Abstract
Valeriana jatamansi Jones is an aromatic medicinal herb and important alternative to V. officinalis, which is utilized for medicinal purposes in China and India and also as spices in India. Bioactive ingredients of V. jatamansi vary in different regions. However, no information is currently available on influence of genotype and environmental factors in the volatile compounds, especially when germplasms and planting locations need to be selected. Based on the results of SNP and volatile constituents from GC-MS analysis, this study found various genotypes and chemotypes of V. jatamansi for wild plants from seven regions in China and common-garden samples; correlations between genotype and chemotype were revealed for the plants. Two distinct populations (PX, FY) were distinguishable from five others (GJ, YL, SY, DD, DY) according to their genotypes and volatile profiles, the consistency of which was observed showing that genotype could significantly influence chemotype. Wild populations and common-garden samples were also separated in their volatile profiles, demonstrating that environmental factors strongly affected their chemotypes. Compounds contributing to the discrimination were identified as discriminatory compounds. This investigation has explored and provided essential information concerning the correlation between genotype and chemotype as well as environmental factors and chemotype of V. jatamansi in some regions of China. Feasible plantation and conservation strategies of V. jatamansi could be further explored based on these results.
Keywords: Valerianae Jatamansi Rhizoma et Radix, Valeriana jatamansi, genotype, chemotype, volatile profile, wild population, common-garden sample
Introduction
Biogenesis of essential oil occurs widely across the plant kingdom and is important for plant physiology in terms of metabolism and preset developmental differentiation programme of the synthesizing tissue (Chappell, 1995; Sangwan et al., 2001). As an essential part of plant metabolism, the biosynthesis of essential oil may depend on both the genetic backgrounds and environmental effects (Chappell, 1995; Sangwan et al., 2001). Commercially, these volatile plant metabolites have already found extensive cosmetic and therapeutic applications (Sangwan et al., 2001).
Valeriana jatamansi Jones is a valuable plant species with volatile components utilized as a herbal medicine. Valerianae Jatamansi Rhizoma et Radix is indexed in Chinese Pharmacopeia (Part 1) in the 1977, 2010, and 2015 editions as a traditional Chinese medicine for gastrointestinal diseases and anxiety. It is prepared from naturally dried rhizome and root of perennial herb V. jatamansi Jones, which is widely distributed throughout temperate Himalayan region and southwestern areas of China (Ming et al., 1997; Jugran et al., 2013). Many phytochemicals including iridoids, sesquiterpenoids, and essential oils have already been identified from this species (Ming et al., 1997; Bhatt et al., 2012; Li et al., 2013). Among them, essential oils commonly used as spices in India had both antioxidant and insecticidal activities whereas iridoids and sesquiterpenoids showed moderate neuroprotective effects and inhibitory activity on acetylcholinesterase (Rana and Sharma, 2000; Xu et al., 2011; Dong et al., 2015), respectively.
For these traditional and newly found pharmaceutical purposes, this medicine's consumption is increasing in these years and the distribution range of crude drugs is expanding. The essential oil composition was explored and was considered to be largely influenced by plant's location of growing (Verma et al., 2011), while some non-volatile components such as the valepotriates are supposed to be similar (Wang et al., 2017). Chinese Pharmacopeia (2015 edition) requires assessments of size, exterior character, water content, total ash, acid-insoluble ash, total extractives, and microscope characteristics together with the TLC profiles of valepotriane (C22H30O8) and acetylvalepotriane (C24H32O10). However, the quality control of this medicinal herb and resource recognition from different areas remain difficult if based solely on the above descriptions. Selection of good germplasms and good agricultural practice (GAP) of V. jatamansi is required, and planting locations is ought to be selected properly. It is well-documented that chemical phenotypes (chemotypes) of several herbal medicines depends on the environment they grow (Wang et al., 2004; Dai et al., 2010a,b). However, whether V. jatamansi possesses different genotypes and how these genotypes and environmental factors affect its chemotype remain to be clarified. To the best of our knowledge, few researches about volatile constituents of V. jatamansi have been published so far.
To address these questions, in current study, type IIB endonucleases restriction-site associated DNA (2b-RAD) approach (Wang et al., 2012) was used to identify the genotypic characteristics for seven wild populations of V. Jatamansi from China. The head-space of SHS-GC-MS analysis would cover all volatile components including not only essential oils but also components with low boiling points, water-soluble volatile components and/or oxidation-prone components (Molina-Calle et al., 2017). Therefore, the composition of volatile compounds was also analyzed as plant chemotype here for all wild populations together with the common-garden samples originating from them. Dependence of the chemotypes on the genotypes and environmental factors was further evaluated for Valerianae Jatamansi Rhizoma et Radix.
Materials and methods
Plant materials
Although V. jatamansi is widely distributed throughout temperate Himalayan region and southwestern areas of China, the origin plants of Valerianae jatamansi Rhizoma et Radix used for medicinal purposes are traditionally collected in Yunnan and Guizhou provinces. To clarify whether different genotypes and chemotypes were present, seven wild populations of V. jatamansi were collected from seven different locations in Yunnan and Guizhou provinces to cover traditional sampling areas plus environmental locations. These included samples from Gejiu (GJ) and Fuyuan (FY) in Yunnan Province, Leishan Yongle (YL), Leishan Dadi (DD), Suiyang (SY), Panxian (PX), and Duyun (DY) in Guizhou Province (Table 1, Supplemental Figure 1).
Table 1.
Information of selected regions plant were sampled.
| Population | Voucher | Season | Location | Longitude | Latitude | Altitude/m | Slope | Biotope |
|---|---|---|---|---|---|---|---|---|
| PX | WT01 | Oct. 2015 | Panxian, Guizhou | 104°24′ | 25°40′ | 1946 | North | Mingled forest |
| FY | FY01 | Oct. 2015 | Fuyuan, Yunnan | 104°13′ | 25°34′ | 1939 | North | Farmland margins |
| GJ | GJ01 | Aug. 2015 | Gejiu, Yunnan | 103°12′ | 23°23′ | 2530 | Southeast | Hillside meadows |
| DD | LS0202 | Sep. 2015 | Dadi, Leishan, Guizhou | 108°08′ | 26°08′ | 1381 | South | Hillside meadows |
| DY | FY01 | Oct. 2015 | Duyun, Guizhou | 107°36′ | 26°18′ | 918 | Northwest | Mingled forest |
| YL | LS0102 | Sep. 2015 | Yongle, Leishan, Guizhou | 108°09′ | 26°11′ | 1056 | East | Farmland margin |
| SY | SY02 | Sep. 2015 | Suiyang, Guizhou | 107°13′ | 27°59′ | 1030 | Northwest | Farmland margin |
In each population, the leaves of 10 plants were collected randomly with spacing greater than 5 m, dried with allochroic silica gel and preserved in normal temperature for genetic analysis. The plant was identified by Professor Zigang Qian, Yunnan University of Traditional Chinese Medicines and a voucher specimen for all the plant material was deposited in The Chinese Medicine Specimen Museum of Yunnan University of Traditional Chinese Medicines for further reference (Table 1).
To avoid seasonal effects on the plant metabolite composition (Xiao et al., 2008), we collected wild samples within a shortest period (practically possible) in autumn according to accumulated experiences of Traditional Chinese Medicine. To take the effects of environmental factors (e.g., altitude, temperature, sunshine, and humidity) into consideration, we also collected the common-garden samples (Yunnan Longfar Pharmaceutical Co. LTD) within 2 days during the same season. Common-garden samples were collected 1 year after transplantation using the same approach as for the corresponding wild populations. The wild plants were also transplanted to the Planting Base in Kunming Unity Township.
For volatile constituent analysis, the rhizomes and roots were removed from the same plants, dried and preserved in the same way described above. Samples were cleaned with brush, cut into pieces with a pair of scissors and ground into powder using SKSI tissue lyser (BiHeng Biotechnology Inc.) at 60 Hz for 30 s with 3 cycles totally. Ground samples were sieved through a 40 mesh sieve and then stored in sample bottles with good sealing property at 4°C before further analysis.
Genetic analysis based on 2b-RAD approach
DNA was extracted by means of AxyPrepTM Multisource Genomic DNA Miniprep Kit 50-prep (AP-MN-BT-GDNA-50G, Axygen, USA). Mass and concentration of DNA were measured using nucleic acid and protein analyzer.
Sequencing libraries with tags from 70 samples of V. jatamansi were built based on 2b-RAD approach (Wang et al., 2012). 5′-NNN-3′ was used to establish libraries for all samples. Paired-end sequencing was conducted for qualified libraries on Hiseq Xten platform.
Tags of individual samples were clustered into allele clusters and mismatch was not allowed. Allele clusters were combined into locus cluster to form reference sequence when two mismatches were allowed. Low-coverage locus cluster formed by sequencing error and high-coverage locus cluster structured by repetitive sequences were excluded based on improved maximum likelihood (iML, whole-genome de novo SNP genotyping method based on mixed Possion distribution model). Threshold was obtained from distribution model of tag numbers. Finally 300,926 tags with BsaXI enzyme cutting sites were obtained.
Original sequences were formed by base calling (raw reads) and were filtered by excluding sequences including N bases and reads with flanking sequences as well as low quality (more than 20% bases with quality less than 20) to get clean data.
Volatile constituent analysis
SHS-GC-MS analysis
Samples were brought to room temperature. Three replicates of powdered samples (about 0.1000–0.1010 g) were put into three different glass vials (20 mL) then sealed with screw-caps having PTFE/silicone septa (17-02-1417-TF, SHIMADZU).
Static headspace equilibration was conducted at 140°C for 20 min with 400 rpm shaking. Then 2.00 mL of headspace gas was injected into injection port and analyzed using QP2010 Ultra Gas Chromatography-Mass Spectrometer (SHIMADZU, Japan) equipped with a Thermo Fisher non-polar TG-5MS column (P/N 26098–2970, 0.32 mm × 30 m × 1 μm); inlet temperature was set to 250°C with split ratio of10:1, helium gas as carrier gas (with a constant flow of 2 mL/min). Mass spectra were recorded with an electron ionization mode (70 eV), a scan range of m/z 50–1,000, ion source temperature of 250°C, transfer line temperature of 280°C, and solvent delay time of 2 min. Under the optimum conditions above, the relative standard deviations (RSD) of all compounds were lower than 20%.
Identification of volatile compounds
The GC-MS raw data were extracted and analyzed using GCMS Solution (Version 2.70). Areas of volatile compounds were calculated automatically with the same integration parameters (slope: 1,000/min, peak width: 2 s, drift: 0 min, smoothing time: 2 s, smoothing width: 2 s). Compounds were identified by automatic retrieval of the mass spectra (NIST 14 and NIST 14s), assisted by comparison between retention indexes from libraries and calculated ones based on experimental retention time of n-alkanes.
Statistical analysis
For genetic analysis, each genotype with markers was assembled head to tail and missing sites were replaced by “–.” Neighbor-joining tree and maximum-likelihood tree were structured using treebest (Version 1.9.2) and MEGA6. Two different analytical methods were employed here to ensure that such clustering were method independent since no single perfect method is available for the time being. Reliability was tested by bootstrap with 1,000 repeats. Genetic differentiation and diversity were presented using Genepop (version 4.2.2).
The compounds, which cover less than 80% of volatile profile of a single population or have signal-to-noise value lower than 10, were excluded from further analysis. Data, based on the areas of volatile compounds, was normalized by mass and scaled using Pareto mode using MateboAnalyst 3.0. Principal Component Analysis (PCA) was conducted for wild populations and common-garden samples separately, where samples outside 95% confidence intervals were excluded.
Clustering analysis was made using MateboAnalyst 3.0 with the distance measurement of Pearson and the Clustering Algorithm of Average. Results were further confirmed with Orthogonal Partial Least Squares-Discriminant Analysis (OPLS-DA) using SIMCA 14.1. S-plots and loadings plots were made to identify discriminatory compounds, verified through Variable Importance in the Projection (VIP) values automatically.
Results and discussion
Genotypic diversity and differentiation for V. jatamansi
F-statistics (Fst) was obtained for all populations and ranged from 0.0226 to 0.2344 (Table 2). Relatively high population differentiation was observed between PX, FY, and other populations with Fst in the range of 0.15–0.25. In contrast, medium inter-group differentiation was observed for PX and FY, for GJ and YL, for GJ and SY with Fst only slightly higher than 0.0500. Other populations shared lower differentiation with each other.
Table 2.
Genetic differentiation (Fst).
| Population | GJ | YL | DD | SY | FY | PX |
|---|---|---|---|---|---|---|
| YL | 0.0536 | |||||
| DD | 0.0437 | 0.0226 | ||||
| SY | 0.0558 | 0.0369 | 0.0338 | |||
| FY | 0.2344 | 0.2310 | 0.2257 | 0.2275 | ||
| PX | 0.2145 | 0.2109 | 0.2049 | 0.2093 | 0.0607 | |
| DY | 0.0439 | 0.0388 | 0.0243 | 0.0376 | 0.2302 | 0.2093 |
Polymorphism Information Content (PIC), Expected Heterozygosity (He), and Observed Heterozygosity (Ho) were also calculated based on the result of SNP (Table 3). Apparently populations of PX and FY had higher PIC, He, and Ho than other populations. This demonstrated they were supposed to own higher genetic diversities.
Table 3.
Genetic diversity.
| Population | He | Ho | PIC |
|---|---|---|---|
| GJ | 0.3650 | 0.5758 | 0.2887 |
| YL | 0.3691 | 0.5855 | 0.2908 |
| DD | 0.3531 | 0.5476 | 0.2800 |
| SY | 0.3661 | 0.5777 | 0.2888 |
| FY | 0.4049 | 0.6846 | 0.3152 |
| PX | 0.4080 | 0.6804 | 0.3179 |
| DY | 0.3499 | 0.5379 | 0.2778 |
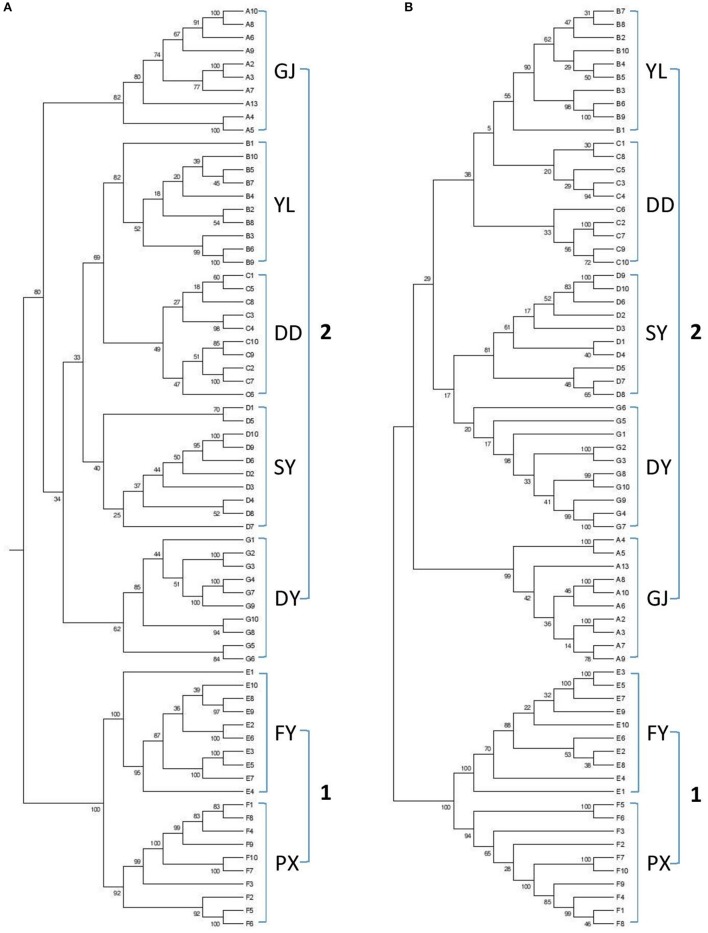
Dendrograms were generated based on DNA sequences for (wild) populations from various regions using reduced-representation sequencing 2b-RAD approach. Two analyses were also conducted from both Neighbor-joining tree (Figure 1A) and Maximum-Likelihood tree (Figure 1B). Results from both analyses clearly showed genetic similarity for populations PX and FY (known as genotype 1 or G1 in the following discussions) and similarity for the rest populations (genotype 2 or G2).
Figure 1.
Dendrograms based on genetic analysis (A: GJ, B: YL, C: DD, D: SY, E: FY, F: PX, G: DY). (A) Neighbor-joining tree, (B) Maximum-Likelihood tree.
Influence of genotype and environment on chemotype of V. jatamansi
Chemotypes of V. jatamansi were evaluated by measuring the composition of volatile compounds with GC-MS method in the aforementioned wild and common garden populations. Sixty-four such volatile compounds were detected and assigned by comparing their retention indices and mass spectral features with the NIST database (Supplemental Table 1). It is apparent that C10 (3-methylvaleric acid), C13 (sabinene), C43 (terpinylacetate), C49 (α-santalene), C50 (β-caryophyllene), C53 (eudesma-3,7(11)-diene), and C58 (α-bulnesene) were abundant volatile compounds in these plants (Supplemental Table 2). It is also interesting to note that many of these volatile metabolites are biosynthetic products terpenoid pathway from geranyl diphosphate and (2E,6E)-farnesyl diphosphate. For instance, C11 (camphene), C13 (sabinene), C14 (β-pinene), C22 (4-R-limonene), and C28 (terpinolene) are metabolites from geranyl diphosphate; C46 (β-elemene), C50 (β-caryophyllene), C52 (S-β-bisabolene), C54 (α-humulene), and C55 (α-patchoulene) are metabolites from (2E,6E)-farnesyl diphosphate (https://www.plantcyc.org/). These 64 compounds are here collectively employed to represent the sample chemotypes, reflecting multiple pathways in the secondary metabolism of this plant.
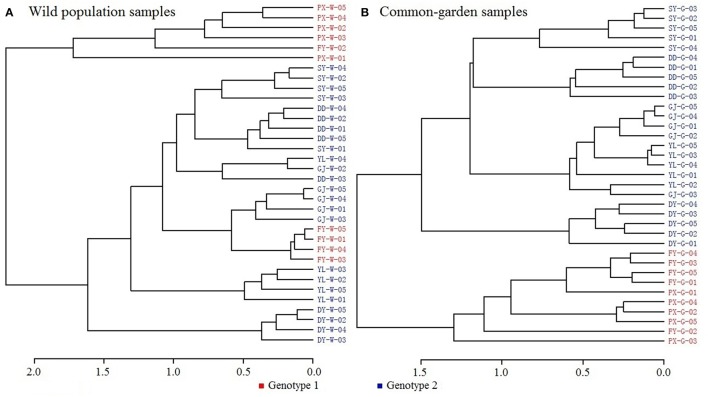
Clustering dendrograms from the volatile constituents (Figure 2A) showed that amongst the wild seven populations, PX was outstandingly different from the rest six populations in terms of chemotypes. This apparently differed from the above genotyping results, where PX and FY clustered but differentiated from the other five populations. This indicates that environmental factors play important roles in establishing chemotypic characteristics. Such notion was further supported by the results that the chemotypes of common garden samples for PX and FY had greater similarities (with closer clustering) than the other five populations (Figure 2B) as shown from the genotypic analysis. This strongly suggests that both genotypes and environmental factors affect the chemotypic characteristics of plants.
Figure 2.
Dendrograms of volatile constituent analysis. (A) Wild population samples, (B) Common-garden samples.
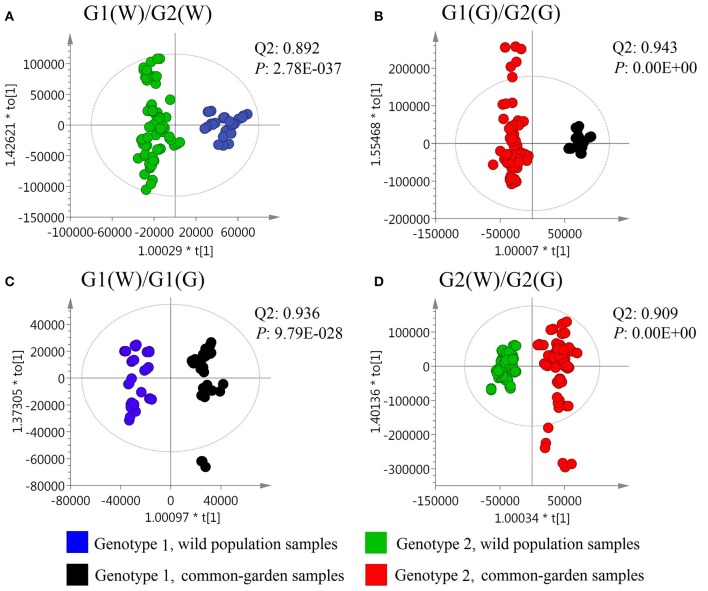
To obtain further details for such chemotypic differences, OPLS-DA of volatile compound composition (chemotypes) was performed for wild and common garden samples in genotype 1 (G1) and 2 (G2) populations. The results showed that G1 and G2 had outstanding chemotypic differences for both wild and common garden samples with Q2 greater than 0.89 whilst p-values smaller than 2.8e–37 in both cases (Figures 3A,B). Furthermore, both G1 and G2 showed significant chemotypic differences between the wild and common garden samples with Q2 greater than 0.9 whilst p-values smaller than 9.8e-28 in both cases (Figures 3C,D).
Figure 3.
OPLS-DA results for the volatile compound composition.
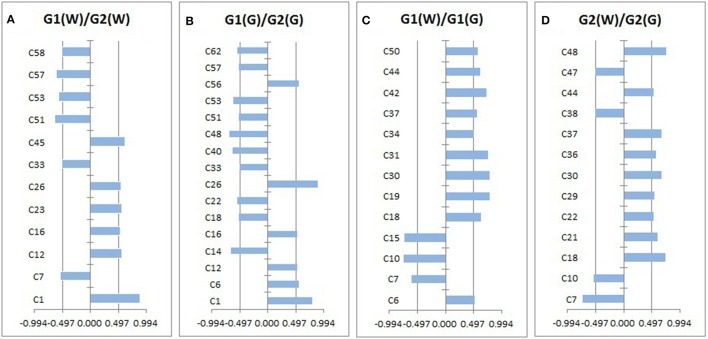
Loadings plots from OPLS-DA (Figure 4, Table 4) showed all volatile compounds having significant inter-group differences between two genotype populations from wild and common garden samples respectively as well as between wild and common garden samples for G1 and G2, respectively. It is worth-noting that common chemotypic features can also be observed in both cases. For both wild and common garden samples, intergroup chemotypic differences are highlighted by significant higher levels of C1 (unknown), C12 (2,4(10)-thujadiene), C16 (1,3,5-trimethylencycloheptan), and C26 (2-acetylpyrrole) but lower levels of C33 (2-ethoxyethyl 3-methylbutanoate), C51 (α-guaiene), C53 (eudesma-3,7(11)-diene), and C57 (2,6,10,10-tetramethylbicyclo[7.2.0]undeca-2,6-diene) in G2 than in G1 (Figures 4A,B). Therefore, these 8 compounds were employable to discriminate G1 and G2. This suggests that two genotypic populations, G1 and G2, have significant difference in their metabolisms with C12, C51, C53 and C57 derived probably from terpenoid pathway. Furthermore, for both G1 and G2, intergroup chemotypic differences are also highlighted by significant higher levels of C18 (isobutyl isovalerate), C30 (3-oxobutan-2-yl 2-methylbutanoate), C37 (unknown), and C44 (pentanoyl pentanoate) but lower levels of C7 (3-methylbut-2-enoic acid) and C10 (3-methylvaleric acid) in common garden samples than in wild samples (Figures 4C,D). These 6 compounds could be used to distinguish the wild populations and common-garden samples (Table 4). This indicates that for both G1 and G2 environmental changes (by transplanting from wild to common gardens) induce significantly in their metabolisms of fatty acids and branch-chain amino acids since these inter-group differential organic acids and esters are mostly β-oxidation metabolites of fatty acids and keto-acids from oxidation of amino acids (Schwab et al., 2008). Moreover, C50 ((–)-β-caryophyllene) has significantly higher levels in common garden samples of G1 than in wild samples of same genotype, indicating that transplantation (or environmental factor) has apparent effects on its (2E,6E)-farnesyl diphosphate metabolism. β-caryophyllene can selectively bind to the cannabinoid receptor type 1 (CB1) and (CB2) thus can be used in the treatment of inflammation, pain, osteoporosis, and atheroscelerosis (Gertsch et al., 2008). It is, therefore, expected that the above effects of genotype, transplantation, or environmental changes on the levels of these compounds may have profound influences on the therapeutic efficiency of this particular herb. Hence planting bases is ought to be selected appropriately for efficacy and quality control purposes.
Figure 4.
The volatile compounds with absolute values of correlation coefficients larger than 0.497 (i.e., significant intergroup differences).
Table 4.
Ranking of the volatile compounds with VIP values larger than 1.00, absolute values of correlation coefficients larger than 0.497, and P-values less than 0.05.
| No. | G1(W)/G2(W) | G1(G)/G2(G) | G1(W)/G1(G) | G2(W)/G2(G) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P-value | CCΔ | VIP | P-value | CC | VIP | P-value | CC | VIP | P-value | CC | VIP | |
| 1 | 0.00E+00 | 0.885 | 0.363 | 0.00E+00 | 0.787 | 0.329 | 7.75E-04 | 0.286 | 0.204 | – | – | – |
| 2 | – | – | – | 2.08E-02 | −0.216 | 0.069 | – | – | – | 4.62E-04 | 0.282 | 0.101 |
| 3 | 3.26E-05 | 0.436 | 0.325 | 8.98E-01 | −0.078 | 0.040 | 4.75E-03 | −0.332 | 0.349 | 7.71E-03 | 0.262 | 0.167 |
| 4 | – | – | – | 6.21E-01 | −0.025 | 0.015 | 1.39E-04 | 0.396 | 0.344 | 2.36E-07 | 0.382 | 0.241 |
| 5 | 2.34E-02 | 0.241 | 0.157 | 1.13E-03 | 0.360 | 0.191 | 5.51E-05 | 0.469 | 0.453 | 2.30E-08 | 0.312 | 0.200 |
| 6 | – | – | – | 7.56E-09 | 0.548 | 0.132 | 1.39E-04 | 0.507 | 0.248 | – | – | – |
| 7 | 1.32E-07 | −0.534 | 1.451 | 2.52E-08 | −0.398 | 0.654 | 1.14E-12 | −0.601 | 2.004 | 0.00E+00 | −0.750 | 1.972 |
| 8 | – | – | – | 8.72E-03 | −0.228 | 0.125 | – | – | – | 7.60E-05 | 0.260 | 0.160 |
| 9 | – | – | – | 1.84E-04 | −0.343 | 0.397 | – | – | – | 2.99E-08 | 0.439 | 0.572 |
| 10 | 4.44E-01 | −0.191 | 1.054 | 2.08E-02 | −0.180 | 0.450 | 5.47E-11 | −0.738 | 4.076 | 3.07E-11 | −0.533 | 2.742 |
| 11 | 1.73E-02 | −0.241 | 0.417 | 1.84E-04 | −0.200 | 0.712 | – | – | – | 2.53E-03 | 0.232 | 0.917 |
| 12 | 1.76E-08 | 0.565 | 0.667 | 7.56E-09 | 0.529 | 0.636 | 5.59E-01 | 0.140 | 0.352 | – | – | – |
| 13 | – | – | – | 2.38E-03 | 0.364 | 0.295 | 1.39E-04 | 0.460 | 0.709 | 4.62E-04 | 0.258 | 0.181 |
| 14 | 7.09E-07 | −0.386 | 0.346 | 6.66E-16 | −0.656 | 0.570 | – | – | – | 4.99E-11 | 0.487 | 0.493 |
| 15 | 1.58E-01 | −0.044 | 0.045 | – | – | – | 5.47E-11 | −0.722 | 0.755 | 0.00E+00 | −0.393 | 0.357 |
| 16 | 1.76E-08 | 0.524 | 0.307 | 1.31E-07 | 0.521 | 0.306 | 2.87E-04 | 0.373 | 0.435 | 2.36E-07 | 0.398 | 0.202 |
| 17 | 6.01E-01 | 0.250 | 0.229 | – | – | – | 1.39E-04 | −0.454 | 0.598 | 2.81E-12 | −0.406 | 0.277 |
| 18 | – | – | – | 9.13E-09 | −0.519 | 0.460 | 1.85E-09 | 0.631 | 0.580 | 0.00E+00 | 0.737 | 0.802 |
| 19 | 1.73E-02 | −0.221 | 0.199 | 5.75E-03 | 0.298 | 0.229 | 1.14E-12 | 0.770 | 1.100 | 1.29E-08 | 0.360 | 0.342 |
| 20 | – | – | – | 1.84E-04 | −0.327 | 0.183 | – | – | – | 2.99E-08 | 0.411 | 0.258 |
| 21 | 1.02E-01 | 0.162 | 0.117 | 2.02E-05 | −0.385 | 0.286 | 3.44E-01 | 0.127 | 0.132 | 9.53E-14 | 0.593 | 0.535 |
| 22 | 2.79E-06 | −0.381 | 0.289 | 6.68E-14 | −0.540 | 0.549 | – | – | – | 2.01E-12 | 0.525 | 0.606 |
| 23 | 1.76E-08 | 0.553 | 0.286 | – | – | – | 1.39E-04 | −0.474 | 0.391 | – | – | – |
| 24 | 1.62E-01 | −0.161 | 0.099 | 2.28E-01 | −0.035 | 0.023 | 3.33E-02 | 0.313 | 0.370 | 5.19E-04 | 0.351 | 0.247 |
| 25 | – | – | – | 2.08E-02 | −0.208 | 0.107 | – | – | – | 4.62E-04 | 0.272 | 0.156 |
| 26 | 1.76E-08 | 0.544 | 0.477 | 0.00E+00 | 0.887 | 0.740 | 1.10E-04 | 0.472 | 0.767 | – | – | – |
| 27 | 1.76E-08 | 0.485 | 0.241 | – | – | – | 1.39E-04 | −0.380 | 0.303 | – | – | – |
| 28 | – | – | – | 2.08E-02 | −0.209 | 0.101 | – | – | – | 4.62E-04 | 0.287 | 0.155 |
| 29 | 2.35E-09 | −0.360 | 0.579 | 1.96E-09 | −0.436 | 1.729 | 5.47E-11 | 0.451 | 1.058 | 0.00E+00 | 0.543 | 2.469 |
| 30 | 1.67E-07 | −0.402 | 0.235 | 2.36E-05 | −0.436 | 0.265 | 1.14E-12 | 0.776 | 0.735 | 0.00E+00 | 0.668 | 0.522 |
| 31 | 4.23E-02 | −0.197 | 0.487 | 6.52E-01 | −0.047 | 0.117 | 0.00E+00 | 0.753 | 2.719 | 2.55E-07 | 0.358 | 1.072 |
| 32 | 1.73E-02 | −0.257 | 0.987 | – | – | – | – | – | – | 2.16E-04 | −0.323 | 1.083 |
| 33 | 1.25E-08 | −0.507 | 1.126 | 1.10E-07 | −0.503 | 0.912 | – | – | – | 6.34E-01 | 0.086 | 0.188 |
| 34 | 1.62E-04 | −0.342 | 0.366 | 1.25E-02 | 0.171 | 0.116 | 1.39E-04 | 0.500 | 0.570 | 1.57E-03 | −0.284 | 0.278 |
| 35 | – | – | – | 2.08E-02 | −0.226 | 0.204 | – | – | – | 4.62E-04 | 0.272 | 0.274 |
| 36 | – | – | – | 1.02E-09 | −0.437 | 0.640 | – | – | – | 0.00E+00 | 0.564 | 0.943 |
| 37 | – | – | – | 6.46E-02 | −0.180 | 0.223 | 1.85E-09 | 0.555 | 1.105 | 0.00E+00 | 0.670 | 0.973 |
| 38 | 2.38E-02 | −0.260 | 0.290 | – | – | – | 1.39E-04 | −0.487 | 0.456 | 1.93E-13 | −0.507 | 0.508 |
| 39 | 8.35E-01 | 0.079 | 0.061 | 4.16E-03 | −0.306 | 0.187 | 8.91E-02 | 0.201 | 0.198 | 4.35E-07 | 0.413 | 0.323 |
| 40 | 1.32E-07 | −0.417 | 1.348 | 6.51E-14 | −0.625 | 2.029 | 9.88E-01 | −0.070 | 0.187 | 1.78E-08 | 0.452 | 1.668 |
| 41 | – | – | – | 2.08E-02 | −0.198 | 0.109 | – | – | – | 4.62E-04 | 0.270 | 0.165 |
| 42 | 1.73E-02 | −0.241 | 0.214 | 6.28E-01 | 0.051 | 0.050 | 1.85E-09 | 0.718 | 1.116 | 2.80E-06 | 0.384 | 0.434 |
| 43 | – | – | – | 8.72E-03 | −0.213 | 0.113 | – | – | – | 7.60E-05 | 0.309 | 0.182 |
| 44 | – | – | – | 1.61E-03 | 0.343 | 0.741 | 5.47E-11 | 0.608 | 2.537 | 2.76E-13 | 0.523 | 1.089 |
| 45 | 7.79E-09 | 0.614 | 0.873 | 3.24E-04 | 0.266 | 0.389 | 1.60E-01 | 0.194 | 0.358 | 7.39E-01 | 0.176 | 0.289 |
| 46 | 3.65E-06 | −0.430 | 1.088 | 7.79E-11 | −0.477 | 1.444 | 1.87E-02 | 0.319 | 0.731 | 1.97E-08 | 0.425 | 1.461 |
| 47 | 3.48E-05 | −0.375 | 0.471 | – | – | – | – | – | – | 4.41E-10 | −0.497 | 0.552 |
| 48 | – | – | – | 6.68E-14 | −0.675 | 0.833 | – | – | – | 0.00E+00 | 0.746 | 1.087 |
| 49 | – | – | – | 1.96E-01 | −0.130 | 0.265 | 1.39E-04 | 0.412 | 0.787 | 7.60E-05 | 0.307 | 0.697 |
| 50 | 7.72E-02 | 0.060 | 0.238 | 2.66E-04 | 0.201 | 0.788 | 4.62E-04 | 0.570 | 3.600 | 9.29E-04 | 0.262 | 1.182 |
| 51 | 1.70E-11 | −0.637 | 2.253 | 4.65E-13 | −0.513 | 2.076 | 3.70E-02 | 0.217 | 0.652 | 3.33E-04 | 0.347 | 1.542 |
| 52 | 1.90E-01 | −0.141 | 0.420 | 1.39E-04 | 0.403 | 1.059 | 7.60E-05 | 0.305 | 1.013 | |||
| 53 | 1.84E-08 | −0.566 | 2.993 | 3.03E-14 | −0.611 | 3.459 | 2.98E-02 | −0.339 | 1.584 | 5.74E-04 | 0.341 | 2.091 |
| 54 | 1.38E-02 | −0.308 | 0.753 | 4.13E-10 | −0.448 | 1.296 | 1.56E-01 | 0.253 | 0.581 | 2.92E-07 | 0.416 | 1.382 |
| 55 | 3.06E-05 | −0.430 | 1.283 | 1.76E-11 | −0.429 | 1.794 | 7.13E-02 | −0.243 | 0.764 | 4.04E-05 | 0.354 | 1.635 |
| 56 | – | – | – | 7.56E-09 | 0.547 | 0.327 | 1.39E-04 | 0.448 | 0.546 | – | – | – |
| 57 | 1.32E-10 | −0.608 | 1.911 | 2.48E-14 | −0.514 | 2.007 | 5.37E-01 | −0.175 | 0.506 | 4.11E-05 | 0.357 | 1.519 |
| 58 | 8.39E-08 | −0.502 | 2.764 | 8.67E-13 | −0.492 | 3.382 | 8.53E-01 | 0.066 | 0.333 | 8.50E-07 | 0.403 | 3.082 |
| 59 | 1.02E-03 | −0.358 | 0.828 | 4.09E-09 | −0.354 | 1.282 | 3.08E-01 | −0.127 | 0.319 | 5.03E-05 | 0.334 | 1.348 |
| 60 | 4.23E-02 | −0.187 | 0.201 | 5.17E-04 | −0.215 | 0.661 | – | – | – | 1.45E-03 | 0.286 | 0.980 |
| 61 | – | – | – | 2.08E-02 | −0.185 | 0.156 | – | – | – | 4.62E-04 | 0.237 | 0.223 |
| 62 | 1.11E-04 | −0.384 | 0.825 | 4.26E-12 | −0.542 | 1.211 | – | – | – | 8.86E-07 | 0.397 | 1.023 |
| 63 | – | – | – | 8.72E-03 | – | 0.233 | – | – | – | 7.60E-05 | 0.311 | 0.354 |
| 64 | – | – | – | 2.08E-02 | −0.165 | 0.189 | – | – | – | 4.62E-04 | 0.255 | 0.323 |
CCΔ, Correlation coefficients; –, No detected in both of two groups; P-value = 0.00E + 00: No detected in one of two groups.
Taking all these together, it is apparent that the chemotypic nature of volatile components in this plant depends on both genotypes and environmental factors with the present of two major genotypic populations. These will be expected to be vital information for understanding physiology of V. jatamansi and for potentially effective commercial applications.
Conclusion
Two genotypicaly different groups were found for V. jatamansi based on genetic analysis and the volatile chemotypes of such groups were not only dependent upon their genotypic features but also environmental factors. When transplanted in common gardens (having similar environment), the two genotypic groups had their own unique composition of volatile constituents. Such genotypic and environmental factors appear to affect the plant secondary metabolites in terpenoid pathway and oxidation metabolism of fatty acids and amino acids.
Common chemotypic differences between two genotypic populations were identifiable for both wild and common garden samples. Eight volatile compounds were discriminatory compounds to differentiate two genotypes in both wild and/or common garden samples. Common chemotypic differences between wild and common garden samples were also observed for both genotypic groups. Six volatile compounds were selectable as discriminatory compounds to differentiate wild and common garden samples for both genotypes. These findings offered better understanding of the correlation of genotype and chemotype for V. jatamansi as well as its genetic and chemical variations in different regions and indicated that both genotyping and chemotyping may need to be considered in breeding new genotypic plants even though the current results themselves may offer little direct guidance at this stage. Nevertheless, selection of good germplasms and proper planting locations of V. jatamansi still need more research come with pharmacodynamic evaluation. It is conceivable that whole metabolomics phenotypes (apart from volatile chemotype) will be of importance for both plant physiology and potential commercial applications.
Author contributions
HX conceived the original project and research plans. XH and ZY collected the samples. XH, SW, and ZS performed experiments. SW, JS, and ZL analyzed the data. HX, SW, and HT wrote paper with all authors participated. All authors agree to be accountable for the content of the work.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Long Xia and Mengxia Zhu for technical assistant of GC/MS.
Footnotes
Funding. The study was supported financially by the National Natural Science Foundation of China (31300281); the Science and Technology entered the Yunnan Special Project of Yunnan Province Key Research and Development Plan (2018IB009); the Science and Technology Research Project of Heilongjiang Province Department of Education (12531781).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01003/full#supplementary-material
Map of the population location. Green marks show wild population, red mark show common-garden.
Volatile compounds identified by automatic retrieval of the mass spectra (NIST 14 and NIST 14s), assisted by retention index.
Volatile compounds in the populations.
References
- Bhatt I. D., Dauthal P., Rawat S., Gaira K. S., Jugran A., Rawal R. S., et al. (2012). Characterization of essential oil composition, phenolic content, and antioxidant properties in wild and planted individuals of Valeriana jatamansi Jones. Sci. Hortic. 136, 61–68. 10.1016/j.scienta.2011.12.032 [DOI] [Google Scholar]
- Chappell J. (1995). Biochemistry and molecular biology of the isoprenoid biosynthetic pathway in plants. Ann. Rev. Plant Physiol. Plant Mol. Biol. 46, 521–547 10.1146/annurev.pp.46.060195.002513 [DOI] [Google Scholar]
- Dai H., Xiao C. N., Liu H. B., Hao F. H., Tang H. R. (2010a). Combined NMR and LC-DAD-MS analysis reveals comprehensive metabonomic variations for three phenotypic cultivars of Salvia miltiorrhiza bunge. J. Proteome Res. 9, 1565–1578. 10.1021/pr901045c [DOI] [PubMed] [Google Scholar]
- Dai H., Xiao C. N., Liu H. B., Tang H. R. (2010b). Combined NMR and LC-MS analysis reveals the metabonomic changes in Salvia miltiorrhiza bunge induced by water depletion. J. Proteome Res. 9, 1460–1475. 10.1021/pr900995m [DOI] [PubMed] [Google Scholar]
- Dong F. W., Yang L., Wu Z. K., Wei G., Zi C. T., Yang D., et al. (2015). Iridoids and sesquiterpenoids from the roots of Valeriana jatamansi Jones. Fitoterapia 102, 27–34. 10.1016/j.fitote.2015.01.021 [DOI] [PubMed] [Google Scholar]
- Gertsch J., Leonti M., Raduner S., Racz I., Chen J. Z., Xie X. Q., et al. (2008). Beta-caryophyllene is a dietary cannabinoid. Proc. Natl. Acad. Sci. U.S.A. 105, 9099–9104. 10.1073/pnas.0803601105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugran A. K., Bhatt I. D., Rawal R. S., Nandi S. K., Pande V. (2013). Patterns of morphological and genetic diversity of Valeriana jatamansi Jones in different habitats and altitudinal range of West Himalaya, India. Flora 208, 13–21. 10.1016/j.flora.2012.12.003 [DOI] [Google Scholar]
- Li Y. D., Wu Z. Y., Li H. M., Li H. Z., Li R. T. (2013). Iridoids from the roots of Valeriana jatamansi. Helv. Chim. Acta 96, 424–430. 10.1002/hlca.201100465 [DOI] [Google Scholar]
- Ming D. S., Yu D. Q., Yang Y. Y., He C. H. (1997). The structures of three novel sesquiterpenoids from Valeriana jatamansi Jones. Tetrahedron Lett. 38, 5205–5208. 10.1016/S0040-4039(97)01112-X [DOI] [Google Scholar]
- Molina-Calle M., Priego-Capote F., Luque de Castro M. D. (2017). Headspace-GC-MS volatile profile of black garlic vs fresh garlic: evolution along fermentation and behavior under heating. LWT Food Sci. Technol. 80, 98–105. 10.1016/j.lwt.2017.02.010 [DOI] [Google Scholar]
- Rana J. C., Sharma B. D. (2000). Naturally occurring spices and aromatic plants of North-West Indian Himalayas and their domestication potential, in Spices and Aromatic Plants: Challenges and Opportunities in the New Century. Contributory Papers. Centennial Conference on Spices and Aromatic Plants (Calicut: Indian Society for Spices; ). [Google Scholar]
- Sangwan N. S., Farooqi A. H. A., Shabih F., Sangwan R. S. (2001). Regulation of essential oil production in plants. Plant Growth Regul. 34, 3–21. 10.1023/A:1013386921596 [DOI] [Google Scholar]
- Schwab W., Davidovich-Rikanati R., Efraim Lewinsohn E. (2008). Biosynthesis of plant-derived flavor compounds. Plant J. 54, 712–732. 10.1111/j.1365-313X.2008.03446.x [DOI] [PubMed] [Google Scholar]
- Verma R. S., Verma R. K., Padalia R. C., Chauhan A., Singh A., Singh H. P. (2011). Chemical diversity in the essential oil of Indian valerian (Valeriana jatamansi Jones). Chem. Biodivers. 8, 1921–1929. 10.1002/cbdv.201100059 [DOI] [PubMed] [Google Scholar]
- Wang C. G., Zheng Z. Q., Deng X. Q., Ma X. J., Wang S. N., Liu J., et al. (2017). Flexible and powerful strategy for qualitative and quantitative analysis of valepotriates in Valeriana jatamansi Jones using high-performance liquid chromatography with linear ion trap Orbitrap mass spectrometry. J. Sep. Sci. 40, 1906–1919. 10.1002/jssc.201601406 [DOI] [PubMed] [Google Scholar]
- Wang S., Meyer E., McKay J. K., Matz M. V. (2012). 2b-RAD: a simple and flexible method for genome-wide genotyping. Nat. Methods 9, 808–810. 10.1038/nmeth.2023 [DOI] [PubMed] [Google Scholar]
- Wang Y. L., Tang H. R., Nicholson J. K., Hylands P. J., Sampson J., Whitcombe I., et al. (2004). Metabolomic strategy for the classification and quality control of phytomedicine: a case study of chamomile flower (Matricaria recutita L.). Planta Med. 70, 250–255. 10.1055/s-2004-815543 [DOI] [PubMed] [Google Scholar]
- Xiao C. N., Dai H., Liu H. B., Wang Y. L., Tang H. R. (2008). Revealing the metabonomic variation of Rosemary extracts using H-1 NMR Spectroscopy and Multivariate Data Analysis. J. Agric. Food Chem. 56, 10142–10153. 10.1021/jf8016833 [DOI] [PubMed] [Google Scholar]
- Xu J., Zhao P., Guo Y. Q., Xie C. F., Jin D. Q., Ma Y. G., et al. (2011). Iridoids from the roots of Valeriana jatamansi and their neuroprotective effects. Fitoterapia 82, 1133–1136. 10.1016/j.fitote.2011.07.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Map of the population location. Green marks show wild population, red mark show common-garden.
Volatile compounds identified by automatic retrieval of the mass spectra (NIST 14 and NIST 14s), assisted by retention index.
Volatile compounds in the populations.