Abstract
Iron as an important element plays crucial roles in various physiological and pathological processes. Iron metabolism behaves in systemic and cellular two levels that usually are in balance conditions. The disorders of the iron metabolism balances relate with many kinds of diseases including Alzheimer’s disease, osteoporosis and various cancers. In systemic iron metabolism that is regulated by hepcidin-ferroportin axis, plasma iron is bound with transferrin (TF) which has two high-affinity binding sites for ferric iron. The generic cellular iron metabolism consists of iron intake, utilization and efflux. During the iron intake process in generic cells, transferrin receptors (TFRs) act as the most important receptor mediated controls. TFR1 and TFR2 are two subtypes of TFRs those bind with iron-transferrin complex to facilitate iron into cells. TFR1 is ubiquitously expressed on the surfaces of generic cells, whereas TFR2 is specially expressed in liver cells. TFR1 has attracted more attention than TFR2 by having diverse functions in both invertebrates and vertebrates. Recently reports showed that TFR1 involved in many kinds of diseases including anemia, neurodegenerative diseases and cancers. Most importantly, TFR1 has been verified to be abnormally expressed in various cancers. Some experimental and clinical drugs and antibodies targeting TFR1 have showed strong anti-tumor effects, herein TFR1 probably become a potential molecular target for diagnosis and treatment for cancer therapy. This paper reviewed the research progresses of the roles of TFR1 in the tumorigenesis and cancer progression, the regulations of TFR1, and the therapeutic effects of targeting TFR1 on many kinds of cancers.
Keywords: Iron, TFR1, TFRC, cancer, cancer targeting drugs
Introduction
Iron as an important element plays crucial roles in various physiological and pathological processes. In all kinds of mammalian cells, iron is indispensable to cell growth and division [1,2], and it predominantly controls the formation of heme- and iron-containing proteins participating in oxygen transport [3-5], energy metabolism [6-8], neurotransmitter generation and release [9,10], synthesis of DNA [11-13], collagen and steroid hormones [14-16], nonspecific resistance, etc. [17]. However, iron concentration must be strictly controlled, as iron is involved in the generation of free radicals in cells [18], a process leading to damage of biomolecules (proteins, lipids, nucleic acids) and in the progression of oxidative stress [19]. Iron metabolism has been reported the close related to cancer progression [20-22]. Disorders of iron metabolism, especially excessive iron acquisition and retention, can induce tumorigenesis and cancer’s growth as well [23,24]. However, high concentration of intracellular iron can make cells in extremely oxidative stress and may induce tumor death. As a novel form of regulated cell death, ferroptosis is typified by lipid peroxidation and relies on iron and reactive oxygen species (ROS). Ferroptosis is morphologically and biochemically different from other known types of cell death [25,26]. Thus, whether iron deprivation or induced iron overload in tumor cells can inhibit tumor growth and cause tumor cell death. A variety of strategies for antitumor therapies have been designed to target intracellular iron [27,28], including utilization of transferrin receptor 1 (TFR1)-mediated cytotoxic drug conjugates and iron chelators. As a membrane protein regulating iron import [29,30], TFR1 is a member of the TFR family that shows nanomolar affinity to transferrin (TF) bound to Fe (III) [31]. The complex of TF-TFR1 is internalized through endocytosis mediated by clathrin, and Fe (III) is disassociated from TF when pH decreases to 5.5. At this pH, apotransferrin and TFR1 are still associated and recycled to cell surface with physiological pH, so the former is released [32,33]. Iron uptake by transferrin receptor is the most important way for cancer cells to absorb iron, thus accumulating evidence has proven that TFR1 participated in tumor onset and progression, and its expression was dysregulated significantly in many cancers [34,35]. The relationship between TFR1 and cancers has been revealed, rendering TFR1 a valuable pharmaceutical target for intervening with cancers [36-39]. Based on these reported studies, in this review will summarize the regulatory effects of TFR1 on tumorigenesis, and the potential therapeutic effects of targeting TFR1 on cancers.
Biological functions and regulations of transferrin receptors
Transferrin receptors (TFRs) encoded by TFRC is a membrane glycoprotein, which can import iron by binding a plasma glycoprotein, transferrin (TF) [40]. TF was first referred to as serum protein, with two specific sites binding Fe (III), so it is an iron source for synthesizing hemoglobin. Meanwhile, TF-bound iron undergoes cellular uptake requiring interaction between this protein and a specific TFR [33,41]. The molecular weight of TFR as a homodimer is 180 kDa [42]. Each monomer contains a TF-binding C-terminal domain, a short N-terminal domain and a single transmembrane domain [33].Transferrin receptors have two subtypes, transferrin receptor 1 (TFR1) and transferrin receptor 2 (TFR2). TFR1 is a homodimeric type II transmembrane glycoprotein that is expressed ubiquitously on the surfaces of most cells while another member of TFRs, TFR2 is mainly expressed in the liver [43,44].
After TF was discovered as the iron source for immature red blood cells synthesizing hemoglobin, TFR1 was first considered as a cell surface receptor by which TF delivered iron to cells [45]. Mammalian TFR1 comprises 760 residue subunits that can be divided into a globular extracellular region, a hydrophobic intramembranous region and the remaining residues within the cytoplasm [33]. Consisting of two monomers, TFR1 is linked by two disulfide bridges, forming a 190 kDa molecule. It is a gatekeeper which regulates iron uptake [46]. Except for mature red cells, almost all cells have TFRs on their surfaces, being most abundant in the placenta, erythron and liver [38]. Human TFR1 has three N-linked and one O-linked oligosaccharide. Appropriate folding and transport of this protein to cell surface are significantly affected by N-linked glycosylation [33].
TFR1 expressions are delicately regulated at many levels, and several genes are involved in the regulation of TFR1 (Table 1). Intracellular iron concentration regulates the TFR gene post-transcriptional regulation by binding iron-regulatory proteins 1 and 2 (IRP1 and IRP2) to the iron response elements in the 5’-untranslated region of TFR transcript [45,47]. IRPs are activated by the deprivation of cellular iron, which is inhibited through iron repletion [48]. IRP activities can be regulated by other iron-independent effectors such as inflammation [49], oxidative stress [50], hypoxia and xenobiotics [51], or such stimuli under pathophysiological conditions [38,52]. Hypoxia induces the transcription of TFR1 gene by binding hypoxia-inducible factors (HIFs) to specific promoter elements [53]. This process can also be activated by an oncogenic transcription factor c-Myc [38]. Thirdly, HFE is implicated in the pathogenesis of disordered toxic and progressive iron overload, i.e. hereditary hemochromatosis [54]. It competes for binding receptor with TF, thus hindering iron uptake [55]. In addition to the compounds mediating TFR1 regulation mentioned above, its transcription is also influenced by some other transcription factors like CREB1 and c-Jun [56] (Figure 1).
Table 1.
Genes involved in the regulation of TFR1
Figure 1.
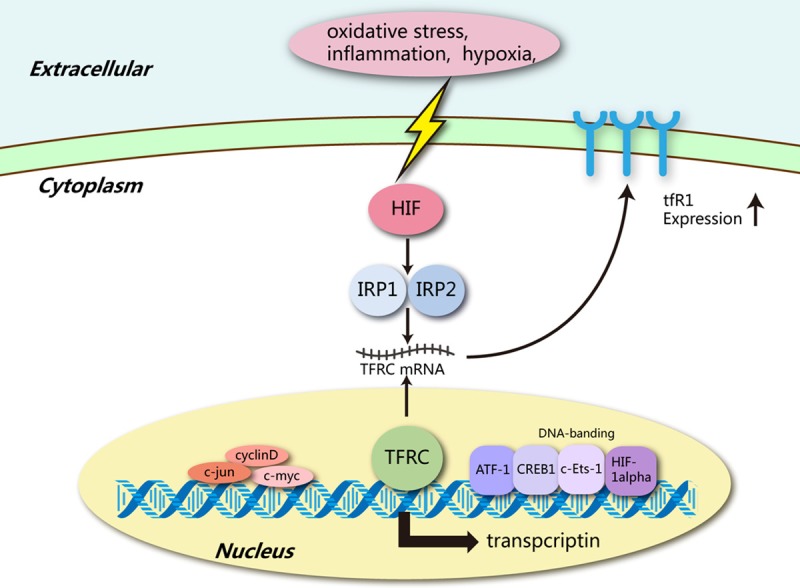
Regulation of TFRC transcription. Oxidative stress, inflammation and hypoxia induce HIF expression. HIF induces binding of IRP1 and IRP2. Binding of IRP1 and IRP2 promotes TFRC transcription. C-Jun, C-Myc and cyclin D expressions also induce TFRC transcription. Meanwhile, CREBBP and EP300 are activated to promote TFRC expression. Moreover, ATF-1, CREB1, c-Ets-1 and HIF-1alpha influence the transcription of TFRC by binding DNA.
Transferrin receptor 1 and cancer
The transformation of normal cells into tumorigenic ones and tumor progression involve complicated processes which are still largely unknown. The changes mainly result from accumulated mutations of some key genes or proteins [64,65], thus damaging the balances of tumor cells growth [66], proliferation [67], death [68], gene transcription [69] and angiogenesis [70]. Tumor cells’ proliferation is enhanced and apoptosis is inhibited by some of the essential signaling pathways varied upon tumorigenesis [71-73], while invasion and metastasis are promoted by others [74,75]. The key roles of transferrin receptors (TFRs) in controlling the above processes have been well demonstrated over the last decade. TFR1 participating in tumor progression is abundantly expressed in liver, breast, lung and colon cancer cells [76-79]. Immunohistochemical findings of TFRC in various tumor tissues showed most cancers displayed moderate to strong cytoplasmic positivity. Carcinoid, prostate and testicular cancer was negative from The Human Protein Atlas [80]. Although the effects of TFRs on cancer pathophysiology have been studied, the expressions of TFR1 in different cancers are inconsistent and the mechanisms by which TFR1 participates in tumor progression remain elusive.
Given that TFR1 is abnormally expressed in various cancers (Figure 2A, 2B), it definitely affects cancer cells’ proliferation [81], migration [82], invasion [83], apoptosis and metastasis [76,84]. Accordingly, several examples showed the regulatory effects of abnormal TFR1 expression on the biological behaviors of cancers (Figure 3).
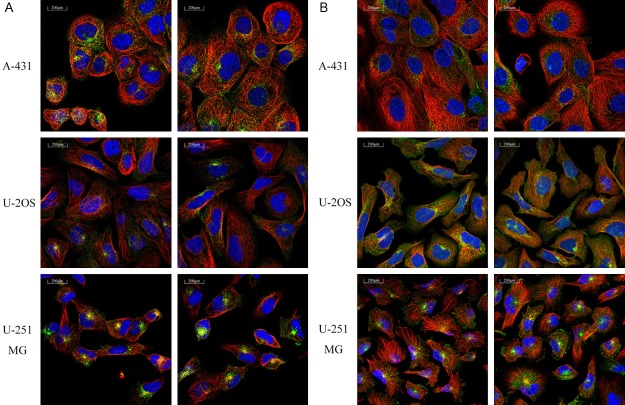
Figure 2.
Location of TFR1 in three cancer cell lines. Immunofluorescence analysis of TFR1 in A431, U2OS and U251 MG was obtained from The Human Protein Atlas database. TFR1 (Green fluorescence) expressed in vesicles (A), endosomes and lysosomes (B).
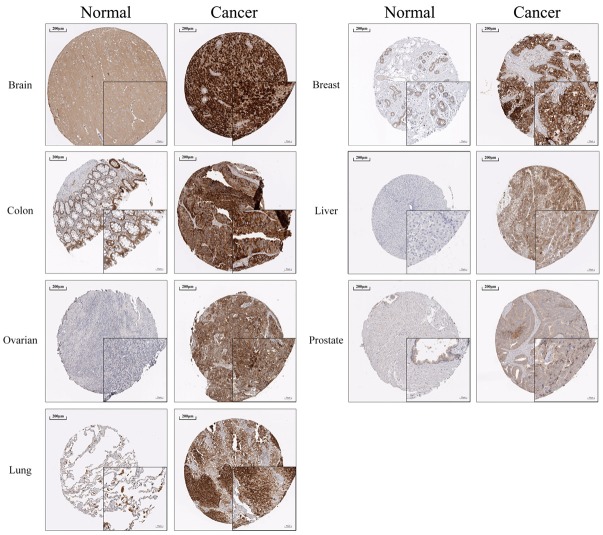
Figure 3.
TFR1 expression in various normal tissues and tumor tissues. Immunohistochemistry analysis of TFR1 in various normal tissues and tumor tissues was obtained from The Human Protein Atlas database. TFR1 is overexpression in multiple tumor tissues compared to normal tissues.
Transferrin receptor 1 in brain cancer
TFR1 participates in regulating the physiology of glioma cells and the progression of brain cancer. Rosager et al. reported that TFR1 was overexpressed in brain cancer [84]. TFR1 mediated ROS formation and iron accumulation, as a crucial downstream effector of corresponding transcription factors facilitating proliferation of glioma and glioma-induced death of neurons [85]. Weston et al. reported that iron was necessary for cell division and cancer pathophysiology was affected by dysregulation of IRPs. Based on the public data from The Cancer Genome Atlas (TCGA), they studied the relationships between the expressions of 61 genes coding iron regulatory proteins (IRPs) in patients with Grade II-III gliomas according to the criteria of World Health Organization and survival. The outcomes were poorer in patients with higher TRF1 expressions, indicating TFR1 played a negative role in the prognosis of glioma [86].
By using the univariate Cox regression model, Yu et al. assessed the prognostic values of genes in two independent datasets of glioblastoma multiforme (GBM). TFR1 was highly expressed at the early stage. Besides being related with clinical outcomes, TFR1 also affected chemoresponse. This reference model potentially allowed identification of new prognostic markers and development of novel therapies [87]. Hence, TFR1 may dominantly mediate the biological behaviors of brain cancer, accurately predict the prognosis of GBM, and help identify new drug targets.
Transferrin receptor 1 in breast cancer
Breast cancer is the most devastating type among females of Western countries and also has the highest incidence in women patients of China [74]. The growth of breast cancer cells requires increasing iron uptake that can be realized by TFR1 over expression [76]. TFR1 has been reported to be overexpressed in human breast cancer [88,89], also as a suitable biomarker for diagnosing and treating cancer patients at the early stage [89]. Based on public microarray datasets consisting of 674 cases of breast cancer, Miller et al. detected the expression of TFR1 gene linked to breast cancer prognosis, and high expression of TFR1 indicated poor prognosis [90]. Singh et al. found that TFR1 expressions in benign and normal lesions were significantly lower than those in invasive carcinoma and premalignant lesions. In the meantime, more TFR1 was expressed in high-grade breast cancer than in other grades [83]. Jiang et al. explored whether breast cancer cells altered the expression of TFRC. The growth of breast cancer was suppressed by regulating the expression of iron transporter genes. Reverse transcription-polymerase chain reaction showed that more TFRC was expressed in MCF-7 cells than in human mammary epithelial MCF-12A cells. Moreover, TFRC antisense oligonucleotides decreased intracellular total iron and TFRC mRNA levels, as well as suppressed 4T1 cell proliferation in culture medium and tumor growth and pulmonary metastasis in a 4T1 mouse model of mammary adenocarcinoma [91]. Wang et al. found that upon breast cancer, IRP2 dominated in iron accumulation. IRP2 overexpression was related with increase of TFR1 and reduction of ferritin heavy chain. Knock-down of IRP2 in human triple-negative breast cancer cells MDA-MB-231 elevated the expression of ferritin heavy chain and reduced that of TFR1, thereby decreasing the labile iron pool and inhibiting the growth of MDA-MB-231 cells in the mammary fat pad of mice [92]. Estrogen receptor (ER)+/progesterone receptor (PR)+ invasive ductal breast cancer accounts for approximately 45% of invasive cases in the United States of America. Buas et al. selected 121 cases and 121 matching controls 12.5 months prior to diagnosis, and divided them equally into testing and training sets. They employed a customized antibody array targeting over 2000 proteins, and found that TFR1 expressions were significantly different (P < 0.05) between cases and matching controls. Additionally, they verified TFR1 as a feasible plasma protein biomarker for ER+/PR+ ductal breast cancer with pre-diagnostic biospecimens [35]. Marques et al. detected TFR1 changes in tumor microenvironment-derived stromal inflammatory cells. TFR1 expressions in tissues of primary breast cancer and axillary lymph nodes were measured by immunohistochemical assay to clarify the iron profiles of lymphocytes, epithelial cells and macrophages. Macrophages and lymphocytes infiltrated primary tumors, and TFR1 expression increased in metastatic lymph nodes, being related to the clinical and pathological markers for poor prognosis (e.g. tumor size and negative hormone receptor status) [93]. Rychtarcikova et al. investigated the deregulation of TFR1 in breast tumor-initiating cells (TICs), managing to find critical genes and proteins related with iron metabolism, which may be applicable to cancer diagnosis or therapy [76]. TFR1 has high expressions in tumor cells, upon iron deficiency in particular. Jian et al. studied the signaling role of TFR1 in cells of breast cancer by using gambogic acid (GA) and a known ligand of TFR1, TF. The sensitivity of TFR1 Tyr (20) phosphomutants to apoptosis mediated by GA was higher. Thus, TFR1 was a signaling molecule indeed, and Tyr (20) phosphorylation by Src resisted apoptosis and boosted the survival of breast cancer cells [94]. Collectively, TFR1 is both a significant independent prognostic marker and a promising therapeutic target for breast carcinoma.
Transferrin receptor 1 in colon cancer
As a prevalent disease, colon cancer has the eighth highest mortality rate among all cancers of adult males at present [95]. The TFR1 levels on cell surface, which modulate the uptake of TF binding iron, are associated with cell proliferation rate [96]. Since cancer cells have higher TFR1 expression than normal cells, it is a potential target for treating cancers. Overexpressed in many types of cancers, TFR1 still exerts unclear effects on colon cancer, needing further studies. Okazaki et al. found that the circadian organization of molecular clock affected TFR1 expression in colon cancer cells of mice and the 24 h rhythm of TFR1 expression may participate in cancer therapies targeting TFR1 [59,96]. The progression of colorectal cancer has been related to high intake of dietary iron and chronic intestinal inflammation. Chua, et al. found high expression of TFR1 activated the IL-6/IL-11-Stat3 signaling pathway in the colon, enhanced DSS-induced proliferation and apoptosis of colon epithelial cells, and aggravated mucosal damage and tumorigenesis [97]. Okazaki et al. observed a 24 h rhythm of IRP2 expression in colon-26 tumor-bearing mice, and IRP2 post-transcriptionally modulated the 24 h rhythm of TFR1 mRNA expression through binding iron-response elements, i.e. RNA stem-loop structures. Moreover, the expression of CLOCK (Delta19) attenuated the proliferation rate of wild-type colon-26 cancer and the time-dependent changes of iron levels in cells. Accordingly, circadian organization regulated iron metabolism to facilitate tumor cell proliferation [59]. Referring to TCGA database, the expressions of IRP2 and TFR1 were evaluated and compared to common mutations in cancers. Compared with the normal colon mucosa, IRP2 had overexpression in colorectal cancer, also being positively correlated with the expression of TFR1 [77]. These results provide a therapeutic target for intervening with colorectal tumorigenesis.
Transferrin receptor 1 in liver cancer
The liver is the most important organ related to iron storage [32]. Thus, liver cancer is closely linked to iron metabolism and expression of TFR1. Iron metabolism is altered upon hepatocellular carcinoma (HCC), which is typified by iron-deficient phenotype and essential to tumor growth. Iron has been suggested as a risk factor mainly in HCC patients with cirrhosis and hereditary haemochromatosis (HH). Beckman et al. found HFE (wild-type HH) protein complexes of TFR. Cys282Tyr and His63Asp, two HFE mutations, augmented the affinity of TFR to TF, thus promoting cellular iron uptake and HCC progression [98]. Holmstrom et al. detected significantly higher mRNA levels of genes participating in uptake of iron, especially TFR1, in HCC. Variations in the expressions of TFR1 in HCCs inferred that bioavailable iron was increasingly required and the iron turnover was high in neoplastic cells [99]. Miwa et al. found that TFRC expression was elevated with increasing cancer stage, and its selective expression in lesions undergoing proliferation indicated that variations in iron homeostasis were involved in the promotion or progression of tumor [100]. By using immunohistochemical assay, Sakurai et al. detected the expressions of TFR1 and TFR2 in tumor and paracancerous normal liver tissues collected from 41 patients with HCC. They also analyzed iron uptake by HCC cells and hepatocytes with iron staining. HCC samples had significantly higher TFR1 expressions than those of normal samples, and such expression was significantly related with the concentrations of serum des-gamma carboxy prothrombin and alpha-fetoprotein [101]. The above results revealed that TFR was expressed responding to iron deficiency in the midst of liver carcinogenesis. Moreover, according to TCGA database, Iryna et al. found that TFRC was overexpressed but microRNA-152 (miR-152) level plummeted in human HCC tissue compared to those in normal liver tissue, suggesting that raised TFRC levels in human HCC cells and tissues may partly be ascribed to the post-transcriptional mechanism that was mediated through miR-152 down-regulation. In short, targeting of TFRC specific to miR-152 may be a selective HCC therapy [78].
Transferrin receptor 1 in ovarian cancer
Ovarian cancer is the fifth most fatal malignancy among females in the USA, and also the most deadly gynecologic type. It is well-documented that TFR1 played key roles in ovarian cancer. Basuli et al. demonstrated that iron metabolism underwent targetable alterations during ovarian cancer. As an iron importer, TFR1 expressions increased in tumor tissues collected from patients with high-grade serous ovarian cancer. Moreover, the expression of TFR1 increased in a TIC model of ovarian cancer [102]. The findings can be exploited therapeutically.
Transferrin receptor 1 in prostate cancer
As a devastating health problem, prostate cancer accounts for 25% of all newly diagnosed cancer cases and approximately 9% of all cancer-related deaths of adult men in the USA annually [74]. However, effects of TFR1 expression and body-iron stores on prostate cancer are still controversial. By using enzymatic immunoassay, Kuvibidila et al. detected serum ferritin and serum transferrin receptor (sTFR) levels in 72 controls and 27 males with newly diagnosed, untreated prostate cancer. The levels of sTFR in males with prostate cancer significantly exceeded those without. However, these changes of sTFR did not correlate with tissue inflammation, tumor stage, or acute-phase proteins [103]. While Johnson et al. detected TFR1 expressions in prostate cancer and normal cells, and reported that the former cells had significantly increased mRNA and protein expressions of TFR1 [104]. Taken together, altered TFR1 expression whether can be a novel biomarker for accurate diagnosis of prostate cancer and prognosis need further study.
Transferrin receptor 1 in lung cancer
TFR1 predominantly mediates the proliferation of lung cancer by regulating the uptake of iron binding TF. Wang et al. demonstrated that TFR1 promoters contained sequences that mediated the transcriptional inhibition depending on cell density. TFR1 expression was affected by lung cancer cell density [105]. Zhu et al. reported that human lung cells SPC-A1 in which more TFR1 was expressed were more sensitive at identical GA concentrations, and TFR1 expression level in tumor tissue, which was quantified by histopathological assay, may predict the sensitivity of lung cancer to treatment with GA [106].
Kukulj et al. reported that TFR1 expression in lung tumor tissue significantly surpassed that in normal lung tissue. The expression in tumor tissue was positively correlated with alpha-globulin level [107]. In addition, epidermal growth factor receptor (EGFR), which drives oncogenesis, binds and modulates subcellular TFR1 distribution through tyrosine kinase activity, thereby being demanded for the import of cellular iron. Accordingly, EGFR can modulate iron homeostasis in cells by redistributing TFR1, so it is necessary for the onset and progression of lung cancer [79].
Transferrin receptor 1 in leukemia
Human leukemias are liquid malignancies characterized by diffuse infiltration of the bone marrow by transformed hematopoietic progenitors [108]. Iron as the most important hematopoietic element plays a key role in leukemia. TFR1 was initially found as an important iron uptake receptor inducing the growth of leukemia cell lines, HL-60 and KG-1 [109]. Many studies have found that TFR1 was upregulated in leukemia [110]. Liu et al. investigated the TFR1 could be a potential marker in the diagnosis of acute leukemia (AL) [111]. Płoszyńska A et al. evaluated TFR1 expression on acute lymphoblastic leukemia (ALL) cells. TFR1 expression was statistically higher on T-lineage leukemias while in the B lineage ALL, a significant difference in TFR1 expression existed between precursor B ALL and mature B-ALL, which showed higher TFR1 expression. TFR1 expression positively correlated with Hgb concentration at diagnosis [112]. In summary, TFR1 could be a good target for leukemia therapy.
Mechanisms by which transferrin receptor 1 affects cancers
Given that TFR1 is widely overexpressed in cancers, the regulatory mechanisms of TFR1 for carcinogenesis are complicated and often interrelated. (Figures 4 and 5) The high expression of TFR1 in tumor cells is mainly to meet the iron requirement of tumor cell proliferation [93,102]. It was reported that TFR1 is a signaling molecule and tyrosine phosphorylation at position 20 by Src enhances anti-apoptosis and potentiates breast cancer cell survival [94]. TFR1 was also reported as a mitochondrial regulator contributed to cancer cell growth via activating JNK signaling pathway [36]. Jeong et al. reported that TFR1 induced the growth of human pancreatic ductal adenocarcinoma (PDAC) by supporting ROS production and mitochondrial respiration in tumor cells. Upregulation of TFR1 expression generated ROS in PDAC cells by inducing oxidative phosphorylation. Moreover, PDAC growth required ROS derived from mitochondria. Furthermore, the sensitivity of PDAC cells to oxidative stress was determined by TFR1 expression. By triggering ROS production and mitochondrial respiration, TFR1 significantly participated in pancreatic cancer growth and survival [34]. Wang et al. found that IRP2 increased TFR1, playing a key role in breast cancer progression. As an early nodal point for iron metabolism changes upon breast cancer, dysregulation of IRP2 may result in unsatisfactory outcomes of some patients [92]. Pham et al. found that sphingosine kinase 1, a lipid kinase catalyzing the production of sphingosine 1-phosphate, could modulate cell proliferation, survival as well as neoplastic transformation by promoting TFR1 expression [113]. Bayeva et al. reported that iron homeostasis was modulated by mammalian target of rapamycin (mTOR) through variations of cellular iron flux and regulation of TFR1 stability. They identified an anti-inflammatory protein, tristetraprolin, as the downstream target of mTOR which bound TFR1 mRNA and facilitated its degradation. Therefore, TFR1 induced carcinogenesis by regulating metabolism, inflammation and iron [114]. Chirasani et al. demonstrated that TFR1-induced accumulation of oxidants altered cellular signaling through inactivation of pRB protein tyrosine phosphatase and p21/cdkn1a, and activation of Akt and mitogen-activated protein kinase. When the cell cycle regulators were inactivated, cells were prone to entry into the S phase. TFR not only affected proliferation, but also facilitated release of glutamate, causing decrease in neuron mass through the mediation by N-methyl-D-aspartate-receptor [85]. O’Donnell et al. proved that TFR1 was a key downstream target for c-Myc, and the expressions of TFR1 in both in vitro and in vivo models of B-cell lymphoma were activated by c-Myc which bound the conserved region of TFR1 directly. Also, inhibiting TFR1 attenuated cell proliferation and induced arrest in the G1 phase without influencing the size of cells. Consistently, expression profiling showed that depletion of TFR1 changed the expressions of cell cycle-regulatory genes. Additionally, increasing TFR1 expression was beneficial to cell growth and significantly boosted in vivo tumor formation mediated by c-Myc. The results mentioned above provide molecular bases for elevated expressions of TFR1 in human tumors, confirming the effects of TFR1 on the network of c-Myc target genes. Targeting TFR1 may be useful for cancer therapy [115].
Figure 4.
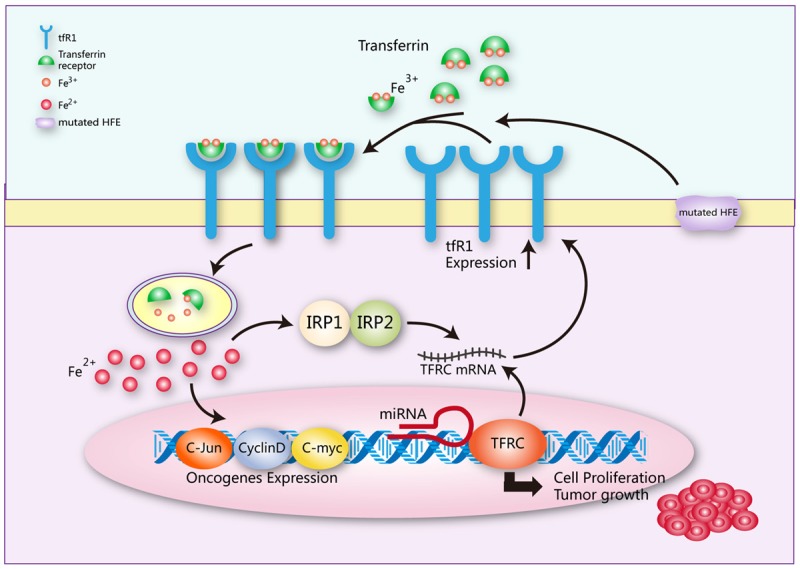
TFR1 and its regulatory effects on tumors Iron concentration changes significantly in cancer cells. TFR1 binds mutated HFE to promote iron intake. Iron concentration influences TFRC post-transcription by regulating the binding of IRP1 and IRP2. Meanwhile, iron concentration affects the activities of c-Jun, cyclin D and C-myc. Some miRNAs also influence TFRC transcription. These bioprocesses contribute to cell progression and tumor growth.
Figure 5.
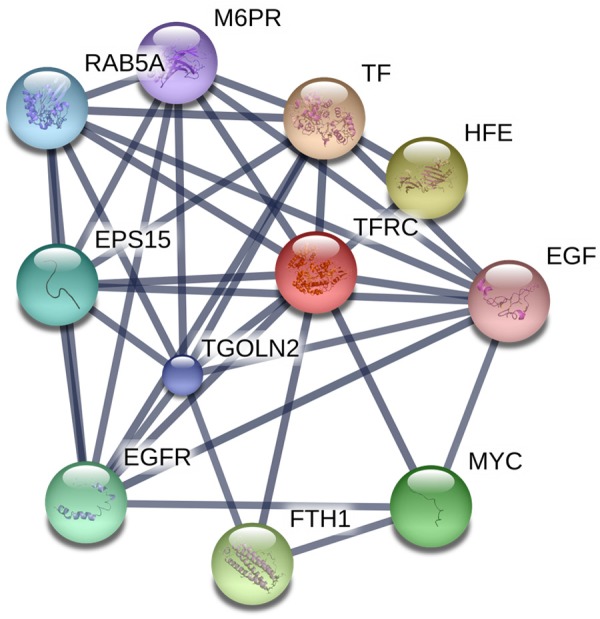
TFR1 interaction network. Based on the search tool of the European Molecular Biology Laboratory for retrieving interacting genes/proteins, an interaction network with confidence levels of > 0.7 is exhibited for genetically interacting, possibly TFR1-related proteins. A thicker line represents a stronger interaction.
Therapeutic potential of targeting TFR1 in cancers
As TFR1 is expressed in many kinds of cancers and significantly involved in tumorigenesis and progression of cancers, it may be feasible to intervene with the progression of cancers by targeting TFR1. As evidenced by currently available studies targeting TFR1, curcumin was among the most successful chemopreventive compounds. Jiao et al. evaluated the influence of curcumin on iron regulatory proteins and TFR1. Both TFR1 and IRP increased responding to curcumin [116]. Yang et al. also found curcumin induced the autophagy and apoptosis of different tumor cells by inhibiting TFR1 expression, inferring that curcumin was a potential TFR1 inhibitor for cancer therapy [117]. Anti-transferrin receptor monoclonal antibody A24 significantly blocks the proliferation of T-cell leukemia cell, induces apoptosis of tumor T lymphocytes from acute T-cell leukemia patients [118,119]. Also, miRNA drug targeting TFR1 has been reported to be a good drug in clinical treatment of leukemia [120]. Moreover, nanomedicine and antibodies targeting TFR1 have been developed for tumor-specific targeted therapy, providing a valuable opportunity for developing eligible TFR1 inhibitors in the field of precision oncology [121,122]. TFR1 is highly expressed in adult T-cell leukemia/lymphoma. Shimosaki et al. developed a novel molecular-targeted therapy against TFR1 to modulate HTLV-1-associated adult T-cell leukemia/lymphoma iron metabolism to inhibit the adult T-cell leukemia/lymphoma. JST-TFR09, an antibody to human TFR1, has great affinity to TFR1 on adult T-cell leukemia/lymphoma cells. It could interfere with binding between TFR1 and TF, inhibited the iron intake of adult T-cell leukemia/lymphoma, which may become a promising therapy for the treatment of adult T-cell leukemia/lymphoma [123].
Conclusions and future directions
We herein reviewed the roles of TFR1 in the onset and progression of cancers, its regulatory effects on tumorigenesis, together with the potential of TFR1-targeted cancer therapies. Regardless of considerable studies concerning TFR1 since it was discovered, several key issues still exist. (Figure 6) Firstly, whether TFR1 interacts with additional signaling pathways or proteins on the cellular level remains elusive, which may be essential to the development of therapies based on TFR1. Being different from the traditional apoptosis and necrosis, the recently discovered ferroptosis is caused by iron-dependent accumulation of lipid peroxides. Cells which are subjected to ferroptotic death suffer from shrinkage of volumes and raised density of the mitochondrial membrane. Iron metabolism plays a curial role in ferroptosis [124]. Ferroptosis induced by erastin or Cys2 deprivation is prevented by silencing TFRC gene, which encodes TFR1 required for the uptake of TF-iron complexes into cells. However, TFR1, an important iron intake receptor in cancer cells, still has unclear functions and mechanisms in ferroptosis. Furthermore, TFR1 expressions are up-regulated in some drug-resistant human cancer cells [45], requiring more in-depth studies though. Last but not least, since drug therapies may suppress TFR1 expression, developing a TFR1-specific reversible antagonist is the only single most effective strategy for both basic research and clinical practice. Notably, tumor progression can be inhibited through magnetic fields that augment the concentrations of TFR1-targeted superparamagnetic iron oxides in tumor tissues, inspiring future cancer therapy [125-127].
Figure 6.
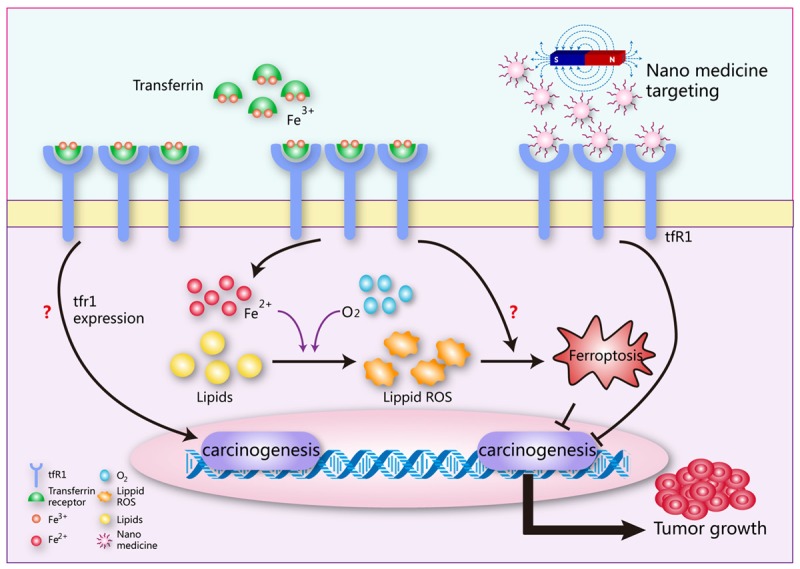
Future directions for TFR1 research in tumor. The direct effects of TFR1 expression on carcinogenesis remains further study. TFR1’s functions and mechanisms in cancer cell ferroptosis are still unclear. Nanomedicine combine with magnetic fields targeting TFR1 will be a potential cancer therapy.
Acknowledgements
We gratefully acknowledge the funding from the Science and Technology Planning Project of Shenzhen of China (JCYJ20170412140904406), and the National Basic Research Program of China (51777171).
Disclosure of conflict of interest
None.
References
- 1.Xue X, Ramakrishnan SK, Weisz K, Triner D, Xie L, Attili D, Pant A, Győrffy B, Zhan M, Carter-Su C, Hardiman KM, Wang TD, Dame MK, Varani J, Brenner D, Fearon ER, Shah YM. Iron uptake via DMT1 integrates cell cycle with JAK-STAT3 signaling to promote colorectal tumorigenesis. Cell Metab. 2016;24:447–461. doi: 10.1016/j.cmet.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torti SV, Torti FM. Iron and cancer: more ore to be mined. Nat Rev Cancer. 2013;13:342–355. doi: 10.1038/nrc3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drakesmith H, Nemeth E, Ganz T. Ironing out ferroportin. Cell Metab. 2015;22:777–787. doi: 10.1016/j.cmet.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bang S, Lee YM, Hong S, Cho KB, Nishida Y, Seo MS, Sarangi R, Fukuzumi S, Nam W. Redox-inactive metal ions modulate the reactivity and oxygen release of mononuclear non-haem iron(III)-peroxo complexes. Nat Chem. 2014;6:934–940. doi: 10.1038/nchem.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ward RJ, Zucca FA, Duyn JH, Crichton RR, Zecca L. The role of iron in brain ageing and neurodegenerative disorders. Lancet Neurol. 2014;13:1045–1060. doi: 10.1016/S1474-4422(14)70117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Čorić I, Mercado BQ, Bill E, Vinyard DJ, Holland PL. Binding of dinitrogen to an iron-sulfur-carbon site. Nature. 2015;526:96–99. doi: 10.1038/nature15246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adak L, Kawamura S, Toma G, Takenaka T, Isozaki K, Takaya H, Orita A, Li HC, TKM S, Nakamura M. Synthesis of aryl c-glycosides via ironcatalyzed cross coupling of halosugars: stereoselective anomeric arylation of glycosyl radicals. J Am Chem Soc. 2017;139:10693–10701. doi: 10.1021/jacs.7b03867. [DOI] [PubMed] [Google Scholar]
- 8.Schlesier J, Rohde M, Gerhardt S, Einsle O. A conformational switch triggers nitrogenase protection from oxygen damage by shethna protein iI (FeSII) J Am Chem Soc. 2016;138:239–247. doi: 10.1021/jacs.5b10341. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh C, Seal M, Mukherjee S, Ghosh DS. Alzheimer’s disease: a heme-aβ perspective. Acc Chem Res. 2015;48:2556–2564. doi: 10.1021/acs.accounts.5b00102. [DOI] [PubMed] [Google Scholar]
- 10.Barañano DE, Snyder SH. Neural roles for heme oxygenase: contrasts to nitric oxide synthase. Proc Natl Acad Sci U S A. 2001;98:10996–11002. doi: 10.1073/pnas.191351298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paul A, Drecourt A, Petit F, Deguine DD, Vasnier C, Oufadem M, Masson C, Bonnet C, Masmoudi S, Mosnier I, Mahieu L, Bouccara D, Kaplan J, Challe G, Domange C, Mochel F, Sterkers O, Gerber S, Nitschke P, Bole-Feysot C, Jonard L, Gherbi S, Mercati O, Ben AI, Lyonnet S, Rötig A, Delahodde A, Marlin S. FDXR mutations cause sensorial neuropathies and expand the spectrum of mitochondrial fe-ssynthesis diseases. Am J Hum Genet. 2017;101:630–637. doi: 10.1016/j.ajhg.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drakesmith H, Prentice A. Viral infection and iron metabolism. Nat Rev Microbiol. 2008;6:541–552. doi: 10.1038/nrmicro1930. [DOI] [PubMed] [Google Scholar]
- 13.Liu X, Theil EC. Ferritins: dynamic management of biological iron and oxygen chemistry. Acc Chem Res. 2005;38:167–175. doi: 10.1021/ar0302336. [DOI] [PubMed] [Google Scholar]
- 14.Cheng Q, Zhang X, Jiang J, Zhao G, Wang Y, Xu Y, Xu X, Ma H. Postmenopausal iron overload exacerbated bone loss by promoting the degradation of type i collagen. Biomed Res Int. 2017;2017:1345193. doi: 10.1155/2017/1345193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu C, Yang F, Fan D, Wang Y, Yu Y. Higher iron bioavailability of a human-like collagen iron complex. J Biomater Appl. 2017;32:82–92. doi: 10.1177/0885328217708638. [DOI] [PubMed] [Google Scholar]
- 16.Tchetina EV, Markova GA, Poole AR, Zukor DJ, Antoniou J, Makarov SA, Kuzin AN. Deferoxamine suppresses collagen cleavage and protease, cytokine, and col10a1 expression and upregulates ampk and krebs cycle genes in human osteoarthritic cartilage. Int J Rheumatol. 2016;2016:6432867. doi: 10.1155/2016/6432867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milto IV, Suhodolo IV, Prokopieva VD, Klimenteva TK. Molecular and cellular bases of iron metabolism in humans. Biochemistry (Mosc) 2016;81:549–564. doi: 10.1134/S0006297916060018. [DOI] [PubMed] [Google Scholar]
- 18.Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascón S, Hatzios SK, Kagan VE, Noel K, Jiang X, Linkermann A, Murphy ME, Overholtzer M, Oyagi A, Pagnussat GC, Park J, Ran Q, Rosenfeld CS, Salnikow K, Tang D, Torti FM, Torti SV, Toyokuni S, Woerpel KA, Zhang DD. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tarangelo A, Dixon SJ. Nanomedicine: an iron age for cancer therapy. Nat Nanotechnol. 2016;11:921–922. doi: 10.1038/nnano.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang YF, Zhang J, Su Y, Shen YY, Jiang DX, Hou YY, Geng MY, Ding J, Chen Y. G9a regulates breast cancer growth by modulating iron homeostasis through the repression of ferroxidase hephaestin. Nat Commun. 2017;8:274. doi: 10.1038/s41467-017-00350-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerins MJ, Ooi A. The roles of NRF2 in modulating cellular iron homeostasis. Antioxid Redox Signal. 2017 doi: 10.1089/ars.2017.7176. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luscieti S, Galy B, Gutierrez L, Reinke M, Couso J, Shvartsman M, Di PA, Witke W, Hentze MW, Pilo BP, Sanchez M. The actin binding protein profilin 2 is a novel regulator of iron homeostasis. Blood. 2017;30:1934–1945. doi: 10.1182/blood-2016-11-754382. [DOI] [PubMed] [Google Scholar]
- 23.Schoenfeld JD, Sibenaller ZA, Mapuskar KA, Wagner BA, Cramer-Morales KL, Furqan M, Sandhu S, Carlisle TL, Smith MC, Abu HT, Berg DJ, Zhang J, Keech J, Parekh KR, Bhatia S, Monga V, Bodeker KL, Ahmann L, Vollstedt S, Brown H, Shanahan Kauffman EP, Schall ME, Hohl RJ, Clamon GH, Greenlee JD, Howard MA, Schultz MK, Smith BJ, Riley DP, Domann FE, Cullen JJ, Buettner GR, Buatti JM, Spitz DR, Allen BG. O2(-) and H2O2-mediated disruption of fe metabolism causes the differential susceptibility of nsclc and gbm cancer cells to pharmacological ascorbate. Cancer Cell. 2017;31:487–500. doi: 10.1016/j.ccell.2017.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung M, Weigert A, Mertens C, Rehwald C, Brüne B. Iron handling in tumor-associated macrophages-is there a new role for lipocalin-2. Front Immunol. 2017;8:1171. doi: 10.3389/fimmu.2017.01171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Latunde-Dada GO. Ferroptosis: role of lipid peroxidation, iron and ferritinophagy. Biochim Biophys Acta. 2017;1861:1893–1900. doi: 10.1016/j.bbagen.2017.05.019. [DOI] [PubMed] [Google Scholar]
- 26.Fearnhead HO, Vandenabeele P, Vanden BT. How do we fit ferroptosis in the family of regulated cell death. Cell Death Differ. 2017;24:1991–1998. doi: 10.1038/cdd.2017.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mai TT, Hamaï A, Hienzsch A, Cañeque T, Müller S, Wicinski J, Cabaud O, Leroy C, David A, Acevedo V, Ryo A, Ginestier C, Birnbaum D, Charafe-Jauffret E, Codogno P, Mehrpour M, Rodriguez R. Salinomycin kills cancer stem cells by sequestering iron in lysosomes. Nat Chem. 2017;9:1025–1033. doi: 10.1038/nchem.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamaï A, Cañeque T, Müller S, Mai TT, Hienzsch A, Ginestier C, Charafe-Jauffret E, Codogno P, Mehrpour M, Rodriguez R. An iron hand over cancer stem cells. Autophagy. 2017;13:1465–1466. doi: 10.1080/15548627.2017.1327104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zanganeh S, Hutter G, Spitler R, Lenkov O, Mahmoudi M, Shaw A, Pajarinen JS, Nejadnik H, Goodman S, Moseley M, Coussens LM, Daldrup-Link HE. Iron oxide nanoparticles inhibit tumour growth by inducing pro-inflammatory macrophage polarization in tumour tissues. Nat Nanotechnol. 2016;11:986–994. doi: 10.1038/nnano.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim SE, Zhang L, Ma K, Riegman M, Chen F, Ingold I, Conrad M, Turker MZ, Gao M, Jiang X, Monette S, Pauliah M, Gonen M, Zanzonico P, Quinn T, Wiesner U, Bradbury MS, Overholtzer M. Ultrasmall nanoparticles induce ferroptosis in nutrient-deprived cancer cells and suppress tumour growth. Nat Nanotechnol. 2016;11:977–985. doi: 10.1038/nnano.2016.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szemraj M, Oszajca K, Szemraj J, Jurowski P. MicroRNA expression analysis in serum of patients with congenital hemochromatosis and age-related macular degeneration (AMD) Med Sci Monit. 2017;23:4050–4060. doi: 10.12659/MSM.902366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crielaard BJ, Lammers T, Rivella S. Targeting iron metabolism in drug discovery and delivery. Nat Rev Drug Discov. 2017;16:400–423. doi: 10.1038/nrd.2016.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aisen P. Transferrin receptor 1. Int J Biochem Cell Biol. 2004;36:2137–2143. doi: 10.1016/j.biocel.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 34.Jeong SM, Hwang S, Seong RH. Transferrin receptor regulates pancreatic cancer growth by modulating mitochondrial respiration and ROS generation. Biochem Biophys Res Commun. 2016;471:373–379. doi: 10.1016/j.bbrc.2016.02.023. [DOI] [PubMed] [Google Scholar]
- 35.Buas MF, Rho JH, Chai X, Zhang Y, Lampe PD, Li CI. Candidate early detection protein biomarkers for ER+/PR+ invasive ductal breast carcinoma identified using pre-clinical plasma from the WHI observational study. Breast Cancer Res Treat. 2015;153:445–454. doi: 10.1007/s10549-015-3554-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Senyilmaz D, Virtue S, Xu X, Tan CY, Griffin JL, Miller AK, Vidal-Puig A, Teleman AA. Regulation of mitochondrial morphology and function by stearoylation of TFR1. Nature. 2015;525:124–128. doi: 10.1038/nature14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreno-Navarrete JM, Novelle MG, Catalán V, Ortega F, Moreno M, Gomez-Ambrosi J, Xifra G, Serrano M, Guerra E, Ricart W, Frühbeck G, Diéguez C, Fernández-Real JM. Insulin resistance modulates iron-related proteins in adipose tissue. Diabetes Care. 2014;37:1092–1100. doi: 10.2337/dc13-1602. [DOI] [PubMed] [Google Scholar]
- 38.Daniels TR, Bernabeu E, Rodríguez JA, Patel S, Kozman M, Chiappetta DA, Holler E, Ljubimova JY, Helguera G, Penichet ML. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim Biophys Acta. 2012;1820:291–317. doi: 10.1016/j.bbagen.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagai K, Nakahata S, Shimosaki S, Tamura T, Kondo Y, Baba T, Taki T, Taniwaki M, Kurosawa G, Sudo Y, Okada S, Sakoda S, Morishita K. Development of a complete human anti-human transferrin receptor C antibody as a novel marker of oral dysplasia and oral cancer. Cancer Med. 2014;3:1085–1099. doi: 10.1002/cam4.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez A, Cacoub P, Macdougall IC, Peyrin-Biroulet L. Iron deficiency anaemia. Lancet. 2016;387:907–916. doi: 10.1016/S0140-6736(15)60865-0. [DOI] [PubMed] [Google Scholar]
- 41.Widera A, Norouziyan F, Shen WC. Mechanisms of TfR-mediated transcytosis and sorting in epithelial cells and applications toward drug delivery. Adv Drug Deliv Rev. 2003;55:1439–1466. doi: 10.1016/j.addr.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 42.Kaplan J. Mechanisms of cellular iron acquisition: another iron in the fire. Cell. 2002;111:603–606. doi: 10.1016/s0092-8674(02)01164-9. [DOI] [PubMed] [Google Scholar]
- 43.Nai A, Lidonnici MR, Rausa M, Mandelli G, Pagani A, Silvestri L, Ferrari G, Camaschella C. The second transferrin receptor regulates red blood cell production in mice. Blood. 2015;125:1170–1179. doi: 10.1182/blood-2014-08-596254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trinder D, Baker E. Transferrin receptor 2: a new molecule in iron metabolism. Int J Biochem Cell Biol. 2003;35:292–296. doi: 10.1016/s1357-2725(02)00258-3. [DOI] [PubMed] [Google Scholar]
- 45.Kazan HH, Urfali-Mamatoglu C, Gunduz U. Iron metabolism and drug resistance in cancer. Biometals. 2017;30:629–641. doi: 10.1007/s10534-017-0037-7. [DOI] [PubMed] [Google Scholar]
- 46.Rienzo M, Schiano C, Casamassimi A, Grimaldi V, Infante T, Napoli C. Identification of valid reference housekeeping genes for gene expression analysis in tumor neovascularization studies. Clin Transl Oncol. 2013;15:211–218. doi: 10.1007/s12094-012-0904-1. [DOI] [PubMed] [Google Scholar]
- 47.Gammella E, Buratti P, Cairo G, Recalcati S. The transferrin receptor: the cellular iron gate. Metallomics. 2017;9:1367–1375. doi: 10.1039/c7mt00143f. [DOI] [PubMed] [Google Scholar]
- 48.Ganz T. Iron homeostasis: fitting the puzzle pieces together. Cell Metab. 2008;7:288–290. doi: 10.1016/j.cmet.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 49.Lazaros G, Antonatou K, Vassilopoulos D. The therapeutic role of interleukin-1 inhibition in idiopathic recurrent pericarditis: current evidence and future challenges. Front Med (Lausanne) 2017;4:78. doi: 10.3389/fmed.2017.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muto Y, Nishiyama M, Nita A, Moroishi T, Nakayama KI. Essential role of FBXL5-mediated cellular iron homeostasis in maintenance of hematopoietic stem cells. Nat Commun. 2017;8:16114. doi: 10.1038/ncomms16114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson SA, Nizzi CP, Chang YI, Deck KM, Schmidt PJ, Galy B, Damnernsawad A, Broman AT, Kendziorski C, Hentze MW, Fleming MD, Zhang J, Eisenstein RS. The IRP1-HIF-2α axis coordinates iron and oxygen sensing with erythropoiesis and iron absorption. Cell Metab. 2013;17:282–290. doi: 10.1016/j.cmet.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghosh MC, Zhang DL, Jeong SY, Kovtunovych G, Ollivierre-Wilson H, Noguchi A, Tu T, Senecal T, Robinson G, Crooks DR, Tong WH, Ramaswamy K, Singh A, Graham BB, Tuder RM, Yu ZX, Eckhaus M, Lee J, Springer DA, Rouault TA. Deletion of iron regulatory protein 1 causes polycythemia and pulmonary hypertension in mice through translational derepression of HIF2α. Cell Metab. 2013;17:271–281. doi: 10.1016/j.cmet.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Udensi UK, Tackett AJ, Byrum S, Avaritt NL, Sengupta D, Moreland LW, Tchounwou PB, Isokpehi RD. Proteomics-based identification of differentially abundant proteins from human keratinocytes exposed to arsenic trioxide. J Proteomics Bioinform. 2014;7:166–178. doi: 10.4172/jpb.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawabata H. Progress in iron metabolism research. Rinsho Ketsueki. 2017;58:1864–1871. doi: 10.11406/rinketsu.58.1864. [DOI] [PubMed] [Google Scholar]
- 55.Barisani D, Conte D. Transferrin receptor 1 (TfR1) and putative stimulator of Fe transport (SFT) expression in iron deficiency and overload: an overview. Blood Cells Mol Dis. 2002;29:498–505. doi: 10.1006/bcmd.2002.0588. [DOI] [PubMed] [Google Scholar]
- 56.Roodman GD. Osteoclasts pump iron. Cell Metab. 2009;9:405–406. doi: 10.1016/j.cmet.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 57.Otero GA, Pliego-Rivero FB, Contreras G, Ricardo J, Fernández T. Iron supplementation brings up a lacking P300 in iron deficient children. Clin Neurophysiol. 2004;115:2259–2266. doi: 10.1016/j.clinph.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 58.Wei H, Ke HL, Lin J, Shete S, Wood CG, Hildebrandt MA. MicroRNA target site polymorphisms in the VHL-HIF1α pathway predict renal cell carcinoma risk. Mol Carcinog. 2014;53:1–7. doi: 10.1002/mc.21917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Okazaki F, Matsunaga N, Okazaki H, Azuma H, Hamamura K, Tsuruta A, Tsurudome Y, Ogino T, Hara Y, Suzuki T, Hyodo K, Ishihara H, Kikuchi H, To H, Aramaki H, Koyanagi S, Ohdo S. Circadian clock in a mouse colon tumor regulates intracellular iron levels to promote tumor progression. J Biol Chem. 2016;291:7017–7028. doi: 10.1074/jbc.M115.713412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marziali G, Perrotti E, Ilari R, Lulli V, Coccia EM, Moret R, Kühn LC, Testa U, Battistini A. Role of Ets-1 in transcriptional regulation of transferrin receptor and erythroid differentiation. Oncogene. 2002;21:7933–7944. doi: 10.1038/sj.onc.1205925. [DOI] [PubMed] [Google Scholar]
- 61.Lok CN, Ponka P. Identification of a hypoxia response element in the transferrin receptor gene. J Biol Chem. 1999;274:24147–24152. doi: 10.1074/jbc.274.34.24147. [DOI] [PubMed] [Google Scholar]
- 62.Biswas S, Tapryal N, Mukherjee R, Kumar R, Mukhopadhyay CK. Insulin promotes iron uptake in human hepatic cell by regulating transferrin receptor-1 transcription mediated by hypoxia inducible factor-1. Biochim Biophys Acta. 2013;1832:293–301. doi: 10.1016/j.bbadis.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 63.Lok CN, Ponka P. Identification of an erythroid active element in the transferrin receptor gene. J Biol Chem. 2000;275:24185–24190. doi: 10.1074/jbc.M000944200. [DOI] [PubMed] [Google Scholar]
- 64.Chromosome 1q21.3 amplification is linked to breast cancer recurrence. Cancer Discov. 2017 doi: 10.1038/nm.4405. [DOI] [PubMed] [Google Scholar]
- 65.Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, Leppa S, Pasanen A, Meriranta L, Karjalainen-Lindsberg ML, Nørgaard P, Pedersen M, Gang AO, Høgdall E, Heavican TB, Lone W, Iqbal J, Qin Q, Li G, Kim SY, Healy J, Richards KL, Fedoriw Y, Bernal-Mizrachi L, Koff JL, Staton AD, Flowers CR, Paltiel O, Goldschmidt N, Calaminici M, Clear A, Gribben J, Nguyen E, Czader MB, Ondrejka SL, Collie A, Hsi ED, Tse E, Au-Yeung RKH, Kwong YL, Srivastava G, Choi WWL, Evens AM, Pilichowska M, Sengar M, Reddy N, Li S, Chadburn A, Gordon LI, Jaffe ES, Levy S, Rempel R, Tzeng T, Happ LE, Dave T, Rajagopalan D, Datta J, Dunson DB, Dave SS. Genetic and functional drivers of diffuse large b cell lymphoma. Cell. 2017;171:481–494. e15. doi: 10.1016/j.cell.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vernieri C, Casola S, Foiani M, Pietrantonio F, de Braud F, Longo V. Targeting cancer metabolism: dietary and pharmacologic interventions. Cancer Discov. 2016;6:1315–1333. doi: 10.1158/2159-8290.CD-16-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 69.Bernardi MP, Ngan SY, Michael M, Lynch AC, Heriot AG, Ramsay RG, Phillips WA. Molecular biology of anal squamous cell carcinoma: implications for future research and clinical intervention. Lancet Oncol. 2015;16:e611–621. doi: 10.1016/S1470-2045(15)00292-2. [DOI] [PubMed] [Google Scholar]
- 70.Ngambenjawong C, Gustafson HH, Pun SH. Progress in tumor-associated macrophage (TAM)-targeted therapeutics. Adv Drug Deliv Rev. 2017;114:206–221. doi: 10.1016/j.addr.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garofalo M, Croce CM. Role of microRNAs in maintaining cancer stem cells. Adv Drug Deliv Rev. 2015;81:53–61. doi: 10.1016/j.addr.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berindan-Neagoe I, Monroig PC, Pasculli B, Calin GA. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64:311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee KM, Giltnane JM, Balko JM, Schwarz LJ, Guerrero-Zotano AL, Hutchinson KE, Nixon MJ, Estrada MV, Sánchez V, Sanders ME, Lee T, Gómez H, Lluch A, Pérez-Fidalgo JA, Wolf MM, Andrejeva G, Rathmell JC, Fesik SW, Arteaga CL. MYC and MCL1 cooperatively promote chemotherapy-resistant breast cancer stem cells via regulation of mitochondrial oxidative phosphorylation. Cell Metab. 2017;26:633–647. e7. doi: 10.1016/j.cmet.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Z, Wu H, Wei Z, Wang X, Shen P, Wang S, Wang A, Chen W, Lu Y. TRPM8: a potential target for cancer treatment. J Cancer Res Clin Oncol. 2016;142:1871–1881. doi: 10.1007/s00432-015-2112-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Augustin HG, Koh GY. Organotypic vasculature: From descriptive heterogeneity to functional pathophysiology. Science. 2017;357 doi: 10.1126/science.aal2379. [DOI] [PubMed] [Google Scholar]
- 76.Rychtarcikova Z, Lettlova S, Tomkova V, Korenkova V, Langerova L, Simonova E, Zjablovskaja P, Alberich-Jorda M, Neuzil J, Truksa J. Tumorinitiating cells of breast and prostate origin show alterations in the expression of genes related to iron metabolism. Oncotarget. 2017;8:6376–6398. doi: 10.18632/oncotarget.14093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Horniblow RD, Bedford M, Hollingworth R, Evans S, Sutton E, Lal N, Beggs A, Iqbal TH, Tselepis C. BRAF mutations are associated with increased iron regulatory protein-2 expression in colorectal tumorigenesis. Cancer Sci. 2017;108:1135–1143. doi: 10.1111/cas.13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kindrat I, Tryndyak V, de Conti A, Shpyleva S, Mudalige TK, Kobets T, Erstenyuk AM, Beland FA, Pogribny IP. MicroRNA-152-mediated dysregulation of hepatic transferrin receptor 1 in liver carcinogenesis. Oncotarget. 2016;7:1276–1287. doi: 10.18632/oncotarget.6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang B, Zhang J, Song F, Tian M, Shi B, Jiang H, Xu W, Wang H, Zhou M, Pan X, Gu J, Yang S, Jiang L, Li Z. EGFR regulates iron homeostasis to promote cancer growth through redistribution of transferrin receptor 1. Cancer Lett. 2016;381:331–340. doi: 10.1016/j.canlet.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 80.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von FK, Forsberg M, Persson L, Johansson F, Zwahlen M, von HG, Nielsen J, Pontén F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 81.Gu Z, Wang H, Xia J, Yang Y, Jin Z, Xu H, Shi J, De Domenico I, Tricot G, Zhan F. Decreased ferroportin promotes myeloma cell growth and osteoclast differentiation. Cancer Res. 2015;75:2211–2221. doi: 10.1158/0008-5472.CAN-14-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ohkuma M, Haraguchi N, Ishii H, Mimori K, Tanaka F, Kim HM, Shimomura M, Hirose H, Yanaga K, Mori M. Absence of CD71 transferrin receptor characterizes human gastric adenosquamous carcinoma stem cells. Ann Surg Oncol. 2012;19:1357–1364. doi: 10.1245/s10434-011-1739-7. [DOI] [PubMed] [Google Scholar]
- 83.Singh M, Mugler K, Hailoo DW, Burke S, Nemesure B, Torkko K, Shroyer KR. Differential expression of transferrin receptor (TfR) in a spectrum of normal to malignant breast tissues: implications for in situ and invasive carcinoma. Appl Immunohistochem Mol Morphol. 2011;19:417–423. doi: 10.1097/PAI.0b013e318209716e. [DOI] [PubMed] [Google Scholar]
- 84.Rosager AM, Sørensen MD, Dahlrot RH, Hansen S, Schonberg DL, Rich JN, Lathia JD, Kristensen BW. Transferrin receptor-1 and ferritin heavy and light chains in astrocytic brain tumors: Expression and prognostic value. PLoS One. 2017;12:e0182954. doi: 10.1371/journal.pone.0182954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chirasani SR, Markovic DS, Synowitz M, Eichler SA, Wisniewski P, Kaminska B, Otto A, Wanker E, Schäfer M, Chiarugi P, Meier JC, Kettenmann H, Glass R. Transferrin-receptor-mediated iron accumulation controls proliferation and glutamate release in glioma cells. J Mol Med (Berl) 2009;87:153–167. doi: 10.1007/s00109-008-0414-3. [DOI] [PubMed] [Google Scholar]
- 86.Weston C, Klobusicky J, Weston J, Connor J, Toms SA, Marko NF. Aberrations in the iron regulatory gene signature are associated with decreased survival in diffuse infiltrating gliomas. PLoS One. 2016;11:e0166593. doi: 10.1371/journal.pone.0166593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu X, Feng L, Liu D, Zhang L, Wu B, Jiang W, Han Z, Cheng S. Quantitative proteomics reveals the novel co-expression signatures in early brain development for prognosis of glioblastoma multiforme. Oncotarget. 2016;7:14161–14171. doi: 10.18632/oncotarget.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pizzamiglio S, De Bortoli M, Taverna E, Signore M, Veneroni S, Cho WC, Orlandi R, Verderio P, Bongarzone I. Expression of iron-related proteins differentiate non-cancerous and cancerous breast tumors. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18020410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Habashy HO, Powe DG, Staka CM, Rakha EA, Ball G, Green AR, Aleskandarany M, Paish EC, Douglas MR, Nicholson RI, Ellis IO, Gee JM. Transferrin receptor (CD71) is a marker of poor prognosis in breast cancer and can predict response to tamoxifen. Breast Cancer Res Treat. 2010;119:283–293. doi: 10.1007/s10549-009-0345-x. [DOI] [PubMed] [Google Scholar]
- 90.Miller LD, Coffman LG, Chou JW, Black MA, Bergh J, D’Agostino R, Torti SV, Torti FM. An iron regulatory gene signature predicts outcome in breast cancer. Cancer Res. 2011;71:6728–6737. doi: 10.1158/0008-5472.CAN-11-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jiang XP, Elliott RL, Head JF. Manipulation of iron transporter genes results in the suppression of human and mouse mammary adenocarcinomas. Anticancer Res. 2010;30:759–765. [PubMed] [Google Scholar]
- 92.Wang W, Deng Z, Hatcher H, Miller LD, Di X, Tesfay L, Sui G, D’Agostino RB, Torti FM, Torti SV. IRP2 regulates breast tumor growth. Cancer Res. 2014;74:497–507. doi: 10.1158/0008-5472.CAN-13-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marques O, Porto G, Rêma A, Faria F, Cruz PA, Gomez-Lazaro M, Silva P, Martins da Silva B, Lopes C. Local iron homeostasis in the breast ductal carcinoma microenvironment. BMC Cancer. 2016;16:187. doi: 10.1186/s12885-016-2228-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jian J, Yang Q, Huang X. Src regulates Tyr (20) phosphorylation of transferrin receptor-1 and potentiates breast cancer cell survival. J Biol Chem. 2011;286:35708–35715. doi: 10.1074/jbc.M111.271585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Carvalho C, Glynne-Jones R. Challenges behind proving efficacy of adjuvant chemotherapy after preoperative chemoradiation for rectal cancer. Lancet Oncol. 2017;18:e354–e363. doi: 10.1016/S1470-2045(17)30346-7. [DOI] [PubMed] [Google Scholar]
- 96.Okazaki F, Matsunaga N, Okazaki H, Utoguchi N, Suzuki R, Maruyama K, Koyanagi S, Ohdo S. Circadian rhythm of transferrin receptor 1 gene expression controlled by c-Myc in colon cancer-bearing mice. Cancer Res. 2010;70:6238–6246. doi: 10.1158/0008-5472.CAN-10-0184. [DOI] [PubMed] [Google Scholar]
- 97.Chua AC, Klopcic BR, Ho DS, Fu SK, Forrest CH, Croft KD, Olynyk JK, Lawrance IC, Trinder D. Dietary iron enhances colonic inflammation and IL-6/IL-11-Stat3 signaling promoting colonic tumor development in mice. PLoS One. 2013;8:e78850. doi: 10.1371/journal.pone.0078850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Beckman LE, Hägerstrand I, Stenling R, Van Landeghem GF, Beckman L. Interaction between haemochromatosis and transferrin receptor genes in hepatocellular carcinoma. Oncology. 2000;59:317–322. doi: 10.1159/000012189. [DOI] [PubMed] [Google Scholar]
- 99.Holmström P, Gåfvels M, Eriksson LC, Dzikaite V, Hultcrantz R, Eggertsen G, Stål P. Expression of iron regulatory genes in a rat model of hepatocellular carcinoma. Liver Int. 2006;26:976–985. doi: 10.1111/j.1478-3231.2006.01316.x. [DOI] [PubMed] [Google Scholar]
- 100.Takahashi M, Shibutani M, Woo GH, Inoue K, Fujimoto H, Igarashi K, Kanno J, Hirose M, Nishikawa A. Cellular distributions of molecules with altered expression specific to the tumor promotion process from the early stage in a rat two-stage hepatocarcinogenesis model. Carcinogenesis. 2008;29:2218–2226. doi: 10.1093/carcin/bgn135. [DOI] [PubMed] [Google Scholar]
- 101.Sakurai K, Sohda T, Ueda S, Tanaka T, Hirano G, Yokoyama K, Morihara D, Aanan A, Takeyama Y, Irie M, Iwata K, Syakado S, Noritomiz T, Yamashita Y, Sakisaka S. Immunohistochemical demonstration of transferrin receptor 1 and 2 in human hepatocellular carcinoma tissue. Hepatogastroenterology. 2014;61:426–430. [PubMed] [Google Scholar]
- 102.Basuli D, Tesfay L, Deng Z, Paul B, Yamamoto Y, Ning G, Xian W, McKeon F, Lynch M, Crum CP, Hegde P, Brewer M, Wang X, Miller LD, Dyment N, Torti FM, Torti SV. Iron addiction: a novel therapeutic target in ovarian cancer. Oncogene. 2017;36:4089–4099. doi: 10.1038/onc.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kuvibidila S, Gauthier T, Warrier RP, Rayford W. Increased levels of serum transferrin receptor and serum transferrin receptor/log ferritin ratios in men with prostate cancer and the implications for body-iron stores. J Lab Clin Med. 2004;144:176–182. doi: 10.1016/j.lab.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 104.Johnson IR, Parkinson-Lawrence EJ, Shandala T, Weigert R, Butler LM, Brooks DA. Altered endosome biogenesis in prostate cancer has biomarker potential. Mol Cancer Res. 2014;12:1851–1862. doi: 10.1158/1541-7786.MCR-14-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang J, Chen G, Pantopoulos K. Inhibition of transferrin receptor 1 transcription by a cell density response element. Biochem J. 2005;392:383–388. doi: 10.1042/BJ20050492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhu X, Zhang H, Lin Y, Chen P, Min J, Wang Z, Xiao W, Chen B. Mechanisms of gambogic acid-induced apoptosis in non-small cell lung cancer cells in relation to transferrin receptors. J Chemother. 2009;21:666–672. doi: 10.1179/joc.2009.21.6.666. [DOI] [PubMed] [Google Scholar]
- 107.Kukulj S, Jaganjac M, Boranic M, Krizanac S, Santic Z, Poljak-Blazi M. Altered iron metabolism, inflammation, transferrin receptors, and ferritin expression in non-small-cell lung cancer. Med Oncol. 2010;27:268–277. doi: 10.1007/s12032-009-9203-2. [DOI] [PubMed] [Google Scholar]
- 108.Ferrando AA, López-Otín C. Clonal evolution in leukemia. Nat Med. 2017;23:1135–1145. doi: 10.1038/nm.4410. [DOI] [PubMed] [Google Scholar]
- 109.Taetle R, Rhyner K, Castagnola J, To D, Mendelsohn J. Role of transferrin, Fe, and transferrin receptors in myeloid leukemia cell growth. Studies with an antitransferrin receptor monoclonal antibody. J Clin Invest. 1985;75:1061–1067. doi: 10.1172/JCI111768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Scott CS, Ramsden W, Limbert HJ, Master PS, Roberts BE. Membrane transferrin receptor (TfR) and nuclear proliferation-associated Ki-67 expression in hemopoietic malignancies. Leukemia. 1988;2:438–442. [PubMed] [Google Scholar]
- 111.Liu Q, Wang M, Hu Y, Xing H, Chen X, Zhang Y, Zhu P. Significance of CD71 expression by flow cytometry in diagnosis of acute leukemia. Leuk Lymphoma. 2014;55:892–898. doi: 10.3109/10428194.2013.819100. [DOI] [PubMed] [Google Scholar]
- 112.Płoszyńska A, Ruckemann-Dziurdzińska K, Jóźwik A, Mikosik A, Lisowska K, Balcerska A, Witkowski JM. Cytometric evaluation of transferrin receptor 1 (CD71) in childhood acute lymphoblastic leukemia. Folia Histochem Cytobiol. 2012;50:304–311. doi: 10.5603/fhc.2012.0040. [DOI] [PubMed] [Google Scholar]
- 113.Pham DH, Powell JA, Gliddon BL, Moretti PA, Tsykin A, Van der Hoek M, Kenyon R, Goodall GJ, Pitson SM. Enhanced expression of transferrin receptor 1 contributes to oncogenic signalling by sphingosine kinase 1. Oncogene. 2014;33:5559–5568. doi: 10.1038/onc.2013.502. [DOI] [PubMed] [Google Scholar]
- 114.Bayeva M, Khechaduri A, Puig S, Chang HC, Patial S, Blackshear PJ, Ardehali H. mTOR regulates cellular iron homeostasis through tristetraprolin. Cell Metab. 2012;16:645–657. doi: 10.1016/j.cmet.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.O’Donnell KA, Yu D, Zeller KI, Kim JW, Racke F, Thomas-Tikhonenko A, Dang CV. Activation of transferrin receptor 1 by c-Myc enhances cellular proliferation and tumorigenesis. Mol Cell Biol. 2006;26:2373–2386. doi: 10.1128/MCB.26.6.2373-2386.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jiao Y, Wilkinson J, Christine PE, Buss JL, Wang W, Planalp R, Torti FM, Torti SV. Iron chelation in the biological activity of curcumin. Free Radic Biol Med. 2006;40:1152–1160. doi: 10.1016/j.freeradbiomed.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 117.Yang C, Ma X, Wang Z, Zeng X, Hu Z, Ye Z, Shen G. Curcumin induces apoptosis and protective autophagy in castration-resistant prostate cancer cells through iron chelation. Drug Des Devel Ther. 2017;11:431–439. doi: 10.2147/DDDT.S126964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Callens C, Moura IC, Lepelletier Y, Coulon S, Renand A, Dussiot M, Ghez D, Benhamou M, Monteiro RC, Bazarbachi A, Hermine O. Recent advances in adult T-cell leukemia therapy: focus on a new anti-transferrin receptor monoclonal antibody. Leukemia. 2008;22:42–48. doi: 10.1038/sj.leu.2404958. [DOI] [PubMed] [Google Scholar]
- 119.Moura IC, Lepelletier Y, Arnulf B, England P, Baude C, Beaumont C, Bazarbachi A, Benhamou M, Monteiro RC, Hermine O. A neutralizing monoclonal antibody (mAb A24) directed against the transferrin receptor induces apoptosis of tumor T lymphocytes from ATL patients. Blood. 2004;103:1838–1845. doi: 10.1182/blood-2003-07-2440. [DOI] [PubMed] [Google Scholar]
- 120.Schaar DG, Medina DJ, Moore DF, Strair RK, Ting Y. miR-320 targets transferrin receptor 1 (CD71) and inhibits cell proliferation. Exp Hematol. 2009;37:245–255. doi: 10.1016/j.exphem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 121.Camp ER, Wang C, Little EC, Watson PM, Pirollo KF, Rait A, Cole DJ, Chang EH, Watson DK. Transferrin receptor targeting nanomedicine delivering wild-type p53 gene sensitizes pancreatic cancer to gemcitabine therapy. Cancer Gene Ther. 2013;20:222–228. doi: 10.1038/cgt.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Daniels-Wells TR, Widney DP, Leoh LS, Martínez-Maza O, Penichet ML. Efficacy of an antitransferrin receptor 1 antibody against aidsrelated non-hodgkin lymphoma: a brief communication. J Immunother. 2015;38:307–310. doi: 10.1097/CJI.0000000000000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shimosaki S, Nakahata S, Ichikawa T, Kitanaka A, Kameda T, Hidaka T, Kubuki Y, Kurosawa G, Zhang L, Sudo Y, Shimoda K, Morishita K. Development of a complete human IgG monoclonal antibody to transferrin receptor 1 targeted for adult T-cell leukemia/lymphoma. Biochem Biophys Res Commun. 2017;485:144–151. doi: 10.1016/j.bbrc.2017.02.039. [DOI] [PubMed] [Google Scholar]
- 124.Yu H, Guo P, Xie X, Wang Y, Chen G. Ferroptosis, a new form of cell death, and its relationships with tumourous diseases. J Cell Mol Med. 2017;21:648–657. doi: 10.1111/jcmm.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhou Z, Song J, Tian R, Yang Z, Yu G, Lin L, Zhang G, Fan W, Zhang F, Niu G, Nie L, Chen X. Activatable singlet oxygen generation from lipid hydroperoxide nanoparticles for cancer therapy. Angew Chem Int Ed Engl. 2017;56:6492–6496. doi: 10.1002/anie.201701181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Aldahoun MA, Jaafar MS, Al-Akhras MH, Bououdina M. Enhanced nanocurcumin toxicity against (PC3) tumor and microbial by using magnetic field in vitro. Artif Cells Nanomed Biotechnol. 2017;45:843–853. doi: 10.1080/21691401.2016.1178137. [DOI] [PubMed] [Google Scholar]
- 127.Liu Z, Zhan X, Yang M, Yang Q, Xu X, Lan F, Wu Y, Gu Z. A magnetic-dependent protein corona of tailor-made superparamagnetic iron oxides alters their biological behaviors. Nanoscale. 2016;8:7544–7555. doi: 10.1039/c5nr08447d. [DOI] [PubMed] [Google Scholar]