- Lin Y. H., Ferguson B. J., Kereszt A., Gresshoff P. M. (2010). Suppression of hypernodulation in soybean by a leaf-extracted, NARK- and Nod factor-dependent, low molecular mass fraction. New Phytol. 185 1074–1086. 10.1111/j.1469-8137.2009.03163.x [DOI] [PubMed] [Google Scholar]
- Lohar D. P., VandenBosch K. A. (2005). Grafting between model legumes demonstrates roles for roots and shoots in determining nodule type and host/rhizobia specificity. J. Exp. Bot. 56 1643–1650. 10.1093/jxb/eri160 [DOI] [PubMed] [Google Scholar]
- Mack K. M. L., Rudgers J. A. (2008). Balancing multiple mutualists: asymmetric interactions among plants, arbuscular mycorrhizal fungi, and fungal endophytes. OIKOS 117 310–320. 10.1111/j.2007.0030-1299.15973.x [DOI] [Google Scholar]
- Martin F. M., Uroz S., Barker D. G. (2017). Ancestral alliances: plant mutualistic symbioses with fungi and bacteria. Science 356:eaad4501. 10.1126/science.aad4501 [DOI] [PubMed] [Google Scholar]
- McGonigle T. P., Miller M. H., Evans D. G., Fairchild G. L., Swan J. A. (1990). A new method which gives an objective measure of colonization of roots by vesicular—arbuscular mycorrhizal fungi. New Phytol. 115 495–501. 10.1111/j.1469-8137.1990.tb00476.x [DOI] [PubMed] [Google Scholar]
- Meixner C., Ludwig-Muller J., Miersch O., Gresshoff P., Staehelin C., Vierheilig H. (2005). Lack of mycorrhizal autoregulation and phytohormonal changes in the supernodulating soybean mutant nts1007. Planta 222 709–715. 10.1007/s00425-005-0003-4 [DOI] [PubMed] [Google Scholar]
- Meixner C., Vegvari G., Ludwig-Müller J., Gagnon H., Steinkellner S., Staehelin C., et al. (2007). Two defined alleles of the LRR receptor kinase GmNARK in supernodulating soybean govern differing autoregulation of mycorrhization. Physiol. Plant. 130 261–270. 10.1111/j.1399-3054.2007.00903.x [DOI] [Google Scholar]
- Mirzaei S., Batley J., El-Mellouki T., Liu S., Meksem K., Ferguson B. J., et al. (2017). Neodiversification of homeologous CLAVATA1-like receptor kinase genes in soybean leads to distinct developmental outcomes. Sci. Rep. 7:8878. 10.1038/s41598-017-08252-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa H., Oka-Kira E., Sato N., Takahashi H., Wu G. J., Sato S., et al. (2010). The receptor-like kinase KLAVIER mediates systemic regulation of nodulation and non-symbiotic shoot development in Lotus japonicus. Development 137 4317–4325. 10.1242/dev.058891 [DOI] [PubMed] [Google Scholar]
- Mohd-Radzman N. A., Laffont C., Ivanovici A., Patel N., Reid D. E., Stougaard J., et al. (2016). Different pathways act downstream of the peptide receptor CRA2 to regulate lateral root and nodule development. Plant Physiol. 171 2536–2548. 10.1104/pp.16.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandi D., Sagan M., Prado-Vivant E., Duc G. (2000). Influence of genes determining supernodulation on root colonization by the mycorrhizal fungus Glomus mosseae in Pisum sativum and Medicago truncatula mutants. Mycorrhiza 10 37–42. 10.1007/s005720050285 [DOI] [Google Scholar]
- Mortier V., De Wever E., Vuylsteke M., Holsters M., Goormachtig S. (2012). Nodule numbers are governed by interaction between CLE peptides and cytokinin signaling. Plant J. 70 367–376. 10.1111/j.1365-313X.2011.04881.x [DOI] [PubMed] [Google Scholar]
- Mortier V., Den Herder G., Whitford R., Van De Velde W., Rombauts S., D’haeseleer K., et al. (2010). CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol. 153 222–237. 10.1104/pp.110.153718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J. (2003). Artificial infection by endophytes affects growth and mycorrhizal colonisation of Lolium perenne. Funct. Plant Biol. 30 419–424. 10.1071/FP02189 [DOI] [PubMed] [Google Scholar]
- Oka-Kira E., Tateno K., Miura K. I., Haga T., Hayashi M., Harada K., et al. (2005). klavier (klv), A novel hypernodulation mutant of Lotus japonicus affected in vascular tissue organization and floral induction. Plant J. 44 505–515. 10.1111/j.1365-313X.2005.02543.x [DOI] [PubMed] [Google Scholar]
- Okamoto S., Kawaguchi M. (2015). Shoot HAR1 mediates nitrate inhibition of nodulation in Lotus japonicus. Plant Signal. Behav. 10:e1000138. 10.1080/15592324.2014.1000138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S., Ohnishi E., Sato S., Takahashi H., Nakazono M., Tabata S., et al. (2009). Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol. 50 67–77. 10.1093/pcp/pcn194 [DOI] [PubMed] [Google Scholar]
- Okamoto S., Shinohara H., Mori T., Matsubayashi Y., Kawaguchi M. (2013). Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat. Commun. 4:2191. 10.1038/ncomms3191 [DOI] [PubMed] [Google Scholar]
- Omacini M., Eggers T., Bonkowski M., Gange A. C., Jones T. H. (2006). Leaf endophytes affect mycorrhizal status and growth of co-infected and neighbouring plants. Funct. Ecol. 20 226–232. 10.1111/j.1365-2435.2006.01099.x [DOI] [Google Scholar]
- Pedrotti L., Mueller M. J., Waller F. (2013). Piriformospora indica root colonization triggers local and systemic root responses and inhibits secondary colonization of distal roots. PLoS One 8:e69352. 10.1371/journal.pone.0069352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid D. E., Ferguson B. J., Gresshoff P. M. (2011a). Inoculation-and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Mol. Plant Microbe Interact. 24 606–618. 10.1094/MPMI-09-10-0207 [DOI] [PubMed] [Google Scholar]
- Reid D. E., Ferguson B. J., Hayashi S., Lin Y. H., Gresshoff P. M. (2011b). Molecular mechanisms controlling legume autoregulation of nodulation. Ann. Bot. 108 789–795. 10.1093/aob/mcr205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid D. E., Nadzieja M., Novak O., Heckmann A. B., Sandal N., Stougaard J. (2017). Cytokinin biosynthesis promotes cortical cell responses during nodule development. Plant Physiol. 175:pp.00832.2017. 10.1104/pp.17.00832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K., Nohara Y. (2009). Soybean (Glycine max [L.] Merr.) shoots systemically control arbuscule formation in mycorrhizal symbiosis. Soil Sci. Plant Nutr. 55 252–257. 10.1111/j.1747-0765.2009.00358.x [DOI] [Google Scholar]
- Sakamoto K., Ogiwara N., Kaji T. (2013). Involvement of autoregulation in the interaction between rhizobial nodulation and AM fungal colonization in soybean roots. Biol. Fertil. Soils 49 1141–1152. 10.1007/s00374-013-0804-8 [DOI] [Google Scholar]
- Sasaki T., Suzaki T., Soyano T., Kojima M., Sakakibara H., Kawaguchi M. (2014). Shoot-derived cytokinins systemically regulate root nodulation. Nat. Commun. 5:4983. 10.1038/ncomms5983 [DOI] [PubMed] [Google Scholar]
- Schaarschmidt S., Gresshoff P. M., Hause B. (2013). Analyzing the soybean transcriptome during autoregulation of mycorrhization identifies the transcription factors GmNF-YA1a/b as positive regulators of arbuscular mycorrhization. Genome Biol. 14:R62. 10.1186/gb-2013-14-6-r62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel E., Journet E. P., De Carvalho-Niebel F., Duc G., Frugoli J. (2005). The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 58 809–822. 10.1007/s11103-005-8102-y [DOI] [PubMed] [Google Scholar]
- Schnabel E. L., Kassaw T. K., Smith L. S., Marsh J. F., Oldroyd G. E., Long S. R., et al. (2011). The ROOT DETERMINED NODULATION1 gene regulates nodule number in roots of Medicago truncatula and defines a highly conserved, uncharacterized plant gene family. Plant Physiol. 157 328–340. 10.1104/pp.111.178756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrihari P. C., Sakamoto K., Inubushi K., Akao S. (2000). Interaction between supernodulating or non-nodulating mutants of soybean and two arbuscular mycorrhizal fungi. Mycorrhiza 10 101–106. 10.1007/s005720000064 [DOI] [Google Scholar]
- Solaiman Z. M., Senoo K., Kawaguchi M., Imaizumi-Anraku H., Akao S., Tanaka A., et al. (2000). Characterization of mycorrhizas formed by glomus sp. on roots of hypernodulating mutants of Lotus japonicus. J. Plant Res. 113 443–448. 10.1007/PL00013953 [DOI] [Google Scholar]
- Soyano T., Kawaguchi M. (2014). “Systemic regulation of root nodule formation,” in Advances in Biology and Ecology of Nitrogen Fixation, ed. Ohyama T. (Rijeka: InTech; ). [Google Scholar]
- Sun X., Wang G.-L. (2011). Genome-wide identification, characterization and phylogenetic analysis of the rice LRR-Kinases. PLoS One 6:e16079. 10.1371/journal.pone.0016079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara M., Magori S., Soyano T., Okamoto S., Yoshida C., Yano K., et al. (2013). Too much love, a novel Kelch repeat-containing F-box protein, functions in the long-distance regulation of the legume-Rhizobium symbiosis. Plant Cell Physiol. 54 433–447. 10.1093/pcp/pct022 [DOI] [PubMed] [Google Scholar]
- van Noorden G. E., Ross J. J., Reid J. B., Rolfe B. G., Mathesius U. (2006). Defective long-distance auxin transport regulation in the Medicago truncatula super numeric nodules mutant. Plant Physiol. 140 1494–1506. 10.1104/pp.105.075879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierheilig H. (2004). Further root colonization by arbuscular mycorrhizal fungi in already mycorrhizal plants is suppressed after a critical level of root colonization. J. Plant Physiol. 161 339–341. 10.1078/0176-1617-01097 [DOI] [PubMed] [Google Scholar]
- Vierheilig H., Coughlan A. P., Wyss U., Piché Y. (1998). Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl. Environ. Microbiol. 64 5004–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierheilig H., Garcia-Garrido J., Wyss U., Piché Y. (2000a). Systemic suppression of mycorrhizal colonization of barley roots already colonized by AM fungi. Soil Biol. Biochem. 32 589–595. 10.1016/S0038-0717(99)00155-8 [DOI] [Google Scholar]
- Vierheilig H., Maier W., Wyss U., Samson J., Strack D., Piché Y. (2000b). Cyclohexenone derivative- and phosphate-levels in split-root systems and their role in the systemic suppression of mycorrhization in precolonized barley plants. J. Plant Physiol. 157 593–599. 10.1016/S0176-1617(00)80001-2 [DOI] [Google Scholar]
- Vierheilig H., Steinkellner S., Khaosaad T., Garcia-Garrido J. M. (2008). “The biocontrol effect of mycorrhization on soilborne fungal pathogens and the autoregulation of the AM symbiosis: one mechanism, two effects?” in Mycorrhiza: State of the Art, Genetics and Molecular Biology, Eco-Function, Biotechnology, Eco-Physiology, Structure and Systematics, ed. Varma A. (Berlin: Springer; ), 307–320. [Google Scholar]
- Wall L. G., Huss-Danell K. (1997). Regulation of nodulation in Alnus incana-Frankia symbiosis. Physiol. Plant. 99 594–600. 10.1111/j.1399-3054.1997.tb05362.x [DOI] [Google Scholar]
- Wei Z., Wang J., Yang S., Song Y. (2015). Identification and expression analysis of the LRR-RLK gene family in tomato (Solanum lycopersicum) Heinz 1706. Genome 58 121–134. 10.1139/gen-2015-0035 [DOI] [PubMed] [Google Scholar]
- Wyss P., Mellor R. B., Wiemken A. (1990). Mutants of soybean (Glycine max) unable to suppress nodulation in the presence of nitrate retain the ability to suppress mycorrhization in the presence of phosphate. J. Plant Physiol. 136 507–509. 10.1016/S0176-1617(11)80045-3 [DOI] [Google Scholar]
- Xu C., Liberatore K. L., Macalister C. A., Huang Z., Chu Y.-H., Jiang K., et al. (2015). A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 47 784–792. 10.1038/ng.3309 [DOI] [PubMed] [Google Scholar]
- Yamamoto E., Karakaya H. C., Knap H. T. (2000). Molecular characterization of two soybean homologs of Arabidopsis thaliana CLAVATA1 from the wild type and fasciation mutant11The sequence data reported in this paper have been submitted to the GenBank data libraries under the accession numbers AF197946 for GmCLV1A and AF197947 for GmCLV1B. Biochim. Biophys. Acta 1491 333–340. 10.1016/S0167-4781(00)00061-0 [DOI] [PubMed] [Google Scholar]
- Zan Y., Ji Y., Zhang Y., Yang S., Song Y., Wang J. (2013). Genome-wide identification, characterization and expression analysis of populusleucine-rich repeat receptor-like protein kinase genes. BMC Genomics 14:318. 10.1186/1471-2164-14-318 [DOI] [PMC free article] [PubMed] [Google Scholar]