Abstract
Hepatocellular carcinoma (HCC) is one of the most prevalent and lethal cancers. It has been demonstrated that aberrant expression of miRNAs plays an important role in HCC development. Here, we observed decreased miR-125b expression status in tumor samples from HCC patients, and the five years survival rate of HCC patients with low miR-125b expression is poor. By using bioinformatics prediction tools combining with luciferase reporter assay, we identified that miR-125b can suppress the expression of SIRT6 by directly targeting the seed-matching region of its 3’UTR. Based on the analysis via TCGA and clinical samples data, the expression of SIRT6 showed negatively correlated with the expression of mir-125b. After knocking-out the expression of SIRT6 through CRISPR/Cas9, HCC cells showed the decreased cell viability and invasiveness, which had the similar function upon the overexpression of the miR-125b. The function induced by overexpression of miR-125b can be rescued by the restoration of SIRT6. Further experiments demonstrated that the HCC cells showed the significant cellular senescence and apoptosis upon overexpression of miR-125b or knockout SIRT6, which is in accordance with the compromised cell malignancy. Thus, we conclude that, by targeting SIRT6, miR-125b can function as a tumor suppressor to induce the cellular senescence and apoptosis in hepatocellular carcinogenesis and could provide a novel insight for HCC treatment.
Keywords: microRNA, SIRT6, hepatocellular carcinoma, CRISPR/Cas9, senescence
Introduction
Hepatocellular carcinoma (HCC) is the most lethal and frequent type of liver cancer in adults. The mortality of HCC is surprisingly high comparing with the overall cancer incidence. The accumulating evidences have been addressed for the cause of this disease, including the high risk factors like infection of HBV or HCV, exposure to toxins such as alcohol or aflatoxin, the fatty liver diseases, the aberrant gene expression situations, and deregulation of cellular growth signaling pathways [1], however, the molecular pathogenesis of HCC is still not completely clear and needs further studies.
MicroRNAs are short, endogenous non-coding RNA molecules which regulate gene expression in multiple aspects. Post-transcriptional repression of target gene through Argonaute protein family involved RNA-induced silencing complex by base pairing to the 3’UTR of target gene mRNA is considered as the principle function of microRNA. The aberrant expression of microRNA is deeply associated with many diseases and is one of the critical reasons for the initiation, development and prognosis of cancers [2]. Mature miR-125b originates from two precursors: pre-miR-125b-1 (located at chr11q24.1) and pre-miRNA-125b-2 (located at chr21q21.1) [3]. Mounting evidences have proven a role for miR-125b in proliferation, apoptosis and cellular differentiation. The underlying molecular and cellular mechanisms are complex according to the different targets that miR-125b might regulate and different signaling pathways that miR-125b might be involved [4].
SIRT6 is one of the NAD+-dependent Sirtuin family members. The Sirtuin protein family has seven members (SIRT1-SIRT7), which have the different enzymatic activity, subcellular localization, and target specificity and involved in regulation of different biological processes [5-7]. SIRT6 is located in the nucleus and plays a key role in genome stability [8-11], transcriptional regulation [12], metabolism [7,13], inflammation [14,15], and the organismal lifespan. SIRT6 has the protein deacetylase activity, which can deacetylate H3K9ac or H3K56ac and is required for transcriptional repression on NF-κB [16], C-JUN [17], MYC [13], and hypoxia-inducible factor-1α (HIF-1α) [18] mediated pathways. Moreover, SIRT6 can interact with SNF2H [19], CtIP [20] and PARP1 [9] to regulate double strand break (DSB) repair process. SIRT6 konckout mice displayed severe progeroid syndrome, profound hypoglycaemia and lifespan only for four weeks [21].
In recent years, numerous reports implied that the aberrant microRNA expression has the significant correlation with HCC. MiR-125b is frequently down-regulated in HCC patients, and its tumor suppressor role has been defined in several studies [22]. To further understand the mechanism of miR-125b functions in HCC, we screened the potential targets of miR-125b and the significant negative correlation between miR-125b and SIRT6 was found. In addition, we discovered that the high SIRT6 expressions occurred in HCC patients accompanied with miR-125b down-regulation. Furthermore, we addressed the role of miR-125b and SIRT6 on HCC development and revealed the underlying mechanisms of SIRT6 knockdown induced cellular senescence and apoptosis upon miR-125b overexpression. Thus, our study highlight a novel perspective on HCC treatment.
Materials and methods
Human liver cancer tissue samples and cells
Hepatocellular carcinoma and adjacent (control) tissue specimens from 20 patients were used for the study. Written informed consent was obtained from all study participants. This study was approved by the Ethics Committee of Peking University.
HEK293T, HepG2 and Sk-hep-1 cells were grown in DMEM with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA) supplemented with 10% (v/v) FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin (Sigma-Aldrich).
CRISPR/Cas9 knockout cell lines
Both SIRT6 and miR-125b knockout cell lines were generated via CRISPR/Cas9 method. First, sgRNA sequences were designed via CRISPR designer at http://crispr.mit.edu/. Then, the LentiCRISPRv2 plasmid containing sgRNA was cotransfected with viral packaging plasmids (psPAX2 and pMD2G) into HEK293T cells. The medium was changed after six hours of transfection, and viral supernatant was filtered through 0.45-mm strainer 42 hours later. After viral supernatant infection of target cells, 1 mg/mL puromycin was used for cell selection for 2 weeks.
The sgRNA sequences targeting pre-miR-125b-1, pre-miR-125b-2 and exon 4 of human SIRT6 gene are shown.
Pre-miR-125b-1: 5’-ACCGTTTAAATCCACGGGTT-3’. Pre-miR-125b-2: 5’-CTCTTGGGACCTAGGCGGAG-3’. SIRT6: 5’-TACGTCCGAGACACAGTCGT-3’.
Plasmids
To generate wild-type (WT) reporter, the SIRT6 3’-UTR containing miR-125b binding site was amplified and inserted into the psiCHECK-2 vector. For mutated reporter (Mut), we used the overlap PCR method to generate mutated SIRT6 3’-UTR containing mutated miR-125b binding motif. After amplification, the resulting fragment was inserted into the psiCHECK-2 vector. The SIRT6 3’-UTR was amplified with following primers: 5’-CCGCTCGAGCCAGGGTGCTTGGGGA-3’ (forward), and 5’-ATAAGAATGCGGCCGCGCAAGAAAGAAATTGTTTTTATTG-3’ (reverse). And the middle overlap primer: 5’-GAGCACTCAAACTCTGAGAGCTGTGCTCCA-3’ (forward), and 5’-TGGAGCACAGCTCTCAGAGTTTGAGTGCTC-3’ (reverse).
To generate miR-125b overexpression construct, we amplified the pre-miR-125b-1 gene locus with a flank about 100 bp and inserted into a pcDNA3.1. The primers used for amplification are following: 5’-CGGGATCCGAAGAAATACCATACCACCT-3’ (forward), and 5’-GCTCTAGAGAGGTATACT CAATCACCTC-3’ (reverse). The cDNA of SIRT6 was amplified and cloned into pcDNA3.1 vector. And the following primers were used. 5’-CGCGGATCCGCCGCCATGTCGGTGAATTACGCGGC-3’ (forward), and 5’-CCGCTCGAGTCAGCTGGGGACCGCCTTGG-3’ (reverse). All expression constructs were checked by Sanger sequencing.
Western blot
Proteins were analyzed by Western blotting according to the standard methods. Visualization was performed using ECL (Thermo Fisher Scientific, #32106). The antibodies used were commercially obtained: anti-b-actin (Santa Cruz Biotechnology, sc-47778, with 1:10,000 dilution), anti-SIRT1 (Santa Cruz technology, sc-74504, with 1:1,000 dilution), anti-SIRT2 (Santa Cruz technology, sc-20966, with 1:1,000 dilution), anti-SIRT3 (Cell Signaling Technology, #5490S, with 1:1,000 dilution), anti-SIRT4 (Santa Cruz technology, sc-135797, with 1:1,000 dilution), anti-SIRT5 (Cell Signaling Technology, #8782, with 1:1,000 dilution), anti-SIRT6 (Cell Signaling Technology, #12486, with 1:1,000 dilution), anti-SIRT7 (Cell Signaling Technology, #5360S, with 1:1,000 dilution), anti-P16 (Abcam, ab51243, with 1:1,000 dilution), anti-Caspase 3 (Santa Cruz technology, sc-7148, with 1:1,000 dilution) and anti-Bax (Cell Signaling Technology, #2772S, with 1:1,000 dilution).
qRT-PCR
Total RNA, including miRNA, was extracted from cell/tissue with TRIzol reagent (Sigma), and RNA was reverse transcribed into cDNA by qPCR RT Kit (FSQ-101 TOYOBO). The gene expression level was determined with a qPCR SYBR Green Master Mix (Q131-03 Vazyme) using the 7500 Fast Real-time PCR System (Applied Biosystems). MiR-125b-5p expression was assessed by qRT-PCR with the following primers: 5’-GGGTCCCTGAGACCCTAAC-3’ (forward), and 5’-CAGTGCGTGTCGTGGAGT-3’ (reverse). And a stem-loop primer, 5’-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTCACAA-3’, was used for miR-125b-5p reverse transcription. The control primers (U6) were 5’-CTCGCTTCGGCAGCACA-3’ (forward) and 5’-AACGGTTCACGAATTTGCGT-3’ (reverse). SIRT6 mRNA expression was assessed by qRT-PCR with the following primers: 5’-CCAAGTTCGACACCACCTTT-3’ (forward), and 5’-CGGACGTACTGCGTCTTACA-3’ (reverse). The control primers (β-actin) were 5’-CGGGACCTGACTGACTACCTC-3’ (forward) and 5’-ATTGGAACGATACAGAGAAGATT-3’ (reverse). The relative fold expression of the target, normalized to the corresponding control, was calculated by the comparative Ct methods.
Immunohistochemistry
For immunohistochemistry, formalin preserved tissue samples were dehydrated using ethanol and embedded in paraffin. Then formalin-fixed paraffin-embedded tissue was cut in sections of 5 μm thickness for SIRT6 visualization. The paraffin-embedded tissue samples were immunoassayed using the immunohistochemistry detection kit (Zsbio, Beijing, China, #PV-9001) by following the manufacturer’s instructions. SIRT6 (Cell Signaling Technology, #12486, with 1:50 dilution) was stained with DAB (Zsbio, Beijing, China, #ZLI-9018) and counterstained with hematoxylin. Brown-stained cells were considered positive. Stained sections were observed and imaged under a microscopy (Eclipse TS100, Nikon).
Luciferase assay
For luciferase report assay, cells were seeded in 6-well plates at a density of 1×105 cells per well. The cells were cotransfected with luciferase reporters, either wild-type or mutant SIRT6 3’UTR, in combination with miR-125b-5p mimics or negative control by using Lipofectamine 2000. After 48 hours, the firefly and renilla luciferase activity were mearsured and analyzed according to the manufacture’s instruction (Promega). The microRNA mimic was acquired from Ribobio (Guangzhou, China).
Colony formation assay
For colony formation assay, cells were seeded in 6-well cell culture plates (2000 cells per well). Cells were cultured for two weeks, and the medium was kept fresh. Then colonies were fixed with 4% paraformaldehyde and stained with 0.2% crystal violet for 30 mins. The number of colonies with more than 1 mm diameters was counted.
Wound healing assay
The cells were seeded into 6-well cell culture plates and cells overgrew in the dish around 24 hours. Then, a wound track was introduced by scraping the cell monolayer with a sterile yellow plastic tip. Cell migration was observed and imaged by microscopy (Eclipse TS100, Nikon) in different time points.
Transwell assay
For the transwell assay, the inserting chambers (Corning, #3422) coated with MaxGelTM ECM (Sigma-Aldrich, #E0282) were inserted into 24-well culture plate and placed in the cell culture hood for 2 hs at 37°C. Then cells were seeded onto the chambers in the upper wells in serum-free medium, and the lower wells were placed with normal growth medium. After 48 hs, the non-invading cells and Matrigel matrix were gently removed with a cotton swab. The invasive cells attached to the lower surfaces of the chamber membranes were stained with Crystal Violet, counted and photographed by microscopy (Eclipse TS100, Nikon).
SA-β-gal assay
Cellular senescence was assayed via the measurement of β-galactosidase (SA-β-gal) activity with the SA-β-gal staining kit (Cell Signaling Technology, #9860) in accordance with the manufacturer’s instructions. Cells were seeded in 6-well cell culture plate and overgrew around 24 hours. Then, the growth medium was removed and the cells were washed three times with 1 ml phosphate buffered saline (PBS) and fixed with 1 mL of fixative solution for 15 mins at room temperature. After the fixative solution was removed, cells were washed three times with PBS and incubated with the staining solution mix overnight at 37°C in a CO2-free cell culture hood. Staining cells were counted and photographed by microscopy (Eclipse TS100, Nikon).
Statistical analysis
Data are presented as means ± Standard Deviation (SD). Statistical comparisons were determined by two-tailed Student’s t-test. P values of less than 0.05 were considered statistically significant.
Results
MiR-125b is aberrantly expressed in HCC and is associated with sirtuin family
Aberrant miR-125b expression is associated with multiple diseases, and the differential expression status might imply the role of miR-125b involved in HCC carcinogenesis. In order to address the miR-125b expression in HCC, we acquired 20 pairs of clinical HCC samples to examine miR-125b levels by qPCR (Figure 1A). The expression levels of miR-125b were downregulated in HCC tissues compared to the adjacent non-tumor tissues, which is consistent with the TCGA clinical data (Figure 1B). Additionally, the miR-125b downregulated patients have the worse overall survival rates [23] (Figure 1C).
Figure 1.
MiR-125b is aberrantly expressed in HCCs and is associated with sirtuin family. A. The miR-125b expression level in clinical samples was analyzed by qPCR. B. miR-125b is differentially expressed in HCC patients and HTSeq-Counts data acquired from TCGA database, which is shown with a logarithmic conversion (log2counts). Unpaired t-test. C. miR-125b gene is related with the overall survival rate of HCC patients. Red line: high expression, blue line: low expression. Data is acquired from TCGA database and analyzed via LinkedOmics bioinformatics [23]. D. Schematic illustrations showing exact position and numbers of miR-125b binding sites of certain sirtuin family members. E. The predicted binding site of miR-125b in the 3’UTR of SIRT2, SIRT3, SIRT5 SIRT6 and SIRT7 mRNA is analyzed via TargetScan bioinformatics [24]. F. Western blots shows the protein level of SIRT1-SIRT7 in cells transfected with miR-125b mimic or control mimic in HepG2 cell lines. G. The expression of SIRT1-SIRT7 in HCC patients. The results, based on TCGA database, were acquired from GEPIA bioinformatics [25].
To find the potential targets of miR-125b that might be involved in HCC carcinogenesis, we searched the candidate targets by using the bioinformatics tools [24]. Interestingly, among all 7 members of sirtuin family, we found that 5 members, including SIRT2, SIRT3, SIRT5, SIRT6 and SIRT7, have miR-125b binding sites (Figure 1D, 1E). Both SIRT3, SIRT5, SIRT6 and SIRT7 were suppressed under miR-125b mimic transfection in HepG2 cell lines (Figure 1F). Additionally, SIRT6 and SIRT7 has been found deregulated in HCC according to the analysis on TCGA database via GEPIA bioinformatics [25] (Figure 1G).
SIRT6 is the target of miR-125b and is up-regulated in HCCs
We conducted a luciferase assay to confirm the inverse correlation between miR-125b and SIRT6. The 3’UTR of SIRT6 was cloned into luciferase reporter vector (WT-vector) and the resulting vector was co-transfected with miR-125b mimic into 293T cells. The luciferase activity was significantly decreased after dual-transfection with mimic and reporter vector. To investigate whether the seed region of 3’UTR is critical for miR-125b to induce the translation suppression. The mutation was introduced into the seed sequence, which was unable to bind to the predicted miR-125b binding site. The luciferase activities showed no differences after co-transfection with mimic and Mut-vector (Figure 2A). The relationship between miR-125b and SIRT6 was also determined via western blot analysis, which showed that the endogenous SIRT6 protein levels were significantly suppressed upon the overexpression of miR-125b in 293T cells (Figure 2B). In addition, the upregulated expression levels of SIRT6 were also verified by qPCR, western blot and IHC (Immunohistochemistry) assays on clinical HCC samples and TCGA data (Figure 2C-F). Notably, the expression levels of SIRT6 in HCC are negatively correlated with miR-125b. Along with the fact that the patients with high levels of SIRT6 have poor 5 years overall survival rate (Figure 2G), we postulated that miR-125b-SIRT6 regulatory axis might have clinical significance.
Figure 2.
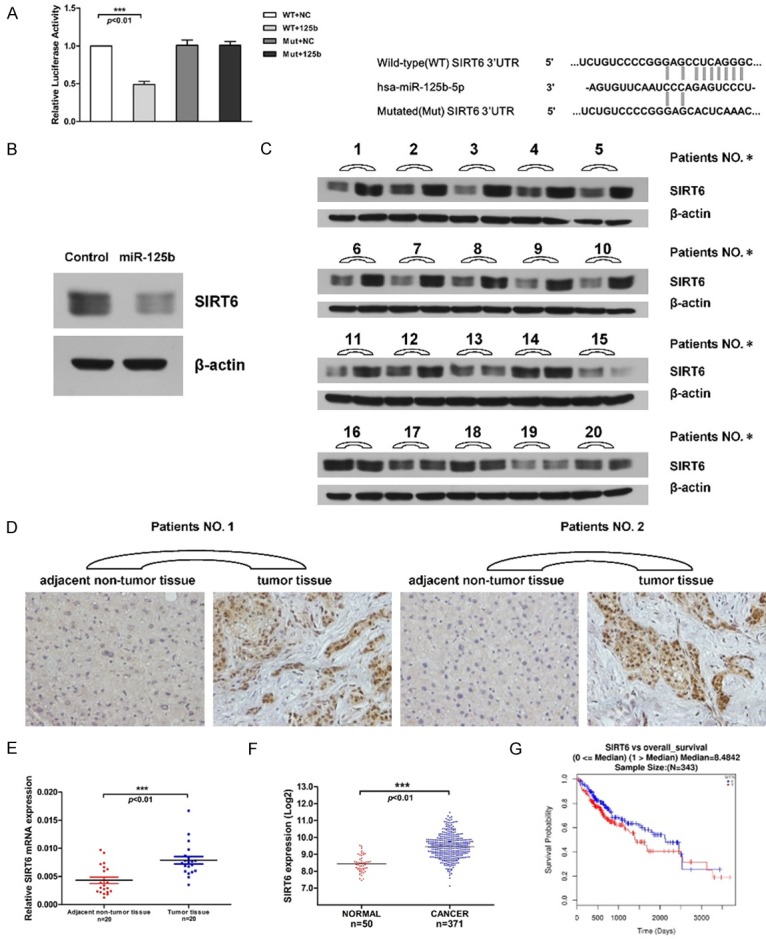
SIRT6 is the target of miR-125b and is up-regulated in HCCs. A. Dual-luciferase reporter assays. Sequences encoding wild-type and mutated fragments, mutated sequence, shown in the right panel, of the SIRT6 3’-UTR were inserted into a luciferase reporter plasmid. MiR-125b mimic was co-transfected with reporter plasmid into 293T cells. Relative renilla luciferase expression was normalized to firefly luciferase. B. Western blots shows the SIRT6 protein level in cells transfected with miR-125b mimic or control mimic. C. SIRT6 protein in tissue samples were measured by western blotting (adjacent non-tumor tissue (left lane) versus tumor tissue (right lane)). β-actin was used as loading control. D. Immunohistochemistry (IHC) shows the SIRT6 expression in adjacent non-tumor tissues (left) and tumor tissues (right). E. The level of SIRT6 mRNA in tissue samples was detected by qRT-PCR. Data were analyzed using 2-ΔCT method in tissue samples and normalized to the expression of β-actin. F. SIRT6 expression in HCC patients and HTSeq-Counts data acquired from TCGA database, which is shown with a logarithmic conversion (log2counts). Unpaired t-test. G. SIRT6 gene is related with the overall survival rate in HCC patients. Red line: high expression, blue line: low expression. Data is acquired from TCGA database and analyzed via Linked Omics bioinformatics [23].
The SIRT6 and miR-125b knockout induced by CRISPR/Cas9 affects the cell proliferation
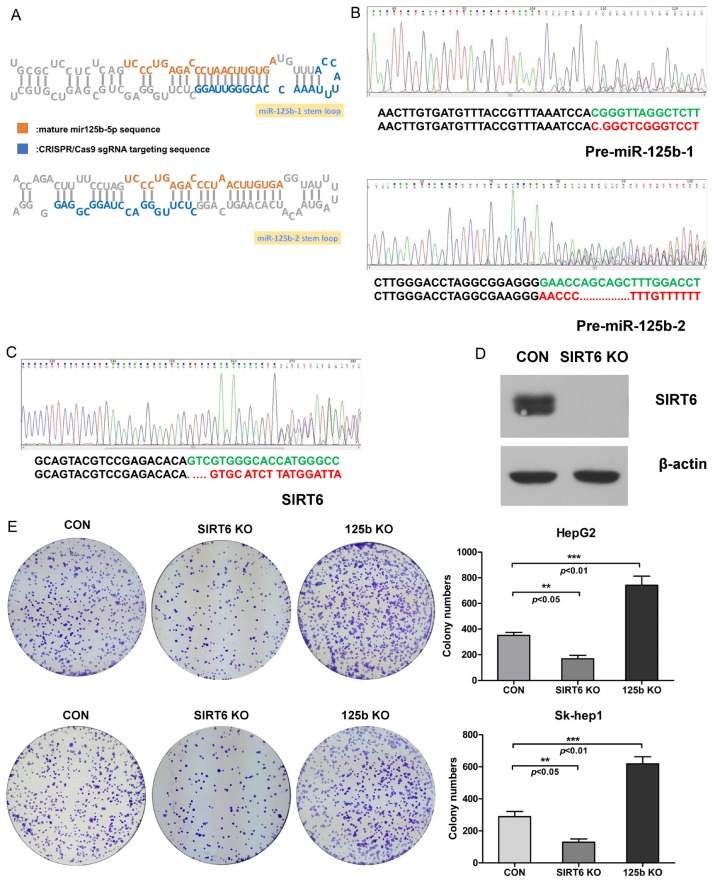
To investigate the role of miR-125b and SIRT6 in HCC carcinogenesis, we generated both miR-125b and SIRT6 knockout (125b KO and SIRT6 KO) HCC cell lines via CRISPR/Cas9 technique. Considering the generation of mature microRNA relies on a series processing on pre-microRNA stem-loop structure, the sgRNA was designed on the stem-loop sequence in order to destroy the spatial conformation (Figure 3A). The genome sequencing results confirmed the miR-125b gene was successfully eliminated (Figure 3B). The SIRT6 knockout cell lines were also generated through CRISPR/Cas9 (Figure 3C) and the SIRT6 expression was completely eliminated in SIRT6 knockout cell lines (Figure 3D). Using these cells, we performed the colony formation assay to examine whether the cell proliferation has been affected. We found the diminished cell proliferation rate in SIRT6 KO cell lines. Whereas, the 125b KO cell lines showed the increased cell proliferation rate (Figure 3E). These observations indicated that miR-125b and SIRT6 affect cell proliferation in an opposite way.
Figure 3.
The SIRT6 and miR-125b knockout induced by CRISPR/Cas9 affects the cell proliferation. A. Schematic illustration on miR-125b knockout design. The orange character represents the mature miR-125b sequence and blue character represents the sgRNA sequence. B. Genome sequencing on pre-miR-125b-1 and pre-miR-125b-2. The expression of miR-125b is successfully eliminated by CRISPR/CAS9 via two sgRNAs targeting both the genomic region of pre-miR-125b-1 and pre-miR-125b-2 separately. C. Genome sequencing on exon 4 of SIRT6. D. The SIRT6 expression in SIRT6 knockout HepG2 cell lines induced by CRISPR/Cas9 was determined via western blot assay. E. The cell proliferation of miR-125b and SIRT6 knockout cell lines was investigated by colony formation assay. Upper lane: HepG2 cell lines. Lower lane: Sk-hep1 cell lines.
MiR-125b-SIRT6 axis affects HCC cell migration
We further examined whether miR-125b and SIRT6 might affect HCC cell migration. Along with the SIRT6 and miR-125b knockout cell lines, the miR-125b overexpression and SIRT6 overexpression (125b OE and SIRT6 OE) HCC cell lines were generated in order to give a comprehensive understanding on the cell function. Through wound scratch assay, we found that miR-125b OE and SIRT6 KO cell lines have the larger wound closure areas comparing to the controls and even larger than that formed by 125b KO and SIRT6 OE cell lines (Figure 4A, 4B). When we reintroduced SIRT6 into 125b OE cell lines, the weakened migration ability was recovered along with the narrowed wound area (Figure 4C). These results indicated that miR-125b and SIRT6 are involved in the HCC cell migration and the migration induced by SIRT6 can be neutralized by miR-125b.
Figure 4.
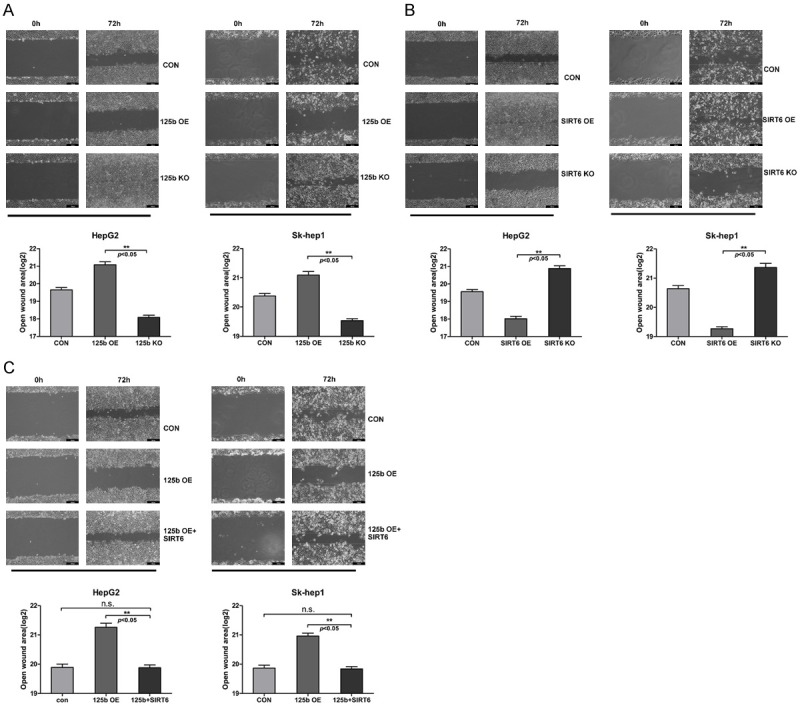
MiR-125b-SIRT6 axis affects the cell migration of HCC cells. A. The effect of miR-125b on the migration of HepG2 and Sk-hep-1 cell lines. A representative photograph from three independent experiments is shown one. The plot shows the results of wound healing assay as mean ± SD of open wound area. Statistical analysis was performed using t-test. B. The effect of SIRT6 on the migration of HepG2 and Sk-hep-1 cell lines. C. The cell migration ability of 125b overexpression HepG2 and Sk-hep-1 cell lines upon SIRT6 transfection.
MiR-125b-SIRT6 axis affects HCC cell invasion
Next, we conducted transwell assay to evaluate the influence of miR-125b and SIRT6 on HCC invasion. Comparing with 125b KO and SIRT6 OE cell lines, the invasive cell numbers of 125b OE and SIRT6 KO cell lines were significantly decreased (Figure 5A, 5B). Reintroducing SIRT6 into 125b OE cell lines was able to restore the decreased invasive cell numbers as expected (Figure 5C). Accordingly, this evidence is in line with the conclusion on the cell proliferation and migration.
Figure 5.
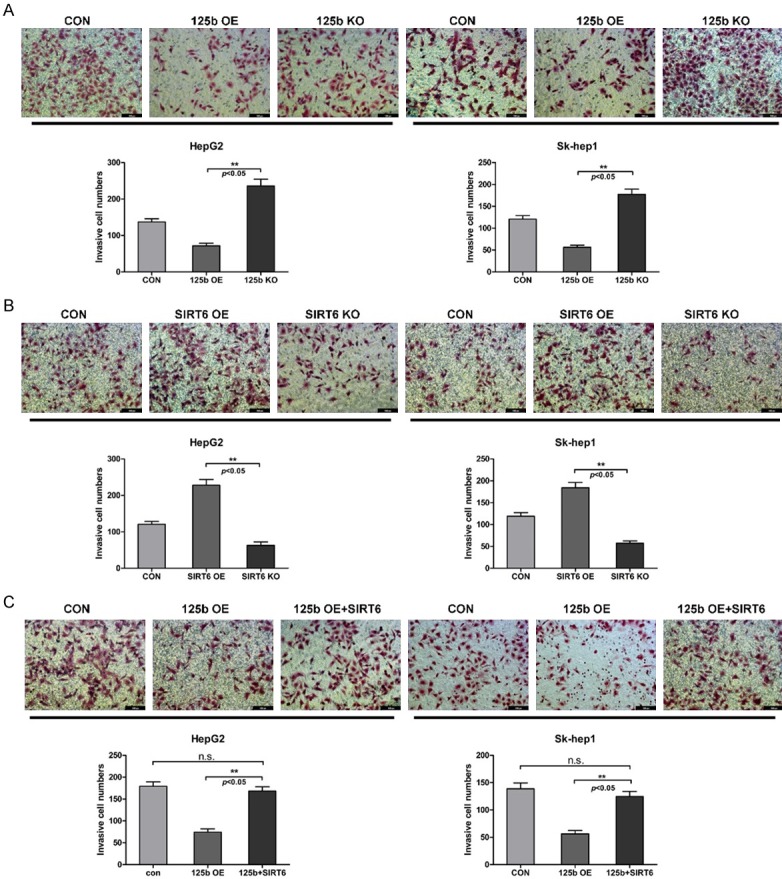
MiR-125b-SIRT6 axis affects the cell invasion on HCC cells. A. The cell invasion ability of miR-125b on HepG2 and Sk-hep-1 cell lines via transwell assay. The invasive cells stained by crystal violet were shown in a representative photograph. The plot data showing the invasive cell counts were presented as the mean ± SD, n=3. Statistical analysis was performed using t-test. B. The cell invasion ability of SIRT6 on HepG2 and Sk-hep-1 cell lines. C. The cell invasion ability of 125b overexpression HepG2 and Sk-hep-1 cell lines upon SIRT6 transfection.
MiR-125b induces HCC senescence and apoptosis via SIRT6 inhibition
We further investigated the associated function of SIRT6 to explain the phenotype change caused by these two genes. Regarding of the function of SIRT6 and the previous study, we speculated that the lost function of SIRT6 could induce cellular senescence and apoptosis of the HCC cells. Therefore, we performed β-GAL staining assays and found that both miR-125b OE and SIRT6 KO cell lines have the increased β-GAL positive cell proportion, and the β-GAL positive cell proportion was decreased in miR-125b overexpression cell lines upon co-transfection with SIRT6 (Figure 6A, 6B). Moreover, cellular senescence associated protein P16 and apoptosis associated protein Bax and Caspase 3 were significantly increased upon SIRT6 knockout and miR-125b overexpression, whereas these effects were eliminated upon SIRT6 restoration. In summary, we found that the inhibition of SIRT6 induced by miR-125b is able to trigger the cell senescence and apoptosis and inhibit the malignancy of HCCs.
Figure 6.

MiR-125b induces HCC senescence and apoptosis via inhibiting SIRT6. A. Senescence-associated β-galactosidase staining was used to exhibit senescence-like phenotypes in SIRT6 knockout HepG2 and Sk-hep-1 cell lines. Cells with positive staining were shown in a representative photograph. Positive staining cell counts were presented as the mean ± SD, n=3. Statistical analysis was performed using t-test. B. SA-β-gal staining assay showing the cell senescence rate in miR-125b overexpression cells and miR-125b overexpression HepG2 and Sk-hep-1 cells upon SIRT6 transfection. C. Western blots shows the SIRT6, P16, Bax and Caspase3 protein level in cells of each group as treated above. β-actin was used as loading control.
Discussion
HCC is one of the most prevalent lethal cancers worldwide. The carcinogenesis of HCC has already been studied for many years and recognized as a complex process. General treatment in clinical practice has severe shortages, along with tremendous pain and economic burden, and the mortality remains high [26,27]. Lack of the molecular and cellular knowledge on the initiation and development of HCC is the huge obstacle for diagnosis and treatment. Recently, microRNA related therapeutic methods showed big advantage and bright application prospection, especially on diagnose and drug discovery [28,29]. Exploring the molecular and cellular mechanism of microRNA involved in carcinogenesis is the key to developing novel HCC treatment. Mounting evidences have proven a role for miR-125b in proliferation, apoptosis and cellular differentiation. The underlying molecular and cellular mechanisms are complex according to the different targets that miR-125b might regulate and different signaling pathways that miR-125b might be involved [4]. Also, miR-125b can be either oncogenes or tumor suppressors depending on its targets. The role as an oncogene or a tumor suppressor of miR-125b can be changed in one cancer type to another, and its expression also varies in different cancers [4,22]. In this study, we confirmed that miR-125b was significant down-regulated in HCC tumor samples. And miR-125b down-regulation in HCC patients is associated with a poor five-year survival rate (Figure 1C). The bioinformatics prediction showed that SIRT6 is a direct target of miR-125b as long as it can bind to the 3’UTR of SIRT6 to suppress SIRT6 mRNA translation, and this prediction is verified with dual-luciferase assay. Additionally, patients with high level of SIRT6 showed poor five-year survival rate (Figure 2G). Thus, the reciprocal correlation of miR-125b and SIRT6 is not only exist in molecular level, but also in clinical relevance.
As a posttranscriptional regulator, the functions of microRNA are deeply associated with its target genes. SIRT6 acts like a double-edge sword in carcinogenesis [6]. In bladder and prostate cancers, SIRT6 was suppressed via E2F-1 pathway [30]. In colorectal cancer, SIRT6 suppresses Myc-related genes expression and glycolytic pathway, and SIRT6 downregulation accelerates tumor progression and leads to poor prognosis [13]. In pancreatic ductal adenocarcinoma, SIRT6 suppression can cause histone hyperacetylation at the Lin28b promoter and upregulate Lin28b expression, which inhibits Let7 expression. Therefore, HMGA2, IGF2BP1 and IGF2BP3, and targets of Let7 were upregulated to enhance cancer progression and metastasis [31]. Several studies also reported SIRT6 overexpression in breast cancer, ovarian cancer, retinoblastoma, lymphocytic leukemia, squamous cell carcinoma, and peripheral blood of patients with head and neck squamous cell carcinoma [12]. One of the important functions of SIRT6 is the regulation of ageing. SIRT6 depletion leads to H3K9 hyperacetylation in telomere, which triggered end-to-end chromosomal fusions and compromised WRN occupancy at telomeres, thus caused cellular senescence [32]. Additionally, in NF-κB signaling, hyperactive NF-κB signaling initiated via SIRT6 deficiency may contribute to premature and normal aging [16]. On the other hand, SIRT6 related apoptosis has been proved as an important reason for certain diseases [33,34]. Here, through colony formation, wound healing and transwell assays, we addressed SIRT6 as an oncogene to accelerate cancer progression, proliferation and metastasis in HCC, and this feature is opposite to miR-125b. Subsequently, the SIRT6 knockout is able to induce the cellular senescence and apoptosis, which has similar effect upon miR-125b overexpression. Additionally, miR-125b can reverse SIRT6 oncogenic ability via the cellular senescence and apoptosis induced by SIRT6 suppression. Thus, the cellular senescence and apoptosis induced by SIRT6 repression is the underlying mechanism of the malignancy elimination of HCC cells induced by miR-125b overexpression. However, the role of SIRT6 in HCC we defined isn’t in line with some other groups’ findings. By investigating a publicly available cancer microarray database and their recently published HCC database, Marquardt et al. [35] found that the decreased SIRT6 expression occurred in HCC specimens, this discrepancy might happened due to different investigation methods and different HCC population, our observations on this issue are in accordance with Ran and colleagues [36]. Be that as it may, we believe either observations can be reasonable. As for transition progress of normal to HCC, SIRT6 may serve as tumor suppressor in maintaining homeostasis and genomic stability, thus SIRT6 suppression accelerates cancer progress, once cancer reached advanced stage, it may harness the ability of SIRT6, e.g. anti-apoptosis and anti-senescence, to establish a new balance which favors carcinogenesis.
In fact, we found that miR-125b is capable of targeting lots of sirtuin family members, this insinuates miR-125b might have strong correlation with sirtuin family and even have an evolutionary conserved manner to some extent. On the basis of our analysis on clinical data, we found that the suppression of SIRT6 and SIRT7 occurred in HCC [25] (Figures 1G, 2C-F). Kim et al. [37] found that the up-regulation of SIRT7 in HCC potentiates tumorigenesis by cell cycle acceleration and protein synthesis enhancement. Yet the role of SIRT6 in HCC, anti-senescence and anti-apoptosis, we addressed here is more like losing brake. Both SIRT6 and SIRT7 act as oncogene in HCC and are downstream target of miR-125b, this makes miR-125b as a potent tumor suppressor, and loss function of miR-125b is considered a critical process upon HCC development.
Taken together, our findings elucidated that miR-125b attenuates HCC carcinogenesis by targeting SIRT6-mediated cellular senescence and apoptosis. Our study uncovered a new understanding of how miR-125b affects carcinogenesis of HCC and may represent a novel therapeutic targets for the treatment of HCC.
Acknowledgements
This work was supported by National Natural Science Foundation of China (81270427, 81471405, 81321003, and 81671389).
Disclosure of conflict of interest
None.
References
- 1.Dhanasekaran R, Limaye A, Cabrera R. Hepatocellular carcinoma: current trends in worldwide epidemiology, risk factors, diagnosis, and therapeutics. Hepat Med. 2012;2012:19–37. doi: 10.2147/HMER.S16316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 3.Shaham L, Binder V, Gefen N, Borkhardt A, Izraeli S. MiR-125 in normal and malignant hematopoiesis. Leukemia. 2012;26:2011–8. doi: 10.1038/leu.2012.90. [DOI] [PubMed] [Google Scholar]
- 4.Sun YM, Lin KY, Chen YQ. Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol. 2013;6:6. doi: 10.1186/1756-8722-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–95. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalkiadaki A, Guarente L. The multifaceted functions of sirtuins in cancer. Nat Rev Cancer. 2015;15:608–624. doi: 10.1038/nrc3985. [DOI] [PubMed] [Google Scholar]
- 7.Chang HC, Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab. 2014;25:138–145. doi: 10.1016/j.tem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 9.Mao Z, Hine C, Tian X, Meter MV, Au M, Vaidya A, Seluanov A, Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Z, Zhang L, Zhang W, Meng D, Zhang H, Jiang Y, Xu X, Meter MV, Seluanov A, Gorbunova V. SIRT6 rescues the age related decline in base excision repair in a PARP1-dependent manner. Cell Cycle. 2015;14:269–276. doi: 10.4161/15384101.2014.980641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao Z, Tian X, Van Meter M, Ke Z, Gorbunova V, Seluanov A. Sirtuin 6 (SIRT6) rescues the decline of homologous recombination repair during replicative senescence. PProc Natl Acad Sci U S A. 2012;109:11800–11805. doi: 10.1073/pnas.1200583109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lerrer B, Gertler AA, Cohen HY. The complex role of SIRT6 in carcinogenesis. Carcinogenesis. 2016;37:108–18. doi: 10.1093/carcin/bgv167. [DOI] [PubMed] [Google Scholar]
- 13.Sebastián C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, Cosentino C, Greenson JK, MacDonald AI, McGlynn L, Maxwell F, Edwards J, Giacosa S, Guccione E, Weissleder R, Bernstein BE, Regev A, Shiels PG, Lombard DB, Mostoslavsky R. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151:1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van GF, Gallí M, Gueydan C, Kruys V, Prevot PP, Bedalov A, Mostoslavsky R, Alt FW, De ST, Leo O. Intracellular NAD levels regulate tumor necrosis factor protein synthesis in a sirtuindependent manner. Nat Med. 2009;15:206–10. doi: 10.1038/nm.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauer I, Grozio A, Lasigliè D, Basile G, Sturla L, Magnone M, Sociali G, Soncini D, Caffa I, Poggi A. The NAD+-dependent histone deacetylase SIRT6 promotes cytokine production and migration in pancreatic cancer cells by regulating Ca2+ responses. J Biol Chem. 2012;287:40924–37. doi: 10.1074/jbc.M112.405837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, Mccord RA, Ongaigui KC, Boxer LD, Chang HY. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sundaresan NR, Vasudevan P, Zhong L, Kim G, Samant S, Parekh V, Pillai VB, Ravindra PV, Gupta M, Jeevanandam V, Cunningham JM, Deng CX, Lombard DB, Mostoslavsky R, Gupta MP. The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat Med. 2012;18:1643–1650. doi: 10.1038/nm.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T. The histone deacetylase SIRT6 regulates glucose homeostasis via Hif1α. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinezpastor B, Giacosa S. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell. 2013;51:454–68. doi: 10.1016/j.molcel.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329:1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Barjoseph Z, Cohen HY. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–21. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 22.Banzhaf-Strathmann J, Edbauer D. Good guy or bad guy: the opposing roles of microRNA 125b in cancer. Cell Commun Signal. 2014;12:30. doi: 10.1186/1478-811X-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956–D963. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 27.Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62:394–9. doi: 10.3322/caac.21161. [DOI] [PubMed] [Google Scholar]
- 28.van der Ree MH, van der Meer AJ, de Bruijne J, Maan R, van Vliet A, Welzel TM, Zeuzem S, Lawitz EJ, Rodriguez-Torres M, Kupcova V, Wiercinska-Drapalo A, Hodges MR, Janssen HL, Reesink HW. Long-term safety and efficacy of microRNA-targeted therapy in chronic hepatitis C patients. Antiviral Res. 2014;111:53–59. doi: 10.1016/j.antiviral.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 29.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu M, Seto E, Zhang J. E2F1 enhances glycolysis through suppressing Sirt6 transcription in cancer cells. Oncotarget. 2015;6:11252–11263. doi: 10.18632/oncotarget.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kugel S, Sebastián C, Fitamant J, Ross KN, Saha SK, Jain E, Gladden A, Arora KS, Kato Y, Rivera MN. SIRT6 suppresses pancreatic cancer through control of Lin28b. Cell. 2016;165:1401–1415. doi: 10.1016/j.cell.2016.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michishita E, Mccord RA, Berber E, Kioi M, Padillanash H, Damian M, Cheung P, Kusumoto R, Kawahara TLA, Barrett JC. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Xie JJ, Jin MY, Gu YT, Wu CC, Guo WJ, Yan YZ, Zhang ZJ, Wang JL, Zhang XL. Sirt6 overexpression suppresses senescence and apoptosis of nucleus pulposus cells by inducing autophagy in a model of intervertebral disc degeneration. Cell Death Dis. 2018;9:56. doi: 10.1038/s41419-017-0085-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu M, Liang K, Zhen J, Zhou M, Wang X, Wang Z, Wei X, Zhang Y, Sun Y, Zhou Z. Sirt6 deficiency exacerbates podocyte injury and proteinuria through targeting Notch signaling. Nat Commun. 2017;8:413. doi: 10.1038/s41467-017-00498-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marquardt JU, Fischer K, Baus K, Kashyap A, Ma S, Krupp M, Linke M, Teufel A, Zechner U, Strand D, Thorgeirsson SS, Galle PR, Strand S. Sirtuin-6-dependent genetic and epigenetic alterations are associated with poor clinical outcome in hepatocellular carcinoma patients. Hepatology. 2013;58:1054–1064. doi: 10.1002/hep.26413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J, Ran RK, Chen Y, Zhang ZZ, Tao NN, Ren JH, Zhou L, Tang H, Chen X, Chen K. SIRT6 overexpression potentiates apoptosis evasion in hepatocellular carcinoma via BCL2-associated X protein-dependent apoptotic pathway. Clin Cancer Res. 2016;22:3372. doi: 10.1158/1078-0432.CCR-15-1638. [DOI] [PubMed] [Google Scholar]
- 37.Kim JK, Ji HN, Jung KH, Eun JW, Bae HJ, Min GK, Chang YG, Shen Q, Park WS, Lee JY. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57:1055–1067. doi: 10.1002/hep.26101. [DOI] [PubMed] [Google Scholar]