CD133, a widely known marker of cancer stem cells, was recently found in extracellular vesicles. However, the mechanisms underlying CD133 translocation to the extracellular space remain largely unknown.
KEYWORDS: ubiquitination, CD133, glycosylation, Tsg101, extracellular vesicles, cell migration
ABSTRACT
CD133, a widely known marker of cancer stem cells, was recently found in extracellular vesicles. However, the mechanisms underlying CD133 translocation to the extracellular space remain largely unknown. Here we report that CD133 is monoubiquitinated. Ubiquitination occurs primarily on complex glycosylated CD133. The lysine 848 residue at the intracellular carboxyl terminus is one of the sites for CD133 ubiquitination. The K848R mutation does not affect CD133 degradation by the lysosomal pathway but significantly reduces CD133 secretion by inhibiting the interaction between CD133 and tumor susceptibility gene 101 (Tsg101). Furthermore, knockdown of the E3 ubiquitin protein ligase Nedd4 largely impairs CD133 ubiquitination and vesicle secretion. Importantly, CD133-containing vesicles are taken up by recipient cells, consequently promoting cell migration. The K848R mutation reduces cell migration induced by CD133. Taken together, our findings show that monoubiquitination contributes to CD133 vesicle secretion and promotes recipient cell migration. These findings provide a clue to the mechanisms of CD133 secretion and cancer stem cell microenvironment interactional effects.
INTRODUCTION
CD133, a 120-kDa pentaspan transmembrane glycoprotein, has been used as a cell surface marker of normal stem cells and cancer stem cells (CSCs) from a broad range of tissue types (1–4). A series of studies have shown that CD133 promotes tumorigenesis, angiogenesis, self-renewal, and cell migration (5–8). The mechanisms by which cellular CD133 contributes to carcinogenic ability have been demonstrated. For example, our previous study showed that the interaction between CD133 and p85 activates the phosphatidylinositol 3-kinase (PI3K)/Akt pathway and promotes tumorigenesis (9). In addition, CD133 interacts directly with a tubulin deacetylase, HDAC6, and suppresses cancer cell differentiation (10). These observations indicate that cellular CD133 plays an oncogenic role in tumor occurrence.
In addition to specific localization in plasma membrane protrusions (11), CD133 is also released into various body fluids. For instance, CD133-containing vesicles have been detected in murine and human cerebrospinal fluids (CSF), seminal fluids, urine, and saliva (12–15). There are two major classes of extracellular CD133 vesicles, namely, P2- and P4-type vesicles (13). In nonepithelial cells, P4-type vesicle secretion is associated with the endocytic-exocytic pathway (16). Notably, in glioblastoma patients, the level of P4-type vesicles is greatly increased (17). In recent years, cumulative evidence has shown that extracellular vesicle (EV) trafficking is involved in important biological processes, such as embryogenesis, immune responses, and cancer progression, as well as in intercellular communication (18–22). It has been reported that secretion of CD133-containing vesicles is increased during cell differentiation (16). CD133 vesicle release might avoid asymmetric inheritance and potentially balance stem cell proliferation and differentiation (23). However, the molecular mechanisms of CD133 trafficking to the extracellular space remain largely unknown. Understanding these mechanisms might allow us to manipulate the fate of cancer stem cells, thus opening novel modalities for stem cell-based therapies.
Posttranslational modifications affect protein interactivity, stability, and subcellular localization (24–26). CD133 trafficking is linked to its posttranslational modification. For example, N-glycosylation directly regulates CD133 trafficking to the plasma membrane without affecting total CD133 levels (27). Similarly, ATase 1 and ATase 2 mediate CD133 lysine acetylation and further regulate its membrane trafficking (28). These observations led us to the hypothesis that posttranslational modifications are involved in CD133 protein trafficking.
Cumulative evidence has shown that ubiquitination directs membrane protein trafficking and contributes to membrane protein internalization and degradation (29). The endosomal sorting complex required for transport (ESCRT) receives the ubiquitinated cargo and drives it into multivesicular bodies (MVBs), thereby playing a key role in exosome formation. Tumor susceptibility gene 101 protein (Tsg101), a component of ESCRT, is relocated from endosomes to the plasma membrane, where it interacts with arrestin domain-containing protein 1 (ARRDC1) (30). This interaction mediates the release of microvesicles. Surprisingly, CD133 released from membrane structures into saliva is ubiquitinated (15). However, whether ubiquitination is involved in regulating CD133 release is not clear.
To further understand the mechanisms by which CD133 is translocated to the extracellular space, we examined the contribution of ubiquitination to CD133 vesicle secretion. Here we report that CD133 is monoubiquitinated. Additionally, ubiquitination occurs primarily on complex glycosylated CD133. We also demonstrate that the lysine 848 residue at the intracellular carboxyl terminus is one of the sites of CD133 ubiquitination. A K848R mutation does not affect CD133 degradation by the lysosomal pathway but significantly reduces CD133 secretion by inhibiting the binding between CD133 and Tsg101. Furthermore, knockdown of the E3 ubiquitin protein ligase Nedd4 largely impairs CD133 ubiquitination and vesicle secretion. Importantly, CD133-containing vesicles are taken up by recipient cells, consequently promoting cell migration. Taken together, our findings show that monoubiquitination contributes to CD133 vesicle secretion and promotes recipient cell migration. This finding contributes to our understanding of the mechanisms and functions of CD133 secretion.
RESULTS
CD133 can be monoubiquitinated.
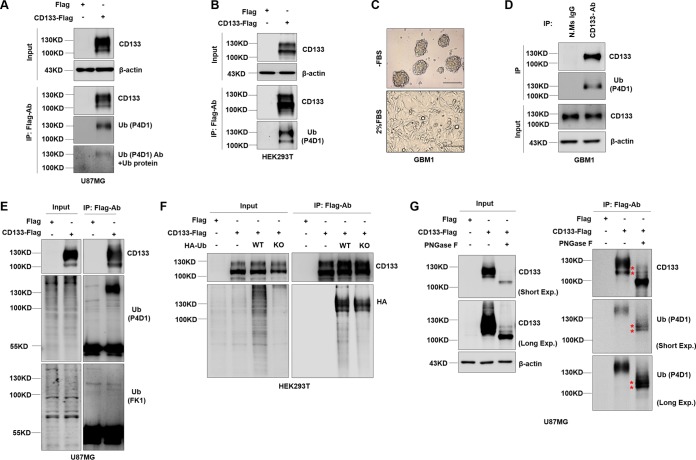
Ubiquitin modification influences cargo trafficking by serving as a signal in the secretory and endocytic pathways (31–34). To identify whether the CD133 protein is ubiquitinated, we ectopically overexpressed CD133-Flag in U87MG cells or HEK293T cells (by lentivirus infection in U87MG cells and by transient transfection in HEK293T cells). Interestingly, there were two bands with different molecular masses (lower band, above 100 kDa; and upper band, approximately 130 kDa) specific for CD133, as shown by Western blotting (Fig. 1A, B, and E to G). The CD133 protein was enriched by immunoprecipitation (IP) assay. A ubiquitin antibody (Ub-P4D1) recognizing both mono- and polyubiquitin was used to detect the presence of ubiquitination. The major band, with a molecular mass of 130 kDa, corresponded to the ubiquitinated CD133 form (Fig. 1A and B). Furthermore, primary GBM1 tumor cells derived from tumor sphere cultures were verified by differentiation experiments (Fig. 1C). Western blotting of immunoprecipitation assay products showed that endogenous CD133 in cultured GBM1 sphere cells could be ubiquitinated (Fig. 1D). Together the results showed that endogenous and exogenous CD133 could be ubiquitinated. To determine the type of CD133 ubiquitination, we used a range of antibodies. The Ub-P4D1 antibody (recognizing both poly- and monoubiquitinated proteins) showed a positive reaction to CD133 by IP-Western blotting, while the Ub-FK1 antibody (recognizing poly-Ub) did not (Fig. 1E) (34). This result indicated that CD133 was monoubiquitinated. To further examine the type of CD133 ubiquitination, two ubiquitin-expressing plasmids were transfected together with the CD133-expressing plasmid into HEK293T cells (a hemagglutinin [HA]-tagged ubiquitin knockout construct [Ub-KO] was unable to cause polyubiquitination, while HA-tagged wild-type ubiquitin [Ub-WT] was able to cause both mono- and polyubiquitination). Western blotting of the immunoprecipitation assay products showed that there was no significant difference between the Ub-WT and Ub-KO lanes (Fig. 1F), indicating that CD133 was monoubiquitinated. We also observed CD133 ubiquitination after treatment with peptide-N-glycosidase (PNGase F), which cleaves N-glycosylated branches (Fig. 1G). This showed that there were two positions on the protein with molecular weights of >100 kDa at which ubiquitination was detected after PNGase F deglycosylation (Fig. 1G, asterisks), indicating that CD133 was multiply monoubiquitinated but that the ratio of total CD133 ubiquitination was very low. Collectively, the data show that CD133 can be monoubiquitinated.
FIG 1.
CD133 undergoes monoubiquitination. (A, E, and G) U87MG cells were infected with a lentivirus expressing Flag (control) or CD133-Flag. (A) CD133 expression was analyzed by Western blotting; β-actin was blotted as a loading control. IP analysis using Flag-M2 was performed to determine the ubiquitination of CD133. Ubiquitin protein was added to neutralizing the primary ubiquitin antibody (bottom panel). (B) HEK293T cells were transfected with a Flag (control) or CD133-Flag plasmid, and IP analysis was performed to determine the ubiquitination of CD133. β-Actin was blotted as a loading control. (C) A differentiation assay was performed. GBM1 cells were cultured in 2% FBS medium for 7 days, and the characteristics of CSCs were evaluated. Representative images of GBM1 cells during the differentiation assay are shown. Bars = 150 μm. (D) Endogenous CD133 from GBM1 sphere culture cells was precipitated by use of CD133 antibody. Normal mouse IgG antibody was used as a negative control. Ubiquitination of CD133 protein was detected by Western blotting; β-actin was blotted as an internal control. (E) CD133 from U87MG cells expressing CD133-Flag was precipitated by use of a Flag antibody. Ub-P4D1 antibody and Ub-FK1 antibody were used to detecting the type of CD133 ubiquitination. (F) HEK293T cells coexpressing CD133-Flag and either the HA-Ub-WT or HA-Ub-KO plasmid. CD133 ubiquitination was detected by Western blotting following co-IP assays. (G) U87MG cells expressing CD133-Flag were subjected to IP with an anti-Flag antibody, and CD133 protein was treated with PNGase F for deglycosylation and then immunoblotted with anti-CD133 or anti-Ub (P4D1) antibodies. Whole-cell lysates were analyzed by immunoblotting, with CD133 and β-actin as input. All results were collected from three independent experiments. N.Ms IgG, normal mouse IgG; IP, immunoprecipitation; Ub-WT, wild-type ubiquitin; Ub-KO, all 7 ubiquitin lysines were mutated to arginine; Exp., exposure.
Ubiquitination occurs primarily on complex glycosylated CD133.
The CD133 protein contains nine potential N-linked glycosylation sites within its putative extracellular domains (35). N-glycosylation of CD133 contributes to its epitope recognition and membrane localization (27). Given that there were two bands specific for CD133 (Fig. 1) but that the ratio of ubiquitination was very low, we hypothesized that the 130-kDa band contained mostly nonubiquitinated glycosylated CD133 and a small amount of ubiquitinated CD133. Therefore, we examined the relationship between ubiquitination and N-glycosylation of CD133. Treatment of CD133 protein with PNGase F, which cleaves between the innermost GlcNAc and asparagine residues of high-mannose, hybrid, and complex oligosaccharides, resulted in the disappearance of both bands and the appearance of a single band with a molecular mass of 100 kDa (Fig. 2A, lane 2). In parallel, we treated purified CD133 with endoglycosidase H (endo H), which cleaves within the chitobiose core of high-mannose and some hybrid oligosaccharides from N-linked glycoproteins, and only the lower band (above 100 kDa) disappeared, shifting to 100 kDa (Fig. 2A, lane 4). These changes in molecular mass of CD133-specific bands suggested that the 130-kDa band was complex glycosylated CD133 and that the band above 100 kDa was high-mannose glycosylated CD133. Phaseolus vulgaris leucoagglutinin (PHA-L) and concanavalin A (ConA), recognizing β-1,6-GlcNAc N-glycans and high-mannose N-glycans, respectively, were also used to distinguish between complex and high-mannose glycosylation (36). Western blotting showed that the 130-kDa CD133 band reacted positively to PHA-L detection, which suggested that this CD133 form was the complex glycosylated form (Fig. 2, red arrows). The minor band (above 100 kDa) was positive for ConA detection, indicating that the CD133 form in this band was of the high-mannose glycosylated type (Fig. 2, blue arrows). Interestingly, while both glycosylated types of CD133 reacted positively to ubiquitin antibody detection, complex glycosylated CD133 was the major type to be ubiquitinated (Fig. 2A, bottom panel). Of course, complex glycosylated CD133 was the form with the highest stable expression in U87MG cells (Fig. 2B, red arrows). Taken together, these results indicate that complex glycosylated CD133 is the major type to be ubiquitinated.
FIG 2.
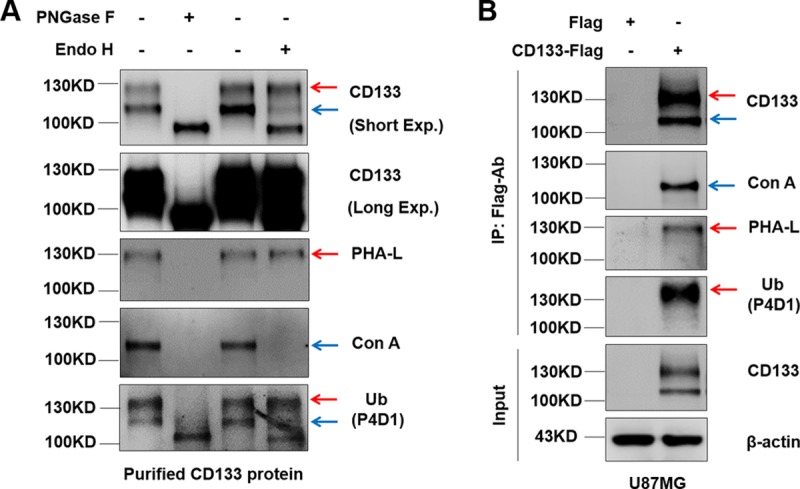
Ubiquitination occurs primarily on complex glycosylated CD133. (A) HEK293T cells were transiently transfected with a Flag (control) or CD133-Flag plasmid. IP methods were performed to purify CD133 protein. PNGase F and endo H were applied for deglycosylation of CD133. PHA-L and ConA were used to examine complex glycosylated CD133 and high-mannose glycosylated CD133, respectively. (B) U87MG cells were used to stably express Flag or CD133-Flag. CD133 was precipitated using anti-Flag antibody. Complex glycosylated CD133 and high-mannose glycosylated CD133 were monitored by use of PHA-L and ConA, respectively. Red arrows indicate complex glycosylated CD133. Blue arrows indicate high-mannose glycosylated CD133. All results were collected from three independent experiments. Exp., exposure; IP, immunoprecipitation.
The lysine 848 residue at the intracellular carboxyl terminus is a site for CD133 ubiquitination.
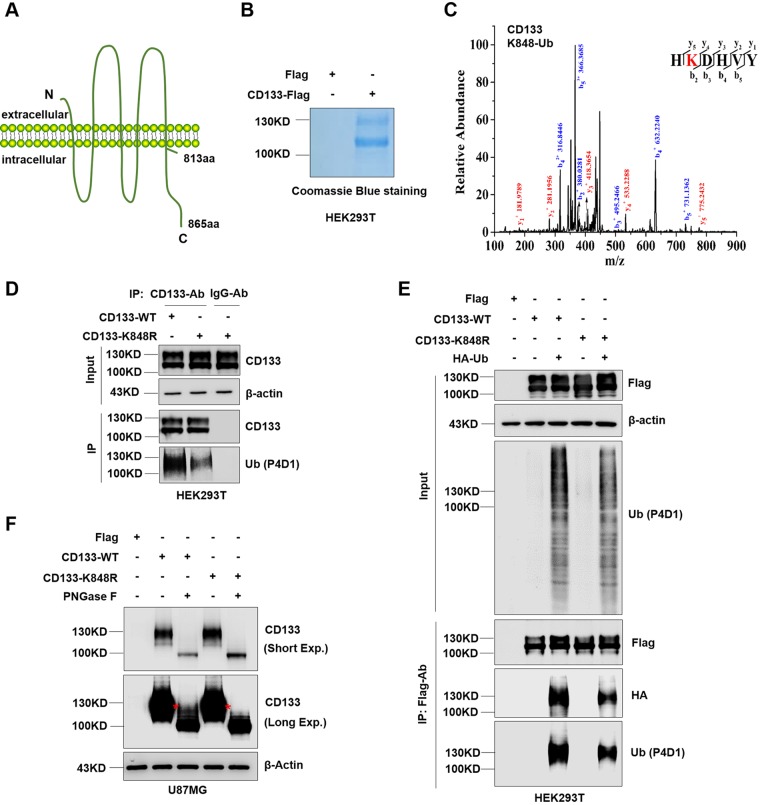
CD133 is a five-transmembrane glycoprotein with a cytoplasmic tail (Fig. 3A) (37). To determine the ubiquitination site of complex glycosylated CD133 (130 kDa), immunoprecipitation followed by tandem mass spectrometry (IP-MS) was performed (Fig. 3B). Lysine 848 was shown to be ubiquitinated (Fig. 3C). Next, to confirm the specific site for CD133 ubiquitination, lysine 848 was mutated to arginine. Western blotting showed a significant decrease in ubiquitination on the CD133-K848R mutant (Fig. 3D). We conformed this result by coexpression of HA-Ub together with CD133-WT or CD133-K848R, followed by IP-Western blotting, which showed that the K848R mutation reduced CD133 ubiquitination (Fig. 3E). We also deglycosylated the CD133-WT and CD133-K848R proteins by use of PNGase F and found that the K848R mutation did prevent the appearance of the protein with a molecular weight of >100 kDa after PNGase F deglycosylation (Fig. 3F, asterisks). Thus, these results show that the lysine 848 residue is a site for CD133 ubiquitination.
FIG 3.
Complex glycosylated CD133 is ubiquitinated at Lys848. (A) Proposed structural model of CD133. (B) Purity of CD133 protein from HEK293T cells, determined by Coomassie blue staining. (C) MS analysis showed complex glycosylated CD133 (≈130 kDa) to be ubiquitinated at Lys848. The multiple lines are the fragment ions that confirm K848 as the ubiquitination site. (D) The K848R mutant or wild-type (WT) plasmid was expressed in HEK293T cells, and immunoprecipitation was performed using a CD133 antibody. Normal mouse IgG antibody was used as a negative control. CD133 ubiquitination was detected by Western blotting; β-actin was blotted as a loading control. (E) Flag-tagged CD133-WT or CD133-K848R was coexpressed with HA-Ub in HEK293T cells, followed by IP-Western blot analysis. (F) U87MG cells were used to stably express Flag, CD133-WT, or CD133-K848R. Cell lysates were treated with PNGase F for deglycosylation and then subjected to Western blotting. β-Actin was blotted as a loading control. All results were collected from three independent experiments. aa, amino acids; MS, mass spectrometry; IP, immunoprecipitation; Exp., exposure.
Lys848 ubiquitination does not affect lysosomal degradation of CD133.
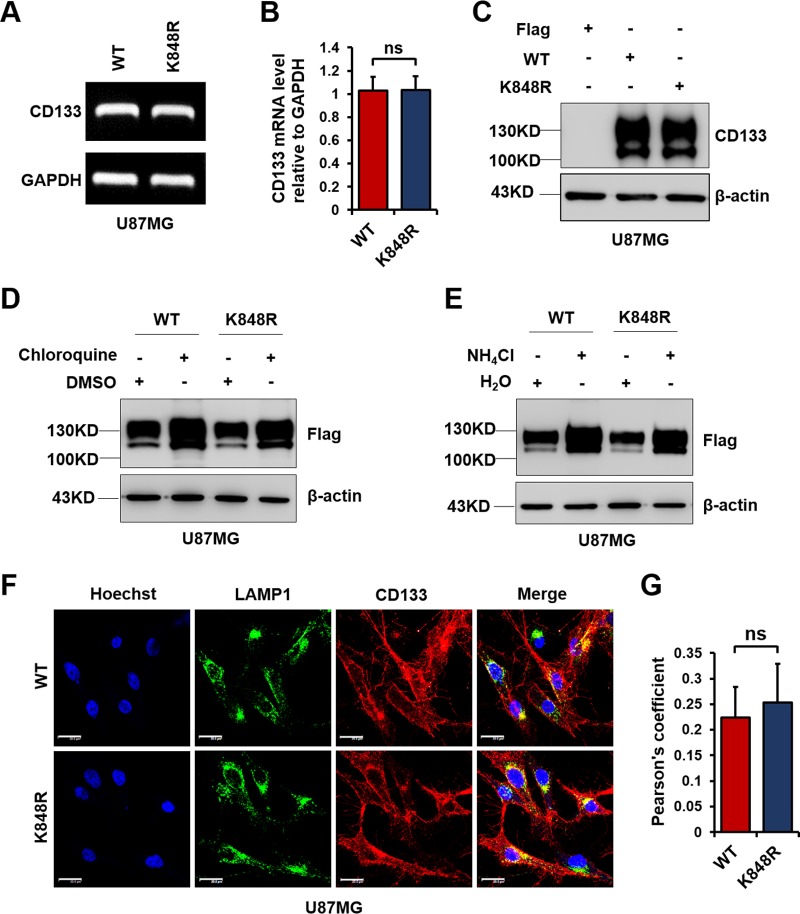
It is known that ubiquitination directs membrane protein trafficking and contributes to membrane protein degradation (29). CD133 is reportedly degraded by the lysosomal pathway (38). It has also been reported that monoubiquitination directs membrane protein internalization and degradation (39). To investigate the effect of Lys848 ubiquitination on CD133 lysosomal degradation, we first overexpressed CD133-WT and CD133-K848R in U87MG cells. The level of CD133 mRNA was examined by reverse transcription-PCR (RT-PCR). The K848R mutation did not affect CD133 at either the mRNA level (Fig. 4A and B) or the protein level (Fig. 4C). Chloroquine and ammonium chloride are widely used to inhibit the lysosomal degradation pathway (40). As shown in Fig. 4D and E, similar to CD133-WT, CD133-K848R was also degraded by the lysosomal pathway. We also observed the lysosomal localization of CD133 by confocal microscopy, which showed that both CD133-WT and CD133-K848R colocalized with the lysosomal marker LAMP1 (Fig. 4F and G). Thus, Lys848 ubiquitination does not prevent CD133 degradation by the lysosomal pathway.
FIG 4.
K848R mutation does not change CD133 degradation by the lysosomal pathway. U87MG cells were infected with a lentivirus containing CD133-WT or CD133-K848R. (A) Relative CD133 mRNA levels were determined by RT-PCR. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. (B) Results are presented as means and SD (n = 3). Statistical analysis was performed using the Student t test (ns, no significance). (C) Protein levels of CD133-WT and CD133-K848R were detected by Western blotting; β-actin was blotted as a loading control. (D) Cells were treated with 50 μM chloroquine or dimethyl sulfoxide (DMSO) for 12 h. The levels of CD133 were determined by Western blotting; β-actin was blotted as a loading control. (E) Cells were treated with 10 mM NH4Cl or H2O for 12 h. The levels of CD133 were determined by Western blotting; β-actin was blotted as a loading control. (F) An immunofluorescence assay was performed to examine the colocalization of CD133 (red) and LAMP1 (green). Bars = 20 μm. (G) Quantification of LAMP1-CD133 colocalization in confocal microscopy images by use of Pearson's coefficients. The fluorescence intensity signals in selected regions with similar areas were measured. Data are presented as means and SD for results from three independent experiments (n ≥ 20 cells) analyzed by the Student t test (ns, no significance).
Lys848 ubiquitination is critical for CD133 secretion.
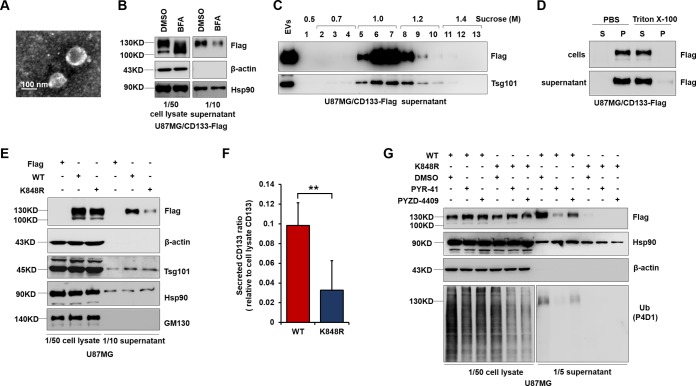
Ubiquitination influences cargo trafficking in the secretory and endocytic pathways (29). CD133 is also found in extracellular vesicles (EVs). To address whether Lys848 ubiquitination is associated with CD133 EV secretion, we harvested extracellular vesicles from the cell culture supernatant and performed transmission electron microscopy (Fig. 5A). This revealed the presence of small vesicles with diameters of approximately 50 to 150 nm in our EV preparations. Western blotting showed that inhibition of protein transport by brefeldin A (BFA; an inhibitor of protein transport) blocked CD133 secretion (Fig. 5B) (16, 41). To confirm the presence of CD133 in EVs, Western blotting of sucrose density gradient fractions was undertaken. Both CD133 and the EV marker Tsg101 were detected (Fig. 5C). Furthermore, EVs were resuspended in phosphate-buffered saline (PBS), with or without 1% Triton X-100, and lysed for 2 h. CD133 was found to be completely soluble in Triton X-100 after centrifugation for 1 h at 100,000 × g, while being insoluble in PBS, as previously described for CD133 vesicles (16). Cells were lysed in parallel under identical conditions (Fig. 5D). These results indicated that CD133 was present in the EVs. The K848R mutation significantly decreased the ratio of released CD133 to cell lysate CD133 (Fig. 5E and F). To further confirm the effect of ubiquitination on CD133 secretion, PYR-41 and PYZD-4409, two inhibitors of the ubiquitin-activating enzyme E1, were used to inhibit total ubiquitination (42, 43). CD133 release was blocked after treatment with PYR-41 or PYZD-4409 (Fig. 5G). Interestingly, extracellular CD133 showed only one high-molecular-mass band, of approximately 130 kDa (Fig. 5C to E and G). Together these results showed that mutation of K848 reduced CD133 EV secretion.
FIG 5.
K848R mutant inhibits CD133 release into EVs. (A) EVs were isolated from the medium of CD133-expressing U87MG cells and verified by transmission electron microscopy. (B) CD133 release into the supernatant was blocked following treatment with 20 μM brefeldin A. (C) Western blotting of sucrose gradient fractions confirmed the presence of CD133 together with the EV marker Tsg101 in purified EVs. (D) EVs were resuspended in PBS alone or in PBS containing 1% Triton X-100 and lysed for 2 h, followed by centrifugation for 1 h at 100,000 × g. The resulting supernatants (S) and pellets (P) were analyzed by Western blotting. (E to G) U87MG cells were infected with a lentivirus expressing CD133-WT or CD133-K848R. (E) EVs were verified by Western blotting of CD133 and known vesicular proteins (Hsp90 and Tsg101). A nonvesicular protein from the Golgi apparatus (GM130) was used as a control. Both cell lysates and EVs were harvested from 5 × 106 cells. (F) Quantification of EV CD133 relative to cell lysate CD133. Data are presented as means and SD for results from three independent experiments. Statistical analysis was performed using the Student t test (**, P < 0.01). (G) Western blotting demonstrated that treatment with 20 μM PYR-41 or 20 μM PYZD-4409 inhibited ubiquitination and CD133 secretion. Hsp90 and β-actin were used as loading controls. Both cell lysates and EVs were harvested from 5 × 106 cells. All results were collected from three independent experiments. EVs, extracellular vesicles.
Lys848 ubiquitination-dependent interaction between CD133 and Tsg101 regulates CD133 secretion.
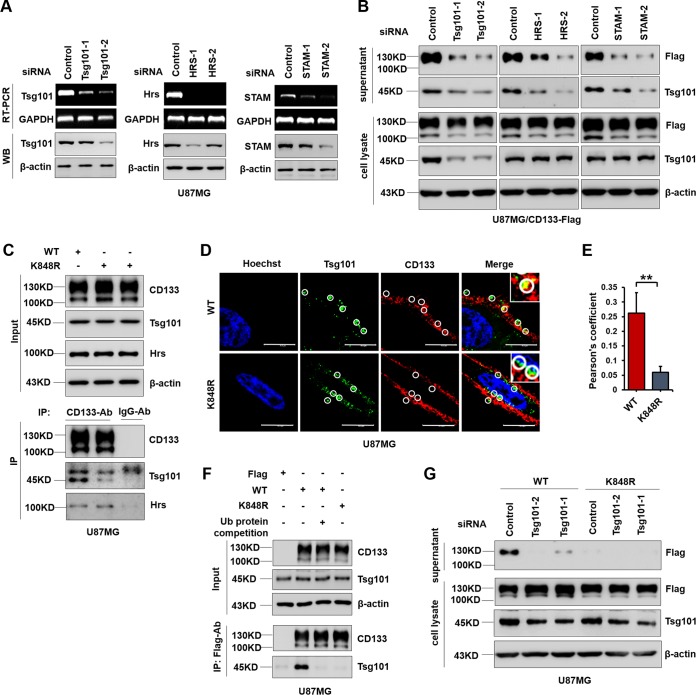
The ESCRT machinery plays a key role in several cellular processes, during which ubiquitin-tagged proteins enter endosomes via the formation of vesicles (44, 45). Three ESCRT members, Hrs, STAM, and Tsg101, can interact with ubiquitinated protein cargos and promote multivesicular body (MVB) formation, thereby playing a significant role in promoting exosome secretion (46–48). To examine the effect of ESCRT on secretion of ubiquitinated CD133, further study was conducted by silencing these ESCRT components by use of small interfering RNA (siRNA) (Fig. 6A). CD133 secretion was markedly decreased following Tsg101, Hrs, or STAM knockdown (Fig. 6B). Along this line, we next examined whether CD133 interacts with these ESCRT components. Co-IP assays showed that CD133-WT could bind to Tsg101, and mutation of K848 obviously reduced the binding of CD133 to Tsg101 (Fig. 6C). In accordance with this, an immunofluorescence assay also showed that CD133 colocalized with Tsg101 in cells expressing CD133 (Fig. 6D and E). CD133-Tsg101 binding was competitively inhibited by adding ubiquitin, indicating that Tsg101-CD133 binding was ubiquitin dependent (Fig. 6F). Knockdown of Tsg101 significantly diminished CD133 secretion in cells expressing CD133 (Fig. 6G). Taken together, these results show that a ubiquitination-dependent interaction between CD133 and Tsg101 regulates CD133 secretion.
FIG 6.
Lys848 ubiquitination regulates CD133 secretion though binding to Tsg101. (A and B) U87MG cells were infected with a lentivirus expressing CD133-Flag. (A) U87MG cells expressing CD133-Flag were transfected with the indicated siRNA for 24 h. RT-PCR was used to determine mRNA levels. Protein levels were detected by Western blotting (WB); β-actin served as a loading control. (B) EVs and cell lysates were subjected to Western blotting following siRNA transfection for 24 h and serum starvation for 48 h. Both cell lysates and EVs were harvested from 2 × 106 cells. β-Actin was used as a loading control. (C) U87MG cells were infected with a lentivirus expressing CD133-WT or CD133-K848R. Co-IP assays were performed by use of CD133 antibody. Normal mouse IgG antibody was used as a control. (D) An immunofluorescence assay was done to observe the colocalization of CD133 (red) and Tsg101 (green). Representative images from confocal microscopy analysis are shown. Circles represent the randomly selected regions that were Tsg101 positive (green). Bars = 10 μm. (E) Quantification of Tsg101-CD133 colocalization in confocal microscopy images by use of Pearson's coefficients. The fluorescence intensity signals in selected regions with similar areas were measured. Data are presented as means and SD for results from three independent experiments (n ≥ 20 cells) analyzed using the Student t test (**, P < 0.01). (F) Ubiquitin protein (0.2 mg/ml) was added during the IP process, and Western blotting of IP assay products demonstrated ubiquitination-dependent CD133-Tsg101 binding. (G) U87MG cells stably expressing CD133-WT or CD133-K848R were transfected with Tsg101 siRNA for 24 h and then serum starved for 48 h. CD133 was detected by Western blotting. Both cell lysates and EVs were harvested from 2 × 106 cells. All results were collected from three independent experiments. IP, immunoprecipitation.
Nedd4 regulates CD133 ubiquitination and extracellular vesicle secretion.
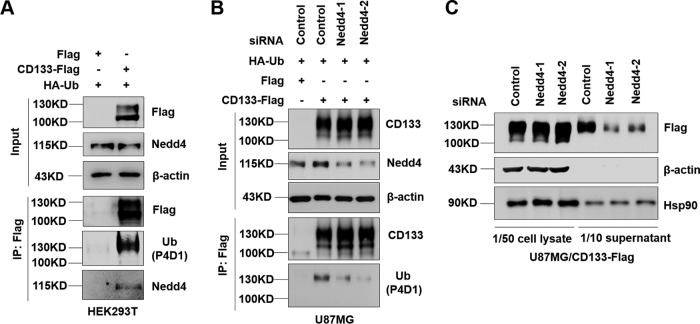
Nedd4, an E3 ligase, plays an important role in identifying and ubiquitinating specific target substrates (49, 50). Nedd4-regulated ubiquitination is also involved in the release of microvesicles and in exosomal secretion (30, 51, 52). Nedd4 is also reported to induce monoubiquitination (53). We next tested the possibility that Nedd4 regulates CD133 ubiquitination and secretion. First, HEK293T cells were transfected with a Flag or CD133-Flag plasmid together with HA-Ub. The binding of Nedd4 to CD133 was observed by Western blotting of co-IP assay products. The results showed that Nedd4 could bind to CD133 (Fig. 7A). We then examined the effect of Nedd4 on CD133 ubiquitination. Knockdown of Nedd4 in U87MG cells expressing CD133-Flag by use of siRNA showed largely decreased CD133 ubiquitination (Fig. 7B). We next explored the effect of Nedd4 on CD133 EV secretion. After knockdown of Nedd4 by siRNA for 24 h and serum starvation for 48 h, the collected EVs were subjected to Western blotting. As expected, CD133 EV secretion was decreased by Nedd4 downregulation (Fig. 7C), accompanied by a small amount of accumulation of intracellular CD133, indicating that Nedd4 regulates CD133 ubiquitination and EV secretion.
FIG 7.
Contribution of Nedd4 to CD133 ubiquitination and secretion. (A) HEK293T cells were transfected with Flag or CD133-Flag in combination with the HA-Ub plasmid. The interaction between Nedd4 and CD133 was examined using a co-IP assay. (B) U87MG cells stably expressing Flag or CD133-Flag were transfected with the HA-Ub plasmid in combination with the indicated siRNA. Nedd4 protein levels and CD133 ubiquitination were analyzed by Western blotting. (C) U87MG cells expressing CD133-Flag were transfected with the indicated siRNA for 24 h and then cultured in FBS-free medium. After 48 h, EVs were harvested from the extracellular medium. Cell lysates and EV lysates were then subjected to Western blotting. Both cell lysates and EVs were harvested from 2 × 106 cells. Hsp90 was used as a loading control. Data are representative of three independent experiments. EVs, extracellular vesicles; IP, immunoprecipitation.
Lys848 mutation reduces cell migration.
Exosomes play a key role in promoting invasive activity and in controlling invasive actin structures (54). We previously reported that CD133-Src interaction promotes cell migration (55). Src also regulates the biogenesis and activity of exosomes and therefore promotes cell migration through an ESCRT-dependent pathway (56, 57). We examined whether the K848R mutation affected cell migration and observed that it reduced cell migration induced by CD133 in a wound-healing assay (Fig. 8A and B) and a Transwell assay (Fig. 8C and D). Thus, Lys848 ubiquitination of CD133 regulates cell migration.
FIG 8.
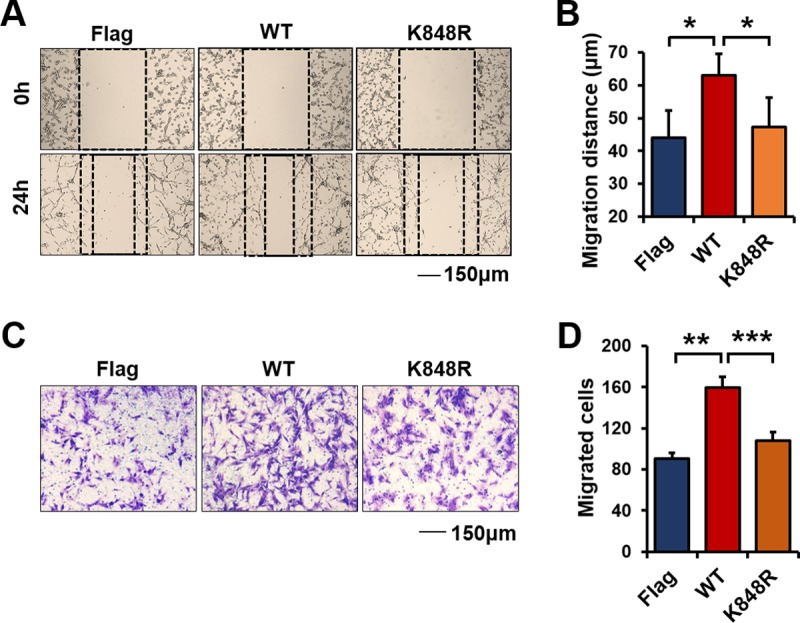
K848R mutation reduces cell migration induced by CD133. U87MG cells were infected with a lentivirus expressing CD133-WT, CD133-K848R, or Flag. (A) Representative images from the wound healing assay. (B) Relative distances of cell migration in the images from panel A were calculated and are presented as means and SD (n = 6). Statistical analysis was performed using the Student t test (*, P < 0.05). (C) Representative images from a Transwell assay of cells expressing CD133-WT, CD133-K848R, or Flag. (D) Cell migration numbers were calculated and are presented as means and SD (n = 3). Statistical analysis was performed using the Student t test (**, P < 0.01; ***, P < 0.001).
CD133 EVs are transferred to recipient cells and facilitate cell migration.
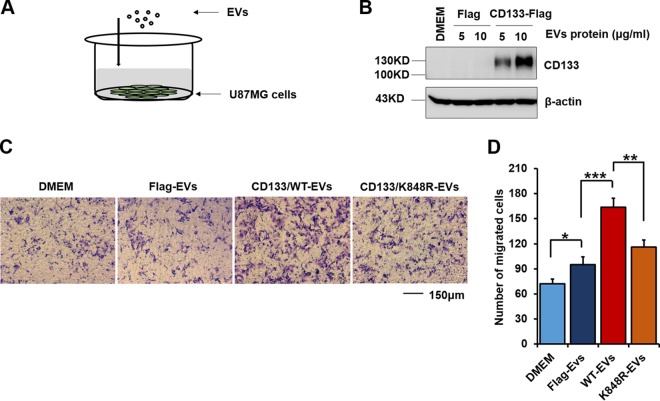
CD133 could be taken up by feeder cells (16), with EVs driving invasive behavior (54). We addressed whether CD133 EVs could be internalized by recipient cells and whether this contributed to cell migration by adding isolated EVs to U87MG cell culture medium (Fig. 9A). Interestingly, after 5 h of incubation, expression of CD133 was observed in recipient cells by use of Western blotting (Fig. 9B). In the Transwell assay, incubation of recipient cells with CD133-WT EVs (10 μg/ml CD133-WT EV protein) promoted recipient cell migration, while adding an equal volume of EVs from CD133-K848R cells showed weakened cell migration (Fig. 9C and D). Collectively, these results show that CD133 EVs are transferred to recipient cells and facilitate cell migration.
FIG 9.
CD133-EVs are taken up by recipient cells and promote cell migration. EVs were harvested from U87MG cells expressing Flag, CD133-WT, or CD133-K848R. (A) Model of U87MG cell incubation with isolated EVs. (B) The indicated EVs were added to the U87MG cell culture medium. After incubation with EVs for 5 h, the cells were washed with PBS, and the expression of CD133 in cell lysates was detected by Western blotting. The expression of β-actin served as a loading control. (C) Representative images from a Transwell assay of cells treated with equal volumes of CD133-WT-, CD133-K848R-, and Flag-EVs. (D) Cell migration numbers were calculated and are presented as means and SD (n = 6). Statistical analysis was performed using the Student t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). EVs, extracellular vesicles.
DISCUSSION
Ubiquitin modification influences cargo trafficking in the secretory and endocytic pathways (29, 31, 32). The present study provides evidence that monoubiquitination contributes to CD133 secretion and demonstrates that Nedd4 is specifically required for CD133 ubiquitination and secretion. Additionally, CD133 undergoes monoubiquitination at its Lys848 residue. Mechanistically, Lys848 ubiquitination regulates CD133 secretion through binding to Tsg101, and mutation of Lys848 significantly blocks CD133 secretion. Importantly, CD133 EVs are taken up by recipient cells and facilitate cell migration. Thus, monoubiquitination at Lys848 of the cancer stem cell marker CD133 regulates its secretion and promotes cell migration.
CD133, a widely known cancer stem cell marker, plays a vital role in tumor progression (5–8). CD133-containing vesicles are reported to be upregulated in cancer (58). CD133-containing vesicles are also released during stem cell differentiation (16, 23). It is therefore apparent that extracellular vesicles are important in relation to the disease. However, the mechanisms underlying CD133 secretion are still largely unknown. We showed in this study that ubiquitin modification contributed to CD133 secretion. We observed that CD133 was monoubiquitinated and demonstrated that the E3 ligase Nedd4 was required for CD133 ubiquitination. Knockdown of Nedd4 by use of siRNA significantly blocked CD133 secretion. We identified Lys848 as a site for CD133 ubiquitination and showed that Lys848 mutation or inhibition of the E1 activating enzyme by use of the PYR-41 or PYZD-4409 inhibitor reduced CD133 ubiquitination and blocked its secretion. These results confirmed the contribution of ubiquitination to the regulation of CD133 secretion.
Cargo selection during MVB sorting is dependent on several endosomal protein complexes, such as ESCRT. Several ESCRT components contain ubiquitin-binding domains (UBDs) that may act as receptors for ubiquitinated proteins. For instance, ESCRT-0, comprising Hrs and STAM, contains Ub-interacting motifs (UIMs) required for MVB sorting of ubiquitinated cargo. Tsg101, an ESCRT-I component which contains a Ub E2 variant domain (UEV), has been implicated in MVB cargo selection. Tsg101 also mediates the release of microvesicles from the plasma membrane through its interaction with ubiquitinated proteins (30). In this study, we found that Tsg101 bound to CD133 and that this binding was disturbed by either Lys848 mutation or ubiquitin protein competitive binding, indicating that ubiquitination mediated CD133 secretion that was at least partly dependent on CD133-Tsg101 binding. However, Lys848 mutation did not affect the binding of CD133 and Hrs. We speculated that Hrs may bind to CD133 in a K848 ubiquitination-independent manner. We must acknowledge that Lys848 is not the only monoubiquitination site, because CD133 is still ubiquitinated in the K848R mutant. The roles of other monoubiquitination sites in CD133 secretion should be examined in the future. Nevertheless, monoubiquitination at Lys848 of CD133 facilitates its secretion.
An interesting finding of our study is that ubiquitination is related to the degree of CD133 N-glycosylation. We demonstrated that ubiquitination occurred primarily on complex glycosylated CD133. One possible implication of this is that different types of N-glycosylation regulate CD133 cell surface distribution. For example, complex N-glycosylation also determines cell surface recognition (27). Another possibility is that N-glycosylation alters the interaction between CD133 and other proteins intracellularly. For example, glycosylation site mutation of CD133 reduces the binding of CD133 to β-catenin (35). However, because our study found that only a small proportion of complex glycosylated CD133 was ubiquitinated, we cannot exclude the possibility of a glycosylation-dependent specific ligand-CD133 receptor interaction regulating CD133 ubiquitination. The mechanisms underlying the effects of glycosylation on CD133 ubiquitination and EV secretion should be explored further in the future.
In summary, we have identified ubiquitination-mediated transfer of CD133 into the extracellular environment and recipient cells. Lys848 ubiquitination of CD133 drives CD133-containing vesicles toward the extracellular space by means of Tsg101 binding. Importantly, CD133 EVs are taken up by recipient cells and promote cell migration. Our results demonstrate a mechanism for CD133 secretion and provide a clue to understanding the mechanisms of cell-cell intercommunication.
MATERIALS AND METHODS
Reagents and antibodies.
Endoglycosidase H and peptide-N-glycosidase F were obtained from New England BioLabs (Beverly, MA). PCR reagents and the DpnI enzyme were obtained from TaKaRa. Pfx DNA polymerase, Lipofectamine 2000 transfection reagent, and Dulbecco's modified Eagle's medium (DMEM) were obtained from Invitrogen. Fetal bovine serum (FBS) was obtained from Biological Industries. A protease inhibitor mixture was obtained from Roche Applied Science. PYR-41 and PYZD-4409 were obtained from Med Chem Express (MCE). Brefeldin A, anti-Ub (P4D1) antibody, anti-LAMP1 antibody, anti-Hrs antibody, anti-STAM antibody, anti-GM130 antibody, anti-HA antibody, and anti-Flag antibody were obtained from Cell Signaling Technology (Danvers, MA). Anti-FK1 (polyubiquitin) antibody was obtained from Bio-Rad. Anti-Tsg101 antibody and anti-Nedd4 antibody were obtained from Abcam. Phaseolus vulgaris leucoagglutinin (PHA-L) and concanavalin A (ConA) were obtained from Vector Laboratories. Anti-Hsp90 antibody was obtained from Santa Cruz Biotechnology. Donkey anti-rabbit antibody–Alexa Fluor 488 and donkey anti-mouse antibody–Alexa Fluor 594 were obtained from Invitrogen. Mouse antiactin antibody, Hoechst 33258 dye, and the Flag peptide were obtained from Sigma. Protein G-agarose was obtained from Roche Life Science. Mouse anti-CD133 (W6B3C1) antibody was obtained from Miltenyi Biotec. Horseradish peroxidase (HRP)-conjugated secondary antibodies and normal mouse IgG antibody were obtained from Santa Cruz Biotechnology.
Cell culture.
HEK293T cells, U87MG cells, and wild-type or mutant CD133-overexpressing cells were cultured in DMEM containing 10% FBS, 50 mg/ml streptomycin, and 100 U/ml penicillin at 37°C in a humidified 5% CO2 incubator. For serum starvation, cells were washed three times with PBS and cultured in DMEM without FBS for 24 to 48 h. GBM1 cells were isolated from primary surgical GBM biopsy specimens in accordance with protocols approved by the Fudan University Institutional Review Boards. GBM1 cells were cultured in DMEM–F-12 medium containing B27 supplement lacking vitamin A (Invitrogen), 2 μg/ml heparin (Sigma), 20 ng/ml epidermal growth factor (EGF; Chemicon), and 20 ng/ml fibroblast growth factor 2 (FGF-2; Chemicon) for a short period before treatment and analysis. For examination of the differentiation capacity of GBM1, GBM1 cells were plated onto polylysine-coated coverslips in DMEM containing 2% FBS for 7 days.
Western blotting.
Cells were lysed with buffer containing 0.5% sodium dodecyl sulfate (SDS), 5% mercaptoethanol, and 1% protease inhibitor mixture. The process of Western blotting was performed as described previously (9). The dilution ratios of primary antibodies were as follows: CD133 (W6B3C1), 1:500; Ub-P4D1, 1:1,000; Ub-FK1, 1:2,000; PHA-L, 1:500; ConA, 1:500; HA, 1:1,000; Tsg101, 1:2,000; Hsp90, 1:2,000; GM130, 1:1,000; Nedd4, 1:2,000; Flag, 1:1,000; Hrs, 1:1,000; STAM, 1:1,000; and β-actin, 1:5,000.
Plasmids, transfection, and lentivirus production and infection.
The cDNA for CD133 was amplified by PCR and cloned into the pRRLSIN.cPPT.PGK vector to create full-length CD133 with a Flag tag (9). The Lys848 residue of CD133-Flag was mutated to arginine by use of PCR, the DpnI enzyme, and the following primers: CD133 (K848R) sense, CGAGATCATGTATATGGTATTCACAATCCT; and CD133 (K848R) antisense, ATATACATGATCTCGATGATAACCATTATT. The pRK5-HA-Ub (WT) and pRK5-HA-Ub (KO) plasmids were provided by Ronggui Hu (59). All the plasmids were verified by DNA sequencing. For ectopic expression of CD133, transient transfection of HEK293T cells was carried out by use of Lipofectamine 2000. Cells were harvested 48 to 72 h after transfection. Lentivirus production and infection were performed as described previously (9).
Immunoprecipitation and CD133 protein purification.
Cells transfected with CD133 or the CD133 site-directed mutant were lysed at 4°C for 2 h by use of lysis buffer (150 mM NaCl, 100 mM Tris [pH 8.0], 0.5% Triton X-100, 1 mM EDTA, protease inhibitor mixture, 1 mM phenylmethylsulfonyl fluoride), and then insoluble materials were removed by centrifugation at 12,000 × g for 10 min. The supernatants of cell lysates were precleared by incubation with protein G-agarose (Roche) at 4°C for 2 h. Anti-Flag antibody-conjugated agarose gel (Flag-M2; Sigma) was incubated with the cell lysates overnight under constant agitation at 4°C. After incubation, the beads with antibody were washed three times in lysis buffer to remove nonspecific binding proteins from the agarose. The collected protein complexes were then analyzed by Western blotting.
To purify the CD133 protein, the lysis and incubation processes were the same as those described above. Flag peptide (100 μg/ml) was used to elute Flag-tagged CD133. The eluate was concentrated to a volume of 20 μl by use of an ultrafiltration tube (Millipore). The purity of enriched CD133 protein was determined by Coomassie blue staining.
Identification of ubiquitination sites by MS.
The identification of ubiquitination sites by MS has been described previously (60). Briefly, CD133 protein was fractionated by 8% SDS-PAGE, and the protein bands were visualized using Coomassie blue staining. The 130-kDa gel lanes were cut into pieces, followed by in-gel trypsin (sequencing grade; Promega) digestion. The peptide samples were analyzed by Q Exactive MS. We analyzed K-ε-GG MS data. Search parameters included the mass tolerance of precursor ions (±2 Da), no enzyme restriction, and a fixed modification of ubiquitinated Lys (114.0429). Only b and y ions were considered during the database match.
Immunofluorescence assay.
Cells were grown on coverslips, fixed with 4% paraformaldehyde (PFA) for 40 min at room temperature, and gently washed three times with PBS. Cells were then blocked with PBS containing 5% normal serum and 0.3% Triton X-100. Cells were sequentially incubated overnight at 4°C with rabbit antibody and mouse monoclonal anti-CD133 (1:40). After washing three times with PBS, cells were incubated with donkey anti-rabbit antibody–Alexa Fluor 488 (1:400) and donkey anti-mouse antibody–Alexa Fluor 594 (1:500) for double immunofluorescence staining. Nuclei were counterstained with Hoechst 33258 (10 g/ml). Immunofluorescence images were collected on a Leica TCS SP5 confocal microscope. For quantification, briefly, fluorescence intensity signals were measured in selected regions with similar areas. Quantification of colocalization was performed for at least 10 cells per experiment, using LAS AF Lite software.
Extracellular vesicle preparation.
Conditioned cell medium was subjected to differential centrifugation at 4°C as follows: 300 × g for 5 min, 500 × g for 5 min, 1,200 × g for 20 min, and 10,000 × g for 30 min. The 10,000 × g supernatant was subjected to filtration with 0.22-μm low-protein-binding Millex-GV filters (Millipore); the filtrate was then concentrated by use of Amicon Ultracel-10K tubes (Millipore) according to the manufacturer's instructions. The concentrate was diluted 1:1 (vol/vol) with PBS and then concentrated by use of Amicon Ultracel-10K tubes (Millipore). The resulting concentrate was resuspended in Laemmli buffer and analyzed by Western blotting.
Electron microscopy.
Transmission electron microscopy was performed according to standard protocols. In brief, EVs were fixed in 100 μl of 2.5% (wt/vol) glutaraldehyde (Sigma) in PBS. The EV preparation was applied to a 200-mesh copper grid supported with Formvar-carbon (ProSciTech, Kirwan, Australia), and the grid was air dried. Grids were washed, negatively stained with 3% saturated aqueous uranyl acetate, and viewed with a transmission electron microscope (CM120; Philips, Netherlands).
Sucrose gradient centrifugation.
The harvested EVs were resuspended in PBS containing 1% protease inhibitor mixture, placed on top of the equilibrium sucrose gradient (0.5 to 1.4 M), and centrifuged at 75,000 × g for 10 h. After centrifugation, fractions of equal volume were collected from the top to the bottom of the gradient and then subjected to Western blotting.
Transient transfection with siRNAs.
U87MG/CD133-Flag cells, seeded at 5.0 × 105 cells per well in a 6-well dish, were grown overnight. The cells were then transiently transfected with 100 nM (each) specific siRNAs by use of Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). siRNAs were designed to target the following specific mRNA sequences: Nedd4 siRNA1, 5′-UAGAGCCUGGCUGGGUUGUUU-3′; Nedd4 siRNA2, 5′-CCAUGAAUCUAGAAGAACA-3′; Hrs siRNA1, 5′-AGAGACAAGUGGAGGUAAA-3′; Hrs siRNA2, 5′-CGACAAGAACCCACACGUC-3′; STAM siRNA1, 5′-UAACUUGGUAUAUAAGGAAAGGGCC-3′; STAM siRNA2, 5′-AUACAUGGAAUACAUCGGAUCUUCG-3′; Tsg101 siRNA1, 5′-CCAGUCUUCUCUCGUCCUA-3′; and Tsg101 siRNA2, 5′-CCUCCAGUCUUCUCUCGUC-3′. A nonspecific (ns) siRNA (5′-UUCUCCGAACGUGUCACGU-3′) was used as a negative control.
Wound healing assay.
In the wound healing assay, cells were seeded in 6-well plates. Scratches were performed by use of a tip when cells were 70 to 80% confluent. PBS was used to remove the nonadherent cells, and the cells were then cultured in medium without FBS. The rate of migration was analyzed by measuring the distance of cell migration in the same view at 0 h and 24 h.
Transwell assay.
Transwell chambers (8 μm; Corning Costar Co., Cambridge, MA) were used according to the manufacturer's instructions. Briefly, after serum starvation for 12 h, cells were resuspended in serum-free medium to a final concentration of 2 × 105/ml. The cell suspension (200 μl) was pipetted into the top chamber, and medium (600 μl) containing 10% FBS was added to the lower chamber. After 12 h of incubation, the cells on the upper side of the membrane were removed with cotton swabs, and cells that migrated to the lower surface were fixed with 4% paraformaldehyde for 30 min and stained with 1% crystal violet in 2% ethanol for 20 min. Cells in 6 randomly selected fields for each section were counted under a microscope.
Statistical analysis.
Statistical analysis was performed by using Microsoft Excel. Comparisons of categorical data were carried out by use of the unpaired two-tailed Student t test. Data are presented as means ± standard deviations (SD). P values of <0.05 were considered statistically significant.
ACKNOWLEDGMENTS
We thank Yalin Huang (Fudan University) for technical assistance with confocal microscopy analysis, Maurizio Pesce for providing the plasmid pRRLSIN.cPPT.hPGK-GFP.WPRE, Bernd Giebel and Denis Corbeil for providing the CD133 plasmid, Haojie Lu (Fudan University) and Liqi Xie (Fudan University) for MS analysis, and Ronggui Hu for providing HA-Ub plasmids.
This work was supported by the National Key R&D Program of China (grant 2016YFA0501303) and the National Natural Scientific Foundation of China (grants 81472724, 31770856, and 81773164). This work was also supported by the Key Laboratory of Glycoconjugate Research, Fudan University, Ministry of Public Health.
We declare that we have no financial and personal relationships with other people or organizations that could inappropriately influence (bias) our work in this study.
F.Y. conducted most of the experiments, analyzed the results, and wrote most of the paper. J.J., Y.W., and Z.A. designed the research. Y.X., X.C., and Y.L. contributed valuable discussions during the study.
REFERENCES
- 1.Yin AH, Miraglia S, Zanjani ED, Almeida-Porada G, Ogawa M, Leary AG, Olweus J, Kearney J, Buck DW. 1997. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood 90:5002–5012. [PubMed] [Google Scholar]
- 2.Wu Y, Wu PY. 2009. CD133 as a marker for cancer stem cells: progresses and concerns. Stem Cells Dev 18:1127–1134. doi: 10.1089/scd.2008.0338. [DOI] [PubMed] [Google Scholar]
- 3.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. 2004. Identification of human brain tumour initiating cells. Nature 432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 4.Richardson GD, Robson CN, Lang SH, Neal DE, Maitland NJ, Collins AT. 2004. CD133, a novel marker for human prostatic epithelial stem cells. J Cell Sci 117:3539–3545. doi: 10.1242/jcs.01222. [DOI] [PubMed] [Google Scholar]
- 5.Yin S, Li J, Hu C, Chen X, Yao M, Yan M, Jiang G, Ge C, Xie H, Wan D, Yang S, Zheng S, Gu J. 2007. CD133 positive hepatocellular carcinoma cells possess high capacity for tumorigenicity. Int J Cancer 120:1444–1450. doi: 10.1002/ijc.22476. [DOI] [PubMed] [Google Scholar]
- 6.Bao S, Wu Q, Sathornsumetee S, Hao Y, Li Z, Hjelmeland AB, Shi Q, McLendon RE, Bigner DD, Rich JN. 2006. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res 66:7843–7848. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- 7.Sansone P, Ceccarelli C, Berishaj M, Chang Q, Rajasekhar VK, Perna F, Bowman RL, Vidone M, Daly L, Nnoli J, Santini D, Taffurelli M, Shih NN, Feldman M, Mao JJ, Colameco C, Chen J, DeMichele A, Fabbri N, Healey JH, Cricca M, Gasparre G, Lyden D, Bonafe M, Bromberg J. 2016. Self-renewal of CD133 (hi) cells by IL6/Notch3 signalling regulates endocrine resistance in metastatic breast cancer. Nat Commun 7:10442. doi: 10.1038/ncomms10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nomura A, Banerjee S, Chugh R, Dudeja V, Yamamoto M, Vickers SM, Saluja AK. 2015. CD133 initiates tumors, induces epithelial-mesenchymal transition and increases metastasis in pancreatic cancer. Oncotarget 6:8313–8322. doi: 10.18632/oncotarget.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei Y, Jiang Y, Zou F, Liu Y, Wang S, Xu N, Xu W, Cui C, Xing Y, Liu Y, Cao B, Liu C, Wu G, Ao H, Zhang X, Jiang J. 2013. Activation of PI3K/Akt pathway by CD133-p85 interaction promotes tumorigenic capacity of glioma stem cells. Proc Natl Acad Sci U S A 110:6829–6834. doi: 10.1073/pnas.1217002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mak AB, Nixon AM, Kittanakom S, Stewart JM, Chen GI, Curak J, Gingras AC, Mazitschek R, Neel BG, Stagljar I, Moffat J. 2012. Regulation of CD133 by HDAC6 promotes beta-catenin signaling to suppress cancer cell differentiation. Cell Rep 2:951–963. doi: 10.1016/j.celrep.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weigmann A, Corbeil D, Hellwig A, Huttner WB. 1997. Prominin, a novel microvilli-specific polytopic membrane protein of the apical surface of epithelial cells, is targeted to plasmalemmal protrusions of non-epithelial cells. Proc Natl Acad Sci U S A 94:12425–12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huttner HB, Janich P, Kohrmann M, Jaszai J, Siebzehnrubl F, Blumcke I, Suttorp M, Gahr M, Kuhnt D, Nimsky C, Krex D, Schackert G, Lowenbruck K, Reichmann H, Juttler E, Hacke W, Schellinger PD, Schwab S, Wilsch-Brauninger M, Marzesco AM, Corbeil D. 2008. The stem cell marker prominin-1/CD133 on membrane particles in human cerebrospinal fluid offers novel approaches for studying central nervous system disease. Stem Cells 26:698–705. doi: 10.1634/stemcells.2007-0639. [DOI] [PubMed] [Google Scholar]
- 13.Marzesco AM, Janich P, Wilsch-Brauninger M, Dubreuil V, Langenfeld K, Corbeil D, Huttner WB. 2005. Release of extracellular membrane particles carrying the stem cell marker prominin-1 (CD133) from neural progenitors and other epithelial cells. J Cell Sci 118:2849–2858. doi: 10.1242/jcs.02439. [DOI] [PubMed] [Google Scholar]
- 14.Pisitkun T, Shen RF, Knepper MA. 2004. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A 101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karbanova J, Laco J, Marzesco AM, Janich P, Vobornikova M, Mokry J, Fargeas CA, Huttner WB, Corbeil D. 2014. Human prominin-1 (CD133) is detected in both neoplastic and non-neoplastic salivary gland diseases and released into saliva in a ubiquitinated form. PLoS One 9:e98927. doi: 10.1371/journal.pone.0098927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bauer N, Wilsch-Brauninger M, Karbanova J, Fonseca AV, Strauss D, Freund D, Thiele C, Huttner WB, Bornhauser M, Corbeil D. 2011. Haematopoietic stem cell differentiation promotes the release of prominin-1/CD133-containing membrane vesicles—a role of the endocytic-exocytic pathway. EMBO Mol Med 3:398–409. doi: 10.1002/emmm.201100147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kucharzewska P, Christianson HC, Welch JE, Svensson KJ, Fredlund E, Ringner M, Morgelin M, Bourseau-Guilmain E, Bengzon J, Belting M. 2013. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc Natl Acad Sci U S A 110:7312–7317. doi: 10.1073/pnas.1220998110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan M, Nickoloff E, Abramova T, Johnson J, Verma SK, Krishnamurthy P, Mackie AR, Vaughan E, Garikipati VN, Benedict C, Ramirez V, Lambers E, Ito A, Gao E, Misener S, Luongo T, Elrod J, Qin G, Houser SR, Koch WJ, Kishore R. 2015. Embryonic stem cell-derived exosomes promote endogenous repair mechanisms and enhance cardiac function following myocardial infarction. Circ Res 117:52–64. doi: 10.1161/CIRCRESAHA.117.305990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis DM. 2007. Intercellular transfer of cell-surface proteins is common and can affect many stages of an immune response. Nat Rev Immunol 7:238–243. doi: 10.1038/nri2020. [DOI] [PubMed] [Google Scholar]
- 20.Thery C, Ostrowski M, Segura E. 2009. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol 9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 21.Muralidharan-Chari V, Clancy JW, Sedgwick A, D'Souza-Schorey C. 2010. Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci 123:1603–1611. doi: 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathivanan S, Ji H, Simpson RJ. 2010. Exosomes: extracellular organelles important in intercellular communication. J Proteomics 73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Dubreuil V, Marzesco AM, Corbeil D, Huttner WB, Wilsch-Brauninger M. 2007. Midbody and primary cilium of neural progenitors release extracellular membrane particles enriched in the stem cell marker prominin-1. J Cell Biol 176:483–495. doi: 10.1083/jcb.200608137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Hung MC. 2001. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol 3:973–982. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 25.Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, Pavletich NP, Carver BS, Cordon-Cardo C, Erdjument-Bromage H, Tempst P, Chi SG, Kim HJ, Misteli T, Jiang X, Pandolfi PP. 2007. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 128:141–156. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grovdal LM, Stang E, Sorkin A, Madshus IH. 2004. Direct interaction of Cbl with pTyr 1045 of the EGF receptor (EGFR) is required to sort the EGFR to lysosomes for degradation. Exp Cell Res 300:388–395. doi: 10.1016/j.yexcr.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Mak AB, Blakely KM, Williams RA, Penttila PA, Shukalyuk AI, Osman KT, Kasimer D, Ketela T, Moffat J. 2011. CD133 protein N-glycosylation processing contributes to cell surface recognition of the primitive cell marker AC133 epitope. J Biol Chem 286:41046–41056. doi: 10.1074/jbc.M111.261545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mak AB, Pehar M, Nixon AM, Williams RA, Uetrecht AC, Puglielli L, Moffat J. 2014. Post-translational regulation of CD133 by ATase1/ATase2-mediated lysine acetylation. J Mol Biol 426:2175–2182. doi: 10.1016/j.jmb.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacGurn JA, Hsu PC, Emr SD. 2012. Ubiquitin and membrane protein turnover: from cradle to grave. Annu Rev Biochem 81:231–259. doi: 10.1146/annurev-biochem-060210-093619. [DOI] [PubMed] [Google Scholar]
- 30.Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. 2012. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci U S A 109:4146–4151. doi: 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukhopadhyay D, Riezman H. 2007. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 32.MacDonald C, Buchkovich NJ, Stringer DK, Emr SD, Piper RC. 2012. Cargo ubiquitination is essential for multivesicular body intralumenal vesicle formation. EMBO Rep 13:331–338. doi: 10.1038/embor.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staub O, Rotin D. 2006. Role of ubiquitylation in cellular membrane transport. Physiol Rev 86:669–707. doi: 10.1152/physrev.00020.2005. [DOI] [PubMed] [Google Scholar]
- 34.Barberon M, Zelazny E, Robert S, Conejero G, Curie C, Friml J, Vert G. 2011. Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc Natl Acad Sci U S A 108:E450–E458. doi: 10.1073/pnas.1100659108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Ren S, Xie L, Cui C, Xing Y, Liu C, Cao B, Yang F, Li Y, Chen X, Wei Y, Lu H, Jiang J. 2015. Mutation of N-linked glycosylation at Asn548 in CD133 decreases its ability to promote hepatoma cell growth. Oncotarget 6:20650–20660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rayon C, Lerouge P, Faye L. 1998. The protein N-glycosylation in plants. J Exp Bot 49:1163–1172. doi: 10.1093/jxb/49.324.1163. [DOI] [Google Scholar]
- 37.Miraglia S, Godfrey W, Yin AH, Atkins K, Warnke R, Holden JT, Bray RA, Waller EK, Buck DW. 1997. A novel five-transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood 90:5013–5021. [PubMed] [Google Scholar]
- 38.Zhou F, Cui C, Ge Y, Chen H, Li Q, Yang Z, Wu G, Sun S, Chen K, Gu J, Jiang J, Wei Y. 2010. Alpha2,3-sialylation regulates the stability of stem cell marker CD133. J Biochem 148:273–280. doi: 10.1093/jb/mvq062. [DOI] [PubMed] [Google Scholar]
- 39.Horak J, Wolf DH. 2001. Glucose-induced monoubiquitination of the Saccharomyces cerevisiae galactose transporter is sufficient to signal its internalization. J Bacteriol 183:3083–3088. doi: 10.1128/JB.183.10.3083-3088.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin H, Shao Q, Igdoura SA, Alaoui-Jamali MA, Laird DW. 2003. Lysosomal and proteasomal degradation play distinct roles in the life cycle of Cx43 in gap junctional intercellular communication-deficient and -competent breast tumor cells. J Biol Chem 278:30005–30014. doi: 10.1074/jbc.M300614200. [DOI] [PubMed] [Google Scholar]
- 41.Lancaster GI, Febbraio MA. 2005. Exosome-dependent trafficking of HSP70: a novel secretory pathway for cellular stress proteins. J Biol Chem 280:23349–23355. doi: 10.1074/jbc.M502017200. [DOI] [PubMed] [Google Scholar]
- 42.Gulia R, Sharma R, Bhattacharyya S. 2017. A critical role for ubiquitination in the endocytosis of glutamate receptors. J Biol Chem 292:1426–1437. doi: 10.1074/jbc.M116.752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu GW, Ali M, Wood TE, Wong D, Maclean N, Wang X, Gronda M, Skrtic M, Li X, Hurren R, Mao X, Venkatesan M, Beheshti ZR, Ketela T, Reed JC, Rose D, Moffat J, Batey RA, Dhe-Paganon S, Schimmer AD. 2010. The ubiquitin-activating enzyme E1 as a therapeutic target for the treatment of leukemia and multiple myeloma. Blood 115:2251–2259. doi: 10.1182/blood-2009-07-231191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katzmann DJ, Babst M, Emr SD. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145–155. doi: 10.1016/S0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 45.Shields SB, Oestreich AJ, Winistorfer S, Nguyen D, Payne JA, Katzmann DJ, Piper R. 2009. ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. J Cell Biol 185:213–224. doi: 10.1083/jcb.200811130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamai K, Tanaka N, Nakano T, Kakazu E, Kondo Y, Inoue J, Shiina M, Fukushima K, Hoshino T, Sano K, Ueno Y, Shimosegawa T, Sugamura K. 2010. Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT-0 protein. Biochem Biophys Res Commun 399:384–390. doi: 10.1016/j.bbrc.2010.07.083. [DOI] [PubMed] [Google Scholar]
- 47.Bache KG, Raiborg C, Mehlum A, Stenmark H. 2003. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J Biol Chem 278:12513–12521. doi: 10.1074/jbc.M210843200. [DOI] [PubMed] [Google Scholar]
- 48.Vardhana S, Choudhuri K, Varma R, Dustin ML. 2010. Essential role of ubiquitin and TSG101 protein in formation and function of the central supramolecular activation cluster. Immunity 32:531–540. doi: 10.1016/j.immuni.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fotia AB, Ekberg J, Adams DJ, Cook DI, Poronnik P, Kumar S. 2004. Regulation of neuronal voltage-gated sodium channels by the ubiquitin-protein ligases Nedd4 and Nedd4-2. J Biol Chem 279:28930–28935. doi: 10.1074/jbc.M402820200. [DOI] [PubMed] [Google Scholar]
- 50.Kawabe H, Neeb A, Dimova K, Young SJ, Takeda M, Katsurabayashi S, Mitkovski M, Malakhova OA, Zhang DE, Umikawa M, Kariya K, Goebbels S, Nave KA, Rosenmund C, Jahn O, Rhee J, Brose N. 2010. Regulation of Rap2A by the ubiquitin ligase Nedd4-1 controls neurite development. Neuron 65:358–372. doi: 10.1016/j.neuron.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nabhan JF, Pan H, Lu Q. 2010. Arrestin domain-containing protein 3 recruits the NEDD4 E3 ligase to mediate ubiquitination of the beta2-adrenergic receptor. EMBO Rep 11:605–611. doi: 10.1038/embor.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Putz U, Howitt J, Doan A, Goh CP, Low LH, Silke J, Tan SS. 2012. The tumor suppressor PTEN is exported in exosomes and has phosphatase activity in recipient cells. Sci Signal 5:ra70. doi: 10.1126/scisignal.2003084. [DOI] [PubMed] [Google Scholar]
- 53.Fukushima T, Yoshihara H, Furuta H, Kamei H, Hakuno F, Luan J, Duan C, Saeki Y, Tanaka K, Iemura S, Natsume T, Chida K, Nakatsu Y, Kamata H, Asano T, Takahashi S. 2015. Nedd4-induced monoubiquitination of IRS-2 enhances IGF signalling and mitogenic activity. Nat Commun 6:6780. doi: 10.1038/ncomms7780. [DOI] [PubMed] [Google Scholar]
- 54.Hoshino D, Kirkbride KC, Costello K, Clark ES, Sinha S, Grega-Larson N, Tyska MJ, Weaver AM. 2013. Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Rep 5:1159–1168. doi: 10.1016/j.celrep.2013.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu C, Li Y, Xing Y, Cao B, Yang F, Yang T, Ai Z, Wei Y, Jiang J. 2016. The interaction between cancer stem cell marker CD133 and Src protein promotes focal adhesion kinase (FAK) phosphorylation and cell migration. J Biol Chem 291:15540–15550. doi: 10.1074/jbc.M115.712976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Imjeti NS, Menck K, Egea-Jimenez AL, Lecointre C, Lembo F, Bouguenina H, Badache A, Ghossoub R, David G, Roche S, Zimmermann P. 2017. Syntenin mediates SRC function in exosomal cell-to-cell communication. Proc Natl Acad Sci U S A 114:12495–12500. doi: 10.1073/pnas.1713433114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tu C, Ortega-Cava CF, Winograd P, Stanton MJ, Reddi AL, Dodge I, Arya R, Dimri M, Clubb RJ, Naramura M, Wagner KU, Band V, Band H. 2010. Endosomal-sorting complexes required for transport (ESCRT) pathway-dependent endosomal traffic regulates the localization of active Src at focal adhesions. Proc Natl Acad Sci U S A 107:16107–16112. doi: 10.1073/pnas.1009471107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marimpietri D, Petretto A, Raffaghello L, Pezzolo A, Gagliani C, Tacchetti C, Mauri P, Melioli G, Pistoia V. 2013. Proteome profiling of neuroblastoma-derived exosomes reveal the expression of proteins potentially involved in tumor progression. PLoS One 8:e75054. doi: 10.1371/journal.pone.0075054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Z, Chen P, Gao H, Gu Y, Yang J, Peng H, Xu X, Wang H, Yang M, Liu X, Fan L, Chen S, Zhou J, Sun Y, Ruan K, Cheng S, Komatsu M, White E, Li L, Ji H, Finley D, Hu R. 2014. Ubiquitylation of autophagy receptor Optineurin by HACE1 activates selective autophagy for tumor suppression. Cancer Cell 26:106–120. doi: 10.1016/j.ccr.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Udeshi ND, Mertins P, Svinkina T, Carr SA. 2013. Large-scale identification of ubiquitination sites by mass spectrometry. Nat Protoc 8:1950–1960. doi: 10.1038/nprot.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]