Abstract
Mineralocorticoids trigger a profibrotic process in the kidney. In mouse cortical collecting duct cells, the present study addressed two main questions: 1) what are microRNAs (miRNAs) and their target genes that are changed by aldosterone? and 2) what do miRNAs, in response to aldosterone, regulate regarding signaling pathways related to fibrosis? A microarray chip assay was done in cells in the absence or presence of aldosterone treatment (10−6 M; 3 days). The candidate miRNAs were identified by the criteria of >30% of fold change among the significantly changed miRNAs (P < 0.05). Twenty-nine miRNAs were upregulated (>1.3-fold), and 27 miRNAs were downregulated (<0.7-fold). Putative target genes of identified miRNAs were associated with 74 Kyoto Encyclopedia of Genes and Genomes pathways. Among them, the wingless-related integration site (Wnt) signaling pathway was highly ranked, where 15 mature miRNAs were observed. These miRNAs were further analyzed by real-time quantitative PCR, and among them, miR-130b-3p, miR-34c-5p, and miR-146a-5p were selected. Through the identification of putative target genes of these three miRNAs, mRNA and protein expression of the Ca2+/calmodulin-dependent protein kinase type II β-chain (Camk2b) gene (a target gene of miR-34c-5p) were found to be increased significantly in aldosterone-treated cells, where fibronectin (FN) and α-smooth muscle actin were induced. When CaMKIIβ small interfering RNA or the miR-34c-5p mimic was transfected, aldosterone-induced FN expression was significantly attenuated, along with reduced CaMKIIβ protein expression. A luciferase reporter assay revealed a decrease of CaMKIIβ translation in cells transfected with miRNA mimics of miR-34c-5p. In conclusion, aldosterone-induced downregulation of miR-34c-5p in the Wnt signaling pathway and a consequent increase of CaMKIIβ expression are likely to be involved in aldosterone-induced fibrosis.
Keywords: aldosterone, CaMKII, microRNA, Wnt signaling
INTRODUCTION
The mineralocorticoid hormone, aldosterone, plays a role in the homeostasis of sodium, potassium, and extracellular fluid volume in the body (20, 25, 44). In addition to its critical role in body fluid homeostasis, aldosterone is associated with inflammatory injury and fibrosis in the kidney, in which renal hypertrophy, tubulointerstitial fibrosis, and glomerulosclerosis are observed (5, 17, 22, 48). The underlying mechanisms have been demonstrated to be associated with production of growth factors and inhibited degradation of ECM (23). Furthermore, aldosterone is known to induce mitochondrial dysfunction through oxidative stress in the renal tubular epithelial cells (61). Consistently, the blockade of the mineralocorticoid receptor (MR) exerted protective effects against renal damage, including podocyte injury, albuminuria, and tubulointerstitial lesions (20, 37, 40, 43).
MicroRNA (miRNA), 20–22 nt in length, is a short, noncoding RNA, which interacts with the 3′-untranslated region (UTR) of mRNAs. miRNAs regulate target mRNAs through an inhibition of translation of target mRNAs or degradation of target mRNAs, which leads to the regulation of protein expression (30). Thus modulation of protein expression via miRNAs in the kidney tubular epithelial cells could affect renal functions. For instance, we have recently identified the miRNAs regulating aquaporin-2 (AQP2) protein expression in the kidney collecting ducts (25, 27). A significant decrease of AQP2 protein expression was observed when mouse cortical collecting duct cells (mpkCCDc14 cells) were transfected with miRNA mimics of miR-32 or miR-137 (27). Moreover, miRNAs were demonstrated to be involved in aldosterone-mediated sodium and potassium transport (14, 15). Additional studies have further revealed that miRNAs are involved in various pathophysiological conditions, e.g., kidney development, renal cell carcinoma, acute kidney injury, chronic kidney disease, and diabetic nephropathy. For instance, downregulation of miRNA-26a in podocytes exacerbated diabetic nephropathy through enhancing the transforming growth factor-β/connective tissue growth factor (TGF-β/CTGF) signaling pathway (28). miR-146a-deficient mice exhibited an early development of glomerulopathy and albuminuria in streptozotocin-induced diabetes mellitus (33), and miR-21-deficient mice were also associated with glomerulopathy in a diabetic condition (32). Several other miRNAs were also reported to inhibit or promote renal cell carcinoma through regulation of cell migration and metastasis (35, 45).
Based on these previous findings, it is hypothesized that altered expression of miRNAs could be involved in a number of pathophysiological conditions in the kidney. It has been demonstrated that mineralocorticoids trigger a profibrotic process in the cells expressing the MR (5). Recent studies demonstrated that renal tubular cells, after injury, could promote fibroblast activation and renal fibrosis (26, 59, 63). Importantly, a recent study for evaluation of expression of mRNA transcripts, along with rat kidney tubule segments using RNA sequencing, showed that the mRNA transcript of MR (gene symbol: Nr3c2) is expressed abundantly along the connecting tubule and collecting ducts (34). Thus aldosterone-mediated changes in the expression of miRNAs and their target genes in the kidney collecting duct epithelial cells would be the subjects of interest. The present study aimed at the following: to 1) examine whether aldosterone induces the expression of the fibrosis marker proteins in mpkCCDc14 cells; 2) profile the aldosterone-regulated miRNAs in mpkCCDc14 cells; 3) identify the signaling pathways where putative target genes of the identified miRNAs are enriched; and 4) examine the roles of identified miRNAs and putative target genes in aldosterone-mediated fibrosis.
MATERIALS AND METHODS
Cell culture.
mpkCCDc14 cells were maintained in a 1:1 mixture of DMEM and Ham’s F-12 medium, containing 60 nM sodium selenite, 5 μg/ml transferrin, 2 mM L-glutamine, 50 nM dexamethasone, 1 nM triiodothyronine, 10 ng/ml epidermal growth factor, 5 μg/ml insulin, 20 mM HEPES, 20 mM D-glucose, 0.1% penicillin/streptomycin solution, and 2% heat-inactivated FBS at 37°C, as previously described (24, 27). All chemicals were purchased from Sigma-Aldrich (St. Louis, MO). Cells were seeded in 12-well plates and grown to 60––70% confluence in culture medium at 37°C in 5% CO2, 95% air atmosphere, and then in FBS-free culture medium for the treatment of TGF-β (5 or 10 ng/ml; 3 days) or aldosterone (10−6 M; 3 or 5 days). Aldosterone was administered every 2 days. The dose of aldosterone treatment was determined from the previous study, showing that administration of a high concentration of aldosterone to the human mesangial cell line attributed to cell apoptosis, accompanied by generation of reactive oxygen species (38).
Animal model of aldosterone infusion.
The animal protocols were approved by the Animal Care and Use Committee of Kyungpook National University (Taegu, Korea). C57BL/6 mice (20–22 g) were obtained from Charles River (Orient Bio, Seongnam, Korea). Mice were allowed ad libitum access to normal diet and water intake. Saline-infused control (n = 7) and aldosterone-infused (n = 7) mice were implanted subcutaneously with osmotic minipumps (model 1002; Durect, Cupertino, CA; saline or 250 μg ⋅ kg−1 ⋅ day−1 sc; 10 days). Aldosterone was dissolved in 100% DMSO (#D2650; Sigma-Aldrich) and then diluted with sterile saline to a final concentration of DMSO of 3% vol/vol. Osmotic minipumps were loaded, according to the manufacturer’s instructions, before implantation subcutaneously on the back of the mice. For the implantation of osmotic minipumps, the mice were anesthetized by isoflurane inhalation. The incision was closed by silk suture, and mice were awakened and returned to normal cages. After 10 days, mice were anesthetized by isoflurane inhalation again, both kidneys were removed, and blood samples were collected from inferior vena cava and rapidly transferred into blood collection tubes containing sodium heparin (BD Vacutainer; Becton Dickinson, Franklin Lakes, NJ). Protein lysates and RNA extracts were prepared from the kidney cortex. Plasma potassium levels were measured by the M420/425 flame photometer (Sherwood Scientific, Cambridge, UK).
Total RNA extraction and microarray analysis.
mpkCCDc14 cells were seeded in six-well plates and treated with aldosterone (10−6 M) on a daily basis for 3 days. Total RNA was purified by the mirVana miRNA Isolation Kit (Ambion; Thermo Fisher Scientific, Waltham, MA), according to the manufacturer’s instruction. Concentrations and purity of total RNA were measured using NanoDrop (Thermo Fisher Scientific). Total RNA (1 μg) was labeled by biotin using the FlashTag Biotin HSR RNA Labeling Kit (Affymetrix; Thermo Fisher Scientific), and miRNA expression was profiled by GeneChip miNRA 4.0 Array (Affymetrix; Thermo Fisher Scientific). Images of the microarray were scanned by the GeneChip Scanner 3000 7G Plus (Affymetrix; Thermo Fisher Scientific), and signal intensity of miRNA expression was analyzed by Expression Console software (version 1.2.1; Affymetrix; Thermo Fisher Scientific).
Computational analysis of signaling pathways and prediction of miRNA target genes.
Prediction of putative target genes of the identified miRNAs was performed using DIANA-mirPath (version 2.0) (54), based on the TargetScan database, using a microT-CDS algorithm (microT > 0.8, and P < 0.05). To identify signaling pathways in which putative target genes of the identified miRNAs were enriched, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were exploited in DIANA-mirPath (http://snf-515788.vm.okeanos.grnet.gr).
Real-time quantitative PCR.
mpkCCDc14 cells were treated with aldosterone (10−6 M; 3 or 5 days) or TGF-β (5 or 10 ng/ml; 3 days), and RNA was prepared by the mirVana miRNA Isolation Kit (Ambion; Thermo Fisher Scientific), according to the manufacturer’s instruction. cDNAs were synthesized using the miScript II RT Kit (Qiagen, Germantown, MD), as per the manufacturer’s protocol. Total RNA (1 μg), isolated from vehicle- or aldosterone-treated cells, was subjected to cDNA synthesis. The relative expression of the identified miRNAs and target genes was determined by real-time quantitative PCR (RT-qPCR), using a miScript SYBR Green PCR Kit (Qiagen) and a QuantiTect SYBR Green PCR Kit (Qiagen), respectively, according to the manufacturer’s instructions. U6 RNA and β-actin mRNA were used as an internal control, and the threshold was set by 0.02 to determine the threshold cycle (Ct) value. The relative miRNA or mRNA expression was calculated by the following formulas: 1) 2−ΔCt, where Ct value presents threshold cycles, and ΔCt = Ct (miRNA) − Ct (U6); 2) 2−ΔCt, where Ct value presents threshold cycles, and ΔCt = Ct (mRNA) − Ct (mRNA of β-actin). RT-qPCR of miRNA and mRNA was carried out using Rotor-Gene-A (Qiagen), and each sample was tested in triplicate. Primers of miRNA were purchased from Qiagen, and primer sequences for mRNA are shown in Table 1.
Table 1.
Primer sequences for real-time quantitative PCR
| Gene | Sense (5′–3′) | Antisense (5′–3′) |
|---|---|---|
| Rock1 | GACTGGGGACAGTTTTGAGAC | GGGCATCCAATCCATCCAGC |
| Skp1a | ATGCCTACGATAAAGTTGCAGAG | TCCATTCCCAAATCTTCCAGC |
| Tbl1xr1 | TCTCATTCTGCGTTTACCTTTGG | GACAGAGACTCGATGGGTC |
| Ppp2r5e | GACGGATTTTCTCGGAAGTCC | GAGGTTGGAACGTCTTTCAGC |
| Plcb1 | GCCCCTGGAGATTCTGGAGT | GGGAGACTTGAGGTTCACCTTT |
| Let1 | TGTTTATCCCATCACGGGTGG | CATGGAAGTGTCGCCTGACAG |
| Camk2b | GCACGTCATTGGCGAGGAT | ACGGGTCTCTTCGGACTGG |
| Prickle1 | ACCTGGAGTATGCTGGCAC | CACAGTGGATTTTTCCATCCTGA |
| Ppp2r5a | ATTGAAGAGCCGCTTTTTAAGCA | TGAGGGTTTTCAGCACATTGT |
| Daam1 | AACTTTGCACTTCAGACAATGGA | CTGGTCCTTTTTCTTGCTACAGT |
| Ppp3r2 | AAATGAGGCCAGCTACCAAAC | CCCGATTTGTCCAAGTCCAG |
| Cxxc4 | CTGCCCGCAGAATCATTCCT | CAGACGCCACAGTTGATGAG |
| Smad4 | ACACCAACAAGTAACGATGCC | GCAAAGGTTTCACTTTCCCCA |
| Nfat5 | CAGCGCCCAATAGTTGGCA | TGCTGGTGAAAAATTGACTGGT |
| Actb | CCTTCTTGGGTATGGAAT | TTGGCATAGAGGTCTTTA |
Semiquantitative immunoblot analysis.
The total cell lysate was obtained in radioimmunoprecipitation assay buffer (10 mM Tris·HCl, 0.15 M NaCl, 1% Nonidet P-40, 1% Na-deoxycholate, 0.5% SDS, 0.02% sodium azide, and 1 mM EDTA, pH 7.4), including proteinase and phosphatase inhibitors (0.4 μg/ml leupeptin, 0.1 mg/ml Pefabloc, 1 mM Na3VO4, 25 mM NaF, and 0.1 μM okadaic acid). Immunoblotting was performed, as previously described (24, 27). Primary antibodies were anti-epithelial sodium channel-α (ENaC-α; 1:1,000, LL766AP) (19), anti-AQP2 (1:5,000, ab3274; MilliporeSigma, Burlington, MA), anti-α-smooth muscle actin (α-SMA; 1:1,000, ab5694; Abcam, Cambridge, MA), anti-fibronectin (anti-FN; 1:5,000, ab2413; Abcam), anti-Ca2+/calmodulin-dependent protein kinase II β (CaMKIIβ; 1:1,000, ab34703; Abcam), and anti-β-actin (1:400,000, A1978; Sigma-Aldrich). Band density was quantitated by ImageJ (NIH, Bethesda, MD), and the densitometry values for each protein were corrected by a densitometry value of β-actin (24, 27).
Transfection of miRNA mimic or CaMKIIβ siRNA.
mpkCCDc14 cells were seeded at 12-well plates. When confluency of the cells reached 50–60% in each well, cells were transfected with Qiagen miScript Inhibitor Negative Control or Qiagen miScript miRNA mimic (Syn-miR-34c-5p), using a Lipofectamine RNAiMAX Reagent (Invitrogen; Thermo Fisher Scientific), as per the manufacturer’s instruction. miRNA mimic (20 nM) was transfected to the cells for 24 h, and aldosterone (10−6 M) was administered for 3 days in serum- and hormone-free medium. Aldosterone was adminmistered every 2 days.
mpkCCDc14 cells were grown in 12-well plates and transfected with small interfering RNA (siRNA), specific for CaMKIIβ (40 nM), or nontarget control siRNA, using the DharmaFECT reagent (Thermo Fisher Scientific). After 24 h of transfection, cells were treated with 10−6 M aldosterone for 3 days in serum- and hormone-free medium. Aldosterone was treated every 2 days.
Luciferase assay.
To examine the direct interaction of miRNA with the target sequence of 3′-UTR of CaMKIIβ mRNA in mpkCCDc14 cells, the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, Madison, WI) was used. The 3′-UTR sequence of mouse CaMKIIβ predicted by DIANA Tools was inserted to the pmirGLO vector using a restriction enzyme (PmeI and XbaI). Dual luciferase assay was carried out, as we previously demonstrated (27). The empty vector or target sequence-inserted vector (0.5 μg) and pRL-SV40 vector (25 ng; Promega) were cotransfected with the 40 nM Syn-miR-34c-5p mimic (Qiagen), using the 0.5 μl Lipofectamine RNAiMAX Reagent to mpkCCDc14 cells seeded on 24-well plates (SPL Life Sciences, Seoul, Korea). After 24 h, luciferase activity was analyzed, according to the manufacturer’s instructions. Luminescence was measured using TriStar2 S LB 942 (Berthold Technologies, Bad Wildbad, Germany).
Statistical analysis.
Quantitative data are shown as means ± SE. Comparisons between two groups (Control vs. Aldosterone) were made by unpaired t-test. Comparisons of multiple groups were made by one-way ANOVA, followed by a post hoc Tukey’s Honestly Significant Difference test. A multiple comparisons test was only applied when a significant difference was determined in ANOVA (P < 0.05). P < 0.05 was considered statistically significant.
RESULTS
Increased expression of fibrosis marker proteins in mpkCCDc14 cells exposed to aldosterone.
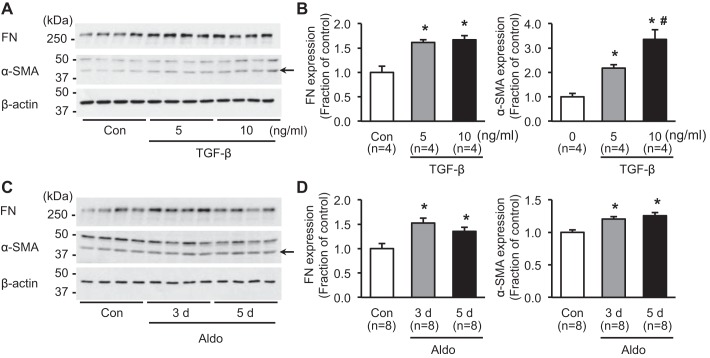
To examine whether aldosterone induces the fibrosis marker proteins in mpkCCDc14 cells, e.g., FN and α-SMA, cells were treated with TGF-β, a key mediator of fibrosis (5 or 10 ng/ml; 3 days) or aldosterone (10−6 M; 3 or 5 days). Semiquantitative immunoblotting demonstrated that protein expression of FN was significantly increased in cells treated with either 5 ng/ml (160 ± 6% of control, P < 0.05) or 10 ng/ml (170 ± 8% of control, P < 0.05; Fig. 1, A and B) of TGF-β. Furthermore, α-SMA protein expression was markedly increased (220 ± 13% and 340 ± 40% of controls, P < 0.05, respectively; Fig. 1, A and B). Importantly, expression of the fibrosis marker proteins was also significantly increased in mpkCCDc14 cells treated with aldosterone (10−6 M) for 3 or 5 days. Immunoblot analysis revealed a significant increase of FN expression (150 ± 10% of control at 3 days; 140 ± 9% of control at 5 days, P < 0.05, respectively) and α-SMA expression (120 ± 4% of control at 3 days; 130 ± 5% of control at 5 days, P < 0.05, respectively; Fig. 1, C and D). The findings indicated that aldosterone per se could induce fibrosis in renal tubular epithelial cells.
Fig. 1.
Expression of fibrosis marker proteins in response to TGF-β or aldosterone (Aldo) treatment in mpkCCDc14 cells. A and B: semiquantitative immunoblotting of fibronectin (FN; ~262 kDa) and α-smooth muscle actin (α-SMA; ~42 kDa; arrow). Densitometric analysis of samples from vehicle-treated control (Con) and TGF-β (5 or 10 ng/ml; 3 days)-treated mpkCCDc14 cells. C and D: semiquantitative immunoblotting of FN (~262 kDa) and α-SMA (~42 kDa; arrow). Densitometric analysis of samples from vehicle-treated control and aldosterone (10−6 M; 3 or 5 days)-treated mpkCCDc14 cells. n, number of cell lysate preparations. *P < 0.05 compared with control group; #P < 0.05 compared with a group of TGF-β treatment (5 ng/ml; 3 days).
Identification of aldosterone-regulated miRNAs in mpkCCDc14 cells.
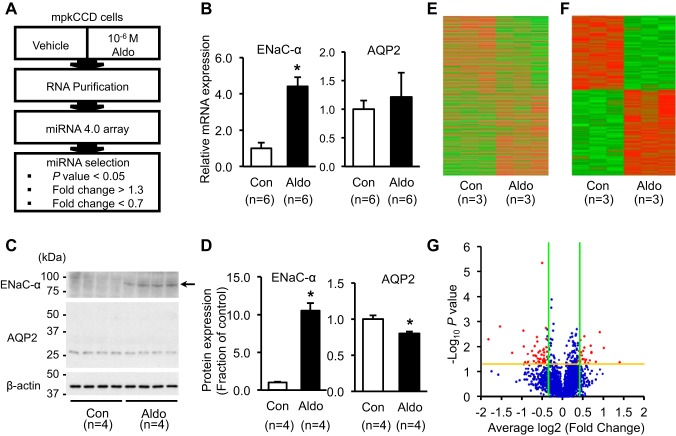
mpkCCDc14 cells were treated with aldosterone (10−6 M) for 3 days (Fig. 2A). The responsiveness of mpkCCDc14 cells to aldosterone treatment was validated by RT-qPCR using a primer specific for the ENaC-α. Consistent with previous studies (9, 44), aldosterone induced a significant increase of the ENaC-α mRNA level (440 ± 51% of vehicle-treated control, P < 0.05), whereas the AQP2 mRNA level was unchanged (Fig. 2B). Consistent with this, protein expression of ENaC-α was significantly increased (1,050 ± 99% of vehicle-treated control, P < 0.05; Fig. 2, C and D), whereas AQP2 protein was decreased after aldosterone treatment (80 ± 2% of vehicle-treated control, P < 0.05; Fig. 2, C and D).
Fig. 2.
Aldosterone (Aldo)-regulated miRNAs and changes of epithelial sodium channel-α subunit (ENaC-α) and aquaporin-2 (AQP2) expression after aldosterone treatment in mpkCCDc14 cells. A: a scheme of the experimental process for examining miRNA expression in mpkCCDc14 cells after aldosterone treatment (10−6 M; 3 days) using the Affymetrix GeneChip miRNA 4.0 array. B: changes of mRNA expression of ENaC-α and AQP2 after aldosterone treatment (10−6 M; 3 days). C and D: changes of protein expression of ENaC-α (~85 kDa; arrow) and AQP2 (deglycosylated; ~29 kDa) after aldosterone treatment (10−6 M; 3 days). In the mpkCCDc14 cells without vasopressin stimulation, basal expression of AQP2 protein is low and variable, and a glycosylated form of AQP2 (between ~35 and 50 kDa) is rarely seen. E and F: heatmap analysis of (E) total miRNAs and (F) selected miRNAs that were significantly changed (P < 0.05) after aldosterone treatment (10−6 M; 3 days). G: volcano plot of fold changes [logarithm to base 2 (log2) value, 1.3-fold; green lines] and probability values (−log10, P = 0.05; yellow line) for miRNAs in the aldosterone-treated group (n = 3) compared with the control group (n = 3). n, number of cell lysate preparation. *P < 0.05 compared with control group.
To identify the miRNAs that were significantly changed in the expression level in mpkCCDc14 cells after aldosterone treatment, a microarray chip assay was performed (Affymetrix GeneChip miRNA 4.0 array containing probes for 1,908 mouse mature miRNAs and 1,255 mouse pre-mature miRNAs). The miRNAs, in response to aldosterone, were identified by the criteria of >30% of fold changes among the significantly changed miRNAs (P < 0.05, n = 3; Fig. 2A). Heatmap analysis revealed the differential expression of either total miRNAs (Fig. 2E) or selected miRNAs that were changed significantly (P < 0.05; Fig. 2F) after aldosterone treatment. The volcano plot also showed the differential expression of total miRNAs, and red dots represented the miRNAs changed >30% of fold changes (Fig. 2G). Fifty-six miRNAs, including mature and pre-mature miRNAs, were identified to be significantly changed after aldosterone treatment. Among them, 29 miRNAs were upregulated, and 27 miRNAs were downregulated (Tables 2 and 3).
Table 2.
Upregulated miRNAs in mpkCCDc14 cells after aldosterone treatment
| Transcript ID | Average Fold Change (Aldo/Con) | P |
|---|---|---|
| mmu-miR-342-5p | 2.641 | 0.041 |
| mmu-miR-676-5p | 2.021 | 0.011 |
| mmu-miR-224-5p | 1.878 | 0.003 |
| mmu-miR-6927-5p | 1.768 | 0.041 |
| mmu-miR-676-3p | 1.635 | 0.018 |
| mmu-miR-181a-2-3p | 1.629 | 0.017 |
| mmu-mir-5099 | 1.608 | 0.009 |
| mmu-miR-6974-5p | 1.590 | 0.035 |
| mmu-miR-130b-3p | 1.556 | 0.044 |
| mmu-miR-181b-1-3p | 1.498 | 0.034 |
| mmu-miR-30b-3p | 1.476 | 0.038 |
| mmu-miR-6373 | 1.464 | 0.046 |
| mmu-miR-378d | 1.449 | 0.004 |
| mmu-mir-200b | 1.449 | 0.011 |
| mmu-miR-331-5p | 1.441 | 0.032 |
| mmu-mir-7037 | 1.422 | 0.015 |
| mmu-miR-1949 | 1.409 | 0.032 |
| mmu-mir-92a-1 | 1.398 | 0.032 |
| mmu-mir-1839 | 1.396 | 0.023 |
| mmu-miR-378b | 1.366 | 0.017 |
| mmu-miR-10b-3p | 1.358 | 0.005 |
| mmu-miR-669h-3p | 1.355 | 0.017 |
| mmu-miR-378a-3p | 1.353 | 0.038 |
| mmu-mir-6387 | 1.351 | 0.041 |
| mmu-miR-7037-5p | 1.344 | 0.022 |
| mmu-miR-7033-5p | 1.335 | 0.050 |
| mmu-miR-378c | 1.327 | 0.015 |
| mmu-mir-501 | 1.325 | 0.025 |
| mmu-miR-6955-5p | 1.310 | 0.036 |
Table 3.
Downregulated miRNAs in mpkCCDc14 cells after aldosterone treatment
| Transcript ID | Average Fold Change (Aldo/Con) | P |
|---|---|---|
| mmu-mir-3107 | 0.697 | 0.021 |
| mmu-mir-7015 | 0.689 | 0.031 |
| mmu-miR-34c-5p | 0.681 | 0.015 |
| mmu-mir-194-2 | 0.678 | 0.047 |
| mmu-miR-7115-3p | 0.677 | 0.041 |
| mmu-miR-7002-5p | 0.675 | 0.032 |
| mmu-miR-7001-3p | 0.665 | 0.017 |
| mmu-mir-6976 | 0.662 | 0.039 |
| mmu-miR-7006-5p | 0.656 | 0.030 |
| mmu-miR-5118 | 0.650 | 0.006 |
| mmu-miR-6956-3p | 0.649 | 0.004 |
| mmu-miR-8112 | 0.639 | 0.024 |
| mmu-miR-6361 | 0.637 | 0.046 |
| mmu-mir-6970 | 0.628 | 0.023 |
| mmu-miR-7088-3p | 0.624 | 0.010 |
| mmu-miR-6416-3p | 0.616 | 0.038 |
| mmu-mir-5131 | 0.610 | 0.022 |
| mmu-miR-6926-3p | 0.594 | 0.015 |
| mmu-miR-1224-3p | 0.557 | 0.040 |
| mmu-miR-99a-5p | 0.553 | 0.020 |
| mmu-miR-7003-5p | 0.546 | 0.048 |
| mmu-miR-1967 | 0.530 | 0.042 |
| mmu-miR-1249-3p | 0.517 | 0.010 |
| mmu-miR-146a-5p | 0.513 | 0.002 |
| mmu-miR-668-3p | 0.423 | 0.018 |
| mmu-miR-3107-3p | 0.345 | 0.002 |
| mmu-miR-6939-5p | 0.284 | 0.005 |
Identification of miRNA target genes and KEGG pathway enrichment.
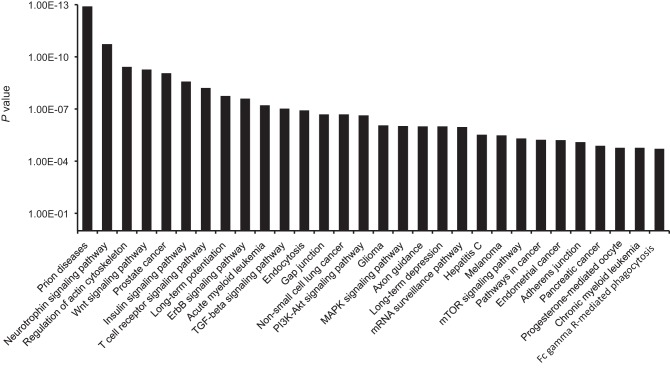
With the use of the DIANA-mirPath tool, we identified KEGG signaling pathways where putative target genes of the identified miRNAs were enriched. Twenty-four out of the identified 56 miRNAs were eligible for the analysis using DIANA-mirPath, based on the microT-CDS algorithm. Seventy-four KEGG pathways were profiled with the criteria of P < 0.05, and microT threshold > 0.8. Thirty top-ranked pathways were demonstrated in Fig. 3. Among them, the wingless-related integration site (Wnt) signaling pathway, represented at the top in the list, was selected for further studies of aldosterone-induced fibrosis, as Wnt/β-catenin signaling has been demonstrated to play a role in the pathogenesis of renal fibrosis (7, 21, 63).
Fig. 3.
Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathways where putative target genes of the identified aldosterone-regulated miRNAs were enriched. KEGG pathways were profiled with the criteria of P < 0.05 and microT threshold > 0.8 using the DIANA-mirPath tool. Thirty top-ranked pathways were demonstrated. Wnt, wingless-related integration site; ErbB, avian erythroblastosis oncogene B; TGF-β, transforming growth factor-β; PI3K-Akt, phosphatidylinositol 3-kinase-PKB; mTOR, mammalian target of rapamycin.
In the Wnt signaling pathway, target genes of miRNAs (seven upregulated, mature miRNAs: miR-342-5p, miR-224-5p, miR-130b-3p, miR-181b-1-3p, miR-30b-3p, miR-1949, and miR-669h-3p; eight downregulated, mature miRNAs: miR-34c-5p, miR-5118, miR-1224-3p, miR-99a-5p, miR-1967, miR-146a-5p, miR-668-3p, and miR-3107-3p) were enriched (Table 4). Thirty-seven putative target genes of these 15 miRNAs were enlisted in Table 4, which were profiled with the criteria of the microT threshold > 0.08.
Table 4.
miRNAs and putative target genes associated with the Wnt signaling pathway
| miRNA | Target Genes | |
|---|---|---|
| Upregulated miRNA | mmu-miR-342-5p | Mapk9, Ctbp2 |
| mmu-miR-224-5p | Ppp2r1b, Psen1, Tcf7l2, Btrc, Wnt9a, Gpc4, Mapk8 | |
| mmu-miR-130b-3p | Rock1, skp1a, Tbl1xr1, Ppp2re, Wnt2b, Plcb1 | |
| mmu-miR-181b-1-3p | Rock1, Csnk1a1, Nfat5, Nratc4 | |
| mmu-miR-30b-3p | Ctbp2 | |
| mmu-miR-1949 | Wif1 | |
| mmu-miR-669h-3p | Rock1, Axin2, Fzd10, Ppp3cb, Plcb1 | |
| Downregulated miRNA | mmu-miR-34c-5p | Lef1, Camk2b, Tbl1xr1, Prickle1, Ppp2r5a, Daam1 |
| mmu-miR-5118 | Camk2g | |
| mmu-miR-1224-3p | Csnk2a1 | |
| mmu-miR-99a-5p | Fzd8 | |
| mmu-miR-1967 | Nfat5 | |
| mmu-miR-146a-5p | Ppp3r2, Cxxc4, Smad4, Nfat5 | |
| mmu-miR-669-3p | Ppp3r2, Camk2g, lrp5, Nfat5 | |
| mmu-miR-3107-3p | Csnk2b |
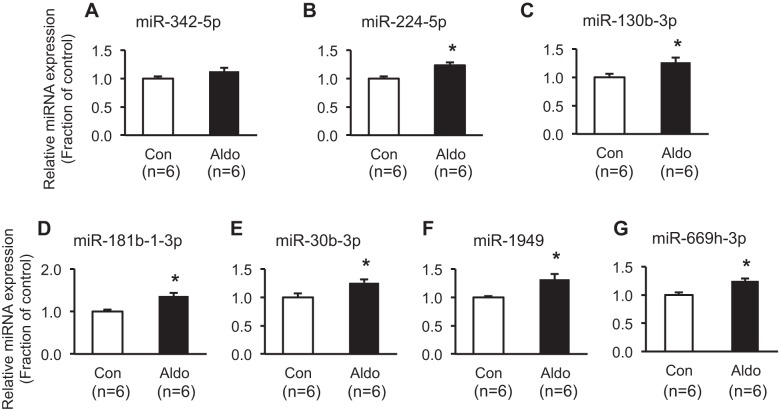
Quantitative analysis of miRNAs enriched in the Wnt signaling pathway.
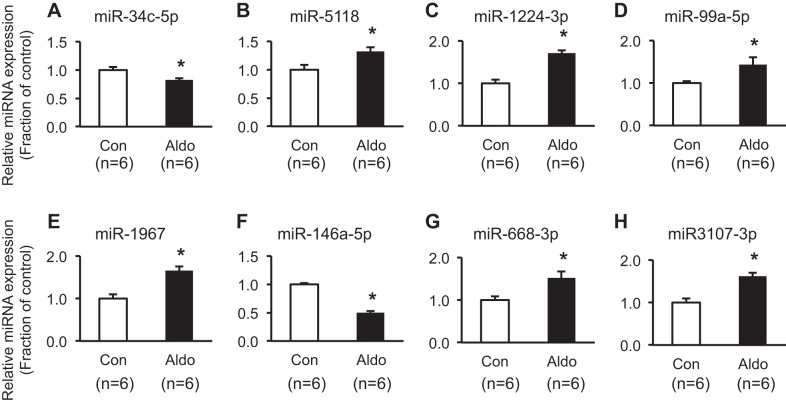
The expression of seven upregulated and eight downregulated mature miRNAs, among 15 miRNAs that were profiled in the Wnt signaling pathway, was further analyzed by RT-qPCR. Six out of seven upregulated miRNAs were found to be significantly increased (miR-224-5p, miR-130b-3p, miR181b-1-3p, miR-30b-3p, miR-1949, and miR-669h-3p; Fig. 4). Moreover, two out of eight downregulated miRNAs were further demonstrated to be markedly decreased (miR-34c-5p and miR-146a-5p; Fig. 5).
Fig. 4.
Real-time quantitative PCR analysis of upregulated miRNAs enriched in the Wnt signaling pathway. Open bars, vehicle-treated mpkCCDc14 cells [control (Con)]; closed bars, aldosterone (Aldo; 10−6 M; 3 days)-treated mpkCCDc14 cells. *P < 0.05 when compared with control. n, number of RNA extract preparations.
Fig. 5.
Real-time quantitative PCR analysis of downregulated miRNAs enriched in the Wnt signaling pathway. Open bars, vehicle-treated mpkCCDc14 cells [control (Con)]; closed bars, aldosterone (Aldo; 10−6 M; 3 days)-treated mpkCCDc14 cells. *P < 0.05 when compared with control. n, number of RNA extract preparations.
miRNAs associated with fibrosis and their target genes.
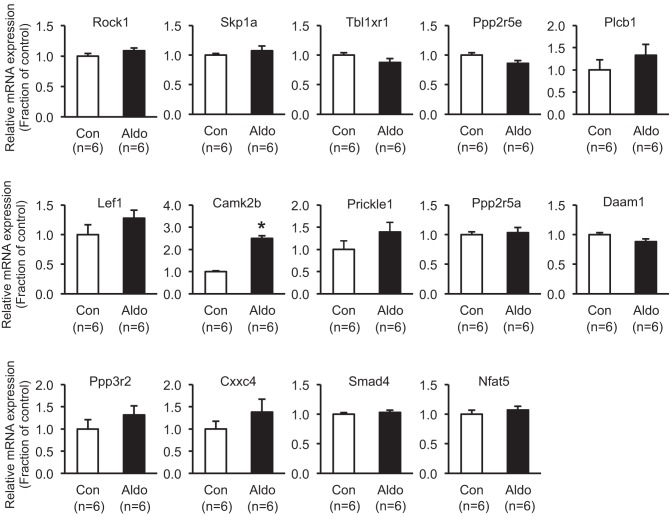
Among miRNAs significantly changed in RT-qPCR analysis (miR-224-5p, miR-130b-3p, miR181b-1-3p, miR-30b-3p, miR-1949, miR-669h-3p, miR-34c-5p, and miR-146a-5p; Figs. 4 and 5), miR-130b-3p, miR-34c-5p, and miR-146a-5p were further selected, based on the previous studies revealing that these three miRNAs were importantly associated with tissue fibrosis in lung, liver, heart, and kidney (2, 3, 11, 12, 16, 41). Putative target genes of these three miRNAs were identified (Table 5), i.e., six target genes (Rock1, skp1a, Tbl1xr1, Ppp2re, Wnt2b, Plcb1) of miR-130b-3p, six target genes (Lef1, Camk2b, Tbl1xr1, Prickle1, Ppp2r5a, Daam1) of miR-34c-5p, and four target genes (Ppp3r2, Cxxc4, Smad4, Nfat5) of miR-146a-5p in Wnt signaling. The changes of all of these target genes were examined by RT-qPCR in mpkCCDc14 cells after aldosterone treatment (10−6 M; 3 days; Fig. 6). Among the 15 target genes (Table 5), mRNA expression of the Camk2b gene was significantly increased in response to aldosterone (10−6 M) treatment for 3 days (250 ± 12% of control, P < 0.05; Fig. 6).
Table 5.
Target genes of miR-130b-3p, miR-34c-5p, and miR-146a-5p
| miRNA | Gene | Accession Number | Protein Description |
|---|---|---|---|
| mmu-miR-130b-3p | Rock1 | NM_009071.2 | Rho-associated coiled-coil containing protein kinase 1 |
| skp1a | NM_011543.4 | S-Phase kinase-associated protein 1A | |
| Tbl1xr1 | NM_030732.3 | Transducin (beta)-like 1 X-linked receptor 1 | |
| Ppp2re | NM_012024.2 | Protein phosphatase 2, regulatory subunit B′, epsilon | |
| Wnt2b | NM_009520.3 | Wingless-type MMTV integration site family, member 2B | |
| Plcb1 | NM_001145830.1 | Phospholipase C, beta 1 | |
| mmu-miR-34c-5p | Lef1 | NM_001276402.1 | Lymphoid enhancer binding factor 1 |
| Camk2b | NM_001174053.1 | Ca2+/calmodulin-dependent protein kinase II, beta | |
| Tbl1xr1 | NM_030732.3 | Transducin (beta)-like 1 X-linked receptor 1 | |
| Prickle1 | NM_001033217.4 | Prickle planar cell polarity protein 1 | |
| Ppp2r5a | NM_144880.4 | Protein phosphatase 2, regulatory subunit B′, alpha | |
| Daam1 | NM_026102.3 | Disheveled associated activator of morphogenesis 1 | |
| mmu-miR-146a-5p | Ppp3r2 | NM_001004025.4 | Protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type II) |
| Cxxc4 | NM_001004367.4 | CXXC finger 4 | |
| Smad4 | NM_008540.2 | SMAD family member 4 | |
| Nfat5 | NM_018823.2 | Nuclear factor of activated T cells 5 |
MMTV, mouse mammary tumor virus.
Fig. 6.
Real-time quantitative PCR analysis of target genes of miR-130b-3p, miR-34c-5p, and miR-146a-5p after aldosterone (Aldo; 10−6 M; 3 days) treatment in mpkCCDc14 cells. *P < 0.05 when compared with control. n, number of RNA extract preparations.
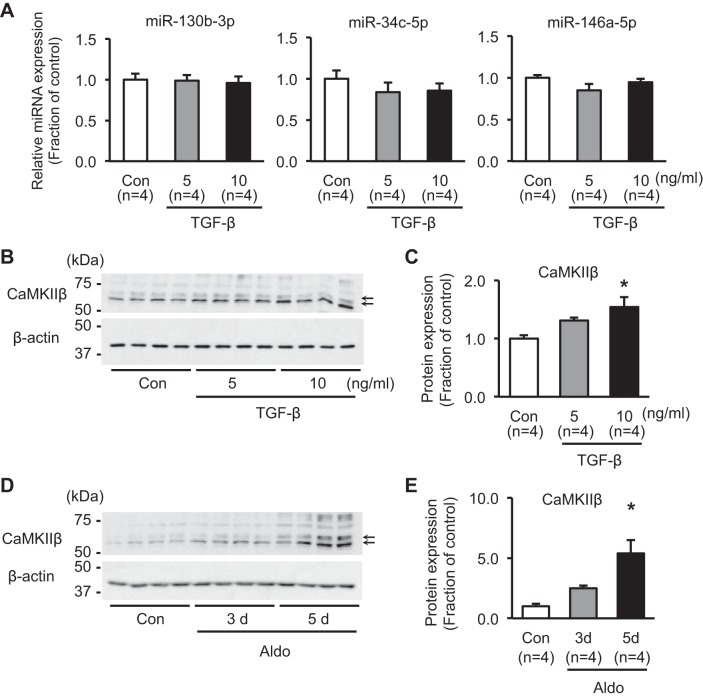
To examine whether the intracellular pathways by which aldosterone induces fibrosis are the same as TGF-β—the key player in development of renal fibrosis—the expression levels of these three miRNAs (miR-130b-3p, -34c-5p, and -146a-5p) were further examined in TGF-β-treated mpkCCDc14 cells (5 or 10 ng/ml; 3 days). Importantly, as shown in Fig. 7A, RT-qPCR demonstrated that miR-130b-3p, -34c-5p, and -146a-5p were unchanged in response to TGF-β. The protein expression of CaMKIIβ—the target of miR-34c-5p—revealed no significant change in response to 5 ng/ml TGF-β treatment for 3 days, whereas it was significantly increased after 10 ng/ml TGF-β treatment for 3 days (150 ± 17% of control, P < 0.05; Fig. 7, B and C). Although aldosterone treatment (10−6 M) for 3 days did not induce a significant change of CaMKIIβ protein expression, its protein expression was fivefold higher after aldosterone treatment (10−6 M) for 5 days (540 ± 110% of control, P < 0.05; Fig. 7, D and E).
Fig. 7.
Changes of miR-130b-3p, miR-34c-5p, miR-146a-5p, and CaMKIIβ expression in mpkCCDc14 cells. A: real-time quantitative PCR analysis of miR-130b-3p, miR-34c-5p, and miR-146a-5p in response to TGF-β treatment (5 or 10 ng/ml; 3 days) in mpkCCDc14 cells. B and C: semiquantitative immunoblotting of CaMKIIβ (~58 and 62 kDa; arrows) in mpkCCDc14 cells treated with TGF-β (5 or 10 ng/ml; 3 days). D and E: semiquantitative immunoblotting of CaMKIIβ (~58 and 62 kDa; arrows) in mpkCCDc14 cells treated with aldosterone (10−6 M; 3 or 5 days). (A) n, number of RNA extract preparations; (C and E) n, number of cell lysate preparation. The predicted size of CaMKIIβ is ~60 kDa, as seen in the kidney cortex tissue samples (see Fig. 10). However, in mpkCCDc14 cells, we noticed that there were 2 main bands (~58 and 62 kDa) that were mainly changed after (D) aldosterone treatment, CaMKIIβ siRNA (see Fig. 8, A and C), and miR-34 mimic treatment (see Fig. 8E). In contrast, bands ~75 kDa were also noted, which were variable. Thus the main bands at ~58 and 62 kDa were selected for semiquantitative immunoblotting. The Camk2b gene is a long gene, based on Ensembl mouse genome information (GRCm38; chromosome 11: 5,969,644–6,066,362), and has at least 10 transcript (splicing variants) coding proteins (479–666 amino acids; UniProt). It has not been clear yet which transcripts are expressed abundantly in each renal tubule segment and mpkCCDc14 cell. *P < 0.05 compared with control group.
Role of CaMKIIβ in aldosterone-induced fibrosis in mpkCCDc14 cells.
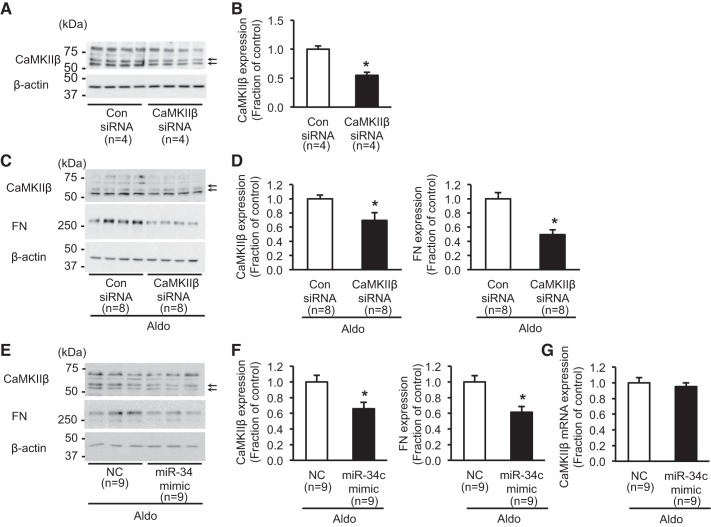
To determine the role of CaMKIIβ in regulation of fibrosis-associated proteins, the changes of protein expression of aldosterone-induced FN were examined in mpkCCDc14 cells with siRNA-mediated CaMKIIβ knockdown. CaMKIIβ expression was significantly decreased in CaMKIIβ siRNA (40 nM)-transfected mpkCCDc14 cells [55 ± 6% of the control siRNA-treated cells in the absence of aldosterone treatment, P < 0.05 (Fig. 8, A and B), or 70 ± 11% of control level in the presence of aldosterone treatment, P < 0.05 (Fig. 8, C and D)]. The difference in the CaMKIIβ protein expression in response to siRNA was likely due to the effect of aldosterone cotreatment under siRNA-mediated knockdown. Importantly, aldosterone (10−6 M; 3 days)-induced protein expression of FN was significantly attenuated in the cells with siRNA-mediated CaMKIIβ knockdown (49 ± 7% of control, P < 0.05; Fig. 8, C and D).
Fig. 8.
Changes of CaMKIIβ and fibronectin (FN) protein expression in mpkCCDc14 cells. A and B: semiquantitative immunoblotting of CaMKIIβ (~58 and 62 kDa; arrows) in mpkCCDc14 cells with CaMKIIβ siRNA-mediated knockdown in the absence of aldosterone treatment. C and D: semiquantitative immunoblotting of CaMKIIβ (~58 and 62 kDa; arrows) and FN (~262 kDa) in mpkCCDc14 cells with siRNA-mediated knockdown of CaMKIIβ (40 nM), followed by aldosterone treatment (10−6 M; 3 days). E and F: semiquantitative immunoblotting of CaMKIIβ (~58 and 62 kDa; arrows) and FN (~262 kDa) in mpkCCDc14 cells transfected with miR-34c-5p mimic (20 nM), followed by aldosterone treatment (10−6 M; 3 days). G: real-time quantitative PCR analysis for the change of CaMKIIβ mRNA expression in mpkCCDc14 cells transfected with the miR-34c-5p mimic (20 nM), followed by aldosterone treatment (10−6 M; 3 days). Aldo, aldosterone; Con siRNA, nontarget control siRNA; NC, miScript Inhibitor Negative Control; n, number of cell lysate preparation. *P < 0.05 when compared with control.
Furthermore, to identify the effect of miR-34c-5p on CaMKIIβ protein expression of its target gene (Camk2b), the miR-34c-5p mimic was transfected to mpkCCDc14 cells. When the miR-34c-5p mimic (20 nM) was transfected to mpkCCDc14 cells, aldosterone-induced protein expression of CaMKIIβ was significantly attenuated (66 ± 8% of control, P < 0.05; Fig. 8, E and F), whereas the mRNA level was unchanged (Fig. 8G). Importantly, consistent with the results observed in the cells with CaMKIIβ siRNA knockdown, aldosterone-induced FN expression was markedly decreased in miR-34c-5p mimic-transfected mpkCCDc14 cells (61 ± 7% of control, P < 0.05; Fig. 8, E and F). Therefore, the results indicated that aldosterone treatment was associated with downregulation of miR-34c-5p and the consequent increase of CaMKIIβ expression, which is likely to play a role in aldosterone-induced fibrosis in kidney collecting duct cells.
Interaction of miR-34c-5p to the target sequence of 3′-UTR of CaMKIIβ mRNA.
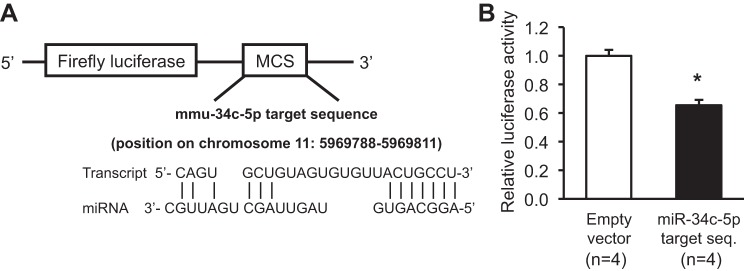
To examine whether miR-34c-5p directly regulates the expression of CaMKIIβ, luciferase assay was performed. The plausible target sequence of miR-34c-5p in 3′-UTR of CaMKIIβ mRNA was analyzed using DIANA Tools and was inserted to the pmirGLO Firefly Luciferase Expression Vector (Fig. 9A). Empty vector or target sequence-inserted vector was transfected to mpkCCDc14 cells with the miR-34c-5p mimic, respectively. When the target sequence-inserted vector was transfected with the miR-34c-5p mimic, firefly luciferase activity was significantly decreased (66 ± 3% of empty vector-transfected control with the miR-34c-5p mimic, P < 0.05; Fig. 9B), indicating that miR-34c-5p could directly bind to 3′-UTR of CaMKIIβ mRNA and hence, regulate CaMKIIβ protein expression.
Fig. 9.
Interaction between miR-34c-5p and 3′-UTR of CaMKIIβ mRNA. A: the target sequence of miR-34c-5p in CaMKIIβ 3′-UTR was inserted to the pmirGLO vector. B: decreased firefly luminescence activity in the cells transfected with the miR-34c-5p target sequence-inserted vector with the miR-34c-5p mimic compared with empty vector-transfected control cells with the miR-34c-5p mimic. *P < 0.05 when compared with the cells transfected with the empty vector and miR-34c-5p mimic. MCS, multiple cloning site; n, number of cell lysate preparation.
A mouse model with aldosterone treatment.
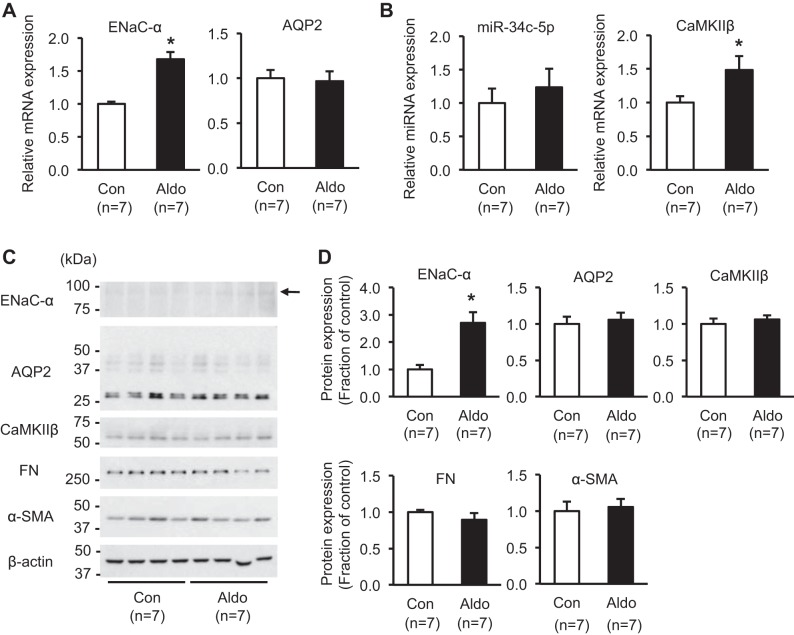
To examine whether aldosterone administration in vivo could also induce the changes of miR-34c-5p and CaMKIIβ expression in the kidney cortex, an animal model was made. Aldosterone (250 μg ⋅ kg−1 ⋅ day−1) was administered subcutaneously to C57BL/6 mice (n = 7) for 10 days via osmotic minipumps. As previously demonstrated (9), the plasma potassium level was significantly decreased to 2.5 ± 0.9 mmol/l in aldosterone-infused mice compared with control (3.4 ± 0.1 mmol/l, P < 0.05). Furthermore, mRNA and protein expression of ENaC-α were significantly increased in the kidney cortex from aldosterone-infused mice (170 ± 11% and 270 ± 40% of control level, respectively, P < 0.05; Fig. 10, A, C, and D), whereas AQP2 mRNA and protein expression were unchanged (Fig. 10, A, C, and D). Consistent with the increased mRNA expression of CaMKIIβ in mpkCCDc14 cells after aldosterone treatment (Fig. 6), CaMKIIβ mRNA expression was significantly increased in the kidney cortex of aldosterone-infused mice (150 ± 20% of control level, P < 0.05; Fig. 10B). In contrast, protein expression of CaMKIIβ was unchanged, and miR-34c-5p expression, FN, and α-SMA showed no significant changes (Fig. 10, B, C, and D).
Fig. 10.
Expression of epithelial sodium channel-α subunit (ENaC-α), aquaporin-2 (AQP2), miR-34c-5p, CaMKIIβ, and fibrosis marker proteins in the kidney cortex from aldosterone-infused mice model. A: real-time quantitative PCR (RT-qPCR) analysis of mRNA expression of ENaC-α and AQP2. B: RT-qPCR analysis of miR-34c-5p and mRNA expression of CaMKIIβ. C and D: semiquantitative immunoblotting of ENaC-α (~85 kDa; arrow), AQP2 (both deglycosylated ~29 kDa and glycosylated ~35–50 kDa), CaMKIIβ (~60 kDa), fibronectin (FN; ~262 kDa), and alpha-smooth muscle actin (α-SMA; ~42 kDa) in the kidney cortex from aldosterone-infused mice in vivo. n, the number of RNA extract preparations or cell or tissue lysate preparation. Aldo, aldosterone. *P < 0.05 when compared with control.
DISCUSSION
Whereas aldosterone-induced glomerular podocyte injury has widely been studied, the underlying mechanisms of aldosterone-induced renal epithelial injury and fibrosis are not fully elucidated (5, 48, 49). Recently, a number of studies have demonstrated that tubule cells trigger interstitial fibrosis, emphasizing the importance of tubule-derived signaling pathways in renal fibrogenesis (26, 59, 63). In this study, we demonstrated that aldosterone induced expression of fibrosis-associated proteins in mpkCCDc14 cells. Changes of miRNA expression in mpkCCDc14 cells after aldosterone treatment were examined by microarray, as we previously demonstrated in kidney collecting duct cells after vasopressin treatment (27). The hierarchical clustering of signaling pathways, in which the putative target genes of identified miRNAs were enriched, was also studied via DIANA-mirPath tool. Among the profiled 74 KEGG pathways, the Wnt signaling pathway was highly ranked, where 37 putative target genes of the 15 identified miRNAs were found (Table 4). In particular, among the 15 identified miRNAs, upregulated miR-130b-3p and downregulated miR-146a-5p and miR-34c-5p were selected for further studies, as these three miRNAs have previously been demonstrated to be involved in tissue fibrosis (3, 4, 41). Among the putative target genes of miR-130b-3p, miR-146a-5p, and miR-34c-5p, the mRNA and protein expression of CaMKIIβ, the products of a target gene of miR-34c-5p (Camk2b), were revealed to be significantly increased by aldosterone treatment. Importantly, when CaMKIIβ siRNA or the miR-34c-5p mimic was transfected into mpkCCDc14 cells, aldosterone-induced FN expression was significantly attenuated, along with decreased CaMKIIβ protein expression, suggesting that miR-34c-5p and CaMKIIβ are involved in aldosterone-induced fibrosis. In addition, a luciferase reporter assay revealed a decrease of CaMKIIβ translation in cells transfected with the miRNA mimic of miR-34c-5p, indicating the direct regulation of miR-34c-5p on mRNA and protein expression of CaMKIIβ.
Aldosterone-induced renal fibrosis.
Evidence showed that aldosterone contributes to the progression of chronic renal disease and fibrosis (10, 48). In addition to its production in the adrenal cortex, local production of aldosterone in the kidney—stimulated by angiotensin II, low salt intake, and hyperglycemia—was demonstrated (58). Combined with the localization of MR in the preglomerular vasculature, mesangial cells, distal tubular epithelial cells, and fibroblasts (10), high aldosterone levels are likely to promote renal injury (10, 48). The tissue injury was also observed in other organs, e.g., the heart, and importantly, anti-aldosterone treatment significantly reduced the mortality in patients with chronic advanced heart failure (47) and chronic advanced heart failure after myocardial infarction (46).
In the kidney, high aldosterone levels are associated with endothelial dysfunction, glomerulosclerosis, renal hypertrophy, and tubulointerstitial fibrosis (42). The proposed underlying mechanisms are production of growth factors and inhibited degradation of the ECM, such as increased proinflammatory factors (reactive oxygen species, monocyte chemoattractant protein 1, IL-6, and IL-1β), growth factors (CTGF and TGF-β), ECM (collagen I and IV and plasminogen activator inhibitor 1), and cell proliferation (6, 42). Consistent with this, we demonstrated that aldosterone induced the fibrosis marker proteins, e.g., FN and α-SMA, in the kidney cortical collecting duct cells. This finding was also supported by the previous findings in the inner medullary collecting duct cells, demonstrating that aldosterone increased the expression of collagen, FN, and CTGF (5). The findings indicate that renal tubular epithelial cells expressing MR are subjected to fibrotic changes, as also shown by a recent study demonstrating the importance of tubule-derived Wnts in kidney fibrosis (63).
Wnt signaling pathway and the role of CaMKII in fibrosis.
We demonstrated that putative target genes of the identified miRNAs were associated with 74 KEGG pathways. Among them, the Wnt signaling pathway was highly ranked. Genome-wide transcriptome analysis previously revealed that novel genes and pathways, including the Wnt signaling pathway, are involved in the development of chronic kidney disease and renal fibrosis (57). Moreover, the Wnt/β-catenin signaling pathway has also been reported to contribute to the development of fibrosis in other organs, such as lung and liver (13, 18). Consistent with this, administration of the Wnt antagonist Dickkopf-1 gene retarded the progression of fibrotic lesions (21).
The Wnt signaling pathways have been associated with TGF-β and epithelial-to-mesenchymal transition in disease conditions. Nineteen Wnt ligands, known in the mouse and human genomes, can mediate their actions through the canonical (β-catenin-dependent) or noncanonical (β-catenin-independent) pathway. In the canonical pathway, activated Wnt stimulus attenuates the degradation of the transcriptional coactivator β-catenin by a multiprotein destruction complex (51, 55). The consequent accumulation of β-catenin in the nucleus interacts with T cell factor/lymphoid enhancer binding factor proteins to regulate gene expression (36). The noncanonical Wnt pathway involves Ca2+ influx and further activation of CaMKII, PKC, and JNK pathways (31, 53).
We demonstrated 15 miRNAs enriched in the Wnt signaling pathway (Table 4), including upregulated miR-130b-3p and downregulated miR-146a-5p and miR-34c-5p, which were previously known to be associated with tissue fibrosis (3, 4, 41). In particular, we found that mRNA and protein expression of CaMKIIβ—the products of a target gene (Camk2b) of miR-34c-5p—were significantly increased by aldosterone treatment. CaMKII is a serine/threonine protein kinase, and it has four different isoforms: α, β, γ, and δ (39). When Ca2+ is released from intracellular stores, it interacts with calmodulin, which can bind to CaMKII (39). It could activate CaMKII by autophosphorylation and acts on a wide range of substrates, including SMAD2 and SMAD4, which are transcription factors downstream of TGF-β mediated signaling (1, 56). In the noncanonical Wnt pathway, Wnt5a exploits Ca2+ as a second messenger. Wnt5a activates PKC, leading to Ca2+ release into cytosol, and released Ca2+ ions interact with calmodulin (8, 29). Recent evidence also indicated the role of Ca2+ channels in the progression of renal fibrosis, since T-type Ca2+ channel blockade blunted the tubulointerstitial fibrosis in rat kidney (52). Importantly, siRNA-mediated CaMKIIβ knockdown significantly attenuated the aldosterone-induced expression of FN in mpkCCDc14 cells, suggesting that CaMKIIβ is likely to play a role in tissue fibrosis. Consistent with this, a recent study revealed that disheveled-1 increased both total CaMKIIδγ and phospho-CaMKIIδγ and aggravated cardiomyopathy, whereas disheveled-1 transgenic mice lacking CaMKIIδγ did not exhibit cardiac hypertrophy, left ventricular dysfunction, and fibrosis (62).
Increased expression of miR-130b-3p and decreased expression of miR-146a-5p and miR-34c-5p.
In the Wnt signaling pathway, we demonstrated that 15 miRNAs were enriched (Table 4). Among them, miR-130b-3p, miR-146a-5p, and miR-34c-5p are known to be involved in tissue fibrosis. For instance, the miR-130/301 family was significantly increased in bleomycin-induced lung fibrosis and carbon tetrachloride-induced liver fibrosis (3). In streptozotocin-induced diabetes mellitus, miR-146a knockout mice showed more severe kidney injury, e.g., exacerbated proteinuria, glomerular hypertrophy, and fibrosis (4). Moreover, administration of miR-34c in a mouse model of unilateral ureteral obstruction attenuated kidney fibrosis and expression of fibrosis markers (41).
Consistent with this, we demonstrated that aldosterone induced expression of fibrosis marker proteins in mpkCCDc14 cells, where miR-130b-3p was upregulated, and miR-146a-5p and miR-34c-5p were downregulated. Since TGF-β is a key mediator in fibrogenesis, and also, aldosterone per se could evoke TGF-β in the MR-dependent manner in the kidney (23), it is important to examine whether the intracellular pathways by which aldosterone induced fibrosis are the same as TGF-β. Interestingly, we observed that mRNA expression of miR-130b-3p, miR-146a-5p, and miR-34c-5p was not changed by TGF-β treatment. Although further studies are needed, the results suggested that miR-130b-3p, miR-146a-5p, and miR-34c-5p are likely to be selectively regulated in aldosterone-induced fibrosis. CaMKIIβ protein expression, however, was induced by both TGF-β and aldosterone, although the induction of CaMKIIβ protein expression was higher after aldosterone treatment. The profibrotic effects of TGF-β are known to be mediated by CTGF, stimulating the proliferation of renal fibroblasts and synthesis of ECM (60). In contrast, a previous study showed that aldosterone increased CTGF gene expression and protein expression, even in the presence of the TGF-β1 neutralizing antibody (20), suggesting that the TGF-β-independent pathway may be present for aldosterone-induced fibrosis. In addition, aldosterone activates its nuclear receptor MR, promoting targeting gene transcription and translation, which may be different from TGF-β. Moreover, a rapid, nongenomic effect of aldosterone was demonstrated in the collecting duct cells (50), suggesting a complexity of aldosterone-induced actions. This issue needs to be investigated in future studies.
Finally, we made an animal model to study whether aldosterone treatment in vivo also induces the changes of miR-34c-5p and CaMKIIβ expression in the kidney. High-dose aldosterone administration for 10 days via osmotic minipumps in mice resulted in hypokalemia and an increase of ENaC-α expression in the kidney cortex, compatible with the aldosterone action in the collecting ducts. Similar to the increase of CaMKIIβ mRNA expression in mpkCCDc14 cells after aldosterone treatment (Fig. 6), its expression in the kidney cortex was significantly induced in mice treated with aldosterone. In contrast, protein expression of CaMKIIβ was unchanged, and miR-34c-5p expression, FN, and α-SMA showed no significant changes after aldosterone infusion for 10 days. Thus the findings suggested that aldosterone-induced renal fibrosis in vivo could be possible on a long-term basis of stimulation.
Conclusions and perspectives.
Tubule cell-derived signaling pathways could play a role in renal fibrogenesis. In the kidney collecting duct cells, aldosterone-induced downregulation of miR-34c-5p in the Wnt signaling and the consequent increase in CaMKIIβ expression could play a role in aldosterone-induced fibrosis. Since Wnt signaling pathways are known to be associated with renal fibrosis, future studies are warranted to examine the role of identified miRNAs and their target genes in in vivo experiments for a better understanding of the pathogenesis and therapeutic targets of aldosterone-mediated renal fibrosis.
GRANTS
This study was supported by the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT and Future Planning, Korea (2014R1A5A2009242 and 2016R1A2B4009365).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
E-J.P., H.J.J., H-J.C., and T-H.K. conceived and designed research; E-J.P. performed experiments; E-J.P., H.J.J., H-J.C., and T-H.K. analyzed data; E-J.P., H.J.J., H-J.C., J-I.C., H-J.P., and T-H.K. interpreted results of experiments; E-J.P. and H.J.J. prepared figures; E-J.P., H.J.J., and H-J.C. drafted manuscript; E-J.P., H.J.J., H-J.C., J-I.C., H-J.P., and T-H.K. edited and revised manuscript; E-J.P., H.J.J., H-J.C., J-I.C., H-J.P., and T-H.K. approved final version of manuscript.
REFERENCES
- 1.Abdel-Wahab N, Wicks SJ, Mason RM, Chantry A. Decorin suppresses transforming growth factor-beta-induced expression of plasminogen activator inhibitor-1 in human mesangial cells through a mechanism that involves Ca2+-dependent phosphorylation of Smad2 at serine-240. Biochem J 362: 643–649, 2002. doi: 10.1042/bj3620643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amrouche L, Desbuissons G, Rabant M, Sauvaget V, Nguyen C, Benon A, Barre P, Rabaté C, Lebreton X, Gallazzini M, Legendre C, Terzi F, Anglicheau D. MicroRNA-146a in human and experimental ischemic AKI: CXCL8-dependent mechanism of action. J Am Soc Nephrol 28: 479–493, 2017. doi: 10.1681/ASN.2016010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertero T, Cottrill KA, Annis S, Bhat B, Gochuico BR, Osorio JC, Rosas I, Haley KJ, Corey KE, Chung RT, Nelson Chau B, Chan SY. A YAP/TAZ-miR-130/301 molecular circuit exerts systems-level control of fibrosis in a network of human diseases and physiologic conditions. Sci Rep 5: 18277, 2015. doi: 10.1038/srep18277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhatt K, Lanting LL, Jia Y, Yadav S, Reddy MA, Magilnick N, Boldin M, Natarajan R. Anti-inflammatory role of microRNA-146a in the pathogenesis of diabetic nephropathy. J Am Soc Nephrol 27: 2277–2288, 2016. doi: 10.1681/ASN.2015010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brem AS, Morris DJ, Ge Y, Dworkin L, Tolbert E, Gong R. Direct fibrogenic effects of aldosterone on normotensive kidney: an effect modified by 11β-HSD activity. Am J Physiol Renal Physiol 298: F1178–F1187, 2010. doi: 10.1152/ajprenal.00532.2009. [DOI] [PubMed] [Google Scholar]
- 6.Brown NJ. Contribution of aldosterone to cardiovascular and renal inflammation and fibrosis. Nat Rev Nephrol 9: 459–469, 2013. doi: 10.1038/nrneph.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuevas CA, Gonzalez AA, Inestrosa NC, Vio CP, Prieto MC. Angiotensin II increases fibronectin and collagen I through the β-catenin-dependent signaling in mouse collecting duct cells. Am J Physiol Renal Physiol 308: F358–F365, 2015. doi: 10.1152/ajprenal.00429.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniels L, Bell JR, Delbridge LM, McDonald FJ, Lamberts RR, Erickson JR. The role of CaMKII in diabetic heart dysfunction. Heart Fail Rev 20: 589–600, 2015. doi: 10.1007/s10741-015-9498-3. [DOI] [PubMed] [Google Scholar]
- 9.de Seigneux S, Nielsen J, Olesen ET, Dimke H, Kwon TH, Frøkiaer J, Nielsen S. Long-term aldosterone treatment induces decreased apical but increased basolateral expression of AQP2 in CCD of rat kidney. Am J Physiol Renal Physiol 293: F87–F99, 2007. doi: 10.1152/ajprenal.00431.2006. [DOI] [PubMed] [Google Scholar]
- 10.Del Vecchio L, Procaccio M, Viganò S, Cusi D. Mechanisms of disease: the role of aldosterone in kidney damage and clinical benefits of its blockade. Nat Clin Pract Nephrol 3: 42–49, 2007. doi: 10.1038/ncpneph0362. [DOI] [PubMed] [Google Scholar]
- 11.Du J, Niu X, Wang Y, Kong L, Wang R, Zhang Y, Zhao S, Nan Y. MiR-146a-5p suppresses activation and proliferation of hepatic stellate cells in nonalcoholic fibrosing steatohepatitis through directly targeting Wnt1 and Wnt5a. Sci Rep 5: 16163, 2015. doi: 10.1038/srep16163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du R, Sun W, Xia L, Zhao A, Yu Y, Zhao L, Wang H, Huang C, Sun S. Hypoxia-induced down-regulation of microRNA-34a promotes EMT by targeting the Notch signaling pathway in tubular epithelial cells. PLoS One 7: e30771, 2012. doi: 10.1371/journal.pone.0030771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edeling M, Ragi G, Huang S, Pavenstädt H, Susztak K. Developmental signalling pathways in renal fibrosis: the roles of Notch, Wnt and Hedgehog. Nat Rev Nephrol 12: 426–439, 2016. doi: 10.1038/nrneph.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edinger RS, Coronnello C, Bodnar AJ, Labarca M, Bhalla V, LaFramboise WA, Benos PV, Ho J, Johnson JP, Butterworth MB. Aldosterone regulates microRNAs in the cortical collecting duct to alter sodium transport. J Am Soc Nephrol 25: 2445–2457, 2014. doi: 10.1681/ASN.2013090931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elvira-Matelot E, Zhou XO, Farman N, Beaurain G, Henrion-Caude A, Hadchouel J, Jeunemaitre X. Regulation of WNK1 expression by miR-192 and aldosterone. J Am Soc Nephrol 21: 1724–1731, 2010. doi: 10.1681/ASN.2009111186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng B, Chen S, Gordon AD, Chakrabarti S. miR-146a mediates inflammatory changes and fibrosis in the heart in diabetes. J Mol Cell Cardiol 105: 70–76, 2017. doi: 10.1016/j.yjmcc.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Greene EL, Kren S, Hostetter TH. Role of aldosterone in the remnant kidney model in the rat. J Clin Invest 98: 1063–1068, 1996. doi: 10.1172/JCI118867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Y, Xiao L, Sun L, Liu F. Wnt/beta-catenin signaling: a promising new target for fibrosis diseases. Physiol Res 61: 337–346, 2012. [DOI] [PubMed] [Google Scholar]
- 19.Hager H, Kwon TH, Vinnikova AK, Masilamani S, Brooks HL, Frøkiaer J, Knepper MA, Nielsen S. Immunocytochemical and immunoelectron microscopic localization of alpha-, beta-, and gamma-ENaC in rat kidney. Am J Physiol Renal Physiol 280: F1093–F1106, 2001. doi: 10.1152/ajprenal.2001.280.6.F1093. [DOI] [PubMed] [Google Scholar]
- 20.Han KH, Kang YS, Han SY, Jee YH, Lee MH, Han JY, Kim HK, Kim YS, Cha DR. Spironolactone ameliorates renal injury and connective tissue growth factor expression in type II diabetic rats. Kidney Int 70: 111–120, 2006. doi: 10.1038/sj.ki.5000438. [DOI] [PubMed] [Google Scholar]
- 21.He W, Dai C, Li Y, Zeng G, Monga SP, Liu Y. Wnt/beta-catenin signaling promotes renal interstitial fibrosis. J Am Soc Nephrol 20: 765–776, 2009. doi: 10.1681/ASN.2008060566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hostetter TH, Ibrahim HN. Aldosterone in chronic kidney and cardiac disease. J Am Soc Nephrol 14: 2395–2401, 2003. doi: 10.1097/01.ASN.0000086472.65806.73. [DOI] [PubMed] [Google Scholar]
- 23.Juknevicius I, Segal Y, Kren S, Lee R, Hostetter TH. Effect of aldosterone on renal transforming growth factor-beta. Am J Physiol Renal Physiol 286: F1059–F1062, 2004. doi: 10.1152/ajprenal.00202.2003. [DOI] [PubMed] [Google Scholar]
- 24.Jung HJ, Kim SY, Choi HJ, Park EJ, Lim JS, Frøkiaer J, Nielsen S, Kwon TH. Tankyrase-mediated β-catenin activity regulates vasopressin-induced AQP2 expression in kidney collecting duct mpkCCDc14 cells. Am J Physiol Renal Physiol 308: F473–F486, 2015. doi: 10.1152/ajprenal.00052.2014. [DOI] [PubMed] [Google Scholar]
- 25.Jung HJ, Kwon TH. Molecular mechanisms regulating aquaporin-2 in kidney collecting duct. Am J Physiol Renal Physiol 311: F1318–F1328, 2016. doi: 10.1152/ajprenal.00485.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang HM, Ahn SH, Choi P, Ko YA, Han SH, Chinga F, Park AS, Tao J, Sharma K, Pullman J, Bottinger EP, Goldberg IJ, Susztak K. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat Med 21: 37–46, 2015. doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JE, Jung HJ, Lee YJ, Kwon TH. Vasopressin-regulated miRNAs and AQP2-targeting miRNAs in kidney collecting duct cells. Am J Physiol Renal Physiol 308: F749–F764, 2015. doi: 10.1152/ajprenal.00334.2014. [DOI] [PubMed] [Google Scholar]
- 28.Koga K, Yokoi H, Mori K, Kasahara M, Kuwabara T, Imamaki H, Ishii A, Mori KP, Kato Y, Ohno S, Toda N, Saleem MA, Sugawara A, Nakao K, Yanagita M, Mukoyama M. MicroRNA-26a inhibits TGF-β-induced extracellular matrix protein expression in podocytes by targeting CTGF and is downregulated in diabetic nephropathy. Diabetologia 58: 2169–2180, 2015. doi: 10.1007/s00125-015-3642-4. [DOI] [PubMed] [Google Scholar]
- 29.Kohn AD, Moon RT. Wnt and calcium signaling: beta-catenin-independent pathways. Cell Calcium 38: 439–446, 2005. doi: 10.1016/j.ceca.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 30.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11: 597–610, 2010. [DOI] [PubMed] [Google Scholar]
- 31.Kühl M, Sheldahl LC, Malbon CC, Moon RT. Ca(2+)/calmodulin-dependent protein kinase II is stimulated by Wnt and Frizzled homologs and promotes ventral cell fates in Xenopus. J Biol Chem 275: 12701–12711, 2000. doi: 10.1074/jbc.275.17.12701. [DOI] [PubMed] [Google Scholar]
- 32.Lai JY, Luo J, O’Connor C, Jing X, Nair V, Ju W, Randolph A, Ben-Dov IZ, Matar RN, Briskin D, Zavadil J, Nelson RG, Tuschl T, Brosius FC III, Kretzler M, Bitzer M. MicroRNA-21 in glomerular injury. J Am Soc Nephrol 26: 805–816, 2015. doi: 10.1681/ASN.2013121274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee HW, Khan SQ, Khaliqdina S, Altintas MM, Grahammer F, Zhao JL, Koh KH, Tardi NJ, Faridi MH, Geraghty T, Cimbaluk DJ, Susztak K, Moita LF, Baltimore D, Tharaux PL, Huber TB, Kretzler M, Bitzer M, Reiser J, Gupta V. Absence of miR-146a in podocytes increases risk of diabetic glomerulopathy via up-regulation of ErbB4 and Notch-1. J Biol Chem 292: 732–747, 2017. doi: 10.1074/jbc.M116.753822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JW, Chou CL, Knepper MA. Deep sequencing in microdissected renal tubules identifies nephron segment-specific transcriptomes. J Am Soc Nephrol 26: 2669–2677, 2015. doi: 10.1681/ASN.2014111067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu GJ, Dong YQ, Zhang QM, Di WY, Jiao LY, Gao QZ, Zhang CG. miRNA-221 promotes proliferation, migration and invasion by targeting TIMP2 in renal cell carcinoma. Int J Clin Exp Pathol 8: 5224–5229, 2015. [PMC free article] [PubMed] [Google Scholar]
- 36.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 17: 9–26, 2009. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matavelli LC, Siragy HM. Reduction of aldosterone production improves renal oxidative stress and fibrosis in diabetic rats. J Cardiovasc Pharmacol 61: 17–22, 2013. doi: 10.1097/FJC.0b013e318274d2ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathew JT, Patni H, Chaudhary AN, Liang W, Gupta A, Chander PN, Ding G, Singhal PC. Aldosterone induces mesangial cell apoptosis both in vivo and in vitro. Am J Physiol Renal Physiol 295: F73–F81, 2008. doi: 10.1152/ajprenal.00435.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Means AR. Regulatory cascades involving calmodulin-dependent protein kinases. Mol Endocrinol 14: 4–13, 2000. doi: 10.1210/mend.14.1.0414. [DOI] [PubMed] [Google Scholar]
- 40.Mejía-Vilet JM, Ramírez V, Cruz C, Uribe N, Gamba G, Bobadilla NA. Renal ischemia-reperfusion injury is prevented by the mineralocorticoid receptor blocker spironolactone. Am J Physiol Renal Physiol 293: F78–F86, 2007. doi: 10.1152/ajprenal.00077.2007. [DOI] [PubMed] [Google Scholar]
- 41.Morizane R, Fujii S, Monkawa T, Hiratsuka K, Yamaguchi S, Homma K, Itoh H. miR-34c attenuates epithelial-mesenchymal transition and kidney fibrosis with ureteral obstruction. Sci Rep 4: 4578, 2014. doi: 10.1038/srep04578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagase M. Activation of the aldosterone/mineralocorticoid receptor system in chronic kidney disease and metabolic syndrome. Clin Exp Nephrol 14: 303–314, 2010. doi: 10.1007/s10157-010-0298-8. [DOI] [PubMed] [Google Scholar]
- 43.Nagase M, Shibata S, Yoshida S, Nagase T, Gotoda T, Fujita T. Podocyte injury underlies the glomerulopathy of Dahl salt-hypertensive rats and is reversed by aldosterone blocker. Hypertension 47: 1084–1093, 2006. doi: 10.1161/01.HYP.0000222003.28517.99. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen J, Kwon TH, Frøkiaer J, Knepper MA, Nielsen S. Maintained ENaC trafficking in aldosterone-infused rats during mineralocorticoid and glucocorticoid receptor blockade. Am J Physiol Renal Physiol 292: F382–F394, 2007. doi: 10.1152/ajprenal.00212.2005. [DOI] [PubMed] [Google Scholar]
- 45.Nishikawa R, Chiyomaru T, Enokida H, Inoguchi S, Ishihara T, Matsushita R, Goto Y, Fukumoto I, Nakagawa M, Seki N. Tumour-suppressive microRNA-29s directly regulate LOXL2 expression and inhibit cancer cell migration and invasion in renal cell carcinoma. FEBS Lett 589: 2136–2145, 2015. doi: 10.1016/j.febslet.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 46.Pitt B, Remme W, Zannad F, Neaton J, Martinez F, Roniker B, Bittman R, Hurley S, Kleiman J, Gatlin M; Eplerenone Post-Acute Myocardial Infarction Heart Failure Efficacy and Survival Study Investigators . Eplerenone, a selective aldosterone blocker, in patients with left ventricular dysfunction after myocardial infarction. N Engl J Med 348: 1309–1321, 2003. doi: 10.1056/NEJMoa030207. [DOI] [PubMed] [Google Scholar]
- 47.Pitt B, Zannad F, Remme WJ, Cody R, Castaigne A, Perez A, Palensky J, Wittes J; Randomized Aldactone Evaluation Study Investigators . The effect of spironolactone on morbidity and mortality in patients with severe heart failure. N Engl J Med 341: 709–717, 1999. doi: 10.1056/NEJM199909023411001. [DOI] [PubMed] [Google Scholar]
- 48.Remuzzi G, Cattaneo D, Perico N. The aggravating mechanisms of aldosterone on kidney fibrosis. J Am Soc Nephrol 19: 1459–1462, 2008. doi: 10.1681/ASN.2007101079. [DOI] [PubMed] [Google Scholar]
- 49.Ritz E, Tomaschitz A. Aldosterone and the kidney: a rapidly moving frontier (an update). Nephrol Dial Transplant 29: 2012–2019, 2014. doi: 10.1093/ndt/gft035. [DOI] [PubMed] [Google Scholar]
- 50.Sheader EA, Wargent ET, Ashton N, Balment RJ. Rapid stimulation of cyclic AMP production by aldosterone in rat inner medullary collecting ducts. J Endocrinol 175: 343–347, 2002. doi: 10.1677/joe.0.1750343. [DOI] [PubMed] [Google Scholar]
- 51.Stamos JL, Weis WI. The β-catenin destruction complex. Cold Spring Harb Perspect Biol 5: a007898, 2013. doi: 10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugano N, Wakino S, Kanda T, Tatematsu S, Homma K, Yoshioka K, Hasegawa K, Hara Y, Suetsugu Y, Yoshizawa T, Hara Y, Utsunomiya Y, Tokudome G, Hosoya T, Saruta T, Hayashi K. T-Type calcium channel blockade as a therapeutic strategy against renal injury in rats with subtotal nephrectomy. Kidney Int 73: 826–834, 2008. doi: 10.1038/sj.ki.5002793. [DOI] [PubMed] [Google Scholar]
- 53.Veeman MT, Axelrod JD, Moon RT. A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev Cell 5: 367–377, 2003. doi: 10.1016/S1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 54.Vlachos IS, Kostoulas N, Vergoulis T, Georgakilas G, Reczko M, Maragkakis M, Paraskevopoulou MD, Prionidis K, Dalamagas T, Hatzigeorgiou AG. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res 40: W498–W504, 2012. doi: 10.1093/nar/gks494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J, Sinha T, Wynshaw-Boris A. Wnt signaling in mammalian development: lessons from mouse genetics. Cold Spring Harb Perspect Biol 4: a007963, 2012. doi: 10.1101/cshperspect.a007963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wicks SJ, Lui S, Abdel-Wahab N, Mason RM, Chantry A. Inactivation of smad-transforming growth factor beta signaling by Ca(2+)-calmodulin-dependent protein kinase II. Mol Cell Biol 20: 8103–8111, 2000. doi: 10.1128/MCB.20.21.8103-8111.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woroniecka KI, Park AS, Mohtat D, Thomas DB, Pullman JM, Susztak K. Transcriptome analysis of human diabetic kidney disease. Diabetes 60: 2354–2369, 2011. doi: 10.2337/db10-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xue C, Siragy HM. Local renal aldosterone system and its regulation by salt, diabetes, and angiotensin II type 1 receptor. Hypertension 46: 584–590, 2005. doi: 10.1161/01.HYP.0000175814.18550.c0. [DOI] [PubMed] [Google Scholar]
- 59.Yang L, Besschetnova TY, Brooks CR, Shah JV, Bonventre JV. Epithelial cell cycle arrest in G2/M mediates kidney fibrosis after injury. Nat Med 16: 535–543, 2010. doi: 10.1038/nm.2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yokoi H, Sugawara A, Mukoyama M, Mori K, Makino H, Suganami T, Nagae T, Yahata K, Fujinaga Y, Tanaka I, Nakao K. Role of connective tissue growth factor in profibrotic action of transforming growth factor-beta: a potential target for preventing renal fibrosis. Am J Kidney Dis 38, Suppl 1: S134–S138, 2001. doi: 10.1053/ajkd.2001.27422. [DOI] [PubMed] [Google Scholar]
- 61.Yuan Y, Chen Y, Zhang P, Huang S, Zhu C, Ding G, Liu B, Yang T, Zhang A. Mitochondrial dysfunction accounts for aldosterone-induced epithelial-to-mesenchymal transition of renal proximal tubular epithelial cells. Free Radic Biol Med 53: 30–43, 2012. doi: 10.1016/j.freeradbiomed.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 62.Zhang M, Hagenmueller M, Riffel JH, Kreusser MM, Bernhold E, Fan J, Katus HA, Backs J, Hardt SE. Calcium/calmodulin-dependent protein kinase II couples Wnt signaling with histone deacetylase 4 and mediates dishevelled-induced cardiomyopathy. Hypertension 65: 335–344, 2015. doi: 10.1161/HYPERTENSIONAHA.114.04467. [DOI] [PubMed] [Google Scholar]
- 63.Zhou D, Fu H, Zhang L, Zhang K, Min Y, Xiao L, Lin L, Bastacky SI, Liu Y. Tubule-derived Wnts are required for fibroblast activation and kidney fibrosis. J Am Soc Nephrol 28: 2322–2336, 2017. doi: 10.1681/ASN.2016080902. [DOI] [PMC free article] [PubMed] [Google Scholar]